Abstract
Colonies of Mycobacterium smegmatis LR222 on iron-limiting (0.1 μM Fe) minimal medium agar fluoresce under UV light due to the accumulation in the cells of the deferri form of the siderophore mycobactin. Two mutants with little or no fluorescence, designated LUN8 and LUN9, were isolated by screening colonies of transposon (Tn611)-mutagenized M. smegmatis. Ferrimycobactin prepared from iron-restricted cells of the wild type had an Rf of 0.62 on high-performance thin-layer chromatography (HPTLC) and a characteristic visible absorption spectrum with a peak near 450 nm. Similar extracts from LUN8 cells contained a small amount of ferrimycobactin with an Rf of 0.58 on HPTLC and an absorption spectrum with the peak shifted to a wavelength lower than that of the wild-type ferrimycobactin. Nuclear magnetic resonance spectroscopy studies suggested that the LUN8 mycobactin may have an altered fatty acid side chain. Mutant strain LUN9 produced no detectable mycobactin. Neither mutant strain produced measurable amounts of excreted mycobactin, although both excreted exochelin (the mycobacterial peptido-hydroxamate siderophore), and both mutants were more sensitive than the wild-type strain to growth inhibition by the iron chelator ethylenediamine-di(o-hydroxyphenylacetic acid). The transposon insertion sites were identified, and sequence analyses of the cloned flanking chromosome regions showed that the mutated gene in LUN9 was an orthologue of the Mycobacterium tuberculosis mycobactin biosynthetic gene mbtE. The mutated gene in LUN8 had homology with M. tuberculosis fadD33 (Rv1345), a gene that may encode an acyl-coenzyme A synthase and which previously was not known to participate in synthesis of mycobactin.
The limited availability of iron in terrestrial and aquatic environments as well as in vertebrate hosts compels most pathogenic and nonpathogenic microorganisms to produce efficient iron acquisition systems, such as the uptake processes that are based on the iron-chelating agents called siderophores (4, 5, 10, 16, 32, 42, 47). With very few exceptions, the mycobacterial species (including both pathogenic and saprophytic forms like Mycobacterium tuberculosis and Mycobacterium smegmatis) synthesize siderophores of the mycobactin class (31, 44). The mycobactins are salicylic acid-derived siderophores containing a hydroxyaryloxazoline cap and two N-hydroxyamides that together form a high-affinity binding site for a ferric iron atom (44). A large number of mycobactins with different alkyl and acyl substituents at five positions on the core of the molecule have been described (44). The mycobactins can be divided into two subgroups based on lipid solubility. In the case of the lipophilic mycobactins (which are the wholly cell-associated form of mycobactin), the N-acyl moiety on the first hydroxylated lysine of the molecule is a long-chain fatty acid (C10 to C20) with a cis double bond (usually at position C-2), while in the more hydrophilic mycobactins (which are excreted from the cells), this acyl moiety is derived from a dicarboxylic acid that may be present as the free acid or the methyl ester (14, 15, 22, 23, 33, 50). The excreted, hydrophilic mycobactins have been called carboxymycobactins (22, 23), exomycobactins (29, 52), or exochelins (14, 50), although Ratledge and Dover (32) have suggested that the exochelin designation be reserved for the excreted petido-hydroxamate siderophores that are found only in saprophytic mycobacteria and which are structurally different from the mycobactins. This convention will be used here.
A cluster of 10 genes (designated mbtA-J) that encodes enzymes for the assembly of mycobactin was identified in the published M. tuberculosis genome sequence (6, 7, 30). Functions have been assigned to the mbt gene products that (together with phosphopantetheinyl transferases) account for the relevant biosynthetic steps to produce mycobactin, although the genes for synthesis of the variable aliphatic side chains and the required acyltransferase for attachment of these units have not been identified (8, 30). Five of the mbt genes form an assembly line of nonribosomal peptide synthetases (mbtB, mbtE, and mbtF) and polyketide synthases (mbtC and mbtD) that activate and elongate the monomers of the mycobactin core. An isochorismate synthase encoded by mbtI and an adenylating enzyme, the MbtA protein, provide activated salicylate. Mycobactin chain initiation is thought to occur by acylation of an aryl carrier domain of the MbtB protein with activated salicylate (29, 30). By gene replacement, the mbtB gene was deleted and an antibiotic marker was inserted, yielding a mutant M. tuberculosis strain that failed to produce mycobactin and which was restricted for growth under low-iron conditions (11). These data link the mbt cluster to biosynthesis of mycobactin in M. tuberculosis, but functional characterization of the remaining genes and their products has yet to be done (11).
The analysis of the mycobactin biosynthetic genes did not reveal associated genes for iron or ferrimycobactin transport. Although there is ample evidence documenting mycobactin-mediated iron acquisition in mycobacteria (reviewed in references 10 and 32), the roles of the excreted (carboxymycobactin-exomycobactin) and the cell-associated form of mycobactin in iron transport are unclear. Uptake of iron from extracellular mycobactin appears to be by facilitated diffusion, and cell-associated mycobactin may serve as a short-term storage mechanism before transfer of iron to the cytoplasm (26, 32). A virulence function for mycobactin was indicated by the finding that deletion and replacement of the mbtB gene with a hygromycin marker impaired growth of the mutant strain in macrophage-like THP-1 cells (11).
Although both excreted and cell-associated mycobactins are produced by the saprophyte M. smegmatis (33), the predominant extracellular siderophore of this organism is exochelin MS, an ornithine-derived trihydroxamate (41). A similar peptido-hydroxamate molecule, exochelin MN, is produced by Mycobacterium neoaurum (40). Genes for biosynthesis, excretion, and uptake of exochelin have been cloned from M. smegmatis and sequenced (13, 51, 52). The genes, named fxuA, fxuB, and fxuC, were proposed to encode the cytoplasmic membrane permease for the transport of ferriexochelin into M. smegmatis (13). Several genes have been shown to be involved in exochelin biosynthesis and export. One (fxbA) was suggested to encode an enzyme that adds a formyl group in the synthesis of exochelin. The others (fxbB, -C, and possibly -D) encode nonribosomal peptide synthetases that likely form a complex with an ABC transporter known as ExiT (or ORF1 and -2) to synthesize and export the peptide siderophore. The exochelins have not been found in pathogenic mycobacteria (32), and the need for both exochelin and mycobactin in saprophytic mycobacteria is unclear. The exochelins may have an important iron acquisition role in the terrestrial and aquatic environments preferred by the exochelin-producing saprophytic mycobacteria.
In this study, we report isolation and characterization of two M. smegmatis mutant strains generated by transposon insertion that are unable to produce (or have greatly diminished production of) mycobactin. The insertion site in one of the mutants (strain LUN9) was in the M. smegmatis orthologue of the mycobactin gene mbtE, while the insertion site for the other mutant (strain LUN8) was in the M. smegmatis orthologue of M. tuberculosis fadD33, a gene that heretofore had not been implicated in mycobactin synthesis. Both mutant strains still produce exochelin, but they were impaired for growth under iron-restricted conditions.
MATERIALS AND METHODS
Bacteria, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli cultures were routinely grown in Luria-Bertani broth or agar medium (27) at 37°C. M. smegmatis cultures were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, Mich.); for plate (TSA) medium, agar (15 g per liter) was added. Antibiotics, when required, were added at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; hygromycin, 200 μg/ml for E. coli and 50 μg/ml for M. smegmatis.
TABLE 1.
Bacterial strains and plasmids
| Designation | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| JM109 | F′ ΔtraD36 lac1q Δ(lacZ)M15 proAB/recA1 endA1 gyrA96 thi hsdR17(rK− mK+) supE44 relA1 Δ(lac-proAB) | Lab stock |
| DH10B | mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK rpsL nupG | Lab stock |
| M. smegmatis strains | ||
| LR222 | Efficient plasmid transformation strain of mc26 | 2 |
| LUN1 | LR222 with Tn611 insertion in exochelin gene fxbC | 52 |
| LUN2 | LR222 with Tn611 insertion in exochelin gene exiT | 52 |
| LUN8 | LR222 with Tn611 insertion in fadD (mycobactin neg.) | This study |
| LUN9 | LR222 with Tn611 insertion in mbtE (mycobactin neg.) | This study |
| Plasmids | ||
| pCG79 | Kanr; temperature-sensitive, E. coli-mycobacterial shuttle plasmid containing Tn611 | 17 |
| pNBV1 | Hyr; E. coli-mycobacterial shuttle vector | 20 |
| pUC18 | Ampr; multiple cloning site vector | Lab stock |
| pCR2.1-TOPO | AT direct cloning vector for PCR products | Invitrogen |
| pCR4Blunt-TOPO | Blunt-end direct PCR cloning vector | Invitrogen |
| Litmus 28 | Ampr; multiple cloning site vector | NEBa |
| pARC6 | pUC18 with 4.4-kb SmaI fragment from LUN9 | This study |
| pWEN3 | pUC18 with 3.5-kb SacI fragment from LUN8 | This study |
| pML222 | pCG79 excised from LUN8 genome by EcoRI digestion | This study |
| pCR-Rv1344 | 3.3-kb PCR product from H37Rv cloned into pCR2.1-TOPO, encodes Rv1344-fadD33-fadE14 | This study |
| pML1344 | Litmus 28 with 3.3-kb EcoRI fragment from pCR-Rv1344 | This study |
| pMLNV1344 | pNBV1 with 3.3-kb HindIII-SpeI insert from pML1344 | This study |
| pCR4-Sm1344 | 3.6-kb PCR product from LR222 cloned into pCR4Blunt-TOPO, encodes acp-fadD-fadE | This study |
| pBLT1344 | pNBV1 with SnaBI insert from pCR4-Sm1344 cloned into the EcoRV site | This study |
New England Biolabs, Inc.
For growth at defined iron concentrations, we used a minimal medium containing (per liter): glucose, 10 g; (NH4)2SO4, 1 g; K2HPO4, 3.5g; KH2PO4, 1.5 g. To lower the trace metal contamination, the medium was treated with Chelex-100 (Bio-Rad Laboratories, Hercules, Calif.) by previously reported methods (1). Before use, the medium was supplemented with 0.1% sterilized Tween 80 and with filter-sterilized solutions of high-purity sulfate salts (Johnson-Matthey, Inc.) of magnesium (830 μM), manganese (40 μM), zinc (0.3 μM), and iron (various concentrations). The glassware was soaked with 0.025 M tetrasodium EDTA overnight to remove contaminating metals and rinsed extensively with laboratory-grade water.
Molecular biology techniques.
Standard molecular biology techniques and procedures were followed as described by Sambrook et al. (37) and Davis et al. (9). Enzymes were used as directed by the manufacturers with the buffers provided. Plasmid DNA from E. coli cells was prepared by the alkaline lysis method (3) and was purified by RNase treatment and phenol extraction. Isolation of plasmid DNA from M. smegmatis (21) was essentially like that from E. coli, i.e., alkaline lysis, except that the cells were first disrupted (beaten for 2 min) by glass beads in a Mini-BeadBeater (Biospec Products, Bartlesville, Okla.).
Introduction of plasmids into M. smegmatis was by electroporation (21). Bacteria suspended in 10% glycerol electroporation buffer with 1 μg of DNA were subjected to a single pulse using the Bio-Rad Gene Pulser (Bio-Rad Laboratories) set at 2.5 kV, 25 μF, with the pulse controller resistance set at infinity. The content of the cuvette was diluted into 5 ml of TSB and incubated at 42°C. After incubation, 100-μl aliquots were plated onto antibiotic-TSA plates; transformants appeared after incubation for 3 days.
Genomic DNA from M. smegmatis cells, disrupted with a Mini-BeadBeater cell disruptor, was isolated with an Easy-DNA kit (Invitrogen Life Technologies, Carlsbad, Calif.) and then was purified by RNase treatment and phenol extraction. Alternatively, genomic DNA was prepared as described elsewhere (49). Cells were lysed with sodium dodecyl sulfate, and proteins were removed by digestion with proteinase K. Cell wall debris and remaining proteins were removed by precipitation with cetyltrimethylammonium bromide, and high-molecular-weight DNA was recovered by isopropanol precipitation. M. tuberculosis DNA was obtained from John T. Belisle and was produced through funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, contract 1-AI-25147 entitled “Tuberculosis Research Materials.”
Southern hybridization probes used in this study were prepared by digesting plasmids carrying the desired gene with the appropriate restriction enzymes, separating the DNA fragments by agarose gel electrophoresis, excising the DNA probe band, and purifying it by phenol extraction. The DNA probe was radiolabeled with [32P]dCTP by the random primer method using the rediprime DNA labeling system (Amersham Biosciences, Piscataway, N.J.). Southern blotting and hybridization were performed according to the instructions of the manufacturer of the transfer membrane (NEN Life Science Products, Boston, Mass.).
Mutagenesis and screening for mycobactin mutants.
Transposon mutagenesis of M. smegmatis LR222 was performed as described by Guilhot et al. (17). Plasmid pCG79 containing Tn611 was introduced into M. smegmatis by electroporation. Transformants were selected at 30°C on TSA plates containing kanamycin. Kanr transformants were grown in TSB at 30°C for 3 days, diluted 1:100 in antibiotic-free medium, and incubated at 42°C for at least 24 h. To screen for mycobactin mutants, dilutions of cultures were plated on low-iron (0.1 μM) minimal medium agar containing 30 μg of kanamycin/ml and supplemented (per liter) with 3 g of sodium glutamate and 20 ml of glycerol. Plates were incubated for 5 days at 42°C. Mycobactin-producing strains fluoresce (18) under UV light (245 nm) on this medium; therefore, colonies were checked for fluorescence and those failing to fluoresce were examined.
Amplification and cloning.
The wild-type acp/fadD/fadE gene cluster was obtained by PCR from M. smegmatis LR222 and M. tuberculosis H37Rv genomic DNA. PCR was carried out using pfuTurbo polymerase (Stratagene, La Jolla, Calif.) in a Bio-Rad Gene Cycler. The oligonucleotide primers (supplied by Invitrogen Life Technologies) were as follows: for M. smegmatis, GTCTGTTTCAAGTCGCCCGG (forward) and ACACCCGACGACATCCCCG (reverse); for M. tuberculosis, GGTGCCCGAGGTCGATTGAAC (forward) and CGACCGTCGGTTGTGTGAGTGG (reverse). PCR products were isolated from agarose and cloned into pCR4Blunt-TOPO (Invitrogen Life Technologies). Constructs were verified as correct by sequencing.
Complementation of strain LUN8.
Plasmids carrying PCR-amplified acp/fadD/fadE DNA from the wild-type M. smegmatis (pBLT1344 and subclones of pBLT1344) or M. tuberculosis (pMLNV1344) were electroporated into mutant strain LUN8, and transformants were selected on appropriate antibiotic-containing media. The transformants were streaked on low-iron (0.1 μM) minimal medium agar containing glycerol and glutamate, and the colonies were observed for fluorescence under UV (245 nm), indicative of mycobactin production. Plasmids were recovered from the desired transformants and analyzed by restriction digestion and electrophoresis to verify the presence of the correct plasmid. Production of mycobactin by fluorescing transformants was confirmed by extraction and the high-performance thin-layer chromatography (HPTLC) procedures described below.
Nucleotide sequencing and analysis.
DNA sequencing was performed manually using the Sequenase system (U.S. Biochemicals, Cleveland, Ohio) and [35S]dATP or [33P]dideoxynucleoside triphosphate with miniprep plasmid DNA. Complete sequences of both strands were obtained by either subcloning or primer walking. Preliminary M. smegmatis sequence data were obtained from The Institute for Genomic Research (TIGR) through their website (http://www.tigr.org).
Siderophore purification and assay.
Cell-associated mycobactin was extracted as previously described (18, 44). Bacteria were grown in high-iron (20 μM iron) minimal medium for 2 days and then subcultured in low-iron (0.1 μM iron) minimal medium without Tween 80 and grown to an optical density at 600 nm of 0.68. The cells recovered by centrifugation were suspended in ethanol. The cell suspension was stirred at room temperature for 24 h. Cell debris was then removed by filtration, and the mycobactin in the filtrate was converted to its ferric complex by the addition of saturated FeCl3 in ethanol. The ferrimycobactin was extracted with chloroform. Excess iron was removed by washing the chloroform layer with water, and the chloroform was collected and evaporated to dryness. The residue was dissolved in methanol, and its absorbance at 450 nm was determined. The amount of ferrimycobactin was calculated using an extinction coefficient (1% solution) of 42.8 (44). Further purification of mycobactin in the crude extracts was done by previously published protocols (31) with slight alteration of volumes of each solvent. The dried mycobactin was dissolved in cyclohexane-butanol (6:1) and then mixed with neutral-grade alumina until all colored material had adsorbed to the alumina. The colored alumina was then washed successively with cyclohexane, mineral spirits, toluene, and finally diethylether. The mycobactin then was eluted from the alumina with chloroform and evaporated to dryness. This provided a siderophore of approximately 80% purity (31).
Carboxymycobactin (exomycobactin) was extracted by the method of Ratledge and Ewing (33). Culture supernatants were filtered through 0.45-μm-pore-size filters, and the pH was adjusted to 3.5 with 2 M H2SO4. Saturated FeCl3 in ethanol was added until a precipitate began to form. This solution was stirred for 1 h and centrifuged. The exomycobactin in the supernatant was extracted into chloroform, which then was washed with water and evaporated. The residue was dissolved in ethanol.
Mycobactin preparations were analyzed spectrophotometrically on a Beckman DU-70 spectrophotometer and by HPTLC on silica gel (Whatman type LHPK) using ethanol-cyclohexane-water-ethyl acetate-acetic acid (5:25:2.5:35:5, by volume) as the developing solvent (33). Mycobactin J obtained from Allied Monitor, Fayette, Mo., was used as a control. The peptido-hydroxamate siderophore exochelin in low-iron culture filtrates was assayed with the liquid chrome azurol S shuttle reagent (39).
EDDA inhibition assay.
To test the effect of iron limitation on the growth of siderophore mutants, ethylenediamine-di(o-hydroxyphenyl acetic acid) (EDDA), a ferric iron chelator, was added at final concentrations from 0 to 300 μg/ml to brain heart infusion agar. Prior to use, the agar was cured for 7 days to allow chelation of iron by EDDA. The EDDA agar was melted by microwaving, and after cooling to 50°C bacterial cells (about 104 to 105 CFU) were added to the agar and mixed. The agar was aliquoted into 12.5-cm2 tissue culture flasks and allowed to solidify with the flask standing on end. The flasks were incubated for 3 to 5 days, and the growth of M. smegmatis was determined.
NMR spectroscopy.
Proton nuclear magnetic resonance (NMR) spectra of mycobactin samples dissolved in methanol-d4 or chloroform-d were recorded and referenced to that of tetramethylsilane on a JEOL Eclipse 400+ spectrometer spinning at 15 Hz at 21 to 25°C in the laboratory of E. Valente, Mississippi College, Clinton, Miss.
Nucleotide sequence accession numbers.
The nucleotide sequences of the M. smegmatis LR222 mbtDE genes and the acp/fadD genes have been assigned the GenBank database accession numbers AY225312 and AF277395, respectively.
RESULTS
Isolation of mycobactin mutants.
M. smegmatis LR222, a strain derived from M. smegmatis mc26 (2), was mutagenized with Tn611, and the insertional library was screened for loss of mycobactin production on low-iron (0.1 μM) minimal medium agar containing glutamate, glycerol, and kanamycin. After 5 days of incubation, colonies were checked for fluorescence under UV light. Mycobactin-producing colonies fluoresce due to the aromatic nucleus of mycobactin that accumulates in the deferri form within iron-restricted cells (18). More than 10,000 colonies were screened, and two potential mycobactin mutants (designated LUN8 and LUN9) that had no or significantly diminished fluorescence under UV light were isolated.
Lack of mycobactin production by strain LUN9 and production of an altered mycobactin by strain LUN8.
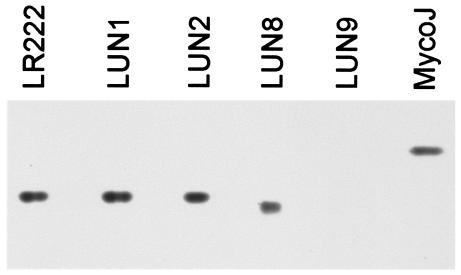
To assess production of cell-associated mycobactin by strains LUN8 and LUN9, ethanolic extracts of cells grown in low-iron (0.1 μM) minimal medium were analyzed by silica gel HPTLC. Strain LUN9 produced no detectable mycobactin, whereas strain LUN8 produced a ferric-chelating molecule that migrated more slowly (Rf, 0.58) than mycobactin produced by the parental, wild-type strain LR222 (Rf, 0.62) (Fig. 1). When the mycobactin preparations from strains LUN8 and LR222 were mixed and chromatographed, separate bands were noted, confirming that the mycobactin-like product from strain LUN8 was distinct from mycobactin produced by the wild type (data not shown). The previously described (52) mutant strains LUN1 and LUN2 that fail to make the peptido-hydroxamate siderophore exochelin also were analyzed and found to produce mycobactin with a wild-type Rf (Fig. 1).
FIG. 1.
HPTLC of cell-associated ferrimycobactins produced by wild-type M. smegmatis strain LR222, by the previously described (52) mutant strains LUN1 and LUN2 that are unable to produce the siderophore exochelin, and by strains LUN8 and LUN9. Mycobactin J was included as a reference.
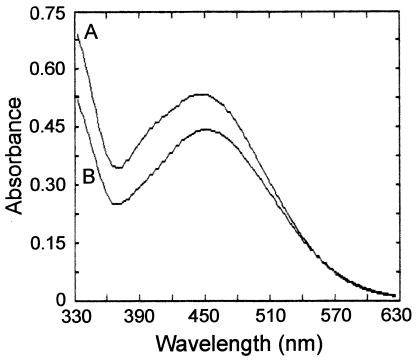
The ferric-chelating material prepared from strain LUN8 was concentrated and purified by adsorption to, and elution from, alumina. Mycobactin from the wild-type strain LR222 was likewise purified. The electronic absorption spectrum of ferrimycobactin from LR222 showed a broad peak with maximum absorbance near 450 nm, characteristic of the ferrimycobactins (Fig. 2). The ferric chelate of the substance produced by strain LUN8 also displayed a broad peak, with the maximum shifted to a wavelength somewhat lower than that of the wild-type ferrimycobactin (Fig. 2). Comparison of the absorption maxima of the identically prepared samples from both strains indicated that strain LUN8 made a mycobactin-like material at about 1/10 the production level of wild-type mycobactin. Preliminary NMR spectroscopy of deferrated mycobactin isolated from both the wild-type and the LUN8 strains depicted molecules with the typical ortho-salicylate aromatic moiety, similar to mycobactin S previously described by Ratledge and Dover (32). The mycobactin from strain LR222 contained an approximately 17-carbon, 2,3-unsaturated aliphatic side chain at the R1 position. Signals from the 2,3-hydrogens at the double bond in the R1 side chain of the wild-type siderophore were seen as a set of correlated signals at δ 6.0 and 6.3 ppm (downfield from standard tetramethylsilane) in one- and two-dimensional proton NMR spectra. These signatures were not seen in the mycobactin produced by the LUN8 strain. Therefore, a saturated alkyl side chain, probably of wild-type carbon length, was predicted to be attached at the R1 position of the mycobactin made by strain LUN8. The resulting alkane group at this position could alter solubility, resulting in the slightly lower Rf value observed for LUN8 mycobactin on HPTLC plates (Fig. 1). The complete structural description of the mycobactin produced by strain LUN8 is in progress and will be the subject of another publication.
FIG. 2.
Absorption spectra of identically prepared samples of ferrimycobactin-like material produced by M. smegmatis mutant strain LUN8 (A) and of ferrimycobactin prepared from wild-type M. smegmatis strain LR222 (B). Wild-type ferrimycobactin was diluted 10-fold for this scan.
To assess production of the excreted form of mycobactin (exomycobactin or carboxymycobactin) by the mutant strains LUN8 and LUN9, chloroform extracts of the cell-free filtrates of low-iron cultures were examined by HPTLC. Excreted mycobactin with a characteristic Rf value of 0.23 was detected in culture filtrates of the wild-type strain LR222 and the previously described exochelin mutants (52) LUN1 and LUN2. Exomycobactin was not detectable in culture filtrates of either strain LUN8 or LUN9, although strain LUN8 may have excreted exomycobactin in amounts below the detection threshold of the assay. Both strains LUN8 and LUN9 excreted the peptido-hydroxamate siderophore exochelin, which was detected by assay of the cell-free filtrates of low-iron cultures with the liquid chrome azurol S shuttle assay of Schwyn and Neilands (39).
Sensitivity of mycobactin mutant strains to iron restriction.
The capacities of the mycobactin mutant strains LUN8 and LUN9 to overcome iron restriction imposed by the iron-chelating agent EDDA were determined and compared to those of the previously described exochelin mutants LUN1 and LUN2 (52). The MIC of EDDA for the wild-type strain LR222 was more than 250 μg of EDDA/ml, whereas the MICs for both strains LUN8 and LUN9 were lowered to approximately 50 μg of EDDA/ml. On the other hand, the MICs for the exochelin mutants LUN1 and LUN2 were essentially identical to that for the wild type, at 250 μg of EDDA/ml. Control studies showed that EDDA inhibition of the M. smegmatis strains was reversed by iron salts, confirming that this inhibition was due to iron restriction. These data suggest that mycobactin synthesis but not exochelin production was necessary to overcome iron limitation caused by EDDA.
Cloning and identification of mycobactin genes.
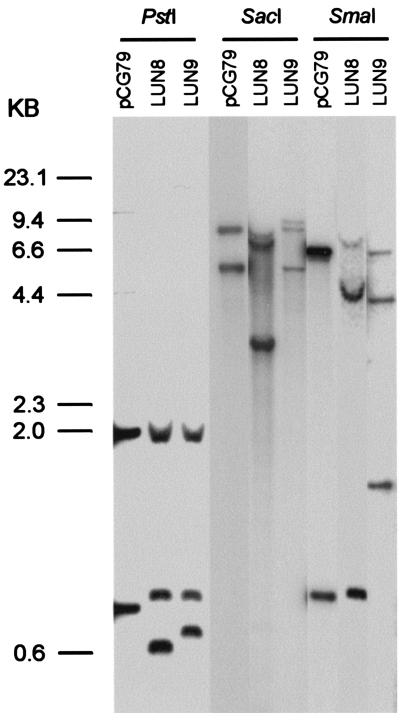
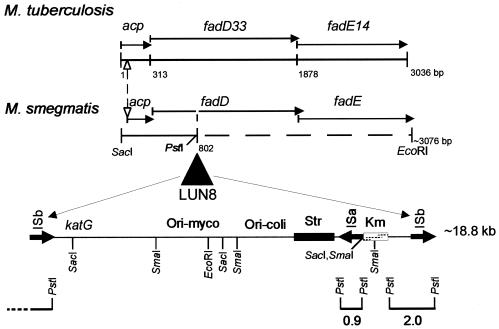
Insertional mutagenesis with Tn611 was confirmed by the previously described strategy (17, 52) of digesting the genomic DNAs from the mutants with PstI and performing Southern hybridizations using the insertion sequence IS6100 as a probe (Fig. 3). Colony blotting was used to clone the transposon-flanking regions of the insertion mutants. Both DNA fragments from the PstI digestion were too short to give enough information for further work. Therefore, we used the enzymes SmaI and SacI to cut the genomic DNA and performed Southern blotting using IS6100 as a probe. The results showed that both mutants had different hybridization patterns from the plasmid control (Fig. 3). By using the same probe, a 3.5-kb SacI fragment (pWEN3) was cloned from LUN8 and a 4.4-kb SmaI fragment (pARC6) was cloned from LUN9. Other fragments seen in the blot were vector sequence. A portion of the LUN8 genome, downstream from the insertion, was cloned by taking advantage of the plasmid vector that was integrated into the chromosome along with Tn611. This integrated vector carries an E. coli origin of replication and resistance genes for kanamycin and streptomycin. By cutting LUN8 DNA with EcoRI, ligating, transforming E. coli cells, and selecting for kanamycin resistance, a plasmid (pML222) was recovered. This plasmid carries the genomic region on the other side of the LUN8 Tn611 insertion from that DNA that yielded pWEN3.
FIG. 3.
Southern hybridization analysis of genomic DNAs isolated from mycobactin mutant strains LUN8 and LUN9 that were digested with PstI, SmaI, and SacI. A 32P-labeled HindIII/PstI internal fragment of IS6100 was used as the probe, and pCG79 was the vector control. The origin of the PstI-generated bands is shown in Fig. 5. Molecular size standards are indicated at the left.
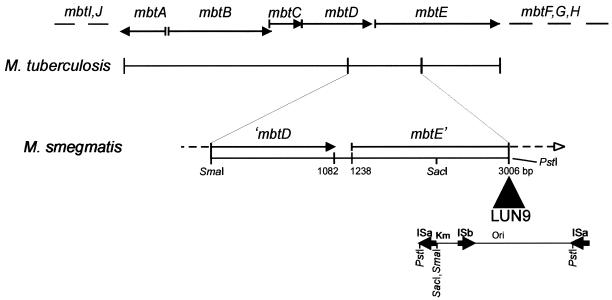
The cloned insertion flanking regions from both LUN8 and LUN9 were sequenced. Nucleotide sequencing showed that the insertion flanking region from LUN9 shared homology with the mycobactin biosynthesis genes mbtD and mbtE of M. tuberculosis (Fig. 4). The mutated gene from LUN8 encoded the N-terminal portion of a protein with homology to M. tuberculosis FadD33, an acyl-coenzyme A (CoA) synthase (Fig. 5). This DNA also encoded an acyl carrier protein (M. tuberculosis Rv1344 orthologue) of unknown function. In addition to the 3′ end of fadD33, the DNA cloned into pML222 encoded an orthologue of the M. tuberculosis FadE14 protein.
FIG. 4.
Physical map of the Tn611 insertion region of M. smegmatis LUN9 compared to the M. tuberculosis mycobactin biosynthetic cluster from nucleotides 2680005 to 2656218. Solid arrows indicate open reading frames for the respective genes. The transposon insertion is shown in the opposite orientation from that in Fig. 5 and, in contrast to the LUN8 insertion, the ISa element was duplicated instead of the ISb element.
FIG. 5.
Physical map of the Tn611 insertion region of M. smegmatis LUN8 compared to the Rv1344(acp)/fadD33/fadE14 sequence from nucleotides 1508968 to 1512003 of M. tuberculosis. The Tn611 insertion location in LUN8 is indicated. Solid arrows indicate open reading frames for the respective genes. The dashed nucleotide line for M. smegmatis represents partially sequenced DNA. PstI digestion hybridization fragments are shown at the bottom of the figure.
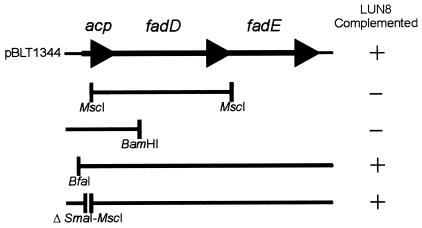
Complementation of LUN8.
Plasmid pBLT1344 carrying PCR-amplified acp/fadD/fadE DNA from the wild-type M. smegmatis strain LR222 and subclones of pBLT1344 were electroporated into strain LUN8, and the resulting transformants were observed for fluorescence under UV (245 nm), indicative of mycobactin production (Fig. 6). Extraction and HPTLC analysis of ferrimycobactin produced by a fluorescing LUN8/pBLT1344 transformant confirmed its production of wild-type levels of mycobactin with a wild-type Rf. Complementation of strain LUN8 by the subclones showed that only those constructs that encoded both of the complete fadD and fadE genes restored mycobactin synthesis in strain LUN8 (Fig. 6). Transformation of strain LUN8 cells with the PCR-amplified M. tuberculosis acp/fadD/fadE orthologues carried on plasmid pMLNV1344 failed to yield fluorescing colonies, indicating that the M. tuberculosis genes were unable to complement the M. smegmatis strain.
FIG. 6.
Complementation of mycobactin expression in mutant M. smegmatis strain LUN8 by plasmid pBLT1344 encoding M. smegmatis acp/fadD/fadE and various subclones. Solid lines below the insert map of pBLT1344 represent the amount of pBLT1344 DNA carried by each subclone. The restriction enzymes used for each construction are indicated.
In silico analysis of mycobactin gene clusters.
Analysis of DNA sequences indicated that the flanking regions from LUN9 had homology with mycobactin genes mbtD and mbtE from M. tuberculosis (Fig. 4). MbtD belongs to the polyketide synthase superfamily of proteins that contain consensus sites for posttranslational phosphopantetheinylation that are involved in acyl group transfer (10, 30). MbtE shares homology with nonribosomal peptide synthetases and has a single amino-acid adenylation domain that activates the lysine residues that are incorporated into mycobactin. From the unfinished M. smegmatis sequence obtained from TIGR, the organization of the mycobactin gene cluster was identical to that of M. tuberculosis with the exception of mbtI and -J (Rv2386c and Rv2385, also called trpE2 and lipK [7]). Whereas mbtI and -J map next to mbtA in M. tuberculosis, these genes were not found to be linked in M. smegmatis. Overall, the proteins encoded in the mycobactin gene cluster displayed a high degree of identity between the two mycobacterial species, from a high of 75.4% for the mbtG orthologues to a low of 49.7% for mbtD.
The flanking regions from LUN8 had homology with an M. tuberculosis acyl carrier protein gene (Rv1344; 52.7% identity) and two genes (fadD33 and fadE14; 62.4 and 76.6% identity, respectively) predicted to be involved in fatty acid degradation. These genes do not map to the mbt gene cluster. The M. tuberculosis acyl carrier protein had an additional 22 amino acids at its N terminus compared to that of the M. smegmatis orthologue. A methionine at the end of this stretch of additional amino acids could serve as a start codon, yielding a product more similar to the one encoded by M. smegmatis. Immediately 5′ of the acp (Rv1344 orthologue) open reading frame of M. smegmatis is a 19-base sequence similar to the consensus IdeR/SirR binding site (19, 45).
DISCUSSION
Mycobactin is an almost ubiquitously occurring siderophore in both pathogenic and saprophytic mycobacteria. The saprophytic forms produce a second siderophore called exochelin (32), but loss of exochelin production did not significantly diminish the MIC of the iron chelator EDDA for M. smegmatis exochelin mutants. However, the necessity for mycobactin in mycobacterial iron metabolism was evidenced by the fivefold decrease in the MIC of EDDA for the mycobactin mutant strains LUN8 and LUN9. The reason for the sensitivity of the mycobactin mutants to iron restriction is unknown.
Analysis of cloned DNA sequences indicated that the regions flanking the transposon insertion in the mycobactin mutant strain LUN9 had homology with mycobactin genes mbtD and mbtE of M. tuberculosis. MbtD belongs to the polyketide synthase superfamily of proteins that contain consensus sites for posttranslational phosphopantetheinylation and may be involved in acyl group transfer (8). MbtE shares homology with nonribosomal peptide synthetases and has a single amino acid adenylation domain that probably activates the lysine residues that are incorporated into mycobactin. Our results are in agreement with those of De Voss et al. (11), who showed that disruption of the M. tuberculosis mbt gene cluster prevented synthesis of the mycobactin core.
The regions flanking the transposon insertion in strain LUN8 have homology with M. tuberculosis genes encoding an acyl carrier protein (Rv1344), an acyl-CoA synthase (fadD33), and an acyl-CoA dehydrogenase (fadE14). These genes, which are not located in the reported mycobactin gene cluster (7), appeared to be necessary for wild-type mycobactin production, an observation not previously reported. The preliminary NMR studies of the mycobactin made by LUN8 suggested that in M. smegmatis the acp/fadD/fadE gene cluster is involved in biosynthesis of the aliphatic side chains attached to the mycobactin nucleus. Because correct side chain synthesis cannot be accomplished in strain LUN8, it produced only a small amount of a mycobactin with an altered side chain. FadD and FadE are predicted, based on sequence, to be involved in lipid degradation (7); these enzymes could initiate modification of a preexisting lipid with subsequent attachment of the lipid to an acyl carrier protein with final transfer to the mycobactin core by an as-yet-to-be-identified acyltransferase (8, 10). Alternatively, the FadD and FadE enzymes could modify the mycobactin side chains after they have been attached by the acyltransferase.
An examination of the putative regulatory region of the M. smegmatis acp gene revealed a consensus “IdeR box,” suggesting a role for the iron-dependent regulatory protein IdeR (38) in the transcriptional control of these genes. Moreover, Rodriguez et al. (36) recently demonstrated by microarray analysis that the expression of the M. tuberculosis Rv1344, fadD33, and fadE14 genes was iron and IdeR repressible. The translational termination and initiation codons for acp-fadD and fadD-fadE were predicted to overlap. These observations and the complementation data from LUN8 transformed with pBLT1344 and its subclones suggest that acp-fadD-fadE comprise an operon with a single polycistronic mRNA under control of the IdeR iron-dependent regulator, making expression of the operon responsive to iron restriction.
Whereas the wild-type M. smegmatis LR222 genes provided the missing function(s) to strain LUN8, transformation of this strain with the M. tuberculosis orthologues failed to restore wild-type mycobactin synthesis. Although there is a high degree of homology between the M. tuberculosis and the M. smegmatis acp/fadD/fadE genes, the lack of complementation of LUN8 by the corresponding M. tuberculosis genes may not be surprising. An explanation may be that the FadD and FadE enzymes from the two species recognize different lipids. Moreover, the aliphatic side chains of the mycobactins are highly variable and species specific (44). M. smegmatis may not produce the particular lipid that is recognized by M. tuberculosis FadD and/or FadE. Wheeler and Ratledge (48) proposed that in vivo-grown M. tuberculosis makes use of the variety and quantity of lipids available within mammalian cells and the tubercle. The many FadD and FadE alleles (36 of each) encoded by the M. tuberculosis genome (7) would suggest a high degree of specificity of each enzyme for its substrate. Various species-specific genes may be used for synthesis and attachment of the variable side chains of mycobactin.
An alternative explanation for the lack of complementation by the M. tuberculosis genes could simply be that the M. smegmatis RNA polymerase does not efficiently initiate transcription from the M. tuberculosis promoter. Although the recognition of promoter sequences appears somewhat conserved among mycobacteria (28), differences in gene expression of cloned mycobacterial DNA have been reported when genes from one species are introduced into another (24, 46). Moreover, Mulder et al. (28) suggested that the validity of studying M. tuberculosis gene expression in other mycobacterial hosts may depend on the particular promoter in question.
A significant amount of evidence has accumulated demonstrating that two of the microbial processes that contribute to M. tuberculosis infection are iron acquisition and lipid metabolism (11, 12, 25, 34, 35, 36, 43). M. tuberculosis strains that are defective in mycobactin synthesis (11) or express a constitutively active iron repressor (25) are attenuated for virulence. Differential display was used to show that fadD33 is underexpressed in the attenuated strain M. tuberculosis H37Ra (35), and strains that are defective in fadD33 expression are inhibited for growth in the liver of BALB/c mice (34). Here we have demonstrated that these two processes, iron acquisition and lipid metabolism, intersect. We have shown that a mutation in the M. smegmatis orthologue of fadD33 disrupts production of mycobactin, resulting in a molecule with an altered side chain. In the pathogenic mycobacteria, the fad genes also may have essential functions in both lipid metabolism and production of mycobactin, a necessary component of the iron acquisition process.
Acknowledgments
This work was supported in part by grants from the Intramural Research Support Program of the University of Mississippi Medical Center (to M.D.L.) and U.S. Public Health Service grant AI35235 from the National Institute of Allergy and Infectious Diseases (NIAID) (to B.R.B.).
Preliminary sequence data were obtained from TIGR through the website at http://www.tigr.org. Sequencing of M. smegmatis by TIGR was accomplished with support from NIAID. We thank J. T. Belisle for providing M. tuberculosis DNA and E. Valente for NMR spectroscopy.
REFERENCES
- 1.Arceneaux, J. E. L., W. B. Davis, D. N. Downer, A. H. Haydon, and B. R. Byers. 1973. Fate of labeled hydroxamates during iron transport from hydroxamate-iron chelates. J. Bacteriol. 115:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beggs, M. L., J. T. Crawford, and K. D. Eisenach. 1995. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J. Bacteriol. 177:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 6.Camus, J.-C., M. J. Pryor, C. Médigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis. Microbiology 148:2967-2973. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garner, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of M. tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics, a manual for genetic engineering. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.De Voss, J. J., K. Rutter, B. G. Schroeder, and C. E. Barry III. 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubnau, E., P. Fontán, R. Manganelli, S. Soares-Appel, and I. Smith. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiss, E. H., S. Y. Yu, and W. R. Jacobs. 1994. Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways. Mol. Microbiol. 14:557-569. [DOI] [PubMed] [Google Scholar]
- 14.Gobin, J., C. H. Moore, J. R. Reeve, Jr., D. K. Wong, B. W. Gibson, and M. A. Horwitz. 1995. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc. Natl. Acad. Sci. USA 92:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gobin, J., D. K. Wong, B. W. Gibson, and M. A. Horwitz. 1999. Characterization of exochelins of the Mycobacterium bovis type strain and BCG substrains. Infect. Immun. 67:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 17.Guilhot, C., I. Otal, I. V. Rompaey, C. Martin, and B. Gicquel. 1994. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 172:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, R. M., and C. Ratledge. 1982. A simple method for the production of mycobactin, the lipid-soluble siderophore, from mycobacteria. FEMS Microbiol. Lett. 15:133-136. [Google Scholar]
- 19.Hill, P., J., A. Cockayne, P. Landers, J. A. Morrissey, C. M. Sims, and P. Williams. 1998. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 66:4123-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, W. R., Jr., G. A. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 22.Lane, S. J., P. S. Marshall, R. J. Upton, and C. Ratledge. 1998. Isolation and characterization of carboxymycobactins as the second extracellular siderophores in Mycobacterium smegmatis. BioMetals 11:13-20. [Google Scholar]
- 23.Lane, S. J., P. S. Marshall, R. J. Upton, C. Ratledge, and M. Ewing. 1995. Novel extracellular mycobactins, the carboxymycobactins from Mycobacterium avium. Tetrahedron Lett. 36:4129-4142. [Google Scholar]
- 24.Levin, M. E., and G. F. Hatfull. 1993. Mycobacterium smegmatis RNA polymerase: DNA supercoiling, action of rifampicin and mechanism of rifampicin resistance. Mol. Microbiol. 8:277-285. [DOI] [PubMed] [Google Scholar]
- 25.Manabe, Y. C., B. J. Saviola, L. Sun, J. R. Murphy, and W. R. Bishai. 1999. Attenuation of virulence in Mycobacterium tuberculosis expressing a constitutively active iron repressor. Proc. Natl. Acad. Sci. USA 96:12844-12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matzanke, B. F., R. Böhnke, U. Möllmann, R. Reissbrodt, V. Schünemann, and A. X. Trautwein. 1997. Iron uptake and intracellular metal transfer in mycobacteria mediated by xenosiderophores. BioMetals 10:193-203. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. (ed.). 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Mulder, M. A., H. Zappe, and L. M. Steyn. 1997. Mycobacterial promoters. Tuber. Lung Dis. 78:211-223. [DOI] [PubMed] [Google Scholar]
- 29.Quadri, L. E. N. 2000. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol. Microbiol. 37:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Quadri, L. E. N., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 31.Ratledge, C. 1984. Metabolism of iron and other metals by mycobacteria, p. 603-627. In G. P. Kubica and L. G. Wayne (ed.), The mycobacteria: a sourcebook. Marcel Dekker, Inc., New York, N.Y.
- 32.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 33.Ratledge, C., and M. Ewing. 1996. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology 142:2207-2212. [DOI] [PubMed] [Google Scholar]
- 34.Rindi, L., L. Fattorini, D. Bonanni, E. Iona, G. Freer, D. Tan, G. Dehb, G. Orefici, and C. Garzelli. 2002. Involvement of the fadD33 gene in the growth of Mycobacterium tuberculosis in the liver of BALB/c mice. Microbiology 148:3873-3880. [DOI] [PubMed] [Google Scholar]
- 35.Rindi, L., N. Lari, and C. Garzelli. 1999. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem. Biophys. Res. Commun. 258:94-101. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schmitt, M. P., M. Predich, L. Doukhan, I. Smith, and R. K. Holmes. 1995. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun. 63:4284-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophore. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 40.Sharman, G. J., D. H. Williams, D. F. Ewing, and C. Ratledge. 1995. Determination of the structure of exochelin MN, the extracellular siderophore from Mycobacterium neoaurum. Chem. Biol. 2:553-561. [DOI] [PubMed] [Google Scholar]
- 41.Sharman, G. J., D. H. Williams, D. F. Ewing, and C. Ratledge. 1995. Isolation, purification and structure of exochelin MS, the extracellular siderophore from Mycobacterium smegmatis. Biochem. J. 305:187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigel, A., and H. Sigel (ed.). 1998. Metal ions in biological systems, vol. 35, p. 37-145. Marcel Dekker, Inc., New York, N.Y.
- 43.Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16:463-496. [DOI] [PMC free article] [PubMed]
- 44.Snow, G. A. 1970. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriol. Rev. 34:99-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao, X., N. Schiering, H.-Y. Zeng, D. Ringe, and J. R. Murphy. 1994. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 46.Timm, J., M. G. Perilli, C. Duez, J. Trias, G. Orefici, L. Fattorini, G. Amicosante, A. Oratore, B. Joris, J. M. FrPre, A. P. Pugsley, and B. Gicquel. 1994. Transcription and expression analysis, using lacZ and phoA gene fusions, of Mycobacterium fortuitum β-lactamase genes cloned from a natural isolate and a high-level β-lactamase producer. Mol. Microbiol. 12:491-504. [DOI] [PubMed] [Google Scholar]
- 47.Weinberg, E. D. 1993. The development of awareness of iron withholding defense. Perspect. Biol. Med. 36:215-221. [DOI] [PubMed] [Google Scholar]
- 48.Wheeler, P. R., and C. Ratledge. 1994. Metabolism of Mycobacterium tuberculosis, p. 353-385. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. American Society for Microbiology, Washington, D.C.
- 49.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 50.Wong, D. K., J. Gobin, M. A. Horwitz, and B. D. Gibson. 1996. Characterization of exochelin of Mycobacterium avium: evidence for saturated and unsaturated and for acid and ester forms. J. Bacteriol. 178:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, S., E. Fiss, and W. R. Jacobs, Jr. 1998. Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J. Bacteriol. 180:4676-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, W., J. E. L. Arceneaux, M. L. Beggs, B. R. Byers, K. D. Eisenach, and M. D. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthetase genes. Mol. Microbiol. 29:629-639. [DOI] [PubMed] [Google Scholar]