Abstract
In human hematopoietic malignancies, RAS mutations are frequently observed. Yet, little is known about signal transduction pathways that mediate KRAS-induced phenotypes in human CD34+ stem/progenitor cells. When cultured on bone marrow stroma, we observed that KRASG12V-transduced cord blood (CB) CD34+ cells displayed a strong proliferative advantage over control cells, which coincided with increased early cobblestone (CAFC) formation and induction of myelomonocytic differentiation. However, the KRASG12V-induced proliferative advantage was transient. By week three no progenitors remained in KRASG12V-transduced cultures and cells were all terminally differentiated into monocytes/macrophages. In line with these results, LTC-IC frequencies were strongly reduced. Both the ERK and p38 MAPK pathways, but not JNK, were activated by KRASG12V and we observed that proliferation and CAFC formation were mediated via ERK, while differentiation was predominantly mediated via p38. Interestingly, we observed that KRASG12V-induced proliferation and CAFC formation, but not differentiation, were largely mediated via secreted factors, since these phenotypes could be recapitulated by treating non-transduced cells with conditioned medium harvested from KRASG12V-transduced cultures. Multiplex cytokine arrays and genome-wide gene expression profiling were performed to gain further insight into the mechanisms by which oncogenic KRASG12V can contribute to the process of leukemic transformation. Thus, angiopoietin-like 6 (ANGPTL6) was identified as an important factor in the KRASG12V secretome that enhanced proliferation of human CB CD34+ cells.
Keywords: Differentiation, Hematopoiesis, Leukemia, Ras, Stem Cell, ANGPTL6, KRAS G12V, Angiopoietin-like 6
Introduction
RAS proteins are small GTPases that control multiple cellular functions like survival, proliferation, differentiation, and cytoskeletal rearrangement (1, 2). Activating mutations in the RAS genes, frequently in codons 12, 13, and 61 prevent the hydrolysis of RAS-GTP and result in constitutive activation of the RAS proteins (2). Three RAS genes exist, NRAS, KRAS, and HRAS, and mutations in all of these have been found in large proportion of solid tumors including cancer of the pancreas (3, 4), thymus (5), colon (6, 7), skin (8), or lung (9, 10). In myeloid leukemias, activating mutations have been found in NRAS and KRAS genes, in particular in acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), and juvenile myelomonocytic leukemia (JMML) (11–13).
To gain insight into the role of KRAS, HRAS, and NRAS proteins in development and malignant transformation many different model systems have been generated. HRas and NRas double knock-out mice displayed normal development and did not show any abnormalities during their life time. However, KRas knock-out mice die between embryonic day 12.5 and the term of gestation due to liver problems and anemia (14). In addition KRas knock-out embryos displayed increased cell death of motor neurons in the medulla and the cervical spinal cord (15). These results show that KRas gene function is essential for normal mouse development, especially for the hematopoietic and central nervous system; while NRas and HRas gene functions are dispensable (16).
Overexpression of oncogenic RAS was studied extensively as well to elucidate the role of RAS in cancer development. Overexpression of HRASG12V in human or mouse fibroblast resulted in a permanent G1 arrest, which was induced by accumulation of p53 and p16. Inactivation of either p53 or p16 prevented RAS-induced growth arrest in these cells (17). When the effect of HRASG12V was studied in erythroid cell differentiation it became clear that HRASG12V mainly blocks terminal erythroid differentiation, and it does not induce apoptosis (18). In addition, KRASG12V overexpression from its endogenous promoter in primary mouse erythroid progenitors induced a terminal differentiation block and upon Epo stimulation the downstream target signaling pathways were hyperactivated (19).
The leukemic potential of KRas, HRas, and NRas was compared using the same transplantation model. Mouse bone marrow cells were transduced with retroviral constructs and transplanted into sublethally irradiated mice. It was found that all of the three oncogenes had the potential to induce myeloid leukemias but their leukemic potential and the phenotype of the disease was different. NRas caused either a CMML- or AML-like disease in the transplanted mice while KRas-transduced BM cells initiated a CMML-like disease. Animals transplanted with HRas-transduced cells developed an AML-like disease similar to NRas, but in the case of HRas the invasiveness of the tumor was higher and the latency of the disease was shorter (20). Transgenic models have been established as well. By using the Mx1-Cre, LSL-KRasG12D mouse model it was observed that overexpression of KRasG12D induced a fatal monocytic myeloproliferative disease in the mice which was similar to CMML and JMML. When signaling pathways were investigated in oncogenic KRas-overexpressing cells, it was found that p-Stat5, p-Erk, and p-S6 levels were abnormal (21). Sabnis et al. investigated the effect of KRasG12D on murine hematopoietic stem and progenitor cells and they found that KRasG12D induced a strong proliferative advantage, increased the fraction of proliferating HSCs, and initiated T-lineage leukemia/lymphoma which was associated with secondary Notch1 mutations. They concluded that MPD-initiating activity was restricted to HSCs in KRasG12D mice and that cooperating mutations appear during cancer progression (22).
Studies to elucidate the role of RAS mutations in human hematopoietic cells have been performed less frequently. Human cord blood (CB)2 CD34+ cells transduced with NRASG13C showed increased expansion in MS5 cocultures and increased myeloid differentiation. Transplantation of NRASG13C-transduced CB CD34+ cells in NOD/SCID mice revealed an increased bone marrow engraftment and higher number of myeloid cells (23). When c-DNA microarray analysis was performed up-regulation of genes encoding cytokines and cycling-dependent kinase inhibitors p16 and p21 were found.
Here, we describe that overexpression of KRASG12V in human CD34+ CB cells induces a strong proliferative advantage coinciding with the formation of early cobblestones on bone marrow stroma. This enhanced proliferation was however transient, because after 3 weeks of culture no progenitors remained and all cells terminally differentiated along the myelomonocytic lineage. The involvement of ERK and p38 MAPK pathways in these phenotypes could be dissected, and interestingly we observed that KRASG12V-induced proliferation and CAFC formation, but not differentiation, were mediated at least in part via secreted factors.
EXPERIMENTAL PROCEDURES
Cell Cultures and Cell Lines
Neonatal CB was collected from healthy full-term pregnancies after informed consent from the obstetrics departments of the Martini Hospital Groningen and the University Medical Center Groningen (UMCG) in The Netherlands. The protocol was approved by the Medical Ethical Committee of the UMCG. Mononuclear cells were isolated using Lymphocyte Separation Medium (PAA Laboratories, Coble, Germany) and CD34+ cells were isolated using the Mini-MACS Separation System (Miltenyi Biotec, Amsterdam, The Netherlands). For MS5 cocultures and LTC-IC assays Gartner's medium was used, consisting of α-modified medium essential media (Fisher Scientific Europe, Emergo, The Netherlands) supplemented with heat-inactivated 12.5% fetal calf serum (Lonza, Leusden, The Netherlands) and heat-inactivated 12.5% horse serum (Invitrogen, Breda, The Netherlands), penicillin and streptomycin (all from PAA Laboratories), 57.2 μm β-mercaptoethanol (Merck Sharp & Dohme BV, Haarlem, The Netherlands) and 1 μm hydrocortisone (Sigma-Aldrich Chemie B.V., Zwijndrecht, The Netherlands). The MEK inhibitor U0126 was from Promega Benelux B.V. Leiden, The Netherlands, the JNK inhibitor SP600125, and the p38 inhibitor SB203580 were obtained from VWR International (Roden, The Netherlands) and were used in concentrations as indicated in the text.
Conditioned medium was collected from MS5 cocultures initiated with either MiNR1 control or KRAS-G12V-transduced CD34+ CB cells. After 1 week of coculture, medium was harvested and filtered with 0.45-mm filters (Millipore B.V., Amsterdam, The Netherlands). The filtered medium was aliquoted and stored at −80 °C. The following antagonizing antibodies were used in the conditioned medium experiments: anti-ANGPTL6 (A12/sc160957, Santa Cruz, Tebu-Bio, Heerhugowaard, The Netherlands), anti-GDF15 (PAB3883, Abnova, Heidelberg, Germany), anti-IGFBP5 (100-05, Peprotech, Tebu-Bio), anti-IL1α (kind gift from Genzyme), anti-IL8 (AB208-NA, R&D, Abindon, UK), and anti-MCP1 (300-04, Peprotech, Tebu-Bio). Concentrations used are indicated in the text.
For CFC assays CD34+ cells or suspension cells from cocultures (1–5 × 103 cells) were plated in duplicate in 35-mm tissue culture dishes containing 1 ml assay medium consisting of methylcellulose (StemCell Technologies, Grenoble, France) supplemented with IMDM (PAA Laboratories, Coble, Germany), 20 ng/ml IL-3, 20 ng/ml IL-6 (both of them from Gist-Brocades, Delft, The Netherlands), 20 ng/ml G-CSF (Rhone-Poulenc Rorer, Amstelveen, The Netherlands), 20 ng/ml c-kit ligand (Amgen, Thousand Oaks), and 6 units/ml erythropoietin (Janssen-Cilag B.V., Tilburg, The Netherlands). After 14 days of culturing colony-forming unit granulocyte-macrophage CFU-GM, burst-forming unit erythroid (BFU-E) and colony-forming unit granulocyte-erythroid-macrophage-megakaryocyte (CFU-GEMM) were scored. For LTC-IC limiting dilution assays cells were plated in the range of 5–1000 cells per well in a 96 well plate in Gartner's medium on MS5. At week five methylcellulose supplemented with the same cytokines as in the CFC assay was added to the wells. Two weeks later wells containing CFCs were scored as positive or negative and LTC-IC frequencies were calculated using ELDA (24).
Retroviral Production and Transduction
Stable PG13 MiNR1 control and PG13 KRAS-G12V retroviral producers were cultured in DMEM (Lonza, Leusden, The Netherlands) supplemented with heat-inactivated 10% fetal calf serum and penicillin/streptomycin. Viral particles for retroviral transduction were collected after 8–12 h of culturing virus producers in hematopoietic progenitor cell growth medium (HPGM) (Lonza, Leusden, The Netherlands). Right before the transduction supernatants were collected and filtered through 0.45-mm filters. 48 h before the first transduction round CB CD34+ cells were pre-stimulated in HPGM supplemented with stem cell factor (SCF; 100 ng/ml), Flt3 ligand (Flt3L; 100 ng/ml; both from Amgen, Thousand Oaks), and thrombopoietin (TPO; 100 ng/ml; Kirin, Japan). Pre-stimulated CB cells were transduced on retronectin-coated plates, (retronectin from Lucron Bioproducts B.V., Gennep, The Netherlands) in 3 consecutive rounds of 8 and 12 h with retroviral supernatant supplemented with the same cytokines and 4 μg/ml Polybrene (Sigma-Aldrich Chemie B.V., Zwijndrecht, The Netherlands). After transduction, NGFR-positive cells were sorted on a MoFlo (Dako Cytomation, Carpinteria, CA).
Flow Cytometry Analysis and Cell Sorting
Antibodies for FACS analysis and for cell sorting were the following: CD34, CD15, CD235a, CD271(NGFR) from BD Bioscience Breda, The Netherlands and CD11b, CD14 antibodies were from BioLegend Europe B.V. Uithoorn, The Netherlands. Cell sorting of transduced cells was performed on the basis of NGFR and the sort was performed on a MoFlo and FACS analysis was performed on a BD FACS Calibur.
Cytokine Multiplex Array
Cytokine levels were determined by the Human 25-Plex panel (Invitrogen, Breda, The Netherlands). The procedure was performed according to the manufacturer's recommendations using conditioned medium from day 5.
Immunoblotting and Cytospins
Cytospins were stained with May-Grunwald-Giemsa staining and images were taken with an Leica DM 3000 microscope (Leica Geosystems B.V., Wateringen, The Netherlands) using a 40 × 1.3 numeric aperture objective. For Western blot analysis we used control and KRAS G12V-transduced CB CD34+ cells, which were cultured in MS5 cocultures for 1 week. Cells were lysed in Laemmli sample buffer and loaded on 10% SDS acrylamide gel. Proteins were transferred to PVDF membranes (Millipore, Etten Leur, The Netherlands) using semidry electroblotting. KRAS antibody was obtained from Tebu Bio BV, Oosterhout The Netherlands, p-JNK, p-p38and p-ERK1/2 antibodies were from Cell Signaling Technology. Secondary antibodies were purchased from Dako Cytomation (Dako Cytomation, Glostrup, Denmark) and they were used in 1:3000 dilutions.
mRNA Analysis
The RNeasy kit (Qiagen Benelux B.V. Venlo, The Netherlands) was used for total RNA isolation. The procedure was performed according to the manufacturer's recommendations from transduced and sorted CB CD34+ cells. For genome-wide expression analysis Illumina (Illumina, Inc., San Diego, CA) BeadChip arrays (Sentrix Human-6; 46,000-probe sets) were used. Hybridization with the arrays was performed according to the manufacturer's instructions. Data were analyzed using the BeadStudio v3 gene expression module (Illumina, Inc.) and Genespring (Agilent, Amstelveen, The Netherlands).
Statistical Analysis
All values are expressed as means ± S.E. Student's t test was used for all other comparisons. Differences were considered statistically significant at p < 0.05.
RESULTS
Expression of KRASG12V in Human CB CD34+ Cells Results in a Transient Proliferative Advantage, the Formation of CAFCs and Myelomonocytic Differentiation
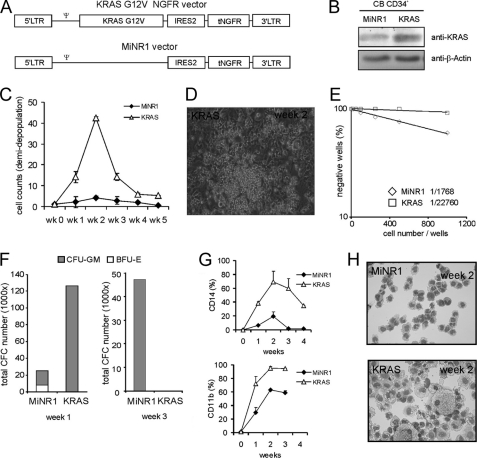
To study KRASG12V-induced phenotypes in human stem and progenitor cells we inserted the KRASG12V gene into the murine stem cell virus (MSCV) retroviral expression vector which contained an encephalomyelocarditis virus-derived internal ribosomal entry site (IRES2) in front of the truncated neural growth factor receptor (MiNR1 vector, Fig. 1A). Stable PG13 virus producer cell lines were generated and CB CD34+ cells were transduced. After transduction cells were sorted and the overexpression level of KRAS was determined by Western blot analysis. KRAS protein expression was increased ∼4-fold in the KRASG12V-transduced cells compared with control MiNR1-transduced cells (Fig. 1B). Transduced and sorted MiNR1 control and KRASG12V cells were used to initiate MS5 cocultures and expansion and differentiation were analyzed weekly. These experiments revealed that KRAS overexpression in CB CD34+ cells induced a massive proliferation within the first weeks but after the second week KRASG12V-transduced cells stopped proliferating. The strong initial expansion in the KRASG12V-transduced cultures coincided with the formation of early cobblestones with high frequency within 5 days after plating (Fig. 1D).
FIGURE 1.
KRASG12V induces a transient growth advantage, early CAFC, and enhanced monocyte/macrophage differentiation. A, schematic representation of the retroviral vectors used in this study. B, cord blood (CB) CD34+cells were transduced with MiNR1 or KRASG12V vectors, cells were harvested after 48 h and KRAS expression levels were measured by Western blotting. C, CB CD34+ cells were transduced with MiNR1 or KRASG12V vectors and cells were cultured in long-term MS5 co-cultures. Data shows cell counts of weekly demidepopulation. The average of three independent experiments is shown. D, to determine stem cell frequencies LTC-IC assays in limiting dilution were performed on MiNR1 and KRASG12V-transduced CB CD34+ cells. E, cobblestone area forming cells (CAFCs) induced by KRASG12V in MS5 cocultures at week 2. F, from each culture at week 1 and week 3, 2000 cells were plated in CFC assays in methylcellulose, colonies were evaluated after 2 weeks and total CFC numbers were calculated based on the cell number in the cocultures. G, FACS analysis on suspension cells from cocultures indicated monocytic differentiation in KRASG12V cocultures. H, MGG-stained cytospins from week 2.
To evaluate the changes in stem cell frequencies induced by KRASG12V, LTC-IC assays were performed in limiting dilution. These experiments indicated that LTC-IC frequencies decreased dramatically upon KRASG12V overexpression (1/1768 in MiNR1 versus 1/22760 in KRASG12V, Fig. 1E). CFC assays revealed that progenitor frequencies were increased 6-fold in KRAS-transduced cells within 1 week after plating on MS5, but this increase was transient. Progenitors were exhausted by week 3 since no CFCs were observed in methylcellulose assays. Also, less BFU-Es were observed in the KRASG12V-transduced cultures compared with MiNR1 controls at week 1 (Fig. 1F). To determine the differentiation profile in the cocultures cell surface markers were measured by FACS analysis. These experiments showed increased CD14 and CD11b expression in the KRASG12V-transduced cultures compared with MiNR1 controls (Fig. 1G). Cytospins from suspension cells of the cocultures confirmed these data and indicated that KRASG12V-transduced cells were all terminally differentiated along the myelomonocytic lineage by week 3 (Fig. 1H).
Inhibition of ERK Impairs Proliferation While Inhibition of p38 Impairs Proliferation and Differentiation in CD34+CB Cells Transduced with KRASG12V
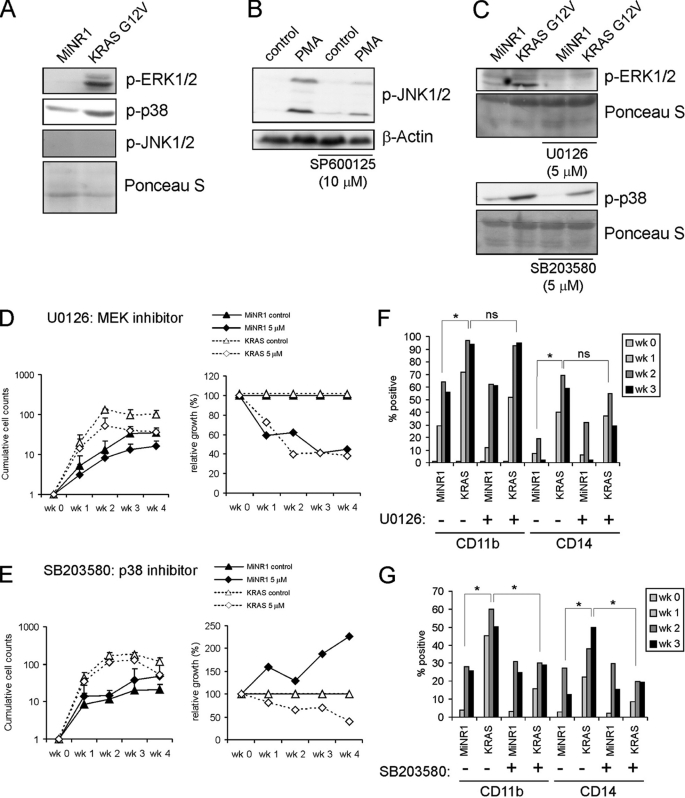
To determine which signaling pathways could be mediating KRASG12V-induced phenotypes we first determined which MAPK signal transduction pathways were activated downstream of KRASG12V by Western blot analysis. These experiments revealed that both the ERK/MAPK and p38/MAPK pathways were activated by KRASG12V in CB CD34+ cells resulting in enhanced phosphorylation levels of ERK1/2 and p38 (Fig. 2A). JNK was not activated by KRASG12V, while PMA did induce phosphorylation of JNK (Fig. 2, A and B). Next, specific inhibitors against MEK (U0126), p38 (SB203580) or JNK (SP600125) were used in order to dissect the involvement of MAPK pathways in KRASG12V-induced phenotypes. Western blot analysis confirmed that the inhibitors effectively downmodulated the activity of the designated pathways (Fig. 2, B and C). To determine the changes in growth, cobblestone formation and differentiation CB CD34+ cells were transduced with either control MiNR1 or KRASG12V vectors. Sorted cells were used to initiate MS5 cocultures which were analyzed for 5 weeks. Inhibition of MEK by U0126 reduced proliferation, both of KRASG12V as well as MiNR1-transduced cells (Fig. 2D). Inhibition of p38 using SB203580 also impaired the expansion of KRASG12V-transduced cells, but to a lesser extent compared with cells treated with the MEK inhibitor (Fig. 2E). Surprisingly, MiNR1 control cells treated with the p38 inhibitor proliferated slightly better compared with controls (Fig. 2E). As expected, no effects were observed on the growth of either MiNR1 control cells or KRASG12V cells treated with the JNK inhibitor SP600125 (data not shown). FACS analysis was used to determine the effects of ERK and p38 on KRASG12V-induced differentiation. As observed earlier, KRASG12V enhanced the differentiation toward CD11b and CD14-positive myelomonocytic cells, which was not affected by inhibition of MEK (Fig. 2F). In contrast, inhibition of p38 by using the inhibitor SB203580 did significantly impair KRASG12V-induced myelomonocytic differentiation (Fig. 2G).
FIGURE 2.
Inhibition of ERK impairs proliferation while inhibition of p38 impairs proliferation and differentiation in CB CD34+ cells transduced with KRASG12V. A, Western blots of MiNR1 and KRASG12V-transduced CB CD34+ cells. B, UT7 cells were treated with PMA for 15 min, and cells were pretreated with the JNK inhibitor SP600125 as indicated after which lysates were prepared for Western blotting. C, Western blots on transduced cells as in A, but now cells were pretreated with the MEK inhibitor U0126 or the p38 inhibitor (SB203580) as indicated. D, proliferation of MiNR1 or KRASG12V cells in MS5 coculture in the presence of MEK inhibitor (U0126). Cultures were demi-depopulated weekly and cumulative cell counts are shown. In the right panels, the relative effects of the inhibitor are shown whereby the untreated MiNR1 and KRASG12V cell counts were normalized to 100% every week. E, As in D, but now MS5 cocultures were performed in the presence of p38 inhibitor (SB203580). F, FACS analysis of cultures described in E. G, FACS analysis of cultures described in E.
Conditioned Medium from KRASG12V-transduced Cells Is Sufficient to Induce Proliferation and Formation of Early CAFCs in CB CD34+ Cells
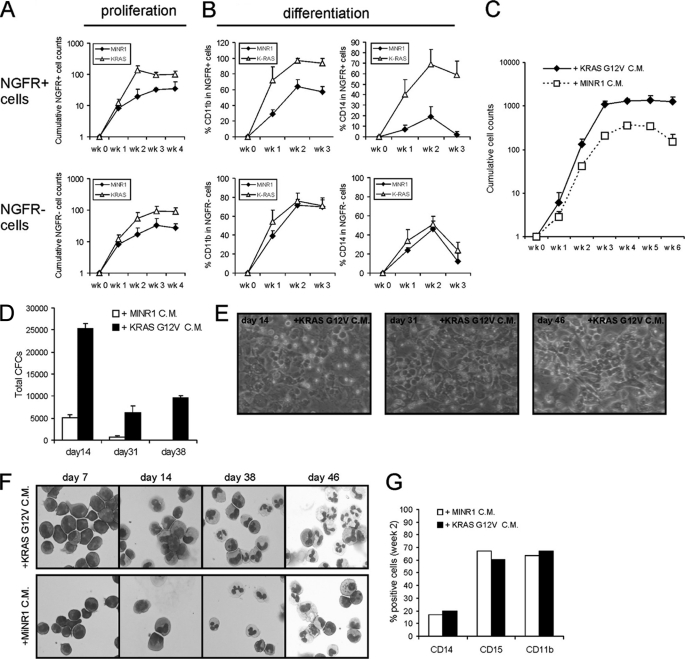
To our surprise, when MS5 cocultures were initiated with non-sorted MiNR1 control and KRASG12V transduced CB CD34+ cells, we observed that the non-transduced cells in KRASG12V-initiated cocultures displayed a proliferative advantage as compared with non-transduced cells in MiNR1-initiated control cocultures. A representative experiment is shown in Fig. 3A, using MiNR1 and KRASG12V-transduced CB cells with initial transduction efficiencies of 52 and 25%, respectively. Cocultures were weekly demi-depopulated, expansion and differentiation of NGFR+ and NGFR− cells were monitored, and cumulative expansion data is shown in Fig. 3A. NGFR+ KRASG12V-transduced cells displayed a strong proliferative advantage over MiNR1 as observed previously. Unexpectedly, the NGFR− cells in KRASG12V-transduced cocultures also displayed a proliferative advantage over NGFR− cells from MiNR1 control cocultures with almost similar kinetics as NGF+ KRASG12V-transduced cells (Fig. 3A, lower panel). In contrast, the myelomonocytic differentiation induced by KRASG12V was only observed in transduced cells, and not in non-transduced NGFR− cells in these cocultures.
FIGURE 3.
Conditioned medium from KRASG12V-transduced cells induces a proliferative advantage and the formation of CAFCs but not myelomonocytic differentiation in freshly isolated CB CD34+ cells. A, MS5 cocultures were initiated with non-sorted, MiNR1, and KRASG12V-transduced CB CD34+ cells. The cumulative expansion of NGFR+ as well as NGFR− populations is shown. B, experiment as in A, but now cultures were analyzed by FACS. C, conditioned medium was collected from MS5 cocultures initiated with MiNR1 and KRASG12V-transduced CB CD34+ cells. Freshly isolated CB CD34+ cells were cultured on MS5 in the presence of MiNR1 and KRASG12V CM as indicated. Cumulative cell counts are shown. D, as in C, but now suspension cells were analyzed for progenitors in CFC assays in methylcellulose. E, representative images of CAFCs of cultures described in C. F, representative cytospins stained with MGG from suspension cells of cultures described in C. G, FACS data of cultures described in C.
To further substantiate these findings, conditioned medium (CM) was harvested from KRASG12V and MiNR1-transduced CB CD34+ cells, which was then used in MS5 cocultures initiated with freshly isolated non-transduced CB CD34+ cells. As shown in Fig. 3C, the KRASG12V CM was sufficient to induce a robust proliferative advantage compared with cells treated with MiNR1 CM. The observed proliferative advantage was also much less transient as compared with directly transduced KRASG12V-transduced CB CD34+ cells. The proliferative advantage coincided with enhanced progenitor frequencies as determined by CFC assays, which persisted for over 6 weeks of coculture (Fig. 3D). Also, CAFCs were formed within 2 weeks of coculture of CB CD34+ cells in the presence of KRASG12V CM, which persisted throughout the culture period (Fig. 3E). In line with what was observed in the non-sorted MS5 coculture experiments, no pronounced acceleration of myelomonocytic differentiation was observed in cultures treated with KRASG12V CM and morphological analysis (Fig. 3F) as well as analysis by FACS (Fig. 3G) revealed normal myeloid differentiation. Thus, these data indicate that a KRASG12V-induced secreted factor strongly contributes to expansion and CAFC phenotypes, while the induction of myelomonocytic differentiation is induced via intrinsic pathways.
Identification of KRASG12V Target Genes in Human CB CD34+ Cells
To identify KRASG12V target genes a genome-wide gene expression profiling study was performed using Illumina BeadChip arrays. CB CD34+ cells were transduced with KRASG12V and MiNR1 control vectors and transduced cells were sorted. RNA was isolated from each sorted population after 24 h, and the RNA was used for hybridization with Illumina Bead Chip arrays. 2258 differentially expressed probesets (>2-fold) upon KRASG12V overexpression were identified, of which 1167 probesets were up-regulated and 1091 were down-regulated (supplemental Table S1). Gene ontology (GO) analysis revealed that the KRASG12V-up-regulated gene list was enriched for genes associating with differentiation, membrane fraction, adhesion, apoptosis, and regulation of the cell cycle (Table 1). The list of up-regulated genes involved in cell cycle regulation included CyclinD1, CyclinD3, p19, and p21. Furthermore, various transcriptional regulators, including CITED2, FOSB, HOXB2, HOXB5, GSK3, JUN, ID2, MAFA, MAFB, MEIS3, and TRIB1 were up-regulated by KRASG12V. Whereas genes associating with myelomonocytic differentiation such as CD14 were also up-regulated, no effects of RASG12V were observed on the expression of GM-CSFR, CSFR, CEBPα, or PU.1. The list of down-regulated genes upon overexpression of KRASG12V was enriched for transcriptional regulators as determined by GO analysis (data not shown), and included HOXA5, HOXA9, NMYC, LEF1, as well as many erythroid-associated genes (supplemental Table S1).
TABLE 1.
GO term annotations of KRAS G12V-upregulated genes in CB CD34+ cells
| GO term | p value | FDR |
|---|---|---|
| GO:0030154∼cell differentiation | 1.96E-08 | 3.75E-05 |
| GO:0005624∼membrane fraction | 1.16E-06 | 1.80E-03 |
| hsa04510:Focal adhesion | 1.48E-05 | 1.85E-02 |
| GO:0006915∼apoptosis | 1.22E-05 | 2.33E-02 |
| GO:0008219∼cell death | 1.29E-05 | 2.47E-02 |
| GO:0048468∼cell development | 1.60E-05 | 3.06E-02 |
| GO:0012501∼programmed cell death | 1.64E-05 | 3.14E-02 |
| GO:0000074∼regulation of progression through cell cycle | 8.47E-05 | 1.62E-01 |
| GO:0051726∼regulation of cell cycle | 9.68E-05 | 1.85E-01 |
| IPR014393:Dual specificity protein phosphatase (MAP kinase phosphatase) | 1.18E-04 | 2.27E-01 |
| GO:0030099∼myeloid cell differentiation | 2.96E-04 | 5.65E-01 |
| GO:0008283∼cell proliferation | 3.75E-04 | 7.16E-01 |
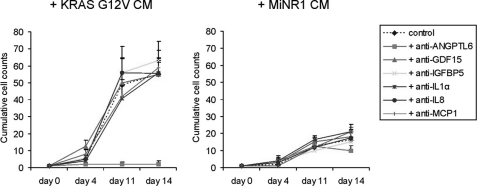
We specifically focused on KRASG12V-up-regulated genes encoding secreted factors. 74 genes were identified that associated with the GO term “secreted”, and a number of these genes is shown in Table 2. Among others, this list included chemokine/cytokine members such as ANGPTL6, CCL2/MCP1, CCL5, IL8, IGFBP5, TNF, and VEGFA. A number of these genes have been previously identified as targets of MEK activation as well (25). To further verify whether up-regulation KRASG12V target genes encoding chemokines/cytokines indeed resulted in actual secretion of these factors, a 25-plex cytokine array analysis was performed on CM from by KRASG12V and MiNR1-transduced CB CD34+ cells. We were able to confirm that the KRASG12V secretome included IL8, IL12p40, CCL2,-3,-4, and 5, and IL1RA. These data are summarized in Table 3. Next, we wished to test whether inhibiting these factors using antagonizing antibodies would be sufficient to impair the enhanced growth and CAFC formation imposed on CB CD34+ cells exposed to KRASG12V-conditioned medium. Polyclonal antibodies against ANGPTL6, GDF15, IGFBP5, IL1a, IL8, and CCL2/MCP1 could be obtained and tested. As shown in Fig. 4, of these tested secreted factors particularly inhibition of ANGPTL6 strongly impaired the enhanced proliferation imposed on CB CD34+ cells by the KRASG12V-conditioned medium. Also the formation of CAFCs upon exposure to KRASG12V-conditioned medium was severely impaired (data not shown). We also determined whether stimulation with only ANGPTL6 would be sufficient to enhance proliferation and CAFC formation, but we only observed a modest increase in expansion of CB CD34+ cells on MS5 cocultures in the presence of up to 4 μg/ml ANGPTL6 (data not shown). Thus, our data argue that ANGPTL6 is required as an essential component of the KRASG12V secretome, but as a single growth factor ANGPTL6 might not be sufficient to impose long-term growth on human stem/progenitor cells.
TABLE 2.
Subset of genes up-regulated by KRASG12D in human CB CD34+ cells associated with the GO term “secreted”
| Gene name | Definition | Fold change |
|---|---|---|
| ANGPTL6 | Angiopoietin-like 6 | 5.1 |
| CCL2/MCP1 | Chemokine (C-C Motif) Ligand 2 | 67.1 |
| CCL23 | Chemokine (C-C Motif) Ligand 23 | 2.3 |
| CCL3L3 | Chemokine (C-C Motif) Ligand 3-like 1 | 4.6 |
| CCL5 | Chemokine (C-C Motif) Ligand 5 | 4.4 |
| COL7A1 | Collagen, Type VII, Alpha 1 | 428.0 |
| COL9A2 | Collagen, Type IX, Alpha 2 | 2.7 |
| ECM1 | Extracellular Matrix Protein 1 | 2.4 |
| ENPP3 | Ectonucleotide Pyrophosphatase/Phosphodiesterase 3 | 2.3 |
| GDF15 | Growth Differentiation Factor 15 | 8.4 |
| IFNA10 | Interferon, Alpha 10 | 2.1 |
| IGFBP5 | Insulin-like Growth Factor-binding Protein 5 | 348.0 |
| IL1RL1 | Interleukin 1 Receptor-like 1 | 2.6 |
| IL8 | Interleukin 8 | 13.2 |
| TNF | Tumor Necrosis Factor (TNF Superfamily, Member 2) | 4.7 |
| VEGFA | Vascular Endothelial Growth Factor | 3.3 |
TABLE 3.
KRAS G12V-induced cytokines and growth factors in human CB CD34+ cells (25-plex multiarray)
| Symbol | MiNR1 | KRAS |
|---|---|---|
| pg/ml | pg/ml | |
| IL-1B | 1,00 | 6,51 |
| IL-1RA | 27,93 | 3757,36 |
| IL-8 | 1,50 | 10010,62 |
| IL-12p40 | 2,00 | 199,83 |
| MIP-1a (CCL3) | 4,00 | 27,24 |
| MIP-1b (CCL4) | 0,40 | 88,59 |
| IP-10 (CXCL10) | 0,00 | 60,71 |
| RANTES (CCL5) | 2,00 | 35,04 |
| MCP-1 (CCL2) | 19,41 | 2145,37 |
FIGURE 4.
The proliferative advantage imposed on CB CD34+ cells by KRASG12V-conditioned medium is impaired by anti-ANGPTL6. Conditioned medium was collected from MS5 cocultures initiated with MiNR1 and KRASG12V-transduced CB CD34+ cells. Freshly isolated CB CD34+ cells were cultured on MS5 in the presence of MiNR1 and KRASG12V CM as indicated. Furthermore, antibodies against Angiopoietin-like 6 (2 μg/ml), GDF15 (2 μg/ml), IGFBP5 (5 μg/ml), IL1α (5 μg/ml), IL8 (2 μg/ml), or MCP1 (5 μg/ml) were added. As controls, cells were left untreated. Cumulative cell counts of a representative experiment are shown.
DISCUSSION
The effects of KRAS mutations on hematopoiesis have been studied extensively in murine model systems, and the effects on proliferation and myeloid commitment have been analyzed in substantial detail. Yet, KRAS-induced phenotypes in the human system have been poorly described, and little insight has been obtained regarding the molecular mechanisms that are involved. Here, we describe that retroviral introduction of KRASG12V in human CB CD34+ resulted in increased proliferation coinciding with the formation of early cobblestone areas underneath the stroma, followed by terminal differentiation along the myelomonocytic lineage. Importantly, we identified the ERK/MAPK pathway as an extrinsic mediator of KRASG12V-induced phenotypes, whereby a growth advantage as well as the capacity to form CAFCs underneath bone marrow stromal cells could be imposed on both transduced cells as well as on non-transduced cells in a paracrine manner. In particular, ANGPTL6 was identified as an important factor in the KRASG12V secretome that mediated enhanced proliferation of human CB CD34+ cells. KRASG12V-induced myelomonocytic differentiation was predominantly regulated via intrinsic pathways mediated via p38/MAPK.
RAS-induced activation of the ERK/MAPK pathway has been linked to enhanced cell cycle progression in various tissues. Among others, this has been associated with an up-regulation of cell cycle genes such as CyclinD1,-2, and -3 (25–27). Indeed, we also observed an up-regulation of these positive cell cycle regulators in KRASG12V-transduced CB CD34+ cells. However, besides these direct effects of the RAS-ERK pathway on the regulation of cell cycle progression, our data now indicate that secreted factors that mediate KRASG12V-induced proliferation in an autocrine/paracrine manner must exist as well. In untransduced cells in non-sorted cocultures, as well as in freshly isolated CB CD34+ cells that were treated with conditioned medium (CM) harvested from KRASG12V-transduced cells, we observed a striking proliferative advantage over cells that had not been cultured in the presence of KRASG12V-expressing cells or CM. Not only the proliferation was enhanced by CM, also the formation of CAFCs was dramatically increased. Experiments whereby KRASG12V-transduced cells were grown in cytokine-driven cultures revealed that the proliferative advantage of KRASG12V-expressing cells compared with controls was not as dominant as observed in MS5 bone marrow stromal cocultures (data not shown). Thus, these data suggest that the formation of CAFCs plays an important role in the autocrine/paracrine proliferative advantage induced by KRASG12V. Genome-wide gene expression profiling allowed the identification of various chemokines, cytokines and growth factors as target genes of KRASG12V, and a number of these were indeed confirmed as being secreted by KRASG12V-transduced cells as determined by 25-plex cytokine arrays. Some of the most pronounced up-regulated factors were CCL2, IGFBP5, ANGPTL6, GDF15, and IL8. In line with our observations, it was recently reported that expression of KRASG12V in various human cell lines up-regulated a number of growth factors and chemokines, including VEGF, IL6, CXCL1, and CXCL8 (IL8) (28). Up-regulation of IL8 and a number of these chemokines by activation of the RAS/ERK pathway was reported by others as well (25, 29). Importantly, deletion of CXCR2, the common receptor for chemokines such as IL8 reduced oncogenic RAS-driven tumorigenesis in mice, indicating that RAS-induced secretion of these factors indeed participates in the process of transformation (28). Nevertheless, we observed that addition of anti-IL8 did not reduce the enhanced proliferation imposed on CB CD34+ by treatment with KRASG12V conditioned medium. Reversely, the addition of IL8 to freshly isolated CB CD34+ cells growth on MS5 was also not sufficient to enhance proliferation or induce CAFC formation. In contrast, the addition of antibodies against angiopoietin-like 6 (ANGPTL6) was sufficient to severely impair KRASG12V CM-induced growth and CAFC formation of CB CD34+ cells. Although little has been revealed about the function of ANGPTL6 in hematopoietic cells, it has been shown that ANGPTL6 is involved in angiogenesis and lipid, glucose, and energy metabolism (30, 31). Also, ANGPTL6 might play a role in wound healing, and might mediate adhesion by interacting with integrin receptors (32). No information is currently available on the effects of ANGTPL6 on hematopoietic stem cells, but it is intriguing that other angiopoietin-like family members have been reported to expand mouse as well as human long-term hematopoietic stem cells (33, 34). While we were able to show that ANGPTL6 is an important component of the KRASG12V secretome, stimulation of CB CD34+ cells with only ANGPTL6 induced a rather modest increase in expansion, suggesting that most likely a multitude of the KRASG12V-secreted factors act in collaboration. Regardless, it remains remarkable that cells that do not express an oncogene are strongly affected via autocrine signaling provided by neighboring cells that express KRASG12V. In this respect it is interesting to note that FOSB was also up-regulated by KRASG12V in CB CD34+ cells. Recently, a number of genes were identified in a stem cell activity screen that could enhance repopulation activity in a non-cell-autonomous manner, and one of these was FOS (35). It will be interesting to determine whether FOS also mediates the expression of KRASG12V-induced chemokines and growth factors.
KRASG12V-induced myelomonocytic differentiation was predominantly mediated via p38/MAPK, and not via the ERK/MAPK pathway. Since our experiments using KRASG12V conditioned medium revealed that only proliferation but not myelomonocytic differentiation was affected, we conclude that the p38/MAPK-mediated differentiation in KRASG12V cells involves activation of intrinsic pathways. The GO term myeloid cell differentiation was significantly enriched in the list of KRASG12V up-regulated genes, and it is possible that a number of these genes are activated via the p38/MAPK pathway. In line with previous observations (19, 36), we find that activation of RAS impairs erythropoiesis, and the reduction of BFU-Es in KRASG12V-transduced cells coincided with a reduction in expression of erythroid genes. The JNK pathway does not appear to play a major role in the observed phenotypes since we did not observe phosphorylation of JNK in KRASG12V-transduced cells and treatment with the JNK inhibitor SP600125 neither affected proliferation or differentiation. Recently, it was reported that HRASG12V can also induce phosphorylation of p38 (26). This induction of p38 activity was attributed to HRASG12V up-regulation of reactive oxygen species (ROS) as treatment with the antioxidant diphenyleneiodonium impaired phosphorylation of p38. Furthermore, the induction of ROS associated with enhanced proliferative capacity of HRASG12V-transduced cells and the increase in proliferation could be counteracted by treatment with antioxidants (26). Whether differentiation of HRASG12V-transduced cells was also affected by treatment with antioxidants is currently unclear. Remarkably, while inhibition of p38 impaired the differentiation, and also to some extent the proliferation of KRASG12V-transduced cells in our studies, we observed that MiNR1 controls cells produced significantly more progeny in bone marrow cocultures. It was reported that ROS-induced p38 activity resulted in stem cell exhaustion and that inactivation of p38 extends the lifespan of HSCs in serial transplantation assays (37). Possibly, immature HSC/MPPs were better maintained in our MiNR1 cultures treated with SB203580 resulting in the production of more progeny. We do not know whether KRASG12V also up-regulates ROS levels in CB CD34+ cells as was observed in HRASG12V-transduced cells where it was associated with enhanced proliferation (26), but it is well possible that the production of ROS induced by RAS initially results in enhanced proliferation, but that ultimately hyperactivation of p38 in KRASG12V-transduced cells induces exhaustion of the stem cell pool as we indeed observed in our studies.
While RAS mutations are frequently observed in human leukemias, it is remarkable that we do not find long-term stem cell self-renewal or transformation phenotypes in our human CB CD34+ model systems. In line with our observations, it was reported that expression of NRASG13C enhanced engraftment in NOD-SCID mice, but could not induce leukemia (23). Also in mouse models, KRAS mutations were able to generate a myeloproliferative disease, but not leukemia (21, 38, 39). Thus, oncogenic RAS might require additional mutations in order to induce overt leukemia, and indeed in a number of model systems it was shown that RAS mutations can effectively cooperate with additional hits (22, 40, 41).
In summary, our data demonstrate that overexpression of KRASG12V in CB CD34+ cells enhances proliferation transiently, increases early cobblestone formation, reduces LTC-IC frequencies and initiates monocytic differentiation. We observed that KRASG12V activates both ERK and p38MAPK pathways but not JNK in CB CD34+ cells. Activation of the ERK pathway correlated with proliferation which was mediated at least in part via secreted factors that can act in an autocrine/paracrine manner, and ANGPTL6 was identified as an important component of the KRASG12V secretome.
Supplementary Material
Acknowledgments
We thank Dr. J. J. Erwich and Dr. A. van Loon and colleagues (Obstetrics Departments of University Medical Centre in Groningen, Martini Hospital Groningen) for collecting cord blood. We would like to acknowledge Johan Bijzet (Dept. of Rheumatology, University Medical Centre in Groningen) for help with the 25-plex cytokine arrays.
This work was supported by grants from the NWO-VENI (2004), NWO-VIDI (2008), and KWF (RUG 2009-4275).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- CB
- cord blood
- FACS
- fluorescent-activated cell sorting
- GO
- gene ontology
- CM
- conditioned medium.
REFERENCES
- 1. Boguski M. S., McCormick F. (1993) Nature 366, 643–654 [DOI] [PubMed] [Google Scholar]
- 2. Bourne H. R., Sanders D. A., McCormick F. (1990) Nature 348, 125–132 [DOI] [PubMed] [Google Scholar]
- 3. Almoguera C., Shibata D., Forrester K., Martin J., Arnheim N., Perucho M. (1988) Cell 53, 549–554 [DOI] [PubMed] [Google Scholar]
- 4. Hruban R. H., van Mansfeld A. D., Offerhaus G. J., van Weering D. H., Allison D. C., Goodman S. N., Kensler T. W., Bose K. K., Cameron J. L., Bos J. L. (1993) Am. J. Pathol. 143, 545–554 [PMC free article] [PubMed] [Google Scholar]
- 5. Lemoine N. R., Mayall E. S., Wyllie F. S., Farr C. J., Hughes D., Padua R. A., Thurston V., Williams E. D., Wynford-Thomas D. (1988) Cancer Res. 48, 4459–4463 [PubMed] [Google Scholar]
- 6. Bos J. L., Fearon E. R., Hamilton S. R., Verlaan-de Vries M., van Boom J. H., van der Eb A. J., Vogelstein B. (1987) Nature 327, 293–297 [DOI] [PubMed] [Google Scholar]
- 7. Breivik J., Meling G. I., Spurkland A., Rognum T. O., Gaudernack G. (1994) Br. J. Cancer 69, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pierceall W. E., Goldberg L. H., Tainsky M. A., Mukhopadhyay T., Ananthaswamy H. N. (1991) Mol. Carcinog. 4, 196–202 [DOI] [PubMed] [Google Scholar]
- 9. Li S., Rosell R., Urban A., Font A., Ariza A., Armengol P., Abad A., Navas J. J., Monzo M. (1994) Lung Cancer 11, 19–27 [DOI] [PubMed] [Google Scholar]
- 10. Mills N. E., Fishman C. L., Rom W. N., Dubin N., Jacobson D. R. (1995) Cancer Res. 55, 1444–1447 [PubMed] [Google Scholar]
- 11. Bos J. L., Verlaan-de Vries M., van der Eb A. J., Janssen J. W., Delwel R., Löwenberg B., Colly L. P. (1987) Blood 69, 1237–1241 [PubMed] [Google Scholar]
- 12. Kalra R., Paderanga D. C., Olson K., Shannon K. M. (1994) Blood 84, 3435–3439 [PubMed] [Google Scholar]
- 13. Onida F., Kantarjian H. M., Smith T. L., Ball G., Keating M. J., Estey E. H., Glassman A. B., Albitar M., Kwari M. I., Beran M. (2002) Blood 99, 840–849 [DOI] [PubMed] [Google Scholar]
- 14. Johnson L., Greenbaum D., Cichowski K., Mercer K., Murphy E., Schmitt E., Bronson R. T., Umanoff H., Edelmann W., Kucherlapati R., Jacks T. (1997) Genes Dev. 11, 2468–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koera K., Nakamura K., Nakao K., Miyoshi J., Toyoshima K., Hatta T., Otani H., Aiba A., Katsuki M. (1997) Oncogene 15, 1151–1159 [DOI] [PubMed] [Google Scholar]
- 16. Esteban L. M., Vicario-Abejón C., Fernández-Salguero P., Fernández-Medarde A., Swaminathan N., Yienger K., Lopez E., Malumbres M., McKay R., Ward J. M., Pellicer A., Santos E. (2001) Mol. Cell Biol. 21, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 18. Zhang J., Socolovsky M., Gross A. W., Lodish H. F. (2003) Blood 102, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J., Liu Y., Beard C., Tuveson D. A., Jaenisch R., Jacks T. E., Lodish H. F. (2007) Blood 109, 5238–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parikh C., Subrahmanyam R., Ren R. (2007) Cancer Research 67, 7139–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Meter M. E., Díaz-Flores E., Archard J. A., Passegué E., Irish J. M., Kotecha N., Nolan G. P., Shannon K., Braun B. S. (2007) Blood 109, 3945–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sabnis A. J., Cheung L. S., Dail M., Kang H. C., Santaguida M., Hermiston M. L., Passegué E., Shannon K., Braun B. S. (2009) PLoS. Biol. 7, e59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen S. W., Dolnikov A., Passioura T., Millington M., Wotherspoon S., Rice A., MacKenzie K. L., Symonds G. (2004) Exp. Hematol. 32, 852–860 [DOI] [PubMed] [Google Scholar]
- 24. Hu Y., Smyth G. K. (2009) J. Immunol. Methods 347, 70–78 [DOI] [PubMed] [Google Scholar]
- 25. Geest C. R., Buitenhuis M., Groot, Koerkamp M. J., Holstege F. C., Vellenga E., Coffer P. J. (2009) Blood 114, 3402–3412 [DOI] [PubMed] [Google Scholar]
- 26. Hole P. S., Pearn L., Tonks A. J., James P. E., Burnett A. K., Darley R. L., Tonks A. (2010) Blood 115, 1238–1246 [DOI] [PubMed] [Google Scholar]
- 27. Shen S., Passioura T., Symonds G., Dolnikov A. (2007) Exp. Hematol. 35, 908–919 [DOI] [PubMed] [Google Scholar]
- 28. O'Hayer K. M., Brady D. C., Counter C. M. (2009) Carcinogenesis 30, 1841–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monticone M., Biollo E., Maffei M., Donadini A., Romeo F., Storlazzi C. T., Giaretti W., Castagnola P. (2008) Mol. Cancer 7, 92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hato T., Tabata M., Oike Y. (2008) Trends Cardiovasc. Med. 18, 6–14 [DOI] [PubMed] [Google Scholar]
- 31. Oike Y., Akao M., Yasunaga K., Yamauchi T., Morisada T., Ito Y., Urano T., Kimura Y., Kubota Y., Maekawa H., Miyamoto T., Miyata K., Matsumoto S., Sakai J., Nakagata N., Takeya M., Koseki H., Ogawa Y., Kadowaki T., Suda T. (2005) Nat. Med. 11, 400–408 [DOI] [PubMed] [Google Scholar]
- 32. Oike Y., Yasunaga K., Ito Y., Matsumoto S., Maekawa H., Morisada T., Arai F., Nakagata N., Takeya M., Masuho Y., Suda T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9494–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang C. C., Kaba M., Iizuka S., Huynh H., Lodish H. F. (2008) Blood 111, 3415–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang C. C., Kaba M., Ge G., Xie K., Tong W., Hug C., Lodish H. F. (2006) Nat. Med. 12, 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deneault E., Cellot S., Faubert A., Laverdure J. P., Fréchette M., Chagraoui J., Mayotte N., Sauvageau M., Ting S. B., Sauvageau G. (2009) Cell 137, 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darley R. L., Hoy T. G., Baines P., Padua R. A., Burnett A. K. (1997) J. Exp. Med. 185, 1337–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y., Suda T. (2006) Nat. Med. 12, 446–451 [DOI] [PubMed] [Google Scholar]
- 38. Braun B. S., Tuveson D. A., Kong N., Le D. T., Kogan S. C., Rozmus J., Le Beau M. M., Jacks T. E., Shannon K. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan I. T., Kutok J. L., Williams I. R., Cohen S., Kelly L., Shigematsu H., Johnson L., Akashi K., Tuveson D. A., Jacks T., Gilliland D. G. (2004) J. Clin. Invest. 113, 528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan I. T., Kutok J. L., Williams I. R., Cohen S., Moore S., Shigematsu H., Ley T. J., Akashi K., Le Beau M. M., Gilliland D. G. (2006) Blood 108, 1708–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J., Wang J., Liu Y., Sidik H., Young K. H., Lodish H. F., Fleming M. D. (2009) Blood 113, 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.