Abstract
Phosphoinositide-dependent kinase 1 (PDK1) is a critical activator of multiple prosurvival and oncogenic protein kinases and has garnered considerable interest as an oncology drug target. Despite progress characterizing PDK1 as a therapeutic target, pharmacological support is lacking due to the prevalence of nonspecific inhibitors. Here, we benchmark literature and newly developed inhibitors and conduct parallel genetic and pharmacological queries into PDK1 function in cancer cells. Through kinase selectivity profiling and x-ray crystallographic studies, we identify an exquisitely selective PDK1 inhibitor (compound 7) that uniquely binds to the inactive kinase conformation (DFG-out). In contrast to compounds 1–5, which are classical ATP-competitive kinase inhibitors (DFG-in), compound 7 specifically inhibits cellular PDK1 T-loop phosphorylation (Ser-241), supporting its unique binding mode. Interfering with PDK1 activity has minimal antiproliferative effect on cells growing as plastic-attached monolayer cultures (i.e. standard tissue culture conditions) despite reduced phosphorylation of AKT, RSK, and S6RP. However, selective PDK1 inhibition impairs anchorage-independent growth, invasion, and cancer cell migration. Compound 7 inhibits colony formation in a subset of cancer cell lines (four of 10) and primary xenograft tumor lines (nine of 57). RNAi-mediated knockdown corroborates the PDK1 dependence in cell lines and identifies candidate biomarkers of drug response. In summary, our profiling studies define a uniquely selective and cell-potent PDK1 inhibitor, and the convergence of genetic and pharmacological phenotypes supports a role of PDK1 in tumorigenesis in the context of three-dimensional in vitro culture systems.
Keywords: AKT PKB, Allosteric Regulation, Cancer Therapy, Enzyme Inhibitors, Phosphatidylinositol-dependent Kinase-1 (PDK1), Phosphatidylinositol 3-Kinase, Protein-Drug Interactions, Protein Kinases, X-ray Crystallography, Biomarker
Introduction
PDK1 (phosphoinositide-dependent kinase-1) was first identified as a protein serine/threonine kinase that linked phosphatidylinositol 3-kinase (PI3K) to AKT (protein kinase B) activation in response to growth factor receptor signaling (1, 2). Growth factor binding to receptor tyrosine kinases (RTKs)3 results in activated PI3K, which phosphorylates the 3′-position of the inositol ring in phosphatidylinositol 4,5-bisphosphate to produce the second messenger phosphatidylinositol 3,4,5-trisphosphate (3). Membrane-bound phosphatidylinositol 3,4,5-trisphosphate recruits AKT to the plasma membrane, where it co-localizes with PDK1 in a pleckstrin homology domain-dependent manner (4–6). The binding of phosphatidylinositol 3,4,5-trisphosphate to AKT induces a conformational shift that alleviates AKT autoinhibition (7) and allows for PDK1-mediated phosphorylation of AKT Thr-308, an event that is absent in both PDK1 null mouse embryonic stem (ES) cells (8) and tissue-specific PDK1 knock-out mice (9). In parallel with the elucidation of the above PI3K/PDK1/AKT signaling cascade, PDK1 has been shown to phosphorylate the conserved threonine/serine residue in the activation loop (T-loop) of about 20 related protein kinases (10). Because this phosphorylation event is a prerequisite for full catalytic activity, PDK1 has been referred to as the “master regulator” of the AGC protein kinase family (10).
The observation that several kinases phosphorylated by PDK1 are positioned in the oncogenic PI3K- or MAPK-signaling pathways and are themselves oncology targets (i.e. AKT, RSK (p90 ribosomal S6 kinase), PKC, and p70S6K (p70 ribosomal S6 kinase)) has prompted the development of small molecule PDK1 inhibitors (11). Because tumor cells often possess pathological activation of the PI3K pathway, pharmacological inhibition of PDK1 is predicted to inhibit oncogenic cellular processes and thus be therapeutically beneficial (11). Consistent with this, several agents targeting components of the PI3K and MAPK pathways are in clinical development, with some showing early signs of activity (12). However, preclinical efficacy studies using pharmacological inhibitors of PDK1 have been hampered by the lack of specific proof-of-concept molecules (11, 13). Thus, gene silencing and expression of functionally impaired and dominant negative mutant forms of PDK1 have frequently been used to probe PDK1 protein function in cells (13–21).
In drug discovery, the cross-validation of cellular phenotypes using both RNA interference (RNAi) and chemical probes are highly valuable because convergent phenotypes provide confidence in conclusions drawn about a protein's biological function and its tractability as a drug target. A critical feature of genetics is the inherent specificity by which point mutations, gene deletion, or knockdown perturbs protein function. By contrast, for a small molecule, it is impossible to comprehensively identify all cellular targets, and pharmacological phenotypes may reflect “off-target” effects of a molecule (22). Although off-target effects are also a potential issue for RNAi, nonspecific gene-silencing effects are routinely controlled for using multiple non-overlapping sequences (15, 23). However, it is important to note that small molecules typically do not alter the expression of their target protein compared with gene silencing, which may disrupt protein complexes or impair protein functional domains that would be unaffected by a drug. Indeed, for many kinases, including PDK1, cellular phenotypes that are independent of the kinase catalytic activity have been reported (14, 24). Taken together, this complexity highlights the importance of combining genetic and chemical approaches for drug target validation studies.
In this report, we provide proof of concept for the value of parallel genetic and chemical approaches focusing on the in vitro characterization of PDK1 function in multiple cancer cell lines. We benchmark candidate PDK1 tool compounds developed at Merck or disclosed in papers and patent applications and document through profiling and x-ray crystallographic studies the existence of an exquisitely selective molecule (compound 7) that inhibits PDK1 in a manner distinct from classical ATP-competitive inhibitors. Using this pharmacological inhibitor and RNAi, we identify anchorage-independent growth and cell migration/invasion as relevant PDK1-dependent assays. We show unambiguously, using a panel of 17 diverse cancer cell lines, that PDK1 inhibition or knockdown does not significantly inhibit cell growth on standard tissue culture plastic. Because the inhibition of monolayer cell growth is a frequently reported read-out for cell potency of PDK1 inhibitors in papers and patents, our findings have significant ramifications for selecting appropriate in vitro assays that can guide the lead optimization of selective PDK1 kinase inhibitors and enable the identification of biomarkers predictive of drug response. Toward this goal, using soft agar colony formation assays, we evaluate the pharmacological inhibition of PDK1 in cancer cell lines and in a panel of 57 primary, patient-derived tumor xenograft lines and identify phospho-PDK1 Ser-241 as a candidate pharmacodynamic (PD) biomarker predictive of efficacy. We also observe that about half of the responsive primary human tumor lines harbor oncogenic RTK mutations, providing a possible patient responder hypothesis for PDK1-targeted therapies.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
All tissue culture reagents were from Invitrogen. Cell lines (ATCC) were grown at 37 °C with 5% CO2 in either Dulbecco's modified Eagle's medium (DMEM) (BT-474, MCF7, KPL-1, T47D, and HCT116), RPMI (A2780 and LS513), α-DMEM (C33a), DMEM/F12K (MDA-MB-231), or F12-K (PC-3) supplemented with 10% (v/v) FBS (Hyclone), 50 units/ml penicillin, and 50 μg/ml streptomycin. Compounds were prepared as 10 mm stock solutions in DMSO and stored in aliquots at −20 °C. The final concentration of DMSO is less than 0.5% (v/v) in all assays. The patient-derived, primary tumor xenograft lines were from Oncotest GmbH (25).
Western Blotting
Cell lysates were prepared using radioimmune precipitation buffer (Pierce) plus protease and phosphatase inhibitor mixtures (Roche Applied Science), analyzed by SDS gel electrophoresis, and transferred to nitrocellulose or PVDF membranes (Invitrogen). Immunoblots were probed with the following antibodies: anti-PDK1, anti-p-PDK1 (p-PDK1) Ser-241, anti-AKT, anti p-AKT Thr-308, anti-p-AKT Ser-473, anti-p-S6K1 Thr-389, anti-p-S6 ribosomal protein (S6RP) Ser-235/236, and anti-β-actin (all from Cell Signaling Technologies (Danvers, MA)) and anti-p-RSK Ser-221 (Invitrogen).
Antisense Molecules and Short Hairpin RNA (shRNA) Sequences
All antisense oligonucleotides were synthesized by Biospring (Frankfurt, Germany) and represent the third generation of gapmer antisense oligonucleotides (26). The PDK1 GeneBloc (GB) and mismatch (mm) molecules have the following sequences: GB1, ugu gaa ATG CCC TTg ccg ugc; mm1, ugu gaa AAG CCG TTg ccg ugc; GB4, caa caa cCT CTT CTC Auc uuc gg; mm4, caa gaa cCA CTT CTC Tuc uuc gc; GB5, aau uuc aCC TTT CAG Aac uuu gu; mm5, aau guc aCC TAT CAT Auc uuu gu underline denotes altered nucleotides in mismatch oligos. The shRNA sequences used are as follows: sh1, 5′-AAG CCA GGG CCA ACT CAT TCG-3′; sh3, 5′-AAT TCC CGG ATA AGC GGA AG G GTT TAT TCA AGA GAT AAA CCC TTC CGC TT A TCC TTT TTG-3′; sh20, 5′-AAT TCC CGA C TC GAA CTC CTT TGA ACT TCA AGA GAG TTC AAA GGA GTT CGA GTC TTT TTG-3′.
Proliferation Assays and Compound Dilution
Cell proliferation reagents ViaLight Plus (Lonza, Rockland, ME) and WST-1 (Roche Applied Science) were used according to the manufacturer's protocol. For the evaluation of compounds 1–7 (Fig. 1), PC-3 cells were seeded into 384-well plates (1000 cells/well) in 36 μl of growth medium and incubated for 24 h. An initial dilution plate was prepared containing 10 mm compounds diluted with DMSO in 10-point half-log serial dilutions. DMSO and staurosporine (200 μm) were utilized as high and low controls, respectively. 5 μl of the serial diluted compounds were then transferred into an intermediate 1:20 dilution plate containing 95 μl of growth medium. After mixing, 4 μl of the resulting medium and compound was transferred into each well of the cell plate (resulting in final compound concentrations ranging from 50 μm to 1.5 nm). After incubation for 72 h, ViaLight lysis reagent (10 μl) was added to each well and incubated at room temperature for 10 min, followed by the addition of ViaLight ATP-monitoring reagent (30 μl). Luminescence was measured (ViewLux plate reader, PerkinElmer Life Sciences), and IC50 values were calculated using the high and low controls for curve fitting. A 96-well plate version of the above protocol was used for the 17-cancer cell line panel (Table 1).
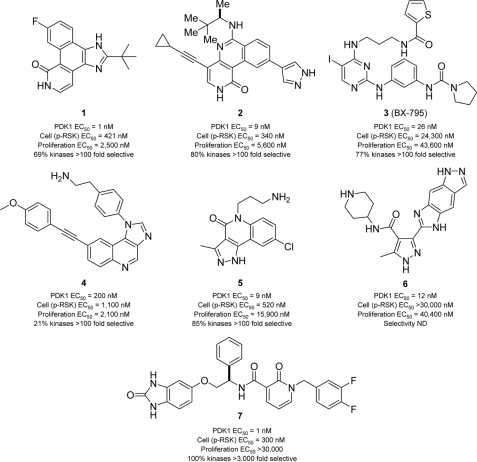
FIGURE 1.
Structure, potency, and selectivity of PDK1 inhibitors. Chemical structures and activity profiles of seven PDK1 inhibitors (compounds 1–7) exemplifying distinct structural classes. Enzymatic potency (PDK1 EC50) was determined using recombinant, purified, full-length human PDK1 enzyme and AKT-Thr-308-tide as substrate (28). Cell biochemical potency (p-RSK EC50) was determined by measuring p-RSK S221 inhibition in PC-3 cells (2 h post-treatment; in-cell Western analysis). Cell viability (proliferation EC50) was determined following 72 h of incubation of PC3 cells with compound (ATP ViaLight). The kinase selectivity data are based on a panel of 82 (compound 1), 183 (compound 5), or >200 protein kinases (compounds 2–4 and 7) and summarizes the percentage of kinases that have an extrapolated EC50 greater than 100 times the compound's EC50 for PDK1 (or 3000 times in the case of compound 7). For instance, 69% of the kinases profiled with compound 1 displayed estimated EC50 values greater than 100 times the EC50 value for PDK1 (see supplemental Table S1 for complete profiles). The selectivity profile was not determined for compound 6 because it did not appear to be cell-potent against p-RSK.
TABLE 1.
Effect of PDK1 and AKT inhibitors on cancer cell proliferation
| Cell line | Origin | EC50a |
|
|---|---|---|---|
| Compound 7 | AKT inhibitor | ||
| μm | |||
| A27801b | Ovary | 8.9 | 0.6 |
| MDA-MB-453c | Breast | 8.1 | 0.144 |
| MDA-MB-231d | Breast | 9.7 | >30 |
| UMC-11 | Lung | 8.2 | >30 |
| T47Dc | Breast | 19 | 0.07 |
| LNCaPb | Prostate | 10.8 | 0.33 |
| BT-474c | Breast | 9.6 | 0.29 |
| NCI-H446b | Lung | 18.5 | 0.6 |
| PC-3b | Prostate | >30 | 1.2 |
| A549d | Lung | >30 | 5 |
| A427d | Lung | >30 | 4.6 |
| NCI-H1437 | Lung | >30 | 2.2 |
| LS-513d | Colon | >30 | 1.9 |
| HCT-116c,d | Colon | >30 | >30 |
| NCI-H1915d | Lung | >30 | >30 |
| U87b | Brain | >30 | >30 |
| MCF7c | Breast | >30 | >30 |
a Calculated from ATP ViaLight data (72 h) using 10-point drug titrations and standard tissue culture plastic and growth medium.
b PTEN-deficient.
c PI3KCA mutant.
d KRAS mutant.
In-cell Western Assay for p-RSK Ser-211 Inhibition
PC-3 cells (30,000 cells/well) were seeded in growth medium (200 μl) in black wall, clear bottom 96-well plates (Corning Glass). The following day, 10-point 3-fold serial dilutions of compounds were transferred into each well and incubated for 2 h. Cells were fixed with 10% (v/v) formalin buffer, permeabilized using PBS supplemented with 0.1% (v/v) Triton X-100 for 15 min at room temperature, and blocked for 2 h with Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE). Next, plates were incubated overnight with a mixture of p-RSK Ser-221 and S6RP primary antibodies (1:100 dilution in Odyssey Blocking Buffer). The following day, plates were washed in PBS, incubated for 2 h with secondary detection antibodies (IRDye 800 CW-conjugated anti-mouse and IRDye 680 CS-conjugated anti-rabbit), washed in PBS, and scanned for infrared signal using the Odyssey Imaging System (LI-COR Biosciences).
Reverse Phase Protein Microarrays
20 nl of denatured protein lysates (10 mm Tris, 100 mm NaCl, 1 mm EDTA, 20 mm Na4P2O7, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate, 2 mm Na3VO4, 1 mm PMSF) were immobilized onto nitrocellulose-coated glass slides (Amersham Biosciences) using an Auchon 2407 microarrayer in 2-fold dilution curves. Arrays were blocked for 2 h at room temperature with casein containing 0.1% Tween 20, followed by blocking of endogenous biotin with avidin (Dako Systems), incubation with primary antibody (1:1000), and finally, incubation with biotinylated secondary antibody (1:5000). Arrays were developed with 3,3′-diaminobenzidine tetrahydrochloride chromogen. Integrated values for each spot were obtained by ImageQuant 3.0. Each assay was performed twice (technical replicas), and on each run, two independent measurements for every phosphorylated protein were obtained (biological replicas). The data were then normalized using the double-z score, and expression level of a housekeeping gene (GAPDH) was followed by averaging across the technical and biological replicas.
Matrigel-based Assays
Network Assay
48-Well plates (flat bottom; Corning Glass) were coated with Matrigel basement membrane matrix (BD Biosciences) according to the manufacturer's protocol. PC-3 cells were seeded at 50,000 cells/well and treated immediately after plating with either drug or vehicle (DMSO). Network formation was captured by contrast microscopy (Zeiss).
Boyden Chamber Assay
Cell invasion was assayed in 24-well Biocoat Matrigel invasion chambers (8 μm; BD Biosciences) according to the manufacturer's protocol. Briefly, 5 × 104 PC-3 or MDA-MB-231 cells were plated in the upper compartment of the chamber in 0.5 ml of serum-free medium with or without drug. Medium in the bottom chamber was supplemented with 10% FBS as a chemoattractant. After 48 h, cells in the bottom chamber were fixed with 4% formaldehyde and Hoechst-stained (Invitrogen), and the total number of migrated cells was quantified using SimplePCI imaging software (Version 6.6).
Spheroid Assay
5000 cells were seeded per well (96-well U-bottom plates) in complete medium supplemented with 2.5% Matrigel and centrifuged for 10 min at 1000 × g to generate single spheroids with homogeneous sizes and morphologies. Compounds were added 24 h later, and spheroid sizes were measured after 5 days using contrast microscopy images.
MCF10A/ErbB2 Acini Formation Assay
Eight-well glass chamber slide (Falcon culture slides) were coated with growth factor-reduced Matrigel (BD Biosciences) and allowed to solidify. Differentiation medium (DMEM/F-12 containing 2% horse serum, 5 ng/ml EGF, 10 μg/ml insulin, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone) containing 2% Matrigel and the MCF10A/ErbB2 cells (5000 cells; 0.4 ml) were added to the top of the solidified layer of Matrigel. The following day and every 4 days, differentiation medium was replaced with or without compound.
Anchorage-independent Growth Assay
For soft agar colony formation assay, trypsinized cells (1 × 104) were suspended in culture medium (Iscove's DMEM containing 0.6% Nobel agar and 20% FBS; 5 ml) and plated on a presolidified layer of soft agar (2.5 ml) in a 6-cm Petri dish. After solidification, culture medium (Iscove's DMEM containing 0.6% Nobel agar and 5% FBS; 2.5 ml) was added on top. An additional 1 ml of Iscove's DMEM was added to retain adequate rehydration. Cells were incubated for 3 weeks at 37 °C. PDK1 inhibitor or vehicle (DMSO) was added to the culture medium at the time of seeding. After 3 weeks, colonies were stained with iodonitrotetrazolium chloride (Sigma) and photographed. The number of macroscopic colonies was determined from images using MetaMorph software (Molecular Devices, Sunnyvale, CA) from three independent plates. The clonogenic assays with patient tumor explants were carried out at Oncotest (25).
RESULTS
Various structural classes of small molecule PDK1 inhibitors have been published in papers and patent applications (11). However, most of these reports disclose limited kinase selectivity data and report highly variable cellular activities (11), emphasizing the need for caution in selecting proof-of-concept PDK1 inhibitors and for the importance of parallel genetic cross-validation of pharmacological phenotypes.
Collection of Small Molecule Inhibitors of PDK1
To enable the pharmacological characterization of PDK1, we first compiled a battery of structurally diverse kinase inhibitors with reported activity toward PDK1 (Fig. 1). These compounds were initially assayed against recombinant purified PDK1 enzyme and characterized in the PC-3 prostatic cancer cell line by monitoring inhibition of proliferation and of p-RSK Ser-221, a bona fide PDK1 substrate (Fig. 1). A parallel high throughput screening campaign (27, 28) complemented our literature search, and seven small molecule inhibitors were selected for further profiling. In brief, compound 1 was identified from a focused kinase library screen using an AKT-derived peptide substrate and full-length PDK1 enzyme (27, 28). It represents the tetracyclic class of pan-Janus kinase inhibitors (29, 30), which we further optimized for selectivity, yielding the tricyclic class of PDK1 inhibitors exemplified by compound 2 (31). Compound 3 is the aminopyrimidine-based PDK1 inhibitor BX-795 reported by Feldman et al. (32). Compound 4 exemplifies the imidazo[4,5-c]quinoline scaffold of BAG956, a dual PDK1/PI3K inhibitor with antitumor activity in vivo (33), and is a chemotype related to NVP-BEZ235, a dual PI3K/mTOR inhibitor currently undergoing clinical trials (34). Compounds 5 and 6 are dual CHK1/PDK1 inhibitors described by Millenium (35) and Vernalis Pharmaceuticals (36), respectively. Compound 7 exemplifies the pyridinonyl-based PDK1 inhibitors described by Sunesis and Biogen Idec (37), which conceptually consist of two fragments tethered by a flexible linker. The activity profiles reported herein are the first attempt to directly compare representative PDK1 inhibitors from various structural classes in an effort to identify suitable tool compounds for in vitro chemical genetic studies.
Compound 7 Is an Exquisitely Selective and Cell-potent PDK1 Inhibitor
Biological characterization of compounds 1–7 revealed a broad range of antiproliferative effects in PC-3 cells. The EC50 values in a 72-h proliferation assay range from 2.1 μm to inactive (>50 μm) for enzymatically equipotent inhibitors, suggesting a possible contribution of “off-target” activities to this phenotype. In general, selectivity represents a key challenge when using pharmacological inhibitors to evaluate the involvement of a particular kinase in cellular processes (22). For example, compound 3 (BX-795) and its analogs have been reported to inhibit cell cycle progression and induce apoptosis (32); however, recent chemical genetic studies show that BX-795 causes G2/M cell cycle arrest in both PDK1 wild type (PDK1+/+) and PDK1 null (PDK1−/−) ES cells (13). Thus, to get a more comprehensive view of non-PDK1 targets, the above compounds were profiled against large commercial panels of distinct protein kinases (supplemental Table S1). For each compound, the degree of selectivity is summarized as the percentage of kinases showing an EC50 value greater than 100-fold of PDK1 EC50 (Fig. 1). Notably, compounds 1–6 inhibit several protein kinases in addition to PDK1, making it difficult to render meaningful conclusions about cell-based functions of PDK1 using these compounds. However, compound 7 displayed greater than 3000-fold selectivity against the 256 kinases tested, making it an attractive tool compound for in vitro cell functional studies.
Compound 7 Binds to the Inactive Kinase Conformation of PDK1 and Induces Conformational Changes Not Previously Reported for AGC Kinases
To understand the molecular basis for the exquisite selectivity of compound 7, we co-crystallized compounds 2 and 7 with PDK1 (Fig. 2a). To date, analysis of a dozen small molecule inhibitor-PDK1 complexes are available in the Protein Data Bank, providing a framework for understanding kinase selectivity in terms of ligand-PDK1 protein interactions (11). The x-ray co-crystal structure of compound 2 (Fig. 2a, left) displays a classical ATP-competitive and kinase-active binding mode (DFG-in). It forms hydrogen bonds to the hinge region of PDK1 (i.e. Ala-161 and Ser-160) that are similar to those reported for BX-517 (close relative of compound 3) (38) and staurosporine (39). In contrast, the high resolution x-ray co-crystal structure of compound 7 (Fig. 2a, right) reveals an inactive kinase conformation similar to the binding mode of imatinib (DFG-out), which is a selective inhibitor of the tyrosine kinase bcr-abl (40). This binding mode is made possible by the DFG residues of the activation loop being folded away from the conformation required for ATP phosphate transfer (41). To our knowledge, this is the first reported example of a DFG-out (i.e. type II) kinase inhibitor for an AGC kinase.
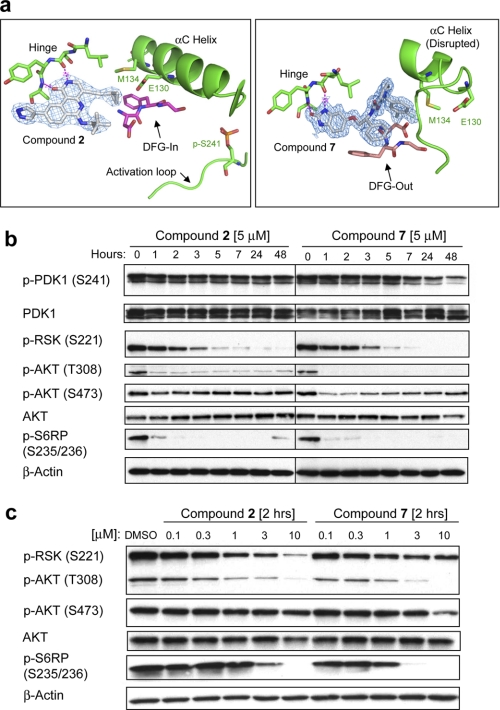
FIGURE 2.
X-ray crystallographic and cell biochemical analysis of compounds 2 and 7 reveal distinct binding modes that differentially impact the phosphorylation state of PDK1 Ser-241. a, co-crystal structures of compounds 2 (2.6 Å resolution; Protein Data Bank code 3NAY) and 7 (1.75 Å resolution; Protein Data Bank code 3NAX). Compound 2 binds to the active, DFG-in conformation of PDK1, like most typical ATP competitive inhibitors (type I). Compound 7 binds in the ATP pocket and induces substantial conformational changes in PDK1, including causing the DFG loop to exist in an “out” conformation (DFG-out) that typifies inactive form kinase inhibitors (type II). Compound 7 additionally disrupts the αC helix, a feature unique to this compound among inactive form kinase inhibitors. The conformational change in the αC helix results in catalytic residue Glu-130 being displaced from the active site. Both compounds show a classical hydrogen bonding pattern to the hinge region. b, time course study. PC-3 cells were incubated with a 5 μm concentration of either compound 2 or compound 7, and cells were collected at the indicated time points for Western blotting analysis. c, dose-response study. PC-3 cells were incubated with either vehicle (DMSO) or the indicated drug concentration and collected 2 h post-treatment for Western blotting analysis.
Conceptually, compound 7 comprises three molecular fragments: a hinge binding group, a linker, and a hydrophobic moiety. The hydrophobic moiety, which occupies a pocket only present in the inactive conformation of PDK1, engages residues in PDK1 that are not conserved broadly within the kinase superfamily. This, together with significant conformational changes in the αC helix (Fig. 2a) which also impact the structural integrity of the PIF pocket, may explain its exquisite kinase selectivity and unique cell biochemical activity in PC-3 cells (see below).
Compound 7 Uniquely Inhibits Phospho-PDK1 Ser-241 Compared with Kinase-active Conformation (Type I) Inhibitors
Western blot analysis of PC-3 cells treated with compounds 2 and 7 revealed potent time- and dose-dependent inhibition of the bona fide PDK1 phosphorylation sites (AKT Thr-308 and RSK Ser-221) as well as the downstream marker S6RP Ser-235/236 (Fig. 2, b and c). This concerted pathway inhibition is consistent with genetic deletion studies, where the loss of PDK1 in mouse ES cells results in decreased kinase activity of AKT, RSK, and p70S6K, the last being the upstream kinase phosphorylating S6RP (8, 42). However, we find that acute small molecule PDK1 inhibition differs from genetic deletion studies in two significant ways.
First, pharmacological PDK1 inhibition showed an acute, transient inhibition of p-AKT Ser-473 (Fig. 2b). This effect was most pronounced for compound 7, which is also 10-fold more potent in the PDK1 enzymatic assay (EC50 = 1 nm) compared with compound 2 (EC50 = 9 nm; Fig. 1). However, prolonged pharmacological inhibition of PDK1 (>24 h), does not lead to a net change in the phosphorylation level of AKT Ser-473, consistent with constitutive PDK1 knock-out studies (8).
Second, for compound 7, we observed a slow time-dependent inhibition of the PDK1 autophosphorylation site (p-PDK1 Ser-241) with about 70% reduction in phosphorylation at 48 h (Fig. 2b). In contrast, compound 2 had no effect on phospho-PDK1 Ser-241 or total PDK1 protein levels (Fig. 2b). Notably, compounds 1–6 are all known (or computationally predicted) to bind PDK1 in a kinase-active conformation (DFG-in mode), and none of these inhibitors induce significant dephosphorylation of p-PDK1 Ser-241 in cells (data not shown). Thus, we speculate that the unique ability of compound 7 to induce PDK1 in a dephosphorylated state results from its inactive kinase binding mode (DFG-out conformation) and note that this binding mode has not previously been reported for AGC kinases.
Selective Pharmacological Inhibition of PDK1 Has Minimal Effect on Two-dimensional Cell Proliferation
Given the potent PDK1 pathway inhibition elicited by compound 7 (Fig. 2), as well as its favorable kinase selectivity profile (>3000 fold; supplemental Table S1), we next screened this inhibitor for antiproliferative effects in a panel of 17 cancer cell lines (Table 1). The cell lines were selected to represent different tumor types (i.e. breast, colon, lung, brain, and prostate) and to cover major signaling abnormalities based on pathway mutational status (i.e. RAS/PI3K/PTEN). To our surprise, compound 7 failed to potently inhibit the growth of plastic-attached monolayer cultures at concentrations achieving >EC90 for cell biochemical pathway inhibition (Table 1). In contrast, several cell lines were sensitive (EC50 < 1 μm) to the highly selective and allosteric naphthyridione-based AKT inhibitor (compound 17 in Ref. 43), including PI3KCA mutant breast cancer lines. Together, these results suggest a difference in potential therapeutic effect of molecules that directly inhibit AKT versus indirectly via PDK1.
PDK1 Knockdown Using either Antisense or RNAi Has Minimal Inhibitory Effect on Standard Plastic-attached Monolayer Cell Growth
RNAi-based gene silencing techniques offer a means to genetically authenticate specific pathway or phenotypic defects attributed to target inhibition by small molecules. To cross-validate the biological effects of compound 7, we first employed GB antisense oligonucleotides (26) to knock down PDK1 in the PTEN-null PC-3 cell line (Fig. 3a). Three independent antisense sequences were used alongside their corresponding mm controls. As expected, each PDK1-targeting antisense molecule reduced phosphorylation of Ser-221 of p90RSK and Thr-308 of AKT, both direct phosphorylation sites of PDK1.
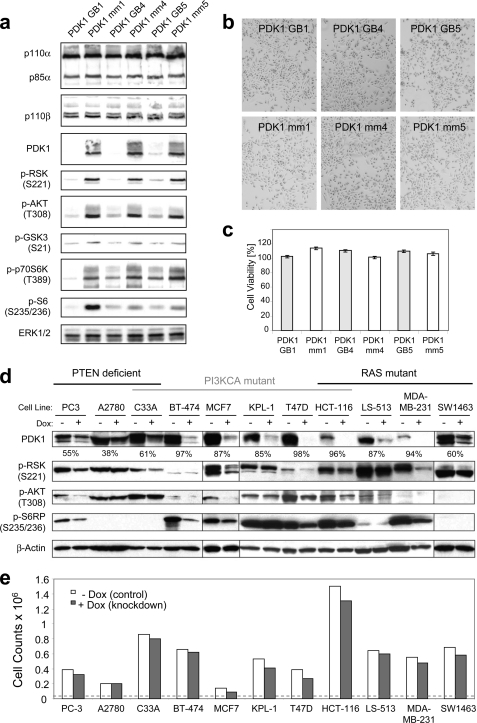
FIGURE 3.
Knockdown of PDK1 using antisense oligonucleotides and inducible RNAi reduces pathway activity but has minimal effect on growth of cancer cell lines. a, PC-3 cells were transfected with antisense RNA oligonucleotides (GB) against PDK1 or mm controls. Cells were harvested at 96 h post-transfection, and lysates were analyzed by Western blotting for target knockdown levels and effects. b, just prior to harvest, the transfected cells were examined for visible growth deficiencies under the microscope. c, in parallel, cell viability (ATP ViaLight) was measured, and the percentage of viability was determined as the ratio of relative light units in the experimental sample over a non-transfected control grown over the same time period (96 h post-transfection). Error bars, S.E. d, a panel of cancer cell lines, grouped by known oncogenic pathway mutations (top), was stably transduced with lentivirus expressing a doxycycline-inducible shRNA targeting PDK1. Following growth in the absence (−) or presence (+) of doxycycline over a 5-day period, transduced cells were harvested and analyzed by Western blotting for the knockdown level of PDK1 as well as for indicators of downstream signaling. The percentage of PDK1 knockdown is given below the PDK1 Western blot. e, cell proliferation assays were performed on the stably transduced cell lines from d by plating the same number of isogenic cells in the presence or absence of doxycycline (Dox) and counting total cells after 5 days.
To determine the functional effects of reduced PDK1 expression and downstream signaling, we transfected cells with PDK1 and control antisense oligonucleotides as above and then allowed cells to proliferate for 96 h. Similar to pharmacological inhibition of PDK1 (compound 7; Table 1), acute knockdown of PDK1 had minimal effect on PC-3 cell growth, as evident from monitoring either cell density and morphology (Fig. 3b) or cell viability (Fig. 3c). To ensure maximum knockdown, the ratio of transfection lipid to DNA was increased, but this resulted in a lipid dose-dependent growth inhibition in both PDK1 and control knockdown cells (supplemental Fig. S1). Notably, the decreased proliferation was more pronounced in PDK1 knockdown samples compared with control samples, possibly due to a combination of PDK1 deficiency and cellular stress induced by the transfection. In line with this observation, pharmacological inhibition of PDK1 in mouse ES cells has little effect on cell growth but can sensitize cells to apoptotic stimuli in response to chemotherapy-induced stress (13).
To eliminate these lipid-derived nonspecific effects, we next used a vector-based doxycycline-inducible shRNA expression system (Tet-on) (44) to screen our cancer cell line panel for a PDK1 knockdown phenotype. Twelve cell lines with putative PI3K/MAPK-activating genotypes were stably transduced with lentivirus containing an shRNA targeting PDK1 (sh1). The ability to reduce PDK1 protein expression in response to doxycycline varied from cell line to cell line, although it approached >90% knockdown in multiple lines (Fig. 3d). Accordingly, signaling nodes downstream of PDK1 were variably affected across the different cell lines, as determined by assessing PDK1-mediated phosphorylation sites on AKT and RSK as well as the downstream marker S6RP. However, we again were unable to detect acute proliferation defects across the panel of cell lines when PDK1 was targeted by shRNA (Fig. 3e), regardless of the knockdown level or the interruption in downstream signaling. The general lack of inhibition of monolayer cell growth under standard tissue culture conditions, either via genetic knockdown or selective small molecule PDK1 targeting, prompted us to explore PDK1 function in alternative assays that capture other cellular responses relevant to cancer.
PC-3 Cells Depend on PTEN Loss for Colony Formation in Soft Agar
Anchorage-independent growth is pivotal to the ability of tumor cells to survive and metastasize in vivo and, under in vitro conditions, allows transformed cells to form colonies in semisolid medium (45). Before evaluating PDK1 function in soft agar colony formation assays, we determined the degree to which PTEN-null PC-3 cells depend on an activated PI3K pathway for their soft agar growth phenotype. For this, PTEN wild type (WT) or catalytically inactive mutant cDNA (C124S) (46) was expressed in PC-3 cells, and stable cell lines were generated. As expected, reconstitution of PTEN WT, but not catalytically inactive PTEN C124S, reduced the phosphorylation levels of AKT and p70S6K (Fig. 4a). Despite this pathway inhibition, however, cell growth on tissue culture plastic was not inhibited. In contrast, when these isogenic PTEN-matched pair cell lines were plated in a soft agar colony-forming assay, PTEN WT expression prevented anchorage independent growth (i.e. >90% inhibition) compared with parental or PTEN C124S reconstituted cells (Fig. 4b). Thus, PC-3 cells depend on PI3K/PTEN signaling for anchorage-independent growth, and the ability of compounds to functionally suppress aberrant PI3K pathway activity can therefore be studied using soft agar colony formation as a quantitative assay.
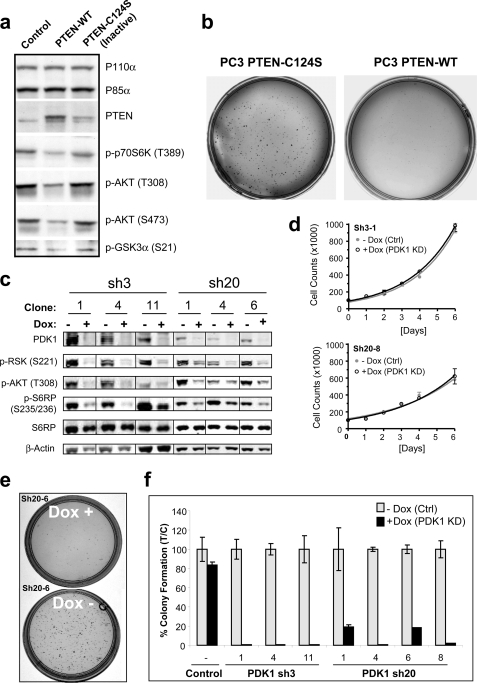
FIGURE 4.
PC-3 cells depend on PTEN loss and PDK1 for growth in soft agar. a, PC-3 cells were stably transfected with empty vector (control) or plasmid encoding either WT PTEN or a catalytically inactive allele of PTEN (C124S) and analyzed by Western blotting using the indicated antibodies. b, in parallel, PTEN-WT and PTEN-C124S cells (5 × 103) were plated on a soft agar bed (6-cm dish) and allowed to form colonies over a 21-day period. c, Western blot analysis of PC-3 cells subcloned from a pool of virally transduced cells with PDK1-targeting shRNA sequences (sh3 and sh20). Cells were lysed after growing for 5 days in the absence (−) or presence (+) of doxycycline. d, growth curves of representative PC-3 subclones monitored through daily cell counts. To ensure maximal PDK1 knockdown at time of plating (day 0), cells were treated with doxycycline (Dox) 5 days prior to the start of the experiment. Error bars, S.E. e, representative soft agar colony formation of the PC3 clonal cell line sh20-6 following 21 days in the continued absence (−) or presence (+) of doxycycline. f, soft agar colony formation was quantified by counting the number of colonies appearing after 21 days of growth on soft agar and represented as the percentage of colonies formed in the presence of doxycycline (T, doxycycline treatment) over control (C, no doxycycline treatment). Non-transduced PC-3 cells were included as added control showing a non-significant effect of doxycycline alone.
PDK1 Knockdown Inhibits Anchorage-independent Growth of PC-3 Cells
Because PC-3 cells proved to rely on PI3K/PTEN signaling (Fig. 4b), we chose that cell line to continue our PDK1 inhibition studies. To further maximize the knockdown efficiency of our lentivirus mediated RNAi screen (Fig. 3a), we tested 30 additional constructs for PDK1 knockdown potential (supplemental Fig. S2). Based on quantitative RT-PCR, two shRNA sequences (sh3 and sh20) showing ∼90% knockdown of mRNA were selected for further knockdown studies (supplemental Fig. S2a).
During the process of identifying sh3 and sh20, we used an inducible lentiviral vector system where shRNA expression is linked to GFP (47). Although virally transduced cells were selected with puromycin to eliminate non-infected cells, flow cytometric cell analysis revealed a heterogeneous population of GFP-positive cells (supplemental Fig. S2b). A small fraction of cells (<10%) did not express GFP in response to doxycycline treatment and thus presumably lacked shRNA expression. To achieve a more homogenous cell population with regard to PDK1 knockdown potential, we therefore single cell-cloned the transduced PC-3 cells (supplemental Fig. S2c). Western blot analysis revealed multiple clonal cell lines for sh3 and sh20 that displayed greater than 95% PDK1 protein knockdown and strong pathway inhibition in response to doxycycline treatment (Fig. 4c). Because the inducible Tet-Krab system affords subcloning of cells in the absence of target knockdown, our approach circumvents the potential risk of negative selection if knockdown were to impair cell growth.
We next characterized the effect of knockdown in these clonal PC-3 cell lines using standard proliferation and soft agar colony-forming assays. Similarly to the PTEN reconstitution experiment (Fig. 4, a and b), PDK1 knockdown did not affect the doubling time of the PC-3 cells when grown as plastic-attached monolayers in medium containing high glucose and 10% serum (Fig. 4d and supplemental Fig. S2d). In contrast, for cells seeded in soft agar, we saw a robust inhibition of colony formation (Fig. 4, e and f) similar to the effect of PTEN reconstitution. Again, the sh3 construct, which affords a more potent PDK1 pathway inhibition compared with sh20 (Fig. 4c), showed the strongest inhibitory effect (>98% efficiency) on colony formation (Fig. 4f).
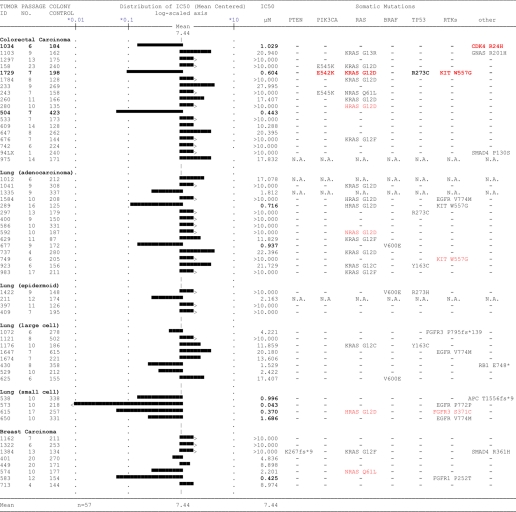
Pharmacological Inhibition of PDK1 Impairs Anchorage-independent Growth in a Subset of Cancer Cells and Primary Tumor Lines
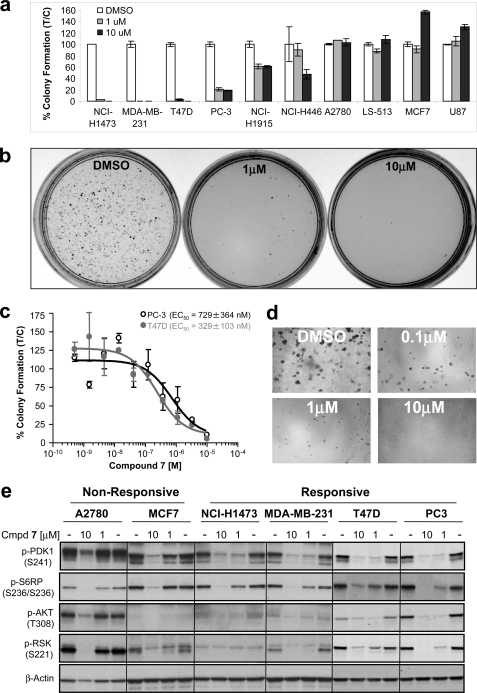
To explore the pathway context for in vitro response to PDK1 inhibition, we next evaluated the ability of compound 7 to inhibit anchorage-independent growth of tumor cells. Of the 17 cell lines listed in Table 1, 10 readily grew in soft agar, and four of these were strongly inhibited by targeting PDK1 (Fig. 5, a and b). Sensitive cell lines included PC-3 cells (mimicking the PDK1 knockdown result), T47D, MDA-MB-231, and NCI-H1437. Using a 96-well miniaturized version of the soft agar assay, IC50 values for compound 7 were determined to be 329 ± 102 nm for T47D and 729 ± 364 nm for PC-3 cells (Fig. 5, c and d). The functional dependence on PDK1 was further validated for the two breast cancer cell lines, with both sh3- and sh20-mediated knockdown resulting in inhibition of soft agar growth. Moreover, removal of doxycycline from the system reversed the phenotype (data not shown). Because none of the PDK1-dependent cell lines shared an obvious common genotype (i.e. PTEN deficiency or PI3K or RAS mutation), we next evaluated drug response in a panel of patient-derived primary human tumor xenografts grown in soft agar. Of the 57 tumor lines, nine were sensitive (EC50 < 1.1 μm) to PDK1 inhibitor treatment (i.e. 16% response rate; compound 7); however, sensitive tumor cells displayed a range of somatic DNA mutations, and inhibitor efficacy did not correlate with any specific pathway aberrations, as judged by somatic tumor mutations in PTEN, PI3K, RAS, or RAF (Table 2). However, small cell lung carcinomas appear more sensitive to PDK1 inhibition (three of four) compared with lung tumor models of different histological subtypes (two of 27) (Table 2). Moreover, looking across all tumor tissue types, RTK mutations (EGFR, KIT, and FGFR3) appeared to sensitize cells to PDK1 inhibition, consistent with recent in vitro findings that PDK1 overexpression can potentiate the effect of RTKs on transformation of MCF10A cells (48).
FIGURE 5.
Inhibition of PDK1 in a subset of cell lines impairs soft agar colony formation. a, the indicated cells were plated on soft agar in the presence of vehicle (DMSO) or compound 7 (1 or 10 μm) and allowed to grow. Colonies were counted after 21 days and scored as a percentage of control colonies (DMSO). Colony formation for each condition was performed in duplicate. Error bars, S.E. b, representative pictures of colony formation assay used for quantification. c, titrations of compound 7 were carried out for PC-3 and T47D cells using a 96-well plate soft agar colony formation assay to determine EC50 values. d, representative dose-response pictures from the 96-well plate soft agar assay analyzed by Isocyte. e, sensitive and resistant cell lines were plated on tissue culture-treated plastic in monolayer and treated with either DMSO or the indicated concentration of phosphoinositide-dependent kinase inhibitor (compound 7). After 48 h, cells were harvested, lysed, and analyzed by Western blotting identifying p-PDK1 Ser-241 as a candidate PD biomarker predictive of drug response.
TABLE 2.
Mean graph analysis based on in vitro IC50 of compound 7 in soft agar colony formation assays using primary, patient-derived, human tumor xenograft lines propagated in nude mice
N.A., not available; fs, frameshift; *, stop codon; ins, insertion. Mutations in red are low confidence assignments.
In an effort to understand the basis for sensitivity to compound 7, we next analyzed the signaling of the responsive and non-responsive cancer cell lines by Western blotting and noted large differences in basal p-AKT and p-RSK levels among cell lines (Fig. 5e). Importantly, pharmacodynamic inhibition of S6RP confirmed the bioavailability of drug in both the sensitive and resistant cell lines. Interestingly, all four sensitive cell lines displayed significant compound-mediated Ser-241 dephosphorylation of PDK1 (1 μm and 48 h post-treatment) compared with the resistant lines (Fig. 5e). Thus, we conclude that efficient modulation of Ser-241 phosphorylation represents a candidate PD biomarker predictive of efficacy.
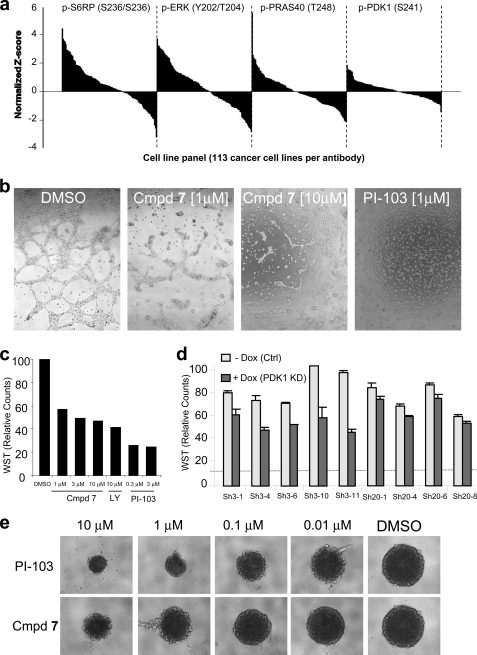
Intrigued by phospho-Ser-241 as a PD biomarker, we next looked at its expression across a large panel of 113 cancer cell lines using a reverse phase protein array (Fig. 6a). Compared with phosphoprotein biomarkers routinely used to access the activation state of the PI3K/MAPK pathway, we discovered that p-PDK1 Ser-241 displayed relatively little variation among tumor lines, supporting the notion that the PDK1 Ser-241 autophosphorylation site is constitutively “on” in cells (10).
FIGURE 6.
PC-3 cells exhibit a strong dependence on PDK1 for network formation and growth on a basement membrane matrix. a, protein expression levels for p-S6RP Ser-235/236, p-ERK Thr-202/Tyr-204, p-PRAS40 Thr-248, and p-PDK1 Ser-241 across a panel of 113 cancer cell lines. Reverse phase protein array values were normalized using the double-z score and the expression level of a housekeeping gene (GAPDH), followed by averaging across the technical and biological replicas. b, PC-3 cells were seeded on a bed of Matrigel and were grown in the presence of vehicle (DMSO), PI3K inhibitor (PI-103 at 1 μm), or PDK1 inhibitor (compound 7 at 1 and 10 μm) for 24 h. Photographs were taken at ×5 magnification. c, PC-3 cell growth on Matrigel in the presence of vehicle or drug was assayed using WST-1 cell proliferation reagent and represented as percentage proliferation. d, PC-3 subclones containing doxycycline-inducible PDK1 knockdown were cultured on a Matrigel bed in the absence or presence of doxycycline, and WST-1 counts for cell proliferation were collected 48 h after seeding. Error bars, S.E. e, PC-3 cells were subjected to a spheroid growth assay in the presence of 2.5% Matrigel and the indicated concentration of either the PI3K inhibitor, PI-103, or compound 7. Photographs were taken following 5 days of growth.
PDK1 Inhibition Impairs Cell-Cell Networks and Growth on Matrigel
We next examined the effect of genetic and pharmacological PDK1 inhibition in cell culture systems where cells are grown either on a Matrigel bed or in spheroids (Fig. 6). Matrigel is an extracellular matrix-containing basement membrane that facilitates growth of cells with increased malignant potential (49). LY294002 has been shown to severely impair cell-cell connections and growth of PC-3 cells in this culture system at concentrations where no antiproliferative effects are seen on regular tissue culture plastic (50). When PC-3 cells were grown on Matrigel, compound 7 induced an intermediate phenotype in the network assay compared with the dual PI3K/mTOR inhibitor PI-103 (Fig. 6, a and b). PDK1 knockdown also showed similar results although less potently (Fig. 6c). Again, PDK1 knockdown levels correlated with the phenotypic effect, with sh3 being more potent than sh20.
PC-3 cells, when seeded in 2.5% Matrigel, form suspension aggregates referred to as spheroids. It has been proposed that such spheroids may more closely resemble the morphology, cell-cell contacts, and diffusion properties of xenografts (51). Treatment for 5 days with compound 7 inhibited the formation of these spheroids in a dose-dependent manner although less potently than the dual PI3K/mTor inhibitor PI-103. Unfortunately, because compound 7 is not suitable for in vivo studies because it is rapidly cleared from the blood (Qliver > hepatic blood flow), the degree by which these in vitro assays predict anti-tumor efficacy in vivo remains unknown.
PDK1 Inhibition Impairs Cell Migration and Invasion
Cancer mortality is due largely to distant metastases, with tumor cell motility and invasion playing crucial roles in the metastatic process. Previous studies using pharmacological inhibitors of PI3K (wortmannin and/or LY294002) indicated that the PI3K/AKT pathway plays a role in regulating cell motility of cancer cell lines (52, 53). Consistent with these findings, PTEN−/− mouse embryonic fibroblasts show increased motility compared with PTEN+/+ mouse embryonic fibroblasts (54), and expression of kinase-dead PDK1 or kinase-dead AKT into PTEN−/− cells inhibits migration (20, 54).
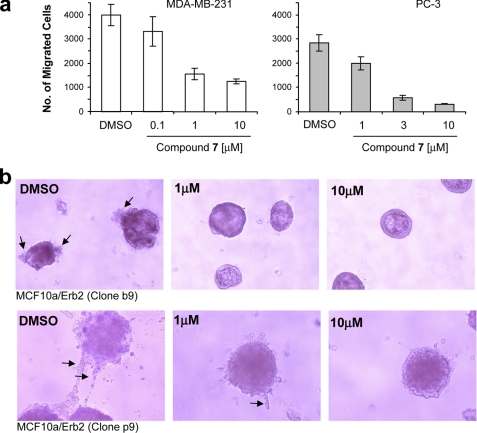
In light of these reports, we next studied the effect of compound 7 on cell motility and invasiveness of the PC-3 prostate and MDA-MB 231 breast cancer cell lines using a Matrigel invasion chamber assay. In this assay, cells must first degrade a reconstituted basement membrane that occludes the pores of the filter and then migrate through these cleared pores to the bottom chamber. The bottom chamber contains the chemoattractant (10% serum). For compound 7, we found a significant dose-dependent inhibition of serum-induced cell invasion for both cell lines (Fig. 7a). Furthermore, inducible PDK1 knockdown showed similar results in both PC-3 and MDA-MB-231 cells, although the effect was less dramatic (2-fold inhibition; data not shown) compared with the inhibition (5–10-fold) observed with compound 7.
FIGURE 7.
PDK1 inhibition reduces cell migration and invasiveness of cancer cells. a, MDA-MB-231 and PC-3 cells were treated with increasing concentrations of compound 7 or DMSO control and were seeded into Matrigel invasion chambers in triplicates. After 48 h, the total number of cells that had migrated into the lower chamber was measured for each condition. Error bars, S.E. b, MCF10A cells engineered to express exogenous ErbB2 (clone b9, low expressing clone; clone p9, high expressing clone) were plated on Matrigel, and their ability to form acini in the presence of DMSO or increasing concentrations of compound 7 was determined after 7 days of inhibitor treatment.
To further evaluate the suggested role of PDK1 in invasion, we looked at phenotypic changes induced by compound 7 on polarized breast epithelial cells. When grown on Matrigel, MCF10A cells form well defined hollow acini (55). Overexpression of mutant ErbB2, which activates both PI3K and MAPK signaling, results in a disturbed morphology (formation of filled multiacini) (55), and ectopic overexpression of PDK1 further potentiates this transformed phenotype (48). Therefore, two clonal MCF10A cell lines, engineered to express mutant ErbB2 and also selected for invasive features (with different degrees of disturbed acini) were incubated with PDK1 inhibitor. In both cell lines, the invasive phenotype (protrusions from acini) was reduced by compound 7, confirming that pharmacological inhibition of endogenous PDK1 indeed may inhibit ErbB2-induced transformation (Fig. 7b, arrows). Interestingly, although activating mutations in PDK1 have not yet been identified from cancer genome sequencing efforts (56), immunohistochemistry studies have revealed that PDK1 may be hyperactivated or overexpressed in a large percentage of invasive human breast cancers based on p-PDK1 Ser-241 quantification (21, 48, 57, 58). This clinical observation, together with the effect of PDK1 inhibition on cell invasion and migration reported herein, suggests that PDK1 inhibitors may find utility as part of a therapeutic strategy to target highly invasive and metastatic cancers.
DISCUSSION
Genetic validation of PDK1 as an oncology drug target has been extensively pursued (13, 19, 20, 57, 59, 60), but supporting pharmacological data has been problematic due to off-target effects of poorly selective compounds (11, 32, 61). Here, we have compared the enzymatic and cell biochemical potency of structurally diverse kinase inhibitors with activity against PDK1. By focusing on kinase selectivity and potency, we uncovered an exquisitely selective PDK1 inhibitor (compound 7), which we characterized in parallel with target knockdown to elucidate the role of PDK1 in cell-based biochemical and functional assays.
Because there are over 500 kinases in the human genome (62), the issue of selectivity for pharmacological inhibitors is critical when studying the cellular function of members of this enzyme family. For example, it can be challenging to obtain specificity of 80% (based on greater than 100-fold selectivity) for prototypical kinase inhibitors (Table 1). Nevertheless, even this degree of selectivity implies that ∼100 additional kinases may be inhibited within a treated cell. Notably, compound 7 exhibits greater than 3000-fold selectivity in a panel of 256 kinases (supplemental Table S1), making it a unique compound for in vitro target efficacy proof-of-concept studies. However, because “off-target” effects cannot formally be excluded for small molecules, parallel genetic knockdown studies were conducted to corroborate compound-induced phenotypes.
To our knowledge, compound 7 is the first example of a molecule that binds to the inactive conformation of an AGC kinase (Fig. 2a). The unique ability of compound 7 to cause PDK1 T-loop dephosphorylation further supports this crystallographic observation, because classical ATP competitive inhibitors (compounds 1–5) do not modulate Ser-241 phosphorylation in cells (Fig. 2b). Notably, in contrast to PDK1 substrate inhibition, which occurs rapidly within a few h following inhibitor treatment, the inhibition of p-PDK1 Ser-241 occurs relatively slowly within 1–2 days (Fig. 2b). Because phosphoproteins like AKT are highly regulated and susceptible to phosphatase activity in cells (Fig. 2b), we speculate that the slow dephosphorylation of p-PDK1 Ser-241 implies that this autophosphorylation site is not subject to tight physiological regulation by protein phosphatases; instead, the dephosphorylation is a direct consequence of the drug-induced conformational changes in the PDK1 enzyme that expose Ser-241 to the solvent and “housekeeping” phosphatase activity. Indeed, this notion is consistent with our reverse phase protein array analysis showing a relatively uniform level of p-PDK1 signal across a large panel of cancer cell lines (Fig. 6a) and with our analysis of PTEN-deficient mouse tumor models where the conditional activation of the PI3K pathway (through PTEN loss) does not lead to changes in PDK1 Ser-241 phosphorylation.
Although targeting the inactive form of tyrosine kinases has been postulated as a strategy to achieve high kinase selectivity, the proposed selectivity benefits have rarely been realized (41). In this example, it is unclear why compound 7 is substantially more selective than compounds 1–6 or compared with other type II kinase inhibitors like imatinib (63), BIRB796 (64), and sorafenib (65) that also bind to a kinase-inactive conformation. Clearly, unique enzyme-inhibitor interactions, made possible by the conformational flip of the DFG residues of the activation loop, provide a plausible explanation for the basis of specificity because compound 7 extends into an “allosteric” conformation-specific hydrophobic pocket adjacent to the ATP-binding pocket. However, our x-ray co-crystal structure also identifies a novel compound-induced disruption of the αC helix, which may contribute to the exquisite selectivity of compound 7. Whether this conformational change in the αC helix is unique to PDK1 or can be leveraged for targeting other AGC kinases is currently unknown.
In the context of cancer, we have defined a role for PDK1 in anchorage-independent cell growth, migration, and invasion. We show using pharmacological PDK1 inhibition that in some cell lines, these cellular processes depended on PDK1 kinase activity. This adds a critical component to existing target validation studies that rely on RNAi-mediated knockdown or ectopic overexpression of PDK1 (19, 21, 48). Because protein scaffolding functions, independent of PDK1 catalytic activity, have been shown to regulate cancer cell motility (19), our pharmacological data address the potential concern that PDK1 knockdown could be disrupting signaling complexes, thereby resulting in phenotypes not mimicked by a small molecule drug.
Notably, compound 7 inhibited the soft agar growth of both cancer cell lines and primary, patient-derived tumor xenograft lines with a response rate of 25% (four of 12) and 16% (nine of 57), respectively (Fig. 7 and Table 2). Interestingly, for the primary tumors, many of drug-responsive lines (five of nine) harbor oncogenic mutations in upstream activating RTKs, providing a biomarker hypothesis that is consistent with recent data suggesting that PDK1 activity can potentiate the transformed phenotype ErbB2 and potentially other RTKs. In contrast, no clear correlation between drug response and genetic mutations emerged from the cancer cell line panel. However, two of the responsive cancer cell lines display low basal levels of p-AKT, which may suggest a dependence on other PDK1 substrates for tumorigenicity, including PKC or SGK3, as proposed by Vasudevan et al. (21). In contrast to that study, we do not find that MCF7 cells, despite their low basal AKT activity (Fig. 5e), depend on PDK1 for soft agar growth and acknowledge that differences in culture conditions and passage numbers may lead to phenotypic variability of MCF7 cells (21). Instead, we identify inhibition of p-PDK1 Ser-241 as a novel candidate biomarker of response based on the correlation between T-loop dephosphorylation and growth inhibition in colony formation assays (Fig. 5e). Because increased levels of p-PDK1 Ser-241 protein have been reported in breast carcinomas (21, 48, 57, 58), targeting PDK1 with molecules like compound 7, which can induce a reduction in PDK1 T-loop phosphorylation, may provide an advantage over prototypical ATP-competitive PDK1 inhibitors (compounds 1–6).
Contrary to PDK1-dependent phenotypes in soft agar and Matrigel-based assays, PDK1 inhibition had minimal effect on plastic-attached monolayer cell growth in all 17 cancer cell lines tested (EC50 > 9 μm for compound 7; Table 1), suggesting that PDK1 activity is not rate-limiting for cell proliferation under standard tissue culture conditions (i.e. medium containing high glucose and 10% serum). Consistent with pharmacological PDK1 inhibition, acute gene silencing of PDK1 also failed to inhibit standard cell proliferation (Fig. 3e). This implies that the antiproliferative effects frequently reported for PDK1 kinase inhibitors in the literature and patent applications are misleading in terms of scoring the potency of PDK1 inhibitors in cells. For example, the antiproliferative effects reported for the celecoxib derivative OSU-03012 in PC-3 cells, which is being studied as a PDK1 inhibitor (66, 67) and is currently undergoing clinical trials (68, 69), is probably due to PDK1-independent effects. In fact, OSU-03012 lacks inhibitory activity in our full-length PDK1 enzymatic assay (28), consistent with another report (11). Likewise, the antiproliferative effects reported for the commonly used PDK1 inhibitor BX-795 are likely misleading. This compound has potent activity against many other anti-cancer drug targets, including Aura A and TBK1 (61), and shows antiproliferative effects in PDK1 knock-out (PDK1−/−) cells (13). Along these lines, derivatives of BX-795 optimized for selectivity possess 10–100-fold higher activity in soft agar growth assays compared with their poor activity under standard tissue culture conditions (70). Notably, PDK1−/− ES cells grow similarly to their wild-type counterparts under standard cell culture conditions, consistent with our observation that the PDK1 enzyme is not rate-limiting for in vitro cell growth of cancer cell lines (13). Interestingly, however, PDK1−/− ES cells display a survival disadvantage over wild-type ES cells when transplanted into nude mice (13) in support of reduced survival in the absence of PDK1 activity.
Unlike pharmacological PDK1 inhibition, six of 17 cell lines were sensitive (EC50 < 1 μm) to the Merck allosteric AKT inhibitor in standard cell proliferation assays (Table 2). Because both inhibitors display single digit nanomolar activity against their respective kinase targets in enzymatic assays, this result suggests a difference in the potential therapeutic effect of molecules that directly inhibit AKT versus indirectly via PDK1. One potential advantage of the allosteric AKT inhibitors may be the dual inhibition of both AKT Thr-308 and Ser-473 compared with PDK1 inhibitors, which only inhibit the steady-state phosphorylation of Thr-308. Indeed, phosphorylation of AKT Ser-473 in response to the mTORC1 inhibitor rapamycin has been highlighted as a potential issue that may limit its single agent activity in the clinic (71).
In summary, preclinical studies using pharmacological inhibitors may help stratify cancer patients and inform the design of clinical trails. Toward this goal, our in vitro studies suggest that potent and selective PDK1 inhibition may find use in oncology via inhibition of non-adherent cell growth, tumor cell migration, and invasion, which all play crucial roles in the metastatic process of malignant neoplasms. In the future, it will be important to study the outcome of in vivo pharmacological inhibition of PDK1 in tumor models, once favorable metabolic and pharmacokinetic properties have been incorporated into selective small molecule proof-of-concept inhibitors.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3NAY and 3NAX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- RTK
- receptor tyrosine kinase
- AGC
- cyclic AMP-dependent, cyclic guanosine 3′,5′-monophosphate-dependent, and protein kinase C
- GB
- GeneBlock
- mm
- mismatch sequence
- ES
- embryonic stem
- T-loop
- kinase activation loop
- PD
- pharmacodynamic
- S6RP
- S6 ribosomal protein
- p-PDK1
- -AKT, -S6K1, and -RSK, phospho-PDK1, -AKT, -S6K1, and -RSK, respectively.
REFERENCES
- 1. Alessi D. R., Deak M., Casamayor A., Caudwell F. B., Morrice N., Norman D. G., Gaffney P., Reese C. B., MacDougall C. N., Harbison D., Ashworth A., Bownes M. (1997) Curr. Biol. 7, 776–789 [DOI] [PubMed] [Google Scholar]
- 2. Anderson K. E., Coadwell J., Stephens L. R., Hawkins P. T. (1998) Curr. Biol. 8, 684–691 [DOI] [PubMed] [Google Scholar]
- 3. Engelman J. A., Luo J., Cantley L. C. (2006) Nat. Rev. Genet. 7, 606–619 [DOI] [PubMed] [Google Scholar]
- 4. Vanhaesebroeck B., Alessi D. R. (2000) Biochem. J. 346, 561–576 [PMC free article] [PubMed] [Google Scholar]
- 5. Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L. C. (2005) Trends Biochem. Sci. 30, 194–204 [DOI] [PubMed] [Google Scholar]
- 6. Ananthanarayanan B., Fosbrink M., Rahdar M., Zhang J. (2007) J. Biol. Chem. 282, 36634–36641 [DOI] [PubMed] [Google Scholar]
- 7. Calleja V., Laguerre M., Parker P. J., Larijani B. (2009) PLoS Biol. 7, e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams M. R., Arthur J. S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D. R. (2000) Curr. Biol. 10, 439–448 [DOI] [PubMed] [Google Scholar]
- 9. Mora A., Lipina C., Tronche F., Sutherland C., Alessi D. R. (2005) Biochem. J. 385, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Mora A., Komander D., van Aalten D. M., Alessi D. R. (2004) Semin. Cell Dev. Biol. 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 11. Peifer C., Alessi D. R. (2008) ChemMedChem 3, 1810–1838 [DOI] [PubMed] [Google Scholar]
- 12. Yuan T. L., Cantley L. C. (2008) Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamgüney T., Zhang C., Fiedler D., Shokat K., Stokoe D. (2008) Exp. Cell Res. 314, 2299–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Higuchi M., Onishi K., Kikuchi C., Gotoh Y. (2008) Nat. Cell Biol. 10, 1356–1364 [DOI] [PubMed] [Google Scholar]
- 15. Bilanges B., Stokoe D. (2005) Biochem. J. 388, 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayascas J. R., Wullschleger S., Sakamoto K., García-Martínez J. M., Clacher C., Komander D., van Aalten D. M., Boini K. M., Lang F., Lipina C., Logie L., Sutherland C., Chudek J. A., van Diepen J. A., Voshol P. J., Lucocq J. M., Alessi D. R. (2008) Mol. Cell. Biol. 28, 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collins B. J., Deak M., Murray-Tait V., Storey K. G., Alessi D. R. (2005) J. Cell Sci. 118, 5023–5034 [DOI] [PubMed] [Google Scholar]
- 18. Haga S., Ozaki M., Inoue H., Okamoto Y., Ogawa W., Takeda K., Akira S., Todo S. (2009) Hepatology 49, 204–214 [DOI] [PubMed] [Google Scholar]
- 19. Pinner S., Sahai E. (2008) Nat. Cell Biol. 10, 127–137 [DOI] [PubMed] [Google Scholar]
- 20. Lim M. A., Yang L., Zheng Y., Wu H., Dong L. Q., Liu F. (2004) Oncogene 23, 9348–9358 [DOI] [PubMed] [Google Scholar]
- 21. Vasudevan K. M., Barbie D. A., Davies M. A., Rabinovsky R., McNear C. J., Kim J. J., Hennessy B. T., Tseng H., Pochanard P., Kim S. Y., Dunn I. F., Schinzel A. C., Sandy P., Hoersch S., Sheng Q., Gupta P. B., Boehm J. S., Reiling J. H., Silver S., Lu Y., Stemke-Hale K., Dutta B., Joy C., Sahin A. A., Gonzalez-Angulo A. M., Lluch A., Rameh L. E., Jacks T., Root D. E., Lander E. S., Mills G. B., Hahn W. C., Sellers W. R., Garraway L. A. (2009) Cancer Cell 16, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moffat J., Sabatini D. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 177–187 [DOI] [PubMed] [Google Scholar]
- 24. Weihua Z., Tsan R., Huang W. C., Wu Q., Chiu C. H., Fidler I. J., Hung M. C. (2008) Cancer Cell 13, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fiebig H. H., Maier A., Burger A. M. (2004) Eur. J. Cancer 40, 802–820 [DOI] [PubMed] [Google Scholar]
- 26. Sternberger M., Schmiedeknecht A., Kretschmer A., Gebhardt F., Leenders F., Czauderna F., Von Carlowitz I., Engle M., Giese K., Beigelman L., Klippel A. (2002) Antisense Nucleic Acid Drug Dev. 12, 131–143 [DOI] [PubMed] [Google Scholar]
- 27. Sun D., Jung J., Rush T. S., 3rd, Xu Z., Weber M. J., Bobkova E., Northrup A., Kariv I. (2010) Comb. Chem. High Throughput Screen. 13, 16–26 [DOI] [PubMed] [Google Scholar]
- 28. Xu Z., Nagashima K., Sun D., Rush T., Northrup A., Andersen J. N., Kariv I., Bobkova E. V. (2009) J. Biomol. Screen. 10, 1257–1262 [DOI] [PubMed] [Google Scholar]
- 29. Thompson J. E., Cubbon R. M., Cummings R. T., Wicker L. S., Frankshun R., Cunningham B. R., Cameron P. M., Meinke P. T., Liverton N., Weng Y., DeMartino J. A. (2002) Bioorg. Med. Chem. Lett. 12, 1219–1223 [DOI] [PubMed] [Google Scholar]
- 30. Lucet I. S., Fantino E., Styles M., Bamert R., Patel O., Broughton S. E., Walter M., Burns C. J., Treutlein H., Wilks A. F., Rossjohn J. (2006) Blood 107, 176–183 [DOI] [PubMed] [Google Scholar]
- 31. Kozina E., Dinsmore C., Siu T., Young J., Northrup A., Altman M., Keenan K. A., Guerin D. J., Jung J. O., Maccoss R. N., Kattar S. (June 24, 2010) U.S. Patent 2010/0160309
- 32. Feldman R. I., Wu J. M., Polokoff M. A., Kochanny M. J., Dinter H., Zhu D., Biroc S. L., Alicke B., Bryant J., Yuan S., Buckman B. O., Lentz D., Ferrer M., Whitlow M., Adler M., Finster S., Chang Z., Arnaiz D. O. (2005) J. Biol. Chem. 280, 19867–19874 [DOI] [PubMed] [Google Scholar]
- 33. Weisberg E., Banerji L., Wright R. D., Barrett R., Ray A., Moreno D., Catley L., Jiang J., Hall-Meyers E., Sauveur-Michel M., Stone R., Galinsky I., Fox E., Kung A. L., Griffin J. D. (2008) Blood 111, 3723–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maira S. M., Stauffer F., Brueggen J., Furet P., Schnell C., Fritsch C., Brachmann S., Chène P., De Pover A., Schoemaker K., Fabbro D., Gabriel D., Simonen M., Murphy L., Finan P., Sellers W., García-Echeverría C. (2008) Mol. Cancer Ther. 7, 1851–1863 [DOI] [PubMed] [Google Scholar]
- 35. Boyle R. G., Imogai H. J., Cherry M., Humphries A. J., Navarro E. F., Owen D. R., Dales N. A., Lamarche M., Cullis C., Gould A. E. (March 31, 2005) Patent WO/2005/028474
- 36. Walmsley D. L., Drysdale M. J., Northfield C. J., Fromont C. (December 21, 2006) Patent WO/2006/134318
- 37. Lind K. E., Lin E. Y., Nguyen T. B., Tangonan B. T., Erlanson D. A., Guckian K., Simmons R. L., Lee W., Sun L., Hansen S., Pathan N., Zhang L. (January 10, 2008) Patent WO/2008/005457
- 38. Islam I., Bryant J., Chou Y. L., Kochanny M. J., Lee W., Phillips G. B., Yu H., Adler M., Whitlow M., Ho E., Lentz D., Polokoff M. A., Subramanyam B., Wu J. M., Zhu D., Feldman R. I., Arnaiz D. O. (2007) Bioorg. Med. Chem. Lett. 17, 3814–3818 [DOI] [PubMed] [Google Scholar]
- 39. Komander D., Kular G. S., Bain J., Elliott M., Alessi D. R., Van Aalten D. M. (2003) Biochem. J. 375, 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dar A. C., Lopez M. S., Shokat K. M. (2008) Chem. Biol. 15, 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y., Gray N. S. (2006) Nat. Chem. Biol. 2, 358–364 [DOI] [PubMed] [Google Scholar]
- 42. Frödin M., Antal T. L., Dümmler B. A., Jensen C. J., Deak M., Gammeltoft S., Biondi R. M. (2002) EMBO J. 21, 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bilodeau M. T., Balitza A. E., Hoffman J. M., Manley P. J., Barnett S. F., Defeo-Jones D., Haskell K., Jones R. E., Leander K., Robinson R. G., Smith A. M., Huber H. E., Hartman G. D. (2008) Bioorg. Med. Chem. Lett. 18, 3178–3182 [DOI] [PubMed] [Google Scholar]
- 44. Urlinger S., Baron U., Thellmann M., Hasan M. T., Bujard H., Hillen W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shoemaker R. H., Wolpert-DeFilippes M. K., Kern D. H., Lieber M. M., Makuch R. W., Melnick N. R., Miller W. T., Salmon S. E., Simon R. M., Venditti J. M. (1985) Cancer Res. 45, 2145–2153 [PubMed] [Google Scholar]
- 46. Myers M. P., Stolarov J. P., Eng C., Li J., Wang S. I., Wigler M. H., Parsons R., Tonks N. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiznerowicz M., Szulc J., Trono D. (2006) Nat. Methods 3, 682–688 [DOI] [PubMed] [Google Scholar]
- 48. Maurer M., Su T., Saal L. H., Koujak S., Hopkins B. D., Barkley C. R., Wu J., Nandula S., Dutta B., Xie Y., Chin Y. R., Kim D. I., Ferris J. S., Gruvberger-Saal S. K., Laakso M., Wang X., Memeo L., Rojtman A., Matos T., Yu J. S., Cordon-Cardo C., Isola J., Terry M. B., Toker A., Mills G. B., Zhao J. J., Murty V. V., Hibshoosh H., Parsons R. (2009) Cancer Res. 69, 6299–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., Brugge J. S. (2001) Nat. Cell Biol. 3, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leenders F., Möpert K., Schmiedeknecht A., Santel A., Czauderna F., Aleku M., Penschuck S., Dames S., Sternberger M., Röhl T., Wellmann A., Arnold W., Giese K., Kaufmann J., Klippel A. (2004) EMBO J. 23, 3303–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ivascu A., Kubbies M. (2006) J. Biomol. Screen. 11, 922–932 [DOI] [PubMed] [Google Scholar]
- 52. Sliva D., Rizzo M. T., English D. (2002) J. Biol. Chem. 277, 3150–3157 [DOI] [PubMed] [Google Scholar]
- 53. Shukla S., Maclennan G. T., Hartman D. J., Fu P., Resnick M. I., Gupta S. (2007) Int. J. Cancer 121, 1424–1432 [DOI] [PubMed] [Google Scholar]
- 54. Liliental J., Moon S. Y., Lesche R., Mamillapalli R., Li D., Zheng Y., Sun H., Wu H. (2000) Curr. Biol. 10, 401–404 [DOI] [PubMed] [Google Scholar]
- 55. Shaw K. R., Wrobel C. N., Brugge J. S. (2004) J. Mammary. Gland. Biol. Neoplasia 9, 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Z., Hou P., Ji M., Guan H., Studeman K., Jensen K., Vasko V., El-Naggar A. K., Xing M. (2008) J. Clin. Endocrinol. Metab. 93, 3106–3116 [DOI] [PubMed] [Google Scholar]
- 57. Xie Z., Yuan H., Yin Y., Zeng X., Bai R., Glazer R. I. (2006) BMC Cancer 6, 77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin H. J., Hsieh F. C., Song H., Lin J. (2005) Br. J. Cancer 93, 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bayascas J. R., Leslie N. R., Parsons R., Fleming S., Alessi D. R. (2005) Curr. Biol. 15, 1839–1846 [DOI] [PubMed] [Google Scholar]
- 60. Garcia-Echeverria C., Sellers W. R. (2008) Oncogene 27, 5511–5526 [DOI] [PubMed] [Google Scholar]
- 61. Clark K., Plater L., Peggie M., Cohen P. (2009) J. Biol. Chem. 284, 14136–14146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 63. Schindler T., Bornmann W., Pellicena P., Miller W. T., Clarkson B., Kuriyan J. (2000) Science 289, 1938–1942 [DOI] [PubMed] [Google Scholar]
- 64. Pargellis C., Tong L., Churchill L., Cirillo P. F., Gilmore T., Graham A. G., Grob P. M., Hickey E. R., Moss N., Pav S., Regan J. (2002) Nat. Struct. Biol. 9, 268–272 [DOI] [PubMed] [Google Scholar]
- 65. Wan P. T., Garnett M. J., Roe S. M., Lee S., Niculescu-Duvaz D., Good V. M., Jones C. M., Marshall C. J., Springer C. J., Barford D., Marais R. (2004) Cell 116, 855–867 [DOI] [PubMed] [Google Scholar]
- 66. Zhu J., Huang J. W., Tseng P. H., Yang Y. T., Fowble J., Shiau C. W., Shaw Y. J., Kulp S. K., Chen C. S. (2004) Cancer Res. 64, 4309–4318 [DOI] [PubMed] [Google Scholar]
- 67. Gao M., Yeh P. Y., Lu Y. S., Hsu C. H., Chen K. F., Lee W. C., Feng W. C., Chen C. S., Kuo M. L., Cheng A. L. (2008) Cancer Res. 68, 9348–9357 [DOI] [PubMed] [Google Scholar]
- 68. Welch S., Hirte H. W., Carey M. S., Hotte S. J., Tsao M. S., Brown S., Pond G. R., Dancey J. E., Oza A. M. (2007) Gynecol. Oncol. 106, 305–310 [DOI] [PubMed] [Google Scholar]
- 69. Edelman M. J., Bauer K. S., Jr., Wu S., Smith R., Bisacia S., Dancey J. (2007) Clin. Cancer Res. 13, 2667–2674 [DOI] [PubMed] [Google Scholar]
- 70. Islam I., Brown G., Bryant J., Hrvatin P., Kochanny M. J., Phillips G. B., Yuan S., Adler M., Whitlow M., Lentz D., Polokoff M. A., Wu J., Shen J., Walters J., Ho E., Subramanyam B., Zhu D., Feldman R. I., Arnaiz D. O. (2007) Bioorg. Med. Chem. Lett. 17, 3819–3825 [DOI] [PubMed] [Google Scholar]
- 71. O'Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. (2006) Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.