Abstract
Mono-ADP-ribosylation is a reversible post-translational modification that can modulate the functions of target proteins. We have previously demonstrated that the β subunit of heterotrimeric G proteins is endogenously mono-ADP-ribosylated, and once modified, the βγ dimer is inactive toward its effector enzymes. To better understand the physiological relevance of this post-translational modification, we have studied its hormonal regulation. Here, we report that Gβ subunit mono-ADP-ribosylation is differentially modulated by G protein-coupled receptors. In intact cells, hormone stimulation of the thrombin receptor induces Gβ subunit mono-ADP-ribosylation, which can affect G protein signaling. Conversely, hormone stimulation of the gonadotropin-releasing hormone receptor (GnRHR) inhibits Gβ subunit mono-ADP-ribosylation. We also provide the first demonstration that activation of the GnRHR can activate the ADP-ribosylation factor Arf6, which in turn inhibits Gβ subunit mono-ADP-ribosylation. Indeed, removal of Arf6 from purified plasma membranes results in loss of GnRHR-mediated inhibition of Gβ subunit mono-ADP-ribosylation, which is fully restored by re-addition of purified, myristoylated Arf6. We show that Arf6 acts as a competitive inhibitor of the endogenous ADP-ribosyltransferase and is itself modified by this enzyme. These data provide further understanding of the mechanisms that regulate endogenous ADP-ribosylation of the Gβ subunit, and they demonstrate a novel role for Arf6 in hormone regulation of Gβ subunit mono-ADP-ribosylation.
Keywords: ADP-ribosylation, G Protein-coupled Receptors (GPCR), Heterotrimeric G Proteins, NAD, Post-translational Modification, Signal Transduction, ADP-ribosylhydrolase, ADP-ribosyltransferase, ARF6, ERK
Introduction
Mono-ADP-ribosylation is a post-translational modification that is catalyzed by bacterial toxins and eukaryotic enzymes and consists of the transfer of the ADP-ribose moiety from β-NAD+ to a specific amino acid of various cellular proteins (1–3). We have previously demonstrated the occurrence of functional mono-ADP-ribosylation of the heterotrimeric G protein β subunit (4, 5).
The heterotrimeric G proteins are the most common transducers for G protein-coupled receptors (GPCRs),4 the largest family of cell-surface receptors. They transduce biological signals from the cell surface to the intracellular compartment (6, 7). In the classical model of G protein signaling, when GPCRs are activated by ligand binding they catalyze the exchange of the tightly bound GDP for GTP on the α subunit of the heterotrimeric G protein; the binding of GTP activates the G protein and the Gα subunit dissociates from the Gβγ dimer. Both the Gα subunit and the Gβγ dimer regulate effector proteins, including adenylyl cyclases, phospholipases, MAPK, and ion channels (see Refs. 8, 9 and references therein). Thus, although the Gβγ dimers were once thought to only be negative regulators of Gα-dependent signaling, they are now well recognized as mediators of receptor signaling in their own right (8, 9).
We have previously demonstrated that a membrane-associated, arginine-specific mono-ADP-ribosyltransferase modifies residue 129 of the Gβ subunit when it is in its active heterodimeric conformation (4, 5, 10). This critical residue is located in the common effector binding surface of the β subunit, and the resulting mono-ADP-ribosylated Gβγ dimer cannot modulate the activity of its effectors adenylyl cyclase, phosphoinositide 3-kinase, and phospholipase C (4, 5). The modified Gβ subunit can then be de-ADP-ribosylated by a cytosolic ADP-ribosylhydrolase (4, 5), thus revealing a cellular ADP-ribosylation/de-ADP-ribosylation cycle that might parallel a functional activation/inactivation cycle of the Gβγ dimer (3–5).
To better understand the physiological relevance of Gβγ mono-ADP-ribosylation, we have studied its hormonal regulation. Our data demonstrate that activation of different GPCRs can lead to either stimulation or inhibition of the endogenous Gβ subunit mono-ADP-ribosylation. Interestingly, stimulation of the gonadotropin-releasing hormone receptor (GnRHR) led to activation of the small G protein Arf6, which in turn inhibited Gβγ dimer mono-ADP-ribosylation. The ADP-ribosylation factors (Arfs) are 20-kDa guanine nucleotide-binding proteins, and they were initially discovered through their ability to allosterically activate the ADP-ribosyltransferase (ART) activity of cholera toxin A1 peptide (CTA1) in host cells (11). Only Arf-GTP (the active, GTP-bound form of the Arfs) can bind effector proteins and activate CTA1 (11). Recently, the crystal structure of a complex between CTA1 and Arf6-GTP was solved (12), revealing that the binding of Arf6-GTP to CTA1 causes major conformational changes to CTA1, which can consequently bind the G protein substrate, Gαs (12). Arf6 is the most divergent member of the Arf family with regard to its sequence, and it constitutes the sole member of the class III Arfs (13). Arf6 localizes to the plasma membrane (14–17) and to some extent to endosomal membranes, where it can act in a wide range of processes, including endocytosis, cytokinesis, and the organization of the actin cytoskeleton (15, 16). Moreover, Arf6 was recently shown to modulate the internalization of various GPCRs (18, 19).
Here, we show that Arf6-GTP modulates the activity of a mammalian ART. We have previously excluded any role of the extracellular, membrane-associated ART enzymes (20) in endogenous Gβ subunit mono-ADP-ribosylation (4). The mammalian intracellular ART enzymes are only now beginning to be identified, and they include two sirtuins and novel members of the poly(ADP-ribose) polymerase (PARP) family (3, 21–23). Some of these PARP enzymes have been proposed to act as cellular mono-ADP-ribosyltransferases, and indeed, this has been demonstrated for PARP10 (24). It is thus possible that novel members of this PARP-like family have an enzymatic activity that modifies the Gβγ dimer and is regulated by Arf6-GTP. The data reported here delineate a novel regulatory pathway that is initiated by GPCR activation and that can modulate G protein signaling through regulation of Gβ subunit mono-ADP-ribosylation.
EXPERIMENTAL PROCEDURES
Cell Culture and Fractionation and Protein Purification
Chinese hamster ovary (CHO) cells were from American Type Culture Collection (ATCC) and were grown as described previously (4). CHO cells stably transfected with the GnRHR were kindly provided by Dr. Roelle (Institute of Pharmacology and Toxicology, Marburg, Germany) and were grown in Ham's F-12 supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 ng/ml streptomycin, and 10% fetal calf serum (FCS) in the presence of 6.25 μg/ml puromycin. CHO cells stably transfected with the adrenergic receptors were kindly provided by Dr. Costa (Istituto Superiore di Sanità, Rome, Italy) and were grown in DMEM/Ham's F-12 (1:1 v/v) supplemented with 34 mg/ml proline, 2 mm l-glutamine, 100 units/ml penicillin, 100 ng/ml streptomycin, and 10% FCS in the presence of 200 μg/ml G418.
Plasma membranes were prepared as described previously (4). Lipofectamine 2000 (Invitrogen) was used to transiently transfect CHO cells, following the manufacturer's instructions. CHO plasma membranes were depleted of Arf6 by incubation with 0.1% v/v Triton X-100 for 30 min at 4 °C, then centrifuged at 13,000 × g, and washed twice with phosphate-buffered saline (PBS). Bovine brain βγ was purified as described previously (5, 25) and concentrated to 2 mg/ml in 50 mm Tris (pH 8.0), 100 mm NaCl, 1 mm DTT, 0.7% CHAPS. The myristoylated Arf6 protein was purified as described previously (26). The Arf1 and Arf6 activities were evaluated using an in vitro ADP-ribosylation assay (see below) that measured their ability to increase the cholera toxin-mediated [32P]ADP-ribosylation of Gαs.
ADP-ribosylation Assay and Immunoblot Analysis
ART activity was measured by following incorporation of radioactive ADP-ribose into membrane proteins, as described previously (4, 10). Briefly, samples (5 μg of plasma membranes) were incubated with 50 μl of ADP-ribosylation buffer (50 mm potassium phosphate buffer (pH 7.4), 5 mm MgCl2, 4 mm dithiothreitol (DTT), 10 μm GTPγS, 700 μm β-NAD+, and 4.5 μCi of [32P]NAD+ (specific activity, 1,000 Ci/mmol) for 60 min at 37 °C, unless otherwise specified. To detect Arf6 ADP-ribosylation, the purified Arf proteins were incubated for 15 min with plasma membranes before the assay. The samples were analyzed by 10% SDS-PAGE, and proteins were electroblotted (4 h at 500 mA) onto nitrocellulose membranes. In all experiments, the filters were exposed to Kodak X-Omat film using an intensifying screen, quantified for the radiolabeled βγ with an InstantImager (Packard Instrument Co.), and then probed with specific primary and peroxidase-conjugated secondary antibodies using chemiluminescence (ECL, Amersham Biosciences). Although this system was previously characterized for the βγ (4, 10), all samples were routinely probed for continuous internal monitoring. The following antibodies were used: an antiserum raised against the C-terminal peptide of the β subunit and an anti-Arf6 monoclonal antibody from Santa Cruz Biotechnology or a monoclonal anti-HA from Covance.
Deribosylation Assay, ARH1 Cloning, and siRNA of ARH1
Following the ADP-ribosylation assay, 32P-labeled membranes were washed twice with 5 mm Tris-HCl (pH 8.0) and incubated for 30 min at 37 °C with ARH1 or ARH3, as indicated (50 ng in 50 μl of 5 mm Tris-HCl (pH 8.0) and protease inhibitors). The analysis of protein samples was performed as described for the ADP-ribosylation assay. ARH1 and ARH3 were purchased from Alexis Biochemicals. Hamster ARH1 was cloned from CHO-DNA by rapid amplification of cDNA ends-PCR. The PCR products were cloned using the TA-cloning technique in the pCR2.1 vector and analyzed by sequencing as follows: forward, 5′-atgggagacatggatggccgggcaccaggt-3′, and reverse, 5′-gataccctgggctacttcaacgggaagtgg-3′. The following four siRNAs were designed with the Dharmacon siDESIGN Center software, on the basis of the nucleotide sequence of the cloned CHO-ARH1: GAAGTACATTGTCCAGTCA, GAACGTGATCAGCTCTATA, GAAGGAACTTGCACACAGA, and TCGTGGAAGCTGACAAATT.
To silence ARH1, CHO cells were transfected with a pool of four siRNAs (200 nm), using Lipofectamine2000. Expression of ARH1 was evaluated by immunoblotting with an anti-ARH1 antibody. ARH1 silencing was visible after 24 h of transfection, and 85% of endogenous ARH1 was knocked down after 72 h.
Cell Permeabilization
Tetanolysin permeabilization of CHO cells was performed as described previously (27), with some modifications. CHO cells were grown in 6-well plates to semi-confluence, washed with 2 ml/well PBS, and then incubated for 10 min at 4 °C with 1 ml/well ice-cold HGI buffer (20 mm PIPES, 2 mm NaATP, 4.8 mm Mg(CH3COO)2, 150 mm potassium glutamate, 2 mm EGTA, 1 mm DTT, KOH to pH 7.0) containing 0.5 μg of tetanolysin. The cells were gently washed with 500 μl of ice-cold HGI buffer and then incubated for the indicated times at 37 °C, with the addition of 1 ml/well HGI buffer containing 4.5 μCi of [32P]NAD and the stimuli as required (1 unit/ml thrombin, 5 nm pertussis toxin, 25 μm Mas7). The cells were then washed twice with PBS and harvested directly into 200 μl of Laemmli sample buffer. Finally, the proteins were resolved by 10% SDS-PAGE and Western blotting (WB). The levels of cell permeabilization were quantified using trypan blue (4 mg/ml).
Determination of Relative Arf6-GTP Levels
The levels of activated Arf6 were determined using a pulldown assay, as described previously (28), with minor modifications. A GST-GGA3 (Golgi-localized, γ-ear-containing Arf-binding protein 3) fusion protein containing the Arf-GTP binding domains of GGA3 was purified from Escherichia coli as described previously (28). The pGEX4T1 vector containing the GST-GGA3(1–226) fusion protein was kindly provided by Dr. P. Chavrier (Institut Curie, Paris, France). GnRHR-CHO cells transiently transfected with FLAG-Arf6 were serum-starved for 4 h and then stimulated with 20 nm GnRH for 15 min. The cells were then lysed with 1 ml of lysis buffer (50 mm Tris-HCl (pH 7.4), 137 mm NaCl, 10 mm MgCl2, 1% Triton X-100, 10% v/v glycerol, plus protease inhibitors). Lysates were cleared by centrifugation at 12,000 × g for 15 min at 4 °C and then snap-frozen in liquid N2 and stored at −80 °C. For each pulldown assay, 50 μg of the GST-GGA3 fusion protein was incubated with lysates in the presence of 0.5% BSA and rotated at 4 °C for 15 min. The samples were pulled down using G-Sepharose beads (Amersham Biosciences) for 1 h at 4 °C. The beads were collected by centrifugation at 700 × g for 5 min and then washed three times with 1 ml of lysis buffer. The protein bound to the resin was then eluted with sample buffer, separated on 10% SDS-PAGE, and analyzed by WB using an anti-Arf6 antibody. The specificity of this assay was confirmed by using GnRH-CHO cell extracts from cells expressing Arf6 as either its GTP-bound form of Arf6(Q67L) or its GDP-bound form of Arf6(T27N). Arf6(Q67L), but not Arf6(T27N), bound GST-GGA3 and was pulled down by the glutathione resin. The plasmids encoding for FLAG-Arf6, FLAG-Arf6(Q67L), and FLAG-Arf6(T27N) were kindly provided by Dr. R. Weigert (National Institutes of Health, Bethesda).
ERK1/2 Assay
The activation status of ERK1/2 was determined as described previously (29) using a mouse anti-p-ERK monoclonal antibody (Cell Signaling) and a rabbit anti-ERK1/2 polyclonal antibody (Santa Cruz Biotechnology).
Statistical Analysis
Data are expressed as means ± S.E. Statistical analyses (Student's t tests) were performed on the means of the replicates within each experimental dataset, and p values were calculated using the GraphPad PRISM software. p < 0.05 was considered significant, unless otherwise indicated.
RESULTS
Mono-ADP-ribosylation of the G Protein β Subunit Is Modulated by GPCRs
We have previously demonstrated that hormones that act on different GPCRs, including thrombin, serotonin, and cholecystokinin, can increase the levels of the ADP-ribosylated β subunit in in vitro ADP-ribosylation assays (5). Here, we have analyzed the mechanisms involved in this hormone modulation.
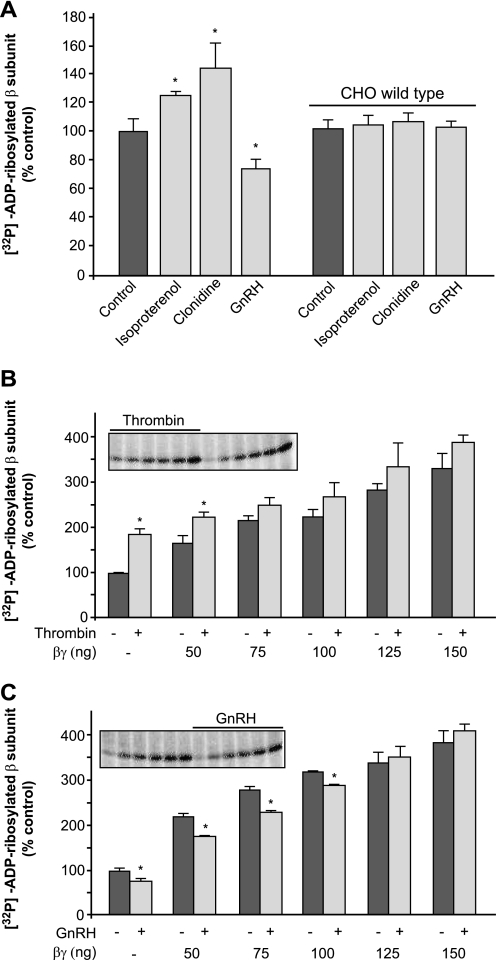
Agonist stimulation of plasma membranes from CHO cells stably transfected with the β2-adrenoreceptor and the α2-adrenoreceptor resulted in increases in β subunit [32P]ADP-ribosylation of 26 ± 3 and 45 ± 16%, respectively (Fig. 1A). This is in line with our previous demonstration that the release of free Gβγ dimer through GPCR activation can lead to mono-ADP-ribosylation of the Gβ subunit (5, 29). However, when we examined Gβ subunit [32P]ADP-ribosylation following stimulation of plasma membranes obtained from CHO cells stably transfected with the GnRHR (GnRHR-CHO cells), a decrease in the levels of modified Gβ subunit was seen (28 ± 6% inhibition, compared with nonstimulated cells; Fig. 1A). Stimulation of plasma membranes from wild-type CHO cells (which do not express α2-adrenoreceptor, β2-adrenoreceptor, and GnRHR) with clonidine, isoproterenol, and GnRH did not modulate this labeling of the Gβ subunit (Fig. 1A), confirming the specificity of this agonist-mediated modulation of Gβ subunit ADP-ribosylation.
FIGURE 1.
G protein β subunit ADP-ribosylation is modulated by GPCR activation. A–C, plasma membrane protein (5 μg) from wild-type CHO cells (A and B) or CHO cells stably transfected with α2-adrenoreceptor (A), β2-adrenoreceptor (A), and GnRHR (A and C) were ADP-ribosylated with [32P]NAD+ for 60 min at 37 °C in the absence (A–C) or presence (B and C) of purified Gβγ dimer and of 10 μm isoproterenol, 15 μm clonidine, 200 nm GnRH, or 10 IU/ml thrombin, as indicated. Fifty ng of purified Gβγ dimer are equivalent to the endogenous Gβ in 5 μg of protein from CHO plasma membranes and correspond to a concentration of 20 nm under these experimental conditions. The solubilized proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes; the counts/min of the [32P]ADP-ribosylated β subunit were quantified by analyzing the filters with an InstantImager, and they are reported as relative (%) to the control (unstimulated plasma membranes). The insets in B and C show images from a typical experiment, as provided by the InstantImager. The data shown represent means (±S.D.) of five independent experiments, each performed in duplicate. *, significantly different from the relevant control (p < 0.01; n = 5).
We then focused on the GnRHR because of its ability to inhibit β subunit [32P]ADP-ribosylation. In parallel to the stimulatory GPCRs, we investigated the thrombin receptor further, which is endogenously expressed in CHO cells (5, 29) and which, when activated, provided the greatest increase in ADP-ribosylation of the β subunit (5). Fig. 1B shows that the thrombin-dependent increase in β subunit [32P]ADP-ribosylation was masked by increasing concentrations of purified bovine brain βγ dimer added to the plasma membranes before the ADP-ribosylation assay (we have previously demonstrated that purified βγ dimer added to the assay mixture is also [32P]ADP-ribosylated (4, 5, 10)). This indicates that the stimulatory effect of thrombin, which is evident in the absence of purified βγ dimers, is due to the release of free βγ from the subset of G proteins coupled to the thrombin receptor, rather than to a thrombin-receptor-mediated activation of the plasma membrane-associated intracellular ART activity. On the contrary, when increasing concentrations of purified bovine brain βγ dimer were added to plasma membranes before the ADP-ribosylation assay in the presence of mastoparan 7, the 2-fold medium stimulation induced by mastoparan 7 was not affected (up to 400 ng of βγ dimer; data not shown). This is in line with mastoparan-7-mediated activation of the ART activity, in agreement with our previous finding that pertussis toxin can only partially block the mastoparan-7-dependent increase in β subunit [32P]ADP-ribosylation (35% inhibition; see data reported in Ref. 5) and in support of our conclusion that thrombin-receptor stimulation did not directly activate the ART enzyme. Moreover, in contrast to the thrombin effect, the GnRH-dependent inhibition of β subunit [32P]ADP-ribosylation was maintained with the addition of free βγ dimer up to 100 ng (Fig. 1C), indicating that activation of the GnRHR can inhibit the plasma membrane-associated ART activity.
Mono-ADP-ribosylation of the G Protein β Subunit Is Induced by Thrombin in Intact Cells and Has a Role in Thrombin Signaling
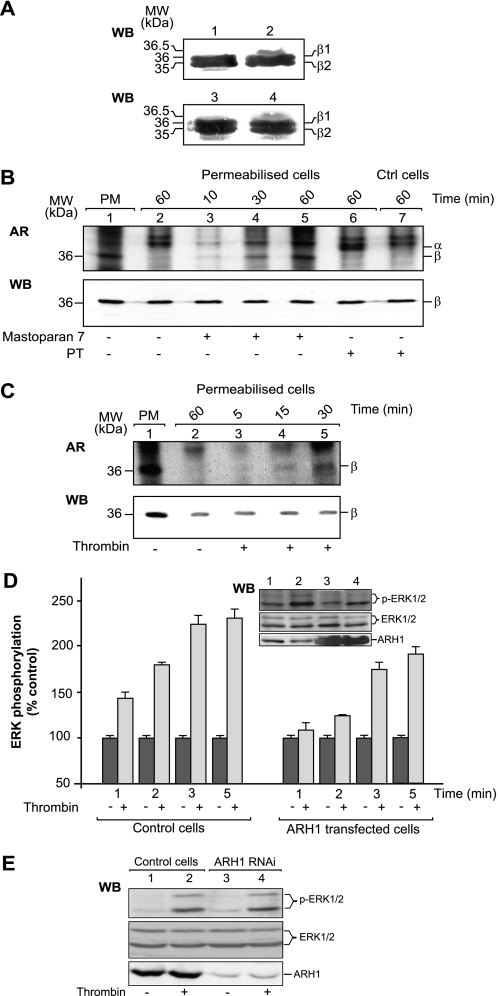
Importantly, thrombin-induced Gβ subunit mono-ADP-ribosylation was confirmed in intact cells (Fig. 2A). Intact CHO cells were stimulated with thrombin, and plasma membranes were then purified and analyzed by WB with a specific anti-Gβ antibody to visualize the ∼500-Da shift in the molecular mass of the Gβ subunit that is due to the addition of an ADP-ribose moiety (Fig. 2A). A limitation of this type of experiment arises from the very low amount of this post-translational modification; indeed, under basal conditions, ∼0.2% of the total cellular Gβ subunit is modified in CHO cells (4, 5). Thus, to detect endogenously ADP-ribosylated β subunit, 150 μg of purified plasma membranes/sample (containing ∼1.5 μg of Gβγ) were analyzed, and SDS-PAGE was performed using long gels for the separation of the unmodified and modified Gβ subunit. In plasma membranes from unstimulated cells (Fig. 2A, lane 1), the antibody revealed two bands, of 36 and 35 kDa, corresponding to the unmodified β1 and β2 subunits, respectively. An additional band of 36.5 kDa was revealed in the plasma membranes purified from thrombin-stimulated cells (Fig. 2A, lane 2). As a further control, we analyzed 150 μg of purified plasma membrane in the in vitro ADP-ribosylation assay, performed under experimental conditions that provided the low level of modification that we expected to occur in vivo upon thrombin stimulation (15 min of incubation with 30 μm NAD; Fig. 2A, lane 4; lane 3 shows the same sample incubated in the absence of NAD). Although the large amounts of protein applied to the gels caused some diffusion and lengthening of the bands, the three bands corresponding to the unmodified and modified β subunits can be resolved, and as expected, the modified β subunit was present at nanogram levels (versus microgram levels of the unmodified β subunit) and was therefore less laterally diffuse than the other two bands. It should also be noted that the protein of 36.5 kDa represents only the ADP-ribosylated β1 subunit; ADP-ribosylation of β2 could not be evaluated due to its co-migration with unmodified β1.
FIGURE 2.
G protein β subunit ADP-ribosylation is stimulated by thrombin in intact and permeabilized cells and can modulate ERK1/2. A, CHO cells were either untreated (lanes 1, 3, and 4) or stimulated with thrombin (1 IU/ml) for 30 min (lane 2), and then plasma membranes were purified and either directly analyzed by WB with an anti-Gβ antibody (lanes 1–3) or subjected to an in vitro ADP-ribosylation assay (150 μg incubated with 30 μm β-NAD+ for 15 min to obtain ∼4–5 ng of modified β subunit) before WB (lane 4). The doublet of β1 and β2 subunits was resolved using extra long gels for protein separation (4, 5). B and C, CHO cells were either untreated (control; B, lane 7) or permeabilized with tetanolysin. In vitro ADP-ribosylated CHO plasma membranes (PM) (5 μg) were used as a further control (B and C, lane 1). Both permeabilized and non-permeabilized cells were incubated with [32P]NAD and with the indicated stimuli (1 unit/ml thrombin, 10 nm pertussis toxin, 25 μm mastoparan 7) for the indicated times. The solubilized proteins were analyzed by autoradiography (AR) and WB with an anti-Gβ antibody. The data shown are representative of 5–10 experiments. D, densitometric quantification and Western blotting (inset) of phosphorylation levels of ERK1/2 in mock-transfected (control cells) and ARH1-overexpressing CHO cells. ERK1/2 activity was evaluated after the indicated times of thrombin (0.5 units/ml) stimulation in control cells or in CHO cells transfected with ARH1. The phosphorylated ERK1/2 was normalized relative to the control (unstimulated cells) total ERK1/2 (%). The data are means (±S.D.) of five independent experiments, each performed in duplicate. The WB in the inset is representative of this ARH1 inhibition as follows: control cells (lanes 1 and 2), cells stimulated with thrombin (lanes 2 and 4; 0.5 units/ml, 5 min), and cells overexpressing ARH1 (lanes 3 and 4). E, WB of a filter labeled with the anti-phospho-ERK1/2 (p-ERK1/2) and anti-ERK1/2 (ERK1/2) antibodies as follows: thrombin stimulation (lanes 2 and 4; 0.5 unit/ml, 5 min) in control cells (lanes 1 and 2) and in cells silenced for ARH1 (lanes 3 and 4). The data are representative of three independent experiments.
The ability of GnRHR stimulation to inhibit β subunit [32P]ADP-ribosylation cannot be evaluated in this system, due to the limitations of this type of experiment. To further investigate hormonal modulation of mono-ADP-ribosylation of the β subunit, we used tetanolysin-permeabilized cells. Tetanolysin is a 55-kDa member of the streptolysin-O family of cholesterol-dependent pore-forming toxins (27), and this system has the advantage of preserving cell structure while allowing [32P]NAD influx (27). Semi-confluent tetanolysin-permeabilized CHO cells were incubated in the presence of [32P]NAD, pertussis toxin (control), mastoparan 7, and thrombin or GnRH. The levels of [32P]ADP-ribosylated β subunit were then analyzed by autoradiography. As expected, pertussis toxin catalyzed the incorporation of [32P]ADP-ribose into a protein with the predicted molecular mass of the Gα subunit in these permeabilized cells but not in the non-permeabilized control cells (Fig. 2B, compare lanes 6 and 7). Moreover, in unstimulated, permeabilized cells (Fig. 2B, lane 2), a weak 32P-labeled band was detected that co-migrated with the in vitro [32P]ADP-ribosylated β subunit (Fig. 2B, lane 1) and that was recognized by the specific anti-β antibody (WB, Fig. 2B, bottom panel, lane 2). As a further control of these experimental conditions, we also evaluated the effects of mastoparan 7. The β subunit labeling significantly increased upon mastoparan 7 addition in a time-dependent manner (Fig. 2B, lanes 3–5). Under the same experimental conditions, thrombin induced an increase in the labeling of the β subunit, with respect to unstimulated cells (Fig. 2C, lanes 3–5). In contrast, incubation of permeabilized GnRHR-CHO cells with [32P]NAD+ revealed only very weak labeling of the β subunit. This was insufficient for the detection of any inhibitory effects due to the stimulation of the GnRHR (data not shown).
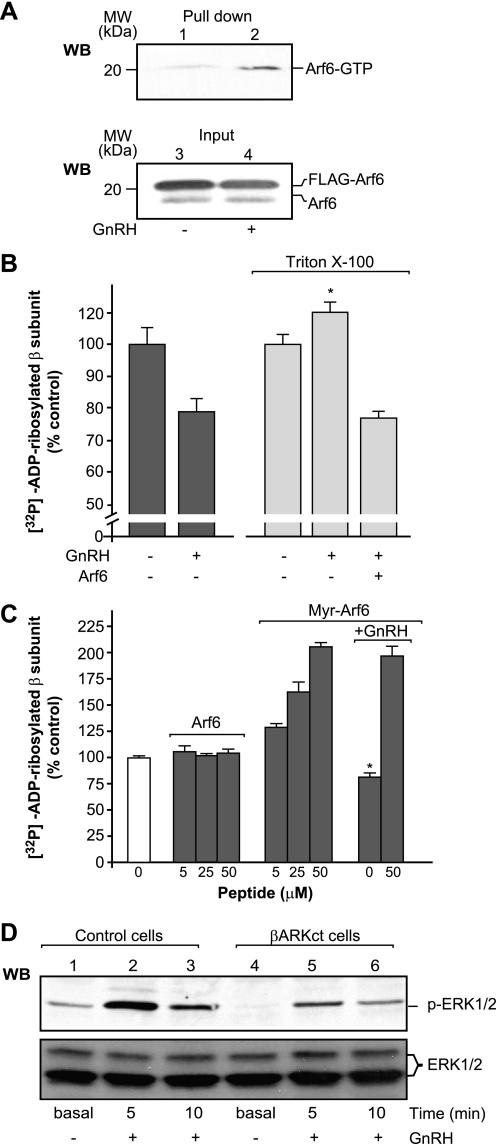
The experiments with both intact and permeabilized cells confirm that Gβ subunit mono-ADP-ribosylation is induced by thrombin stimulation. Thus, we investigated whether ADP-ribosylation of the Gβ subunit has a role in thrombin signaling. We have previously demonstrated that in CHO cells, thrombin-dependent ERK phosphorylation is mediated by the α subunit of the G12/13 and Gq families (29). Here, we initially investigated whether the Gβγ dimers released upon thrombin receptor activation were directly involved in ERK1/2 phosphorylation. When CHO cells were transiently transfected with the βARKct peptide, which is a specific Gβγ-binding peptide that prevents free Gβγ dimers from activating their downstream effectors (30), the levels of thrombin-dependent ERK1/2 phosphorylation were not affected (data not shown). This is because the Gβγ released upon thrombin stimulation was mono-ADP-ribosylated and therefore not directly involved in the ERK1/2 signaling pathway. Under the same experimental conditions, the overexpression of the arginine-specific ADP-ribosylhydrolase ARH1, which regulates the mono-ADP-ribosylation cycle by reversing Gβγ ADP-ribosylation, resulted in decreased thrombin-dependent ERK1/2 phosphorylation (Fig. 2D). This suggests that overexpressed ARH1 can deribosylate Gβγ, which can then reassociate with Gα, favoring the formation of the trimeric, inactive G proteins, and in this way ARH1 can modulate thrombin- and α subunit-dependent ERK1/2 phosphorylation. Moreover, Fig. 2E shows that knockdown of ARH1 using RNA interference (RNAi) did not impair this thrombin-dependent increase in the levels of ERK1/2 phosphorylation. This can be explained considering that when ARH1 is knocked down, Gβγ is “frozen” in the mono-ADP-ribosylated, inactive form. Altogether, these data imply that the ADP-ribosylation/deribosylation cycle induced by thrombin stimulation can modulate Gβγ dimer reassociation with the Gα subunit, and in this indirect way this cycle can modulate thrombin signaling to ERK1/2.
Arf6-GTP Inhibits Gβ Subunit Mono-ADP-ribosylation
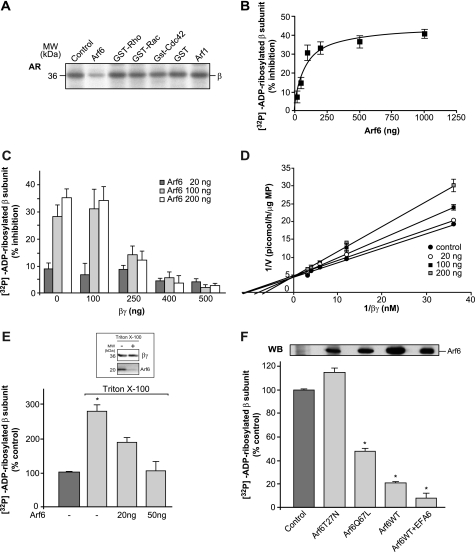
Considering that the Arf proteins were originally named for their ability to function as cofactors for cholera toxin-catalyzed mono-ADP-ribosylation of the αs subunit of trimeric G proteins (11, 12), we explored their ability to modulate endogenous ADP-ribosylation of the Gβ subunit. Plasma membranes from CHO cells were preincubated with purified Arf6 for 15 min at 37 °C and then analyzed in an in vitro [32P]ADP-ribosylation assay. Arf6 caused a marked decrease in [32P]ADP-ribosylation of the β subunit, as compared with the control (Fig. 3A). Under the same experimental conditions, Arf1 and various small G proteins, including Rho, Rac, and Cdc42, had no effects (Fig. 3A). The Ki value for Arf6 evaluated in in vitro [32P]ADP-ribosylation assays was 84 ± 20 nm (Fig. 3B). Moreover, the Arf6-dependent decrease in [32P]ADP-ribosylation of the β subunit was counteracted by increasing concentrations of purified Gβγ (Fig. 3C), thus revealing a competitive mechanism of inhibition (Fig. 3D). These data are the first indication that Arf6 can negatively regulate endogenous ADP-ribosylation of the β subunit. Here, the CHO plasma membranes were treated with 0.1% Triton X-100 (Fig. 3E), as this relatively low detergent concentration does not affect the β subunit, although it is effective for the almost complete solubilization and release of Arf6 (∼90% depletion, as evaluated by WB; Fig. 3E, inset). An almost 3-fold increase in β subunit [32P]ADP-ribosylation was seen using these Arf6-depleted plasma membranes, as compared with untreated plasma membranes (Fig. 3E). Importantly, re-addition of purified, myristoylated Arf6 to the Arf6-depleted plasma membranes was sufficient to restore the level of [32P]ADP-ribosylated β subunit to the level of untreated plasma membranes (Fig. 3E), thus ruling out the possibility of a nonspecific Triton X-100 effect.
FIGURE 3.
Arf6-GTP inhibits G protein β subunit ADP-ribosylation. A, CHO plasma membranes (5 μg) were incubated for 15 min at 37 °C with 1 μg of the indicated purified proteins before the [32P]ADP-ribosylation assay. The data shown are representative of five experiments. B, CHO plasma membranes (5 μg) were incubated for 15 min at 37 °C with the indicated concentrations of purified Arf6 before the [32P]ADP-ribosylation assay. The solubilized proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes, and the [32P]ADP-ribosylated β subunit was quantified by analyzing the filters with the InstantImager, as % inhibition. The data represent means (±S.D.) of six independent experiments, each performed in duplicate. C and D, CHO plasma membranes (5 μg) were incubated for 15 min at 37 °C with the indicated concentrations of purified Arf6 before the [32P]ADP-ribosylation assay, which was performed in the presence of increasing concentrations of purified Gβγ. The solubilized proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes, and the [32P]ADP-ribosylated β subunit was quantified by analyzing the filters with an InstantImager, as % inhibition. The data represent means (±S.D.) of three independent experiments, each performed in duplicate. D shows Lineweaver-Burk plot of these data. E, GnRHR-CHO plasma membranes were left untreated or were treated with 0.1% Triton X-100, and then they were [32P]ADP-ribosylated in the absence or presence of 20 or 50 ng Arf6. Twenty ng of purified Arf6 are equivalent to the endogenous Arf6 in 5 μg of protein from CHO plasma membranes and correspond to a concentration of 20 nm under these experimental conditions. *, significantly different from the relevant control (p < 0.05) and from the sample containing Arf6. The inset shows an immunoblot (representative of at least five independent experiments) of CHO plasma membranes untreated (lane 1) or treated with 0.1% Triton X-100 (lane 2). F, CHO cells were transfected with the indicated constructs for 24 h, permeabilized, and incubated with [32P]NAD for 60 min at 37 °C in the presence of 25 μm mastoparan 7. The inset shows the level of expression of the indicated Arf6 proteins, as performed by immunoblotting with an anti-Arf6 antibody. The samples were analyzed by autoradiography, and the levels of ADP-ribosylated Gβ subunit were determined using an InstantImager (as % control). The data shown represent means (±S.D.) of four independent experiments, each performed in duplicate (n = 4).
This negative regulation of β subunit ADP-ribosylation by Arf6 was then confirmed in tetanolysin-permeabilized cells (Fig. 3F). CHO cells were transiently transfected with either wild-type Arf6 (Arf6WT), a dominant-negative Arf6 (Arf6T27N; which cannot exchange GDP for GTP), or a constitutively active Arf6 (Arf6Q67L; which lacks GTPase activity), or they were co-transfected with Arf6WT and EFA6 (Arf6WT/EFA6), an Arf6-specific exchange factor that exclusively localizes to the plasma membrane (31). The permeabilized transfected cells were incubated with [32P]NAD+ and mastoparan 7 (to increase the basal levels of the [32P]ADP-ribosylated β subunit) and then analyzed for 32P-labeling of the β subunit. Although Arf6WT, Arf6Q67L, and Arf6WT/EFA6 all caused an important decrease in β subunit ADP-ribosylation (ranging from 50 to 90%; Fig. 3F), there was no significant difference between Arf6T27N and the control. In control experiments, CHO cells transiently transfected with wild-type Arf1 (Arf1WT) or the constitutively active or inactive forms of Arf1 (Arf1Q71L and Arf1T31N, respectively) or co-transfected with Arf1 and ARNO, all showed no differences in the labeling of the β subunit when analyzed under the same experimental conditions (data not shown). Thus, it is the activated form of Arf6 that specifically mediates this inhibition of β subunit ADP-ribosylation.
Arf6-GTP Is Mono-ADP-ribosylated by the CHO Plasma Membrane-associated Mono-ADP-ribosyltransferase
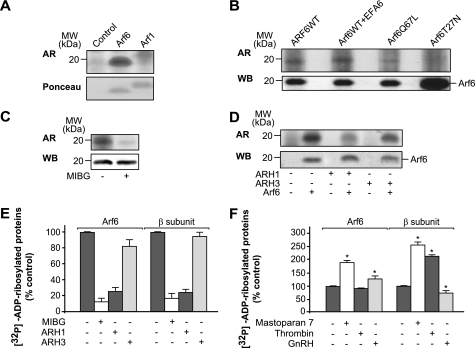
We have provided evidence that Arf6 can act as a competitive inhibitor for the endogenous CHO plasma membrane-associated ART activity. We next evaluated whether Arf6 was ADP-ribosylated by this endogenous ART enzyme. Purified Arf6 was added to CHO plasma membranes and [32P]ADP-ribosylated; labeling of an ∼20-kDa protein that corresponded precisely to the purified Arf6 revealed by Ponceau staining was seen (Fig. 4A). Importantly, Arf6 [32P]ADP-ribosylation was confirmed in tetanolysin-permeabilized CHO cells that had previously been transfected with either Arf6WT or Arf6Q67L or co-transfected with Arf6WT and EFA6. All of these conditions resulted in labeling of an ∼20-kDa protein that was recognized by the anti-Arf6 antibody (Fig. 4B). However, there was no labeling when CHO cells were transfected with Arf6T27N, even when it was expressed at higher levels (Fig. 4B). Thus, only the activated form of Arf6 was ADP-ribosylated, which parallels the ability of the activated form of Arf6 to inhibit β subunit ADP-ribosylation. These data indicate that Arf6 can substitute βγ in the catalytic site of endogenous ART. To provide data to support a model in which the same endogenous ART enzyme catalyzes both β subunit and Arf6 ADP-ribosylation, we evaluated the effects of activators and inhibitors of this reaction on the levels of [32P]ADP-ribosylated Arf6. The Arf6 [32P]ADP-ribosylation was inhibited by the arginine-specific ART inhibitor MIBG (Fig. 4C) (4, 5), as with β subunit ADP-ribosylation (Fig. 4E). We then asked whether [32P]ADP-ribosylated Arf6 can be deribosylated by ARH1. Among the three ARH family members identified, only ARH1 can hydrolyze the ADP-ribose-arginine bond; ARH3 catalyzes hydrolysis of poly(ADP-ribose) and O-acetyl-ADP-ribose, whereas ARH2 does not hydrolyze any known substrate (32, 33). Again, as with the ADP-ribosylated Gβ subunit, [32P]ADP-ribosylated Arf6 can be deribosylated by purified ARH1 (Fig. 4, D and E), although under the same experimental conditions, ARH3 had no effect (Fig. 4D). Altogether, these data demonstrate that mono-ADP-ribosylation of Arf6 occurs on an arginine.
FIGURE 4.
Arf6-GTP is ADP-ribosylated. A, CHO plasma membranes (5 μg) were incubated for 60 min at 37 °C with 1 μg of the indicated purified proteins before the [32P]ADP-ribosylation assay. B, CHO cells were transfected with the indicated constructs for 24 h, permeabilized with tetanolysin, and incubated with [32P]NAD for 60 min at 37 °C. WB shows the levels of expression of the indicated Arf6 proteins, as performed by immunoblotting with an anti-Arf6 antibody. C, CHO plasma membranes (5 μg) were incubated for 60 min at 37 °C with 1 μg of purified Arf6 in the absence or presence of MIBG (200 μm) before the [32P]ADP-ribosylation assay. D, CHO plasma membranes (5 μg) were ADP-ribosylated in the presence of 1 μg of purified Arf6, washed, and then incubated for an additional 30 min at 37 °C in the absence and presence of 50 ng of purified ARH1 or ARH3, as indicated. E and F, levels of [32P]ADP-ribosylated Gβ subunit and Arf6 with the indicated stimuli were measured using an InstantImager, as % control. *, significantly different from the relevant control (p < 0.05). A–F, data are representative of three to five experiments. The solubilized proteins were analyzed by autoradiography (AR) and then probed with an anti-ARF6 antibody. The levels of [32P]ADP-ribosylated Arf6 were measured using an InstantImager, as % control.
Finally, we analyzed the level of Arf6 [32P]ADP-ribosylation in plasma membranes obtained either from wild-type CHO cells stimulated with thrombin or mastoparan 7 or from GnRHR-CHO cells stimulated with GnRH. Fig. 4F shows a mastoparan-7-dependent increase in Arf6 [32P]ADP-ribosylation (85 ± 12%). Thrombin stimulation was not effective on Arf6 [32P]ADP-ribosylation (Fig. 4F). In contrast, an increase in the labeling of Arf6 was seen upon activation of the GnRHR (Fig. 4F; evaluated in the presence of 20 ng of Arf6, a concentration that can be easily revealed by autoradiography but that is not sufficient to inhibit ADP-ribosylation of the β subunit (see also Fig. 3C)). These data led to the conclusion that stimulation of the GnRHR activates Arf6, which can became a substrate of the endogenous mono-ADP-ribosyltransferase.
GnRHR Stimulation Leads to Inhibition of Gβ Subunit Mono-ADP-ribosylation through the Activation of Arf6
Previous studies have demonstrated that certain GPCRs can activate Arf6 (18, 34). We thus evaluated whether activation of the GnRHR can activate Arf6 and whether this activation leads to inhibition of β subunit mono-ADP-ribosylation. To test whether the GnRHR can activate Arf6, we performed a pulldown assay for Arf6-GTP (28) using cell lysates from unstimulated and GnRH-stimulated GnRHR-CHO cells. The pulled down Arf6 was then analyzed by WB using a specific anti-Arf6 antibody. Stimulation of CHO cells with GnRH resulted in a significant increase in the amounts of Arf6-GTP pulled down, as compared with unstimulated CHO cells (Fig. 5A). This thus revealed a previously unknown pathway of Arf6 activation that is initiated by the GnRHR. To determine whether this Arf6 activation by the GnRHR is responsible for the GnRHR-mediated inhibition of β subunit ADP-ribosylation, plasma membranes purified from GnRHR-CHO cells were depleted of endogenous Arf6 with Triton X-100 treatment and then subjected to the in vitro [32P]ADP-ribosylation assay. Under these conditions, the inhibitory effects mediated via GnRHR stimulation were completely lost (Fig. 5B). Importantly, re-addition of purified Arf6 restored the inhibition mediated by the GnRHR (Fig. 5B). Moreover, when a myristoylated peptide corresponding to the N terminus of Arf6 (myrArf6), which inhibits endogenous Arf6 (34), was incubated with the plasma membranes before the ADP-ribosylation assay for 15 min at 37 °C, this induced a dose-dependent increase in the labeling of the β subunit and blocked the GnRHR-dependent inhibition of β subunit mono-ADP-ribosylation (Fig. 5C). Under the same experimental conditions, the corresponding nonmyristoylated peptide and myristoylated coenzyme A were ineffective (Fig. 5C and data not shown).
FIGURE 5.
A, GnRHR-CHO cells were transfected with FLAG-Arf6 and either incubated with control buffer (lanes 1 and 3) or stimulated with 20 nm GnRH for 15 min at 37 °C (lanes 2 and 4). Cell lysates were incubated with GST-GGA3(1–226), which only binds the active form of Arf6. The WB is representative of three independent experiments with both input and pulled down proteins, as performed with an anti-Arf6 antibody. B, GnRHR-CHO plasma membranes were left untreated or were treated with 0.1% Triton X-100, and then they were [32P]ADP-ribosylated in the absence or presence of 20 ng of Arf6 and 200 nm GnRH. *, significantly different from the relevant control (p < 0.05) and from the sample containing Arf6. C, nonmyristoylated (Arf6 p) and myristoylated (myrArf6) Arf6 peptides were incubated with plasma membranes before the ADP-ribosylation assay in the absence or presence of 200 nm GnRH. *, significantly different from the relevant control (white column; p < 0.01). B and C, samples were analyzed by autoradiography, and the levels of ADP-ribosylated Gβ subunit were determined using an InstantImager (as % control), for untreated plasma membranes. The data shown represent means (±S.D.) of four (B) or three (C) independent experiments, each performed in duplicate. D, WB of filter labeled with the anti-phospho-ERK1/2 (p-ERK1/2) and anti-ERK1/2 (ERK1/2) antibodies. ERK1/2 activity was evaluated after the indicated times of 20 nm GnRH stimulation in control cells (lanes 1–3) or in CHO cells transfected with the βARKct peptide (lanes 4–6). The data shown are representative of three independent experiments.
Altogether, these data demonstrate that Arf6 is directly involved in GnRHR-dependent inhibition of β subunit mono-ADP-ribosylation. Thus, it appears that the pathway initiated by agonist stimulation of the GnRHR leads to the activation of Arf6, which in turn inhibits the enzyme responsible for β subunit mono-ADP-ribosylation. The functional consequences of this is that free Gβγ dimers can contribute directly to GnRHR signaling. We have analyzed this possibility by evaluating GnRHR signaling and ERK1/2 phosphorylation.
GnRH stimulation of GnRHR-CHO cells resulted in ERK1/2 phosphorylation (Fig. 5D, lanes 2 and 3), in line with previous data (35). This ERK1/2 phosphorylation was not affected when GnRHR-CHO cells were transiently transfected with Arf6, Arf6T27N, or Arf6Q67L or co-transfected with Arf6WT and EFA6, demonstrating that GnRH-induced activation of ERK1/2 occurs independently of Arf6. In contrast, Gβγ dimers released upon GnRHR activation appear to be directly involved in ERK1/2 phosphorylation, as evaluated in GnRHR-CHO cells transiently transfected with βARKct peptide (Fig. 5D, lanes 4–6), thus suggesting that indeed free and active Gβγ dimers directly contribute to GnRHR signaling.
DISCUSSION
Heterotrimeric G protein-mediated cellular signaling is based on G protein activation/inactivation cycles that occur at the plasma membrane, where GPCRs are stimulated by their ligands in the extracellular environment (36–41). The role of Gβγ dimers in regulating cellular signaling upon agonist stimulation of the GPCRs has become well established over the past 20 years (9, 42). However, despite the crucial role of the Gβγ dimer in cell signaling, the only known mechanism controlling its activity is the intrinsic GTPase activity of the Gα subunit. Gα-GDP binds to free Gβγ dimer to form the inactive trimeric G protein complex. Thus, the mono-ADP-ribosylation cycle appears to represent a mechanism to directly control Gβγ dimer activity independent of the GTPase activity of the Gα subunit.
We have previously demonstrated that the β subunit of free and active Gβγ dimers can undergo post-translational mono-ADP-ribosylation that is catalyzed by an intracellular mono-ADP-ribosyltransferase and that this post-translational modification has a key role in the ability of Gβγ to regulate its effectors (4, 5). Here, we have extended our previous studies to explore how GPCR stimulation by their cognate ligands regulates endogenous mono-ADP-ribosylation of Gβγ dimers.
Although GPCR stimulation of CHO plasma membranes with clonidine, isoproterenol, and thrombin induced an increase in β subunit [32P]ADP-ribosylation, GnRH inhibited this modification, indicating that the active βγ dimers released upon GPCR activation can be differentially modulated, potentially through regulation of the endogenous mono-ADP-ribosyltransferase.
The ability of thrombin to induce β subunit ADP-ribosylation is demonstrated here in three different systems. The first system is an in vitro assay using purified CHO plasma membranes that endogenously express the thrombin receptor. Here, we observe a doubling of β subunit [32P]ADP-ribosylation, which appears to be due to the absence of the cytosolic ADP-ribosylhydrolase under these experimental conditions. The second system is in tetanolysin-permeabilized cells, where the architecture of the cells is preserved. This can thus be considered as a semi-intact system, which provided β subunit [32P]ADP-ribosylation that was detected within 15 min of thrombin stimulation, a time that is compatible with GPCR-mediated signaling regulation. When we evaluated ADP-ribosylhydrolase activity in these permeabilized cells, 25% of the endogenous activity was released into the medium (data not shown), which would result in a partial shift of the β subunit ADP-ribosylation/deribosylation equilibrium toward its ADP-ribosylated form. Finally, in intact cells, we have previously reported that ∼0.2% of the β subunit is mono-ADP-ribosylated under basal conditions (4). We now show that this fraction of modified β subunit can increase upon thrombin stimulation, which induces the 500-Da shift of the β subunit that is a hallmark of ADP-ribosylation. Under these experimental conditions, modification of the β subunit was quantified as ∼0.5% of the total Gβ. This fraction would thus correspond to the βγ pool that is coupled to the thrombin receptor. Therefore, although activation of thrombin receptors leads to activation of the G protein α and βγ subunits, a parallel inactivation of βγ function can occur through its mono-ADP-ribosylation. This can eventually regulate the duration of the βγ signaling independent of the GTPase activity of the α subunit.
Here, we have demonstrated that thrombin-dependent activation of ERK1/2 does not involve Gβγ. However, considering that overexpression of ARH1 inhibited this thrombin-dependent ERK1/2 phosphorylation, we can hypothesize that Gβ mono-ADP-ribosylation induced by thrombin stimulation can modulate Gα subunit-mediated signaling. It remains to estimate the role of this thrombin-dependent Gβ mono-ADP-ribosylation in signaling that is directly regulated by Gβγ. Because it has been previously reported that thrombin activates PI3K and focal adhesion kinase via Gβγ, we have measured both focal adhesion kinase and AKT/PKB phosphorylation, the last of which represents a read-out of PI3K activation. Neither of these pathways was active in CHO cells. Thus the problem of if and how thrombin-induced Gβ mono-ADP-ribosylation regulates Gβγ signaling remains to be defined.
Importantly, a different GPCR can block the enzymatic activity involved in β subunit ADP-ribosylation, the GnRHR. This is mediated through activation of the small GTP-binding protein Arf6 (see Fig. 3 and scheme shown in Fig. 6). This finding was both surprising and of great interest, considering that although the name of the Arf family arose from the ability of ARF1 to stimulate the ART activity of cholera toxin (43, 44), a similar activity on mammalian ARTs has never been shown previously. Here, we provide the first evidence that when activated a member of the Arf family can modulate mammalian ART, which, in turn, can modify Gβγ dimers. Moreover, a further analysis of how Arf6 modulates this mammalian ART led to the demonstration that Arf6 is a substrate of this enzyme. This explains the Arf6 inhibitory effect on Gβ subunit ADP-ribosylation, and importantly, it opens up the possibility that this post-translational modification can also regulate Arf6 function.
FIGURE 6.
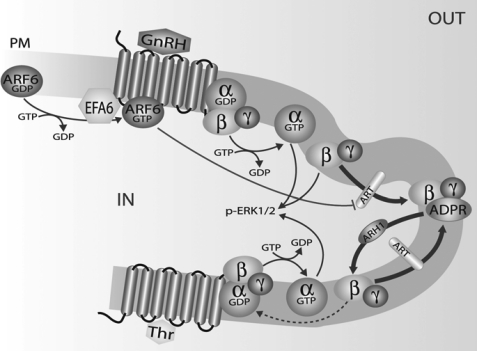
Schematic representation of thrombin and GnRH-induced regulation of mono-ADP-ribosylation of the Gβ subunit. Gβγ released upon thrombin stimulation is mono-ADP-ribosylated. The Gβγ ADP-ribosylation/deribosylation cycle can affect Gβγ dimer reassociation with the Gα subunit, and in this indirect way it can modulate thrombin signaling to ERK1/2. Activation of the GnRHR by its cognate ligand promotes activation of trimeric G proteins and of the small G protein Arf6. Activated Arf6 can in turn inhibit the endogenous intracellular ART enzyme, which catalyzes Gβ subunit mono-ADP-ribosylation, and consequently Arf6 can modulate Gβγ-mediated signaling.
How Arf6 activation is achieved in response to agonist stimulation of a GPCR remains poorly characterized. It has been shown that agonist stimulation of the TPβ receptor results in potent activation of Arf6 and that Gαq mediates this Arf6 activation (45). It is also known that the GnRHR signals mainly through Gq, and thus it is conceivable that once activated, Gq can promote activation of Arf6 through its exchange factor EFA6; this activated Arf6 can, in turn, regulate plasma membrane-associated ART and in this way prolong Gβγ-mediated signaling. An alternative possibility is for direct molecular interactions between the GPCRs and the Arfs. This hypothesis is supported by data in the literature reporting co-immunoprecipitation of some GPCRs and Arfs upon agonist stimulation. Co-immunoprecipitation experiments indicate that GPCRs and Arfs are part of the same complex, but they do not provide a definitive answer to the question of whether the two proteins can directly interact. The hypothesis of direct interactions between GPCRs and Arfs is further supported by the demonstration that both ARF1 and ARF6 can directly interact with an NPXXY domain of several GPCRs and become activated upon receptor stimulation (46, 47). Specifically, the NPXXY motif in the C-terminal tail determines the Arf1 activation, whereas the motif DPXXY, which is present in the GnRHR, is indicative of Arf6 coupling. We have demonstrated that GnRHR stimulation led to Arf6 activation with [35S]GTPγS binding assays using GnRHR-CHO membranes, with Arf6 and its exchange factor added (data not shown). More importantly, there was activation of Arf6 in intact GnRHR-CHO cells using a well characterized GST pulldown assay (28, 48). These experiments demonstrate that the GnRHR in GnRHR-CHO cells leads to activation of Arf6, although whether this is due to a direct interaction between GnRHR and Arf6, to G protein-mediated activation, or to recruitment of a specific guanine nucleotide exchange factor for Arf6 remains to be defined. A consequence of GnRH-mediated activation of Arf6 was inhibition of Gβγ mono-ADP-ribosylation. Because GnRHR did not induce focal adhesion kinase and AKT/PKB phosphorylation in this cell context (our data), we could only investigate the role of Gβγ in ERK1/2 activation. As we measured a decrease in GnRH-induced ERK1/2 phosphorylation in GnRHR-CHO cells transiently transfected with βARKct, this suggests that the Gβγ dimers released upon GnRHR activation are directly involved in ERK1/2 phosphorylation and thus contribute to GnRHR signaling.
To define the role of Gβγ mono-ADP-ribosylation in G protein-mediated signaling, it is crucial to identify the ART involved in this process. Some PARP family members that lack conserved residues crucial for polymer elongation have been proposed to act as cellular mono-ADP-ribosyltransferases, and indeed, this has been demonstrated for PARP10 (24). Future studies will define which of these enzymes is involved in Gβ subunit mono-ADP-ribosylation, and this will be a great help toward a better understanding of the role of Gβ subunit mono-ADP-ribosylation.
Acknowledgments
We thank Francis Schuber (Strasbourg University, Strasbourg, France) for useful discussions and critical reading of this manuscript. We thank Oliviano Martella for technical assistance, Christopher Paul Berrie for editorial assistance, and Elena Fontana and Roberta Le Donne for preparation of figures.
This work was supported in part by Ministero dell'Istruzione, Università e Ricerca, Grants RBIP06LSS2_005 and DM26545.
- GPCR
- G protein-coupled receptor
- Arf
- ADP-ribosylation factor
- ART
- ADP-ribosyltransferase
- GnRHR
- gonadotropin-releasing hormone receptor
- PARP
- poly(ADP-ribose) polymerase
- WB
- Western blot
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Deng Q., Barbieri J. T. (2008) Annu. Rev. Microbiol. 62, 271–288 [DOI] [PubMed] [Google Scholar]
- 2. Seman M., Adriouch S., Haag F., Koch-Nolte F. (2004) Curr. Med. Chem. 11, 857–872 [DOI] [PubMed] [Google Scholar]
- 3. Di Girolamo M., Dani N., Stilla A., Corda D. (2005) FEBS J. 272, 4565–4575 [DOI] [PubMed] [Google Scholar]
- 4. Lupi R., Corda D., Di Girolamo M. (2000) J. Biol. Chem. 275, 9418–9424 [DOI] [PubMed] [Google Scholar]
- 5. Lupi R., Dani N., Dietrich A., Marchegiani A., Turacchio S., Berrie C. P., Moss J., Gierschik P., Corda D., Di Girolamo M. (2002) Biochem. J. 367, 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourne H. R., Sanders D. A., McCormick F. (1991) Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 7. Wettschureck N., Offermanns S. (2005) Physiol. Rev. 85, 1159–1204 [DOI] [PubMed] [Google Scholar]
- 8. Dupré D. J., Robitaille M., Rebois R. V., Hébert T. E. (2009) Annu. Rev. Pharmacol. Toxicol. 49, 31–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smrcka A. V. (2008) Cell. Mol. Life Sci. 65, 2191–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dani N., Stilla A., Marchegiani A., Tamburro A., Till S., Ladurner A. G., Corda D., Di Girolamo M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4243–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsai S. C., Noda M., Adamik R., Chang P. P., Chen H. C., Moss J., Vaughan M. (1988) J. Biol. Chem. 263, 1768–1772 [PubMed] [Google Scholar]
- 12. O'Neal C. J., Jobling M. G., Holmes R. K., Hol W. G. (2005) Science 309, 1093–1096 [DOI] [PubMed] [Google Scholar]
- 13. Maranda B., Brown D., Bourgoin S., Casanova J. E., Vinay P., Ausiello D. A., Marshansky V. (2001) J. Biol. Chem. 276, 18540–18550 [DOI] [PubMed] [Google Scholar]
- 14. Hunzicker-Dunn M., Gurevich V. V., Casanova J. E., Mukherjee S. (2002) FEBS Lett. 521, 3–8 [DOI] [PubMed] [Google Scholar]
- 15. Donaldson J. G. (2003) J. Biol. Chem. 278, 41573–41576 [DOI] [PubMed] [Google Scholar]
- 16. D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 17. Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 18. Claing A. (2004) Biochem. Cell Biol. 82, 610–617 [DOI] [PubMed] [Google Scholar]
- 19. Houndolo T., Boulay P. L., Claing A. (2005) J. Biol. Chem. 280, 5598–5604 [DOI] [PubMed] [Google Scholar]
- 20. Koch-Nolte F., Kernstock S., Mueller-Dieckmann C., Weiss M. S., Haag F. (2008) Front. Biosci. 13, 6716–6729 [DOI] [PubMed] [Google Scholar]
- 21. Otto H., Reche P. A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. (2005) BMC Genomics 6, 139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 23. Liszt G., Ford E., Kurtev M., Guarente L. (2005) J. Biol. Chem. 280, 21313–21320 [DOI] [PubMed] [Google Scholar]
- 24. Kleine H., Poreba E., Lesniewicz K., Hassa P. O., Hottiger M. O., Litchfield D. W., Shilton B. H., Lüscher B. (2008) Mol. Cell 32, 57–69 [DOI] [PubMed] [Google Scholar]
- 25. Sternweis P. C., Robishaw J. D. (1984) J. Biol. Chem. 259, 13806–13813 [PubMed] [Google Scholar]
- 26. Randazzo P. A., Fales H. M. (2002) Methods Mol. Biol. 189, 169–179 [DOI] [PubMed] [Google Scholar]
- 27. Riese M. J., Goehring U. M., Ehrmantraut M. E., Moss J., Barbieri J. T., Aktories K., Schmidt G. (2002) J. Biol. Chem. 277, 12082–12088 [DOI] [PubMed] [Google Scholar]
- 28. Niedergang F., Colucci-Guyon E., Dubois T., Raposo G., Chavrier P. (2003) J. Cell Biol. 161, 1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mariggiò S., Bavec A., Natale E., Zizza P., Salmona M., Corda D., Di Girolamo M. (2006) Cell. Signal. 18, 2200–2208 [DOI] [PubMed] [Google Scholar]
- 30. Hawes B. E., van Biesen T., Koch W. J., Luttrell L. M., Lefkowitz R. J. (1995) J. Biol. Chem. 270, 17148–17153 [DOI] [PubMed] [Google Scholar]
- 31. Franco M., Peters P. J., Boretto J., van Donselaar E., Neri A., D'Souza-Schorey C., Chavrier P. (1999) EMBO J. 18, 1480–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oka S., Kato J., Moss J. (2006) J. Biol. Chem. 281, 705–713 [DOI] [PubMed] [Google Scholar]
- 33. Ono T., Kasamatsu A., Oka S., Moss J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16687–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salvador L. M., Mukherjee S., Kahn R. A., Lamm M. L., Fazleabas A. T., Maizels E. T., Bader M. F., Hamm H., Rasenick M. M., Casanova J. E., Hunzicker-Dunn M. (2001) J. Biol. Chem. 276, 33773–33781 [DOI] [PubMed] [Google Scholar]
- 35. Caunt C. J., Finch A. R., Sedgley K. R., McArdle C. A. (2006) Trends Endocrinol. Metab. 17, 308–313 [DOI] [PubMed] [Google Scholar]
- 36. Gilman A. G. (1987) Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 37. Dessauer C. W., Posner B. A., Gilman A. G. (1996) Clin. Sci. 91, 527–537 [DOI] [PubMed] [Google Scholar]
- 38. Offermanns S., Simon M. I. (1996) Cancer Surv. 27, 177–198 [PubMed] [Google Scholar]
- 39. Bourne H. R. (1997) Curr. Opin. Cell Biol. 9, 134–142 [DOI] [PubMed] [Google Scholar]
- 40. Offermanns S. (2003) Prog. Biophys. Mol. Biol. 83, 101–130 [DOI] [PubMed] [Google Scholar]
- 41. Bourne H. R. (2006) Ernst Schering Found. Symp. Proc. 2, 1–21 [DOI] [PubMed] [Google Scholar]
- 42. Gautam N., Downes G. B., Yan K., Kisselev O. (1998) Cell. Signal. 10, 447–455 [DOI] [PubMed] [Google Scholar]
- 43. Kahn R. A., Gilman A. G. (1986) J. Biol. Chem. 261, 7906–7911 [PubMed] [Google Scholar]
- 44. Lee C. M., Chang P. P., Tsai S. C., Adamik R., Price S. R., Kunz B. C., Moss J., Twiddy E. M., Holmes R. K. (1991) J. Clin. Invest. 87, 1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giguère P., Rochdi M. D., Laroche G., Dupré E., Whorton M. R., Sunahara R. K., Claing A., Dupuis G., Parent J. L. (2006) Cell. Signal. 18, 1988–1994 [DOI] [PubMed] [Google Scholar]
- 46. Mitchell R., McCulloch D., Lutz E., Johnson M., MacKenzie C., Fennell M., Fink G., Zhou W., Sealfon S. C. (1998) Nature 392, 411–414 [DOI] [PubMed] [Google Scholar]
- 47. Robertson D. N., Johnson M. S., Moggach L. O., Holland P. J., Lutz E. M., Mitchell R. (2003) Mol. Pharmacol. 64, 1239–1250 [DOI] [PubMed] [Google Scholar]
- 48. Santy L. C., Casanova J. E. (2001) J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]