Abstract
Arenavirus RNA genomes are initiated by a “prime and realign” mechanism, such that the initiating GTP is found as a single unpaired (overhanging) nucleotide when the complementary genome ends anneal to form double-stranded (ds) RNA panhandle structures. dsRNAs modeled on these structures do not induce interferon (IFN), as opposed to blunt-ended 5′ pppdsRNA. This study examines whether these viral structures can also act as decoys, by trapping RIG-I in inactive dsRNA complexes. We examined the ability of various dsRNAs to activate the RIG-I ATPase (presumably a measure of helicase translocation on dsRNA) relative to their ability to induce IFN. We found that there is no simple relationship between these two properties, as if RIG-I can translocate on short dsRNAs without inducing IFN. Moreover, we found that 5′ pppdsRNAs with a single unpaired 5′ ppp-nucleotide can in fact competitively inhibit the ability of blunt-ended 5′ pppdsRNAs to induce IFN when co-transfected into cells and that this inhibition is strongly dependent on the presence of the 5′ ppp. In contrast, 5′ pppdsRNAs with a single unpaired 5′ ppp-nucleotide does not inhibit poly(I-C)-induced IFN activation, which is independent of the presence of a 5′ ppp group.
Keywords: ATPases, Innate Immunity, Interferon, RNA, RNA Viruses
Introduction
The innate immune system senses RNA virus infections primarily via two constitutively expressed DEX(D/H) box helicases, RIG-I and mda-5 (1). These cytoplasmic sensors act as pattern recognition receptors that respond to two RNA pathogen-associated molecular patterns (PAMPs),2 namely dsRNA (e.g. poly(I)·poly(C) or poly(I-C)) and 5′ pppRNA (2–4)). These RNAs can act as PAMPs because their presence in the cell cytoplasm is thought to be primarily restricted to virus infection. RIG-I, which senses negative-strand RNA virus infections (5), is a large protein composed of three domains as follows: N-terminal tandem CARDs (the effector domain), a central DEX(D/H) box helicase domain that contains a dsRNA-dependent ATPase, and a C-terminal regulatory domain (CTD) that binds 5′ pppdsRNA (6, 7). The CARD domain of quiescent RIG-I is thought to be auto-inhibited by other protein domains and to become activated, i.e. available for interaction with the synonymous domain of IPS-1 (the central adaptor of the pathway to IFNβ activation) via conformational changes in RIG-I induced upon binding of PAMP RNAs to the CTD (8).
In the earliest reports, 5′ pppRNA was thought to act as a PAMP whether the triphosphate was present on ssRNA or dsRNA. More recently, good evidence has been presented that 5′ pppRNA without dsRNA character is not a PAMP (9–11). In the latest descriptions of this signaling pathway, RIG-I is activated and IFN induced only when the CTD interacts with dsRNA that also contains a 5′ ppp-end, at least for relatively short dsRNAs (<100 bp). Both groups found that a stretch of dsRNA near the 5′ ppp-end, as well as the 5′ ppp-end itself, was essential for RIG-I to induce IFN. However, these reports differed about the minimum length of the dsRNA region required and, more importantly in the case of arenaviruses, whether this dsRNA end needed to include the nucleotide carrying the 5′ ppp. Arenaviruses (such as Tacaribe and Junin viruses) contain two ambisense genome segments whose terminal 19 nt are either fully complementary (large segment) or contain two mismatches at positions 6 and 8 (12). These mismatches would create a symmetrical 3-nt bulge when these terminal sequences anneal to form dsRNA. This sequence complementarity and the mismatches presumably reflect the similarity and subtle differences of the various genomic and antigenomic promoters. Remarkably, arenavirus genome synthesis initiates with GTP at position +2 of the template rather than at the precise 3′-end (position +1), and the initiating 5′ pppG is then realigned (as 5′ pppGpCOH) before this primer is extended (13). The net result of this “prime and realign” mechanism of genome initiation is that 5′ pppG is found as an unpaired 5′-nucleotide overhang when the complementary genome ends form dsRNA (see Fig. 5A). Arenavirus RNA genomes are also unusual in that they are found in circular nucleocapsids, presumably due to the annealing of their complementary terminal sequences (14). Using 5′ pppRNA made in vitro and purified so that all dsRNA side products have been removed, we determined that both this 5′-nucleotide overhang and the mismatches within the dsRNA panhandles clearly reduce the ability of these dsRNAs to induce IFN (15). The presence of this unpaired 5′ ppp-nucleotide overhang thus appears to be another way that some viruses use to avoid activating the IFN system.
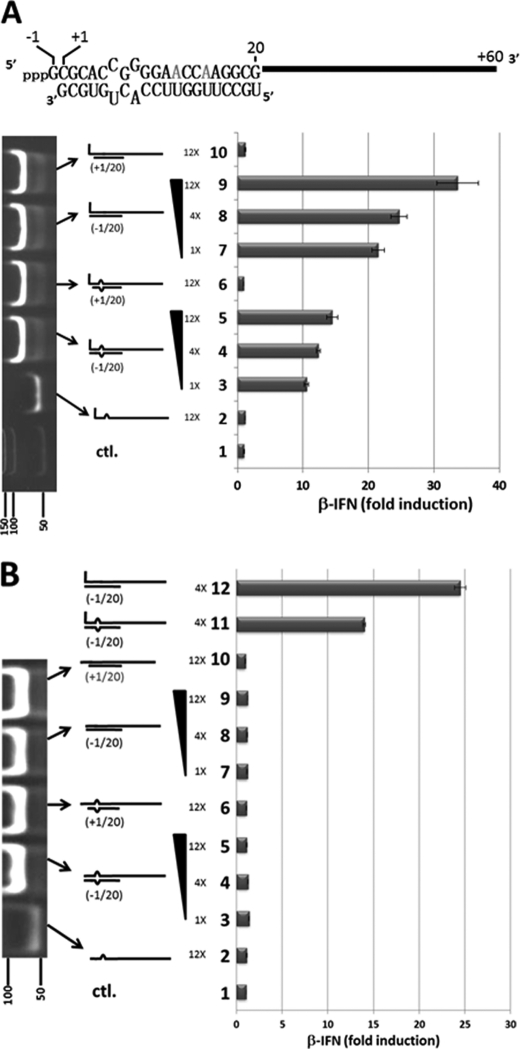
FIGURE 5.
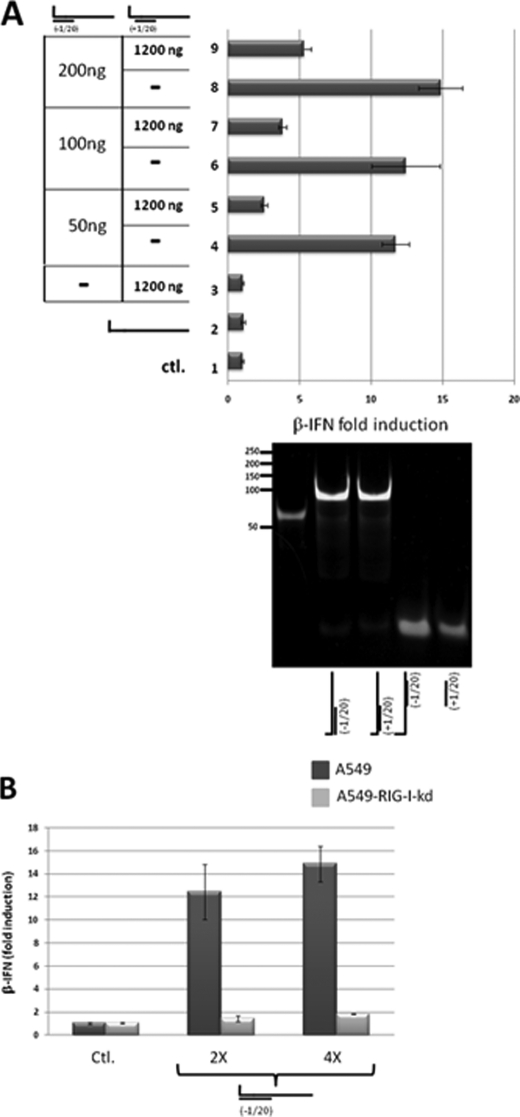
dsRNA-induced activation of the IFNβ promoter. A, parallel cultures of A549 cells were transfected with pIFNβ-(firefly)luciferase and pTK-(Renilla)luciferase. 24 h later, the cultures were transfected in duplicate with either. Lane 1, control; just tRNA. Lane 2, 1200 ng of 5′ pppJun*61 (ssRNA). Lanes 3–5, 100, 400, and 1200 ng of bulged and blunt-ended 5′ pppJun*61/(−1–20). Lane 6, 1200 ng of bulged and overhanging 5′ pppJun*61/(+1–20). Lanes 7–9, 100, 400, and 1200 ng of blunt-ended, nonbulged 5′ pppJun*61/(−1–20). Lane 10, 1200 ng of overhanging 5′ pppJun*61/(+1–20). The vertical panel on the left shows an ethidium bromide-stained, nondenaturing PAGE of the various RNAs transfected; the bottom lane shows dsDNA length markers. tRNA was added so that the total amount of RNA was constant for all transfections. Cell extracts were prepared after a further 20 h, and their luciferase activities were determined. The results are plotted as fold induction relative to the tRNA control. A schematic representation of overhanging and bulged 5′ pppJun*61/(+1–20) is shown above. B, transfection of 5′ OHdsRNAs. Same as A, except as follows: lane 2, 1200 ng of 5′ OHJun*61 (ssRNA). Lanes 3–5, 100, 400, and 1200 ng of bulged and blunt-ended 5′ OHJun*61/(−1–20). Lane 6, 1200 ng of bulged and overhanging 5′ OHJun*61/(+1–20). Lanes 7–9, 100, 400, and 1200 ng of blunt-ended, nonbulged 5′ OHJun*61/(−1–20). Lane 10, 1200 ng of overhanging, nonbulged 5′ OHJun*61/(+1–20). Lane 11, 400 ng of bulged, blunt-ended 5′ pppJun*61/(−1–20). Lane 12, 400 ng of nonbulged, blunt-ended 5′ pppJun*61/(−1–20). ctl., control.
A great deal of attention has been focused on the RIG-I CTD, as the structure of this domain was determined at atomic resolution, and structure/function studies have been carried out (6, 7). More recently, three crystal structures of the CTD bound to 12–14 bp 5′ ppp-dsRNAs were determined (16, 17). The CTD binds to both ends of the self-complementary 5′ pppdsRNAs and contacts the A-form dsRNA primarily through a few nucleotides at the 5′-end, with the α, β, and γ 5′-phosphates surrounded by multiple lysines and anchored in the basic cleft via electrostatic interactions and multiple H-bonds. The CTD also interacts with the duplexes via the stacking of Phe-853 with the terminal base pair, providing additional hydrophobic interaction energy. The modes of RNA recognition in the three structures are remarkably similar despite the very different RNA sequences, suggesting that 5′ pppdsRNA recognition is sequence-independent. RNA binding studies were also carried out, and the binding affinity of the CTD for 5′ pppdsRNA was found to be around 15-fold stronger than for the same 5′ OHdsRNA. The binding affinity for 5′ pppdsRNA with a single unpaired 5′ ppp-nucleotide was not reported.
Although the manner in which the CTD interacts with blunt-ended 5′ pppdsRNA is now well documented, and the role of the N-terminal CARDs in transmitting the signal to IPS-1 is clear, the role of the central helicases/ATPase domain in RIG-I function is less well understood. The ATPase activity of RIG-I is critical for RIG-I function, as a mutation of the ATP-binding site (K270A), like the deletion of the CARDs, eliminates all ability to induce IFN (2). The RIG-I helicase/ATPase was initially found to display typical helicase activity, i.e. to separate the strands of dsRNA (6), but more recently RIG-I was found to translocate on dsRNA without separating the strands (18, 19). Moreover, the N-terminal CARDs were found to strongly suppress this translocation in the absence of a 5′-triphosphate group, and the presence of 5′ pppRNA (tethered to the dsRNA via a ssRNA extension) strongly stimulated the translocation. Although a canonical dsRNA-binding motif is not present in these helicases, a point mutant in the helicase domain predicted to establish contacts with the RNA (Q299A) fails to bind RNA (20), and residual dsRNA-dependent ATPase activity of the isolated RIG-I helicases/ATPase domain points to a dsRNA-interacting site here (7). It also seems intuitive that the RIG-I helicase/ATPase domain binds dsRNA independently of the CTD, as structural studies of helicases bound specifically to double-stranded nucleic acids indicate that the highly conserved amino acids within the ATPase core are essential for the recognition of the double-stranded lattice (21, 22). Thus, one possible scenario for RIG-I action is that this pattern recognition receptor initially binds to any region of dsRNA via its helicase domain binding site and then moves in an ATP-dependent fashion to locate and interact with the dsRNA ends. In the most favorable case for IFN activation induced by relatively short dsRNAs (<100 bp), if a blunt 5′ pppdsRNA end is encountered, this optimally leads to the conformational changes that release the CARDs for interaction with IPS-1 and signaling to the IFN promoter occurs. However, in this scenario where RIG-I first binds to dsRNA independent of their ends, one would expect the arenavirus dsRNA panhandles (that are inactive in inducing IFN) to also act as competitive inhibitors of dsRNAs that can induce IFN, by sequestering RIG-I in inactive complexes. This study examines the binding of various RNAs to RIG-I and their ability to stimulate its ATPase, and it then examines whether 5′ pppdsRNAs whose 5′ ppp-nucleotide is unpaired can in fact competitively inhibit the ability of blunt-ended 5′ pppdsRNA to induce IFN.
EXPERIMENTAL PROCEDURES
Plasmids
pβ-IFN-fl-lucter contains the firefly luciferase gene under the control of the human IFN-β promoter, as described previously (23). pTK-rl-lucter, used as a transfection standard, contains the herpes simplex virus thymidine kinase promoter region upstream of the Renilla luciferase gene (Promega). pEBS-RIG-I expressed full-length RIG-I.
Immunoblotting
Cytoplasmic extracts were prepared using 0.5% Nonidet P-40 buffer. Equal amounts of total proteins were separated by SDS-PAGE and transferred onto Immobilon-P membranes by semidry transfer. The secondary antibodies used were alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Bio-Rad). The immobilized proteins were detected by light-enhanced chemiluminescence (Pierce) and analyzed in a Bio-Rad light detector using Quantity One software. Primary antibody used is a mouse anti-Rig-I (Alexis).
Transfections
100,000 cells were plated into 6-well plates 20 h before transfection with 1.5 μg of pβ-IFN-fl-lucter, 0.5 μg of pTK-rl-lucter, and TransIT-LT1 transfection reagent (Mirus). At 24 h post-transfection, the cells were (or were not) transfected with the indicated RNAs using Trans-messenger transfection reagent (Qiagen) (a total of 1.2–2 μg of RNA was always transfected, the difference always being made up with tRNA. Transfection of tRNA by itself was neutral.). Twenty hours later, cells were harvested and assayed for firefly and Renilla luciferase activity (Dual-Luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of Renilla luciferase.
In Vitro Synthesis of RNA, Purification, and Annealed dsRNA
DNA for T7 RNA polymerase synthesis of 5′ pppJun*61 was prepared by annealing two oligonucleotides as follows: 5′-TAATACGACTCACTATAgcgcaccggggatcctaggcgattttggttacgctataattgtgactgttttctgtttgga (T7 promoter (underlined) and the −1 to +60 Junin 5′ sequence where U residues at position +6, +8, and +19 were replaced by the indicated nucleotides to allow synthesis of single strand RNA in a T7 reaction devoid of U residues) and its complementary oligonucleotide 5′-tccaaacagaaaacagtcacaattatagcgtaaccaaaatcgcctaggatccccggtgcgcTATAGTGAGTCGTATTA. Transcription was performed with 100 pmol of gel-purified dsDNA using T7 MEGAshortscript (Ambion) according to the manufacturer's instructions (no UTP in the reaction). The total T7 transcripts were digested with DNase I and then chromatographed on NucAway Spin columns (Ambion) to remove unincorporated nucleotides and DNA fragments.
DNA for T7 RNA polymerase synthesis of biotinylated RNA was prepared by PCR using the partially complementary primers 5′-TAATACGACTCACTATAgggACACACCACAACCAACCCACAAC-3′ (forward) (start sites are in lowercase type) and 5′-GAAAGAAAGGTGTGGTGTTGGTGTGGTTGTTGTGGGTTGGTTGTGG-3′ (reverse). Transcription was performed with 100 pmol of purified PCR product using T7 MEGAshortscript (Ambion) according to the manufacturer's instructions. Biotinylated RNA1 was synthesized using equal amounts of 5′-biotin-UTP and UTP. The reaction mixture (unpurified 5′ biotinylated ppp-RNA1) was digested with DNase I and then chromatographed on NucAway Spin columns (Ambion) to remove unincorporated nucleotides and DNA fragments.
For further purification of biotinylated 5′ppp-ssRNA, slightly radiolabeled ([α-32P]CTP) T7 biotinylated transcripts were electrophoresed on 10% preparative denaturing gels. The major 54-nucleotide band was excised from the gel, and the RNA was eluted, followed by ethanol precipitation. For RNase III treatment, 50 μg of gel-purified RNA1 was digested with 50 unit of RNase III (Ambion) for 60 min at 37 °C in 40 μl of 50 mm NaCl, 10 mm Tris-HCl, (pH 7.9), 10 mm MgCl2, 1 mm dithiothreitol. The digestion products were then phenol/chloroform-extracted and ethanol-precipitated.
Annealed dsRNA
T7 5′ pppJun*61 (or synthetic 5′ OHJun*6) was mixed with the indicated synthetic complementary 5′OH-oligoribonucleotides in a molar ratio 1:3, in a final volume of 50 μl (300 mm NaCl, 50 mm Tris (pH 7.5), 1 mm EDTA), heated for 1 min at 90°C, and progressively cooled to room temperature. T7 reaction products were checked for dsRNA contamination by RNase III treatment and transfection into A549 cells (15).
Cells
2fTGH, A549, and A549-RIG-I-kd (24) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
5′ ppp-RNA Beads and RNA Pulldown
Streptavidin-agarose beads (Fluka 85881) were pre-equilibrated with Blocking buffer (Base buffer (20 mm Hepes (pH 7.9), 2 mm EDTA, 15% glycerol, 0.05% Nonidet P-40, 50 mm NaCl, 500 unit/ml RNasin (Promega N2515), 0.02 mg/ml tRNA (Roche Applied Science, 10109495001), 1% protease inhibitor mixture (Sigma, P8340) and 2 mm DTT) plus 100 mm NaCl, another 100 units/ml RNasin, 0.1 mg/ml glycogen, and 2.5 mg/ml BSA, for 2 h at 4 °C. Biotinylated RNA was bound to the beads in Base buffer for 2 h at 4 °C. Beads to which 0.5 μg of RNA was added were used for each assay. For “in vitro” RNA pulldown, streptavidin beads to which 500 ng of biotinylated pppRNA had been added were incubated with 1 μg of purified RIG-I (25). After 2 h at 4 °C, the beads were washed three times with Base buffer; SDS protein sample buffer was added, and the beads were analyzed by Western blotting with anti-RIG-I. For “in vivo” RNA pulldown, 5 μg of biotinylated pppRNA were transfected into 2fTGH cells using trans-messenger (Qiagen). 4 h post-transfection, cells were resuspended in 120 μl of sonication buffer (50 mm Hepes-KOH (pH 7.5), 150 mm potassium acetate, 5 mm magnesium acetate, 0.1% digitonin, 2 mm DTT, and protease inhibitors) and sonicated. 30 μl of streptavidin-agarose beads were added and the samples treated as described above.
RESULTS
RNA Binding to RIG-I
The binding of dsRNA to RIG-I was studied primarily using a pulldown assay. Because modified nucleosides were shown to interfere with the ability of 5′ pppRNAs to induce IFN via RIG-I (4), a 54-nt-long RNA1 was designed to contain 46 nt of spacer between the patch of biotinylated uridines at the 3′-end (that anchor the RNA to the bead) and the 5′ ppp “business end” of the RNA, to minimize the effect of these modified nucleosides on RIG-I interaction. This approach would not have been possible with natural RNAs containing arena-like structures.
5′ pppRNA was prepared in vitro with T7 RNA polymerase (T7) in the presence of 5-biotin-UTP, which was incorporated uniquely within the 3′-terminal 8 nucleotides of the DNA-encoded 54′-mer. As described previously, the product of this T7 reaction is mostly double-stranded (virtually all the product is sensitive to RNase III digestion), being composed of the DNA-encoded 54′-mer and complementary RNA fragments (mostly 15–25 nt long), as the DNA-encoded 54′-mer itself acts as a template for complementary RNA synthesis (15). When transfected into cells, these unpurified T7 transcripts are generally very potent inducers of IFN. However, when the 54-nt ssRNA is purified from the mixture, it has lost virtually all its ability to bind to RIG-I, stimulate its ATPase, or induce IFN when transfected into cells, similar to chemically made 5′ pppssRNA (9, 10, 15).
To ensure that this biotinylated, partially dsRNA bound endogenous RIG-I under natural conditions in vivo, the unpurified T7 transcripts were transfected into 2fTGH cells which had or had not been treated with IFN (to increase their endogenous levels of RIG-I). Cell extracts were prepared 4 h post-transfection and incubated with streptavidin beads, and the amount of RIG-I that was bound to the beads after washing was examined by Western blotting. As shown in Fig. 1, IFN treatment strongly increased the endogenous level of RIG-I (∼10-fold; lanes 7 and 8 versus lanes 1 and 2), and RIG-I was efficiently bound to the beads in both cases only when the biotinylated in vitro transcripts had been transfected into the cells (lanes 4 and 6 versus lanes 3 and 5). We next examined the ability of 5′ pppdsRNA whose 5′ ppp-nucleotide was base-paired, or not, to bind to purified, bacterially expressed RIG-I in vitro. The T7 DNA-directed 5′ ppp54′-mer (when purified from the reaction products by denaturing PAGE and RNase III digestion) did not increase the binding of RIG-I to the beads over the background (Fig. 2, lanes 3 and 4 versus lanes 1 and 2), as expected. However, when this 5′ ppp54′-mer was annealed to synthetic 18′-mers complementary to positions 1–18 (5′ppp54′-mer/1–18), 19–36, or 37–54, all three 5′ pppdsRNAs clearly bound RIG-I, albeit 5′ ppp54′-mer/1–18 being the most efficient (Fig. 2, lane 5 versus lanes 6 and 7). Moreover, in competition pulldown assays, where nonbiotinylated RNAs are used to compete with the unpurified biotinylated in vitro transcripts for binding to RIG-I, synthetic 41-bp 5′ OHdsRNA and poly(I-C) were found to compete at least as well as unpurified nonbiotinylated in vitro transcripts for binding to RIG-I (Fig. 3). Thus, all the dsRNAs tested bound to RIG-I, independent of whether they contained a 5′ ppp. These results are consistent with the presence of two dsRNA-binding sites on RIG-I as follows: the CTD, where the presence of a 5′ triphosphate group also contributes to dsRNA binding, and the helicases/ATPase domain, where binding is independent of the RNA ends.
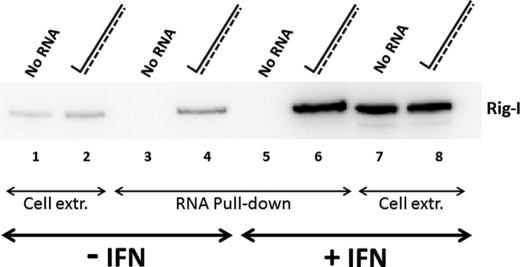
FIGURE 1.
Interaction of RIG-I and in vitro transcribed 5′pppdsRNA1 in vivo. Parallel cultures of 2fTGH cells were treated (or not) with 1000 IU/ml of IFNβ for 12 h and then transfected (or not) with 5 μg of unpurified in vitro transcribed RNA1 containing 5′-biotin-UMP. After 4 h of incubation, cytoplasmic extracts (extr.) were prepared, and equal amounts of extracts were incubated with streptavidin beads. After four cycles of washing, the RIG-I remaining on the beads was determined by Western blotting. The amount of RIG-I present in the extract of 50,000 cells is shown (lanes 1, 2, 7, and 8), as well as that “pulled down” from the extract of 250,000 cells (lanes 3–6).
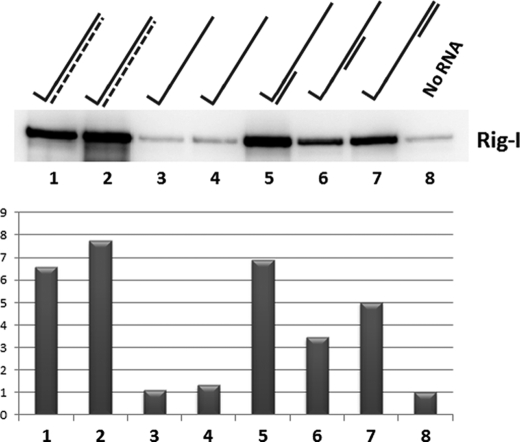
FIGURE 2.
Interaction of RIG-I and purified in vitro transcribed 5′pppRNA1 in vitro. Purified, bacterially expressed RIG-I (1 μg) was incubated with either of two preparations of unpurified in vitro transcribed, biotinylated 5′ pppRNA1 (500 ng, lanes 1 and 2), 500 ng of 5′ pppssRNA1 purified from the above (lanes 3 and 4), or 500 ng of purified 5′ pppssRNA1 annealed with 500 ng of synthetic 5′ OHRNA 18′-mers complementary to position 1–18 (lane 5), 19–36 (lane 6), and 37–54 (lane 7). Streptavidin beads were then added, and the incubation was continued for 10 min. After four cycles of washing, the RIG-I remaining on the beads was determined by Western blotting. Lane 8, no RNA control. The RIG-I bands were quantified (“Experimental Procedures”) and are plotted below. The no RNA control was set at 1.
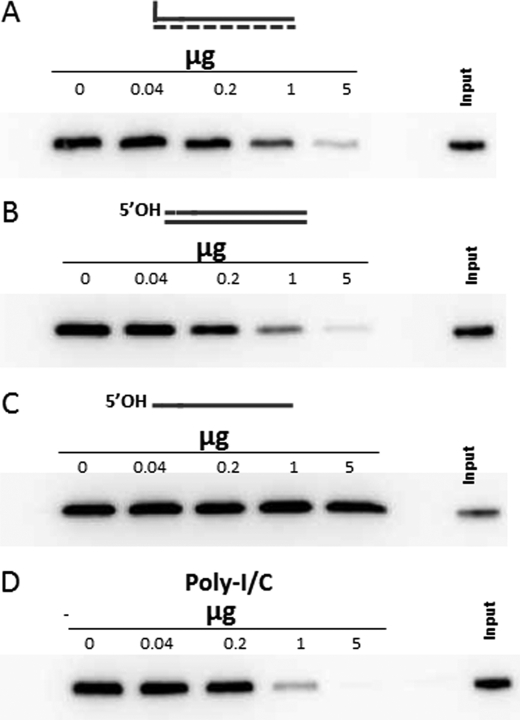
FIGURE 3.
Competition between various RNAs and unpurified in vitro transcribed 5′pppRNA1 for binding to RIG-I. Streptavidin beads to which 0.5 μg of biotinylated pppRNA1 had been added were incubated with 1 μg of RIG-I and increasing amounts of the various competitor RNAs as indicated (A, nonbiotinylated pppRNA1; B, 41-bp 5′ OHdsRNA; C, 41-nt 5′ OHssRNA; D, poly(I-C)). After 2 h at 4°C, the beads were washed three times with base buffer, and the RIG-I remaining on the beads was determined by Western blotting.
dsRNA-dependent ATPase
To investigate the effects of a single 5′ pppnt overhang, or a 3-nt bulge, on the ability of 5′ pppdsRNA to stimulate the RIG-I ATPase, we prepared 5′ pppdsRNAs based on the Junin virus genome panhandles. These 5′ pppdsRNAs were composed of a single-stranded 5′ ppp61′-mer representing the 5′-end of the Junin virus large genome segment (except that the uridines were changed to adenosines or cytidines; Jun*) and chemically synthesized RNAs complementary to positions −1–20 or +1–20. Position −1 corresponds to the overhanging, pseudo-templated 5′ pppG resulting from the prime and realign initiation of genome synthesis; position +1 represents the first nucleotide that is base-paired with the genome 3′-terminal nucleotide (see Fig. 5A). The 5′ ppp61′-mer could be made in vitro as relatively pure ssRNA due to the absence of UTP in the T7 reaction, and annealing with a slight excess of complementary oligoribonucleotides converted all the 5′ ppp61′-mers to dsRNAs (Figs. 5A and 6A).
FIGURE 6.
Effect of dsRNA with an overhanging 5′pppnt on IFNβ activation induced by blunt-ended 5′pppdsRNA. A, parallel cultures of A549 cells were transfected with pIFNβ-(firefly)luciferase and pTK-(Renilla)luciferase. 24 h later, the cultures were transfected in duplicate with either. Lane 1, tRNA control. Lane 2, 1200 ng of 5′ pppJun*61 ssRNA. Lane 3, 1200 ng of overhanging 5′ pppJun*61/(+1–20). Lanes 4, 6, and 8, 50, 100, and 200 ng of blunt-ended, nonbulged 5′ pppJun*61/(−1–20). Lanes 5, 7, and 9, 50, 100, and 200 ng of blunt-ended 5′ pppJun*61/(−1–20) plus 1200 ng of overhanging 5′ pppJun*61/(+1–20). Cell extracts were prepared after a further 20 h, and their luciferase activities were determined. The results are plotted as fold induction relative to the tRNA control. The panel below shows an ethidium bromide-stained, nondenaturing PAGE of the various RNAs transfected. tRNA was added so that the total amount of RNA was constant for all transfections. B, parallel cultures of A549 cells and A549 cells in which RIG-I was knocked down with a constitutively expressed miRNA (A549-RIG-I-kd cells) were transfected with pIFNβ-(firefly)luciferase and pTK-(Renilla)luciferase. 24 h later, the cultures were transfected in duplicate with either 100 (2×) or 200 ng (4×) of nonbulged and blunt-ended 5′ pppJun*61/(−1–20). Cell extracts were prepared after a further 20 h, and their luciferase activities were determined. The results are plotted as fold induction relative to the tRNA control (ctl).
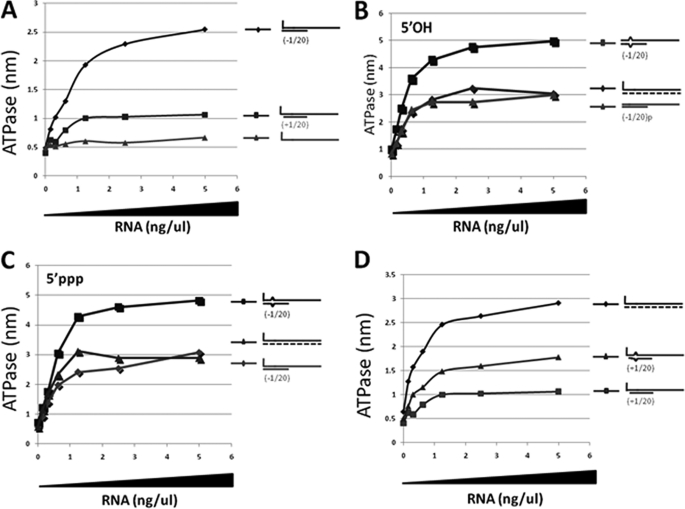
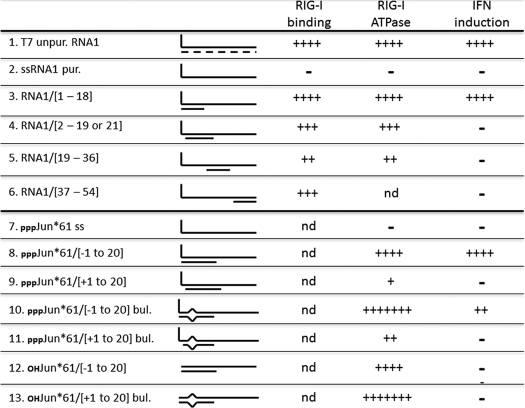
When single-stranded 5′ pppJun*61′-mer, which had little or no ability to stimulate the RIG-I ATPase by itself (Fig. 4A), was annealed with oligoribonucleotides complementary to positions −1–20 or +1–20, both dsRNAs stimulated the ATPase, but the presence of the unpaired 5′ ppp-nucleotide was found to strongly reduce the stimulation (∼4-fold, Fig. 4A), consistent with their relative abilities to activate the IFNβ promoter (Fig. 5A, lane 9 versus lane 10). However, when 5′ pppJun*61/(−1–20) and 5′ OHJun*61/(−1–20) were compared for their stimulation of the ATPase activity (relative to unpurified T7 transcripts), the absence of the 5′-triphosphate group had little or no effect on the ability of this dsRNA to stimulate the ATPase (Fig. 4, B and C). This result is somewhat surprising, as the affinity of the isolated CTD domain for blunt-ended 5′pppdsRNAs is much higher than for 5′OHdsRNAs (16), and because 5′ OHJun*61/(−1–20) has little or no ability to activate the IFNβ promoter (Fig. 5B, lanes 7–9). Even more surprisingly, when the sequences of the complementary oligoribonucleotides were altered so that a symmetrical 3-nt bulge was created at positions 6–8 upon annealing to the 5′ pppJun*61 (similar to the dsRNA panhandles of arenavirus small genome segments), the 5′ pppdsRNA containing the bulge unexpectedly stimulated the ATPase relative to the perfectly base-paired dsRNA (∼2-fold), independent of whether the 5′ pppG was base-paired or unpaired (Fig. 4, C and D). Note that unpurified T7 transcripts were also used to stimulate the ATPase in Fig. 4, B, C, and D, to provide a common point of reference. Given that this 3-nt bulge reduces the ability of blunt-ended 5′ pppdsRNA to activate the IFNβ promoter ∼2-fold (Fig. 5A) and that 5′ OHdsRNA is inactive in this respect, there does not appear to be a simple relationship between the ability of dsRNAs to stimulate RIG-I ATPase activity and to induce IFN. Table 1 summarizes the interactions of RIG-I with the various RNAs.
FIGURE 4.
dsRNA-stimulated RIG-I ATPase activity. Increasing amounts of various dsRNAs (as indicated) were incubated with 200 ng of purified, bacterially expressed RIG-I and 500 μm [lsqb]γ-32P]ATP in 15 μl of 50 mm Tris acetate (pH 6.0), 5 mm DTT, and 1.5 mm MgCl2 for 15 min at 37 °C. The reactions were quenched by adding 3.8 μl of 5 m formic acid. Aliquots (3 ul) of the mixtures were spotted onto PEI TLC plates, and developed with 1 m formic acid, 0.5 m LiCl. 32PO4 release was quantified in a phosphorimager. The results are plotted as a function of RNA concentration (ng/μl). A, blunt-ended 5′ pppJun*61/(−1–20), overhanging 5′ pppJun*61/(+1–20), and 5′ pppJun*61 (ssRNA). B, 5′ OHJun*61/(−1–20) and bulged 5′ OHJun*61/(−1–20) versus unpurified T7 transcripts. C, 5′ pppJun*61/(−1–20) and bulged 5′ pppJun*61/(−1–20) versus unpurified T7 transcripts. D, 5′ pppJun*61/(+1–20) and bulged 5′ pppJun*61/(+1–20) versus unpurified T7 transcripts. The same data set for the +1/20 RNA was used as in A.
TABLE 1.
Interaction of RIG-I with various RNAs
Table lists the names of the various RNAs, the line drawings of their structures, and their relative abilities to bind to RIG-I, to stimulate the ATPase, and to activate the IFN promoter upon transfection into cells.
Short dsRNAs with a Single, Unpaired 5′ ppp-Nucleotide End May Act as RIG-I Decoys
If blunt-ended 5′ pppdsRNA binds directly to the CTD of RIG-I to initiate signaling, 5′ pppdsRNA with a single unpaired 5′ pppnucleotide would compete poorly for this interaction (because of its expected lower CTD affinity), and thus poorly inhibit the activity of blunt-ended 5′ pppdsRNA. On the other hand, if 5′ pppdsRNA binds first to the helicase domain and RIG-I then moves along the dsRNA so that the 5′-ends can interact with the CTD, then 5′ pppdsRNA with a single unpaired 5′ pppnucleotide might compete well for this interaction, as this nonblunt-ended 5′ pppdsRNA does not induce IFN. In this case, 5′ pppdsRNA with a single unpaired 5′ pppnucleotide might sequester RIG-I in an inactive complex.
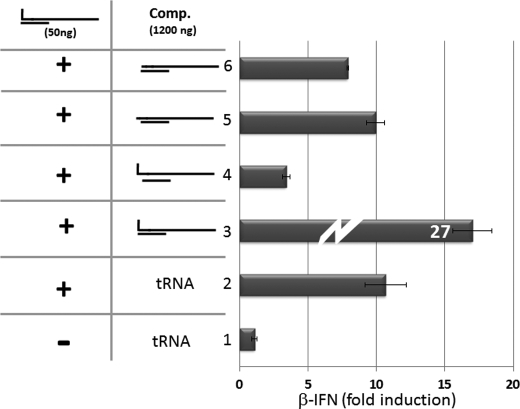
When 1200 ng of single-stranded 5′ pppJun*61′-mer alone was transfected into A549 cells, no activation of the IFNβ promoter was detected (Fig. 6A, top, lane 2), whereas transfection of 50–200 ng of 5′ pppJun*61/(−1–20) was clearly active (lanes 4, 6 and 8). This activation of the IFNβ promoter did not occur in A549 cells is which RIG-I levels had been knocked down (Fig. 6B). Transfection of 1200 ng of 5′ pppJun*61/(+1–20) (whose 5′ pppnucleotide is unpaired) did not activate the IFNβ promoter (Fig. 6B, lane 3). However, transfection of 1200 ng of 5′ pppJun*61/(+1–20) (which mimics the Junin virus panhandles) along with 50–200 ng of blunt-ended 5′ pppJun*61/(−1–20) dsRNA clearly inhibited the ability of the latter to activate the IFNβ promoter (78 to 64% inhibition; Fig. 6B, lanes 5, 7, and 9). This inhibition did not appear to be due simply to high (dsRNA) substrate inhibition, because the transfection of up to 1200 ng of blunt-ended 5′ pppJun*61/(−1–20) continued to increase IFNβ promoter activation (Fig. 5A, lane 9, and Fig. 7, lane 3). Thus, 5′ pppdsRNA with a single unpaired 5′ pppnt inhibits the ability of blunt-ended 5′ pppdsRNA to activate the IFNβ promoter, presumably by competing for RIG-I and sequestering it in an inactive complex. In this case, arenavirus genome 5′ pppdsRNA panhandles would not only fail to induce IFN, they would simultaneously inhibit RIG-I activity.
FIGURE 7.
dsRNA with an overhanging 5′ OHnt does not inhibit IFNβ activation induced by blunt-ended 5′pppdsRNA. Parallel cultures of A549 cells were transfected with pIFNβ-(firefly)luciferase and pTK-(Renilla)luciferase. 24 h later, the cultures were transfected in duplicate with either. Lane 1, tRNA control. Lane 2, 50 ng of blunt-ended 5′ pppJun*61/(−1–20). Lane 3, 50 ng of blunt-ended 5′ pppJun*61/(−1–20) and 1200 ng of blunt-ended 5′ pppJun*61/(−1–20). Lane 4, 50 ng of blunt-ended 5′ pppJun*61/(−1–20) and 1200 ng of nonblunt-ended 5′ pppJun*61/(+1–20). Lane 5, 50 ng of blunt-ended 5′ pppJun*61/(−1–20) and 1200 ng of nonblunt-ended 5′ OHJun*61/(+1–20). Lane 6, 50 ng of blunt-ended 5′ pppJun*61/(−1–20) and 1200 ng of blunt-ended 5′ OHJun*61/(−1–20). Cell extracts were prepared after a further 20 h, and their luciferase activities were determined. The results are plotted as fold induction relative to the tRNA control. tRNA was added so that the total amount of RNA was constant for all transfections.
Interestingly, in contrast to 5′ pppJun*61/(+1–20) dsRNA, transfection of 1200 ng of nonphosphorylated 5′ OHJun*61/(+1–20) dsRNA along with 50 ng of 5′ pppJun*61/(−1–20) dsRNA had little if any ability to inhibit IFN activation induced by the 5′ ppp blunt-ended dsRNA (Fig. 7, lane 5). Moreover, even blunt-ended 5′ OHJun*61/(−1–20) dsRNA had only a modest ability to inhibit 5′ pppJun*61/(−1–20) dsRNA-induced IFN activation (26% inhibition, Fig. 7, lane 6, versus 68%, lane 4), under conditions where addition of the same dsRNA, but bearing a 5′ ppp, roughly doubles the IFN activation (Fig. 7, lane 3). The ability of dsRNA with a single unpaired 5′-nucleotide to inhibit blunt-ended 5′ pppdsRNA activity is thus strongly dependent on the presence of a 5′-triphosphate group. 5′ OHdsRNAs, whether blunt-ended or containing a single unpaired 5′-nucleotide, appear to be very poor competitive inhibitors of blunt-ended 5′ pppdsRNA-induced IFN activation.
Poly(I-C)-induced IFN Activation
In contrast to relatively short dsRNAs, which appear to require a 5′ ppp-blunt end to act as a PAMP, commercial poly(I-C), which is unlikely to contain blunt ends and certainly does not terminate with a 5′ ppp, nevertheless potently induces IFN. This induction occurs exclusively via RIG-I when it is several hundred bp long and, remarkably, primarily via mda-5 when the poly(I-C) is several thousand bp long (25, 26). Moreover, co-infection with Sendai viruses that express a transgenic GFP mRNA plus an mRNA with the complement of the GFP ORF (that are expected to form 700-bp dsRNA with ssRNA tails containing a cap at the 5′-end and a poly(A) tail at the 3′-end) also induces IFN via RIG-I (27). These longer dsRNAs can apparently activate RIG-I without the need for a 5′ pppdsRNA blunt end.
Because the ability of dsRNA with a single unpaired 5′-nucleotide to inhibit blunt-ended 5′ pppdsRNA activity is strongly dependent on the presence of its 5′-triphosphate group (Fig. 7), it was of interest to determine whether this inhibitor was equally active against poly(I-C), whose activity is independent of the presence of a 5′ ppp. As shown in Fig. 8, transfection of 50 ng of our commercial poly(I-C) strongly induces IFN activation (Fig. 8, lane 2), and the co-transfection of 1200 ng of blunt-ended 5′ pppJun*61/(−1–20) dsRNA roughly doubles the IFN activation (lane 3). Co-transfection of 1200 ng of 5′ pppJun*61/(+1–20) dsRNA (with a single unpaired 5′ ppp-nt), in contrast, appears to have no effect; it neither increases nor decreases the activation due to the poly(I-C) (Fig. 8, lane 4). 5′ pppdsRNA with a single unpaired 5′ ppp-nt thus appears to be a selective inhibitor of short blunt-ended 5′ pppdsRNA-induced IFN activation.
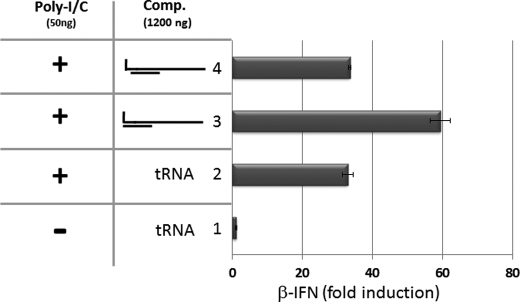
FIGURE 8.
dsRNA with an overhanging 5′ pppnt does not inhibit IFNβ activation induced by poly(I-C). Parallel cultures of A549 cells were transfected with pIFNβ-(firefly)luciferase and pTK-(Renilla)luciferase. 24 h later, the cultures were transfected in duplicate with either. Lane 1, tRNA control. Lane 2, 50 ng of poly(I-C). Lane 3, 50 ng of poly(I-C) and 1200 ng of blunt-ended 5′ pppJun*61/(−1–20). Lane 4, 50 ng of poly(I-C) and 1200 ng of nonblunt-ended 5′ pppJun*61/(+1–20). Cell extracts were prepared after a further 20 h, and their luciferase activities were determined. The results are plotted as fold induction relative to the tRNA control. tRNA was added so that the total amount of RNA was constant for all transfections.
DISCUSSION
RIG-I is a member of the DEX(H/D) box family of ATP-dependent motor proteins (28). Although typically called helicases, many of these enzymes display different functions, such as strand annealing and protein displacement, and in the case of RIG-I translocation along dsRNA. In many cases, their activity is governed by dsRNA-dependent ATP hydrolysis that modulates the protein conformation, thereby converting chemical energy into mechanical movement in a stepwise manner or simply altering the conformation of the protein relative to the RNA. RIG-I appears to carry out both of these functions as follows: (i) it tracks along the phosphodiester backbone of a single strand of the duplex, uniquely in the 5′ to 3′ direction, powered by ATP hydrolysis (18, 19), and (ii) its signaling to the IFNβ promoter requires the interaction of the CTD with a blunt-ended 5′ pppdsRNA (if the dsRNA is relatively short) (9, 15), leading to a proposed conformational change that releases the effector CARD domains to initiate signaling. This latter function again apparently requires ATP, as the K270A RIG-I mutant that cannot bind ATP cannot signal under any conditions, although it can bind dsRNA (20).
This double function of ATP hydrolysis may explain why there is no simple relationship between the ability of various dsRNAs to stimulate the ATPase (Fig. 4) and to activate the IFNβ promoter (Fig. 5), e.g. why blunt-ended 5′ pppdsRNA that contain a bulge increases the ATPase activity relative to perfectly base-paired 5′ pppdsRNAs, whereas having the opposite effect in signaling, and why short 5′ OHdsRNA stimulates the ATPase efficiently but does not induce IFN. Recent single molecular studies have found that RIG-I translocates the entire length of dsRNA in both directions in a robust manner (18). If RIG-I tracks a single strand of the duplex uniquely in the 5′ to 3′ direction, it must switch strands at the end of the duplex to reverse direction. Although highly speculative, the two strands may come apart at the end for RIG-I to switch strands; this would be facilitated by the bulge, and hence the bulge would increase ATPase activity. At the same time, if this “breathing” of the ends interferes with the conformational change in RIG-I that releases the CARDs for downstream signaling, this would decrease IFNβ promoter activation.
We have provided evidence that 20-bp-long 5′ pppdsRNAs with a single unpaired 5′ pppnt (modeled on the Junin virus large genome segment) inhibits the ability of blunt-ended 5′ pppdsRNA to activate the IFNβ promoter when these dsRNAs are co-transfected into cells (Figs. 6 and 7). Remarkably, this inhibition requires the presence of the 5′ ppp group; 5′ OHdsRNAs, whether blunt-ended or containing a single unpaired 5′-nucleotide, appear to be poor competitive inhibitors of blunt-ended 5′ pppdsRNA-induced IFN activation (Fig. 7). The high resolution structures of the CTD bound to 5′ pppdsRNAs have found that the CTD interacts with dsRNA in large part through extensive electrostatic interactions with the 5′ ppp group, consistent with the significantly higher affinity of the CTD for 5′ pppdsRNAs than for 5′ OHdsRNAs. Despite the highly electrostatic nature of CTD/5′ pppdsRNA interactions, both its association and dissociation are relatively slow (17, 29). The unique slow dissociation kinetics of this interaction (t½ = 327 s), as opposed to that of CTD and 5′ OHdsRNA (t½ = 28s), together with the higher affinity of the 5′ pppdsRNA presumably help explain why our 5′ pppdsRNA strongly induces IFN, whereas our 5′ OHdsRNA does not (Fig. 5B, lane 8). It may also help explain why our 5′ OHdsRNAs are such poor competitive inhibitors of 5′ pppdsRNA-induced IFN activation. If RIG-I binds anywhere on dsRNA, and then translocates on the dsRNA to examine its ends, the association of the CTD with a 5′-OH end may not be sufficiently stable so that RIG-I remains associated with (or trapped on) this duplex. One reason why 5′ pppdsRNAs with a single unpaired 5′ pppnt presumably acts as a competitive inhibitor of blunt-ended 5′ pppdsRNA (whereas 5′ OHdsRNAs do not) is that this dsRNA has retained the affinity and half-life of the blunt-ended 5′ pppdsRNA, but this interaction is nonproductive for whatever reason (e.g. because this 5′ pppnt is no longer correctly positioned relative to the following A-form duplex when Phe-857 stacks on a newly reformed blunt end). This RIG-I is then trapped on an inactive 5′ pppdsRNA and is no longer available for productive signaling. This is possibly another way that arenaviruses counteract the innate immune response.
Although the blunt-ended 5′ OHdsRNAs that we have used have not induced IFN upon transfection into cells, Lu et al. (17) have reported that a 27-bp blunt-ended 5′ OHdsRNA does induce IFN. More importantly, they have found that mutation of one of the four basic residues that directly interacts with the 5′ ppp (K861E) abolished the ability of 5′ pppdsRNA to induce IFN but had virtually no effect on that of the 27-bp blunt-ended 5′ OHdsRNA. RIG-I can thus be activated by more than one type of dsRNA, and in more than one way. As mentioned above, poly(I-C), a dsRNA mimetic, strongly induces IFN via RIG-I in a 5′ ppp-independent fashion in A549 cells (24), and this activation is not inhibited by 5′ pppdsRNAs with a single unpaired 5′ pppnt (Fig. 8). The ability of our arenavirus genome dsRNA mimetic to inhibit one type of dsRNA but not another is further evidence that RIG-I recognizes different RNA PAMPs in different ways.
Finally, in contrast to our investigations using defined RNAs based on the known structures of arenavirus genomes, two other groups have extracted the RNAs from polyethylene glycol precipitates of the supernatants of lassa virus and lymphocytic choriomeningitis virus-infected cultures, and both have found that these RNAs induce IFN when transfected into cells (30, 31). However, given that arenavirions are known to include cellular components like ribosomes, they may therefore also contain some antigenome segments and mRNAs as well. The precise nature of the PAMPs present in these extracted RNAs remains to be determined.
This work was supported by a grant from the Swiss National Science Fund.
- PAMP
- pathogen-associated molecular pattern
- dsRNA
- double-stranded RNA
- nt
- nucleotide
- CTD
- C-terminal regulatory domain
- ssRNA
- single-stranded RNA.
REFERENCES
- 1. Takeuchi O., Akira S. (2007) Immunol. Rev. 220, 214–224 [DOI] [PubMed] [Google Scholar]
- 2. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 3. Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 4. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 5. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 6. Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. (2008) Mol. Cell 29, 428–440 [DOI] [PubMed] [Google Scholar]
- 7. Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. (2008) Mol. Cell 29, 169–179 [DOI] [PubMed] [Google Scholar]
- 8. Yoneyama M., Fujita T. (2010) Rev. Med. Virol. 20, 4–22 [DOI] [PubMed] [Google Scholar]
- 9. Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M. C., Besch R., Hopfner K. P., Endres S., Rothenfusser S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schlee M., Hartmann G. (2010) Mol. Ther. 18, 1254–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albariño C. G., Bergeron E., Erickson B. R., Khristova M. L., Rollin P. E., Nichol S. T. (2009) J. Virol. 83, 5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcin D., Kolakofsky D. (1992) J. Virol. 66, 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palmer E. L., Obijeski J. F., Webb P. A., Johnson K. M. (1977) J. Gen. Virol. 36, 541–545 [DOI] [PubMed] [Google Scholar]
- 15. Marq J. B., Kolakofsky D., Garcin D. (2010) J. Biol. Chem. 285, 18208–18216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., Hartmann G., Patel D. J. (2010) Nat. Struct. Mol. Biol. 17, 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu C., Xu H., Ranjith-Kumar C. T., Brooks M. T., Hou T. Y., Hu F., Herr A. B., Strong R. K., Kao C. C., Li P. (2010) Structure 18, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Myong S., Cui S., Cornish P. V., Kirchhofer A., Gack M. U., Jung J. U., Hopfner K. P., Ha T. (2009) Science 323, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Myong S., Ha T. (2010) Curr. Opin. Struct. Biol. 20, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plumet S., Herschke F., Bourhis J. M., Valentin H., Longhi S., Gerlier D. (2007) PLoS ONE 2, e279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matranga C., Pyle A. M. (2010) J. Biol. Chem. 285, 25363–25371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dürr H., Körner C., Müller M., Hickmann V., Hopfner K. P. (2005) Cell 121, 363–373 [DOI] [PubMed] [Google Scholar]
- 23. King P., Goodbourn S. (1994) J. Biol. Chem. 269, 30609–30615 [PubMed] [Google Scholar]
- 24. Marq J. B., Hausmann S., Luban J., Kolakofsky D., Garcin D. (2009) J. Biol. Chem. 284, 25471–25478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hausmann S., Marq J. B., Tapparel C., Kolakofsky D., Garcin D. (2008) PLoS ONE. 3, e3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., Akira S. (2008) J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Strähle L., Marq J. B., Brini A., Hausmann S., Kolakofsky D., Garcin D. (2007) J. Virol. 81, 12227–12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zou J., Chang M., Nie P., Secombes C. J. (2009) BMC Evol. Biol. 9, 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng C., Wu H. (2010) Structure 18, 894–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Habjan M., Andersson I., Klingström J., Schümann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Mühlberger E., Mirazimi A., Weber F. (2008) PLoS ONE 3, e2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou S., Cerny A. M., Zacharia A., Fitzgerald K. A., Kurt-Jones E. A., Finberg R. W. (2010) J. Virol. 84, 9452–9462 [DOI] [PMC free article] [PubMed] [Google Scholar]