Abstract
Resveratrol, a polyphenol compound, is known for its effects on energy homeostasis. With properties of energy sensors mediating effects of calorie restriction, sirtuins are targets of resveratrol. The mammalian sirtuin homolog SIRT1 is a protein deacetylase playing a role in glucose metabolism and islet function. Here, we investigated the effects of resveratrol and possible link with SIRT1 in β-cells. Insulinoma INS-1E cells and human islets were cultured with resveratrol before analyzing their physiological responses. Treatment of INS-1E cells for 24 h with 25 μm resveratrol resulted in marked potentiation of glucose-stimulated insulin secretion. This effect was associated with elevated glycolytic flux, resulting in increased glucose oxidation, ATP generation, and mitochondrial oxygen consumption. Such changes correlated with up-regulation of key genes for β-cell function, i.e. Glut2, glucokinase, Pdx-1, Hnf-1α, and Tfam. In human islets, chronic resveratrol treatment similarly increased both the glucose secretory response and expression of the same set of genes, eventually restoring the glucose response in islets obtained from one type 2 diabetic donor. Overexpression of Sirt1 in INS-1E cells potentiated resveratrol effects on insulin secretion. Conversely, inhibition of SIRT1 achieved either by expression of an inactive mutant or by using the EX-527 inhibitor, both abolished resveratrol effects on glucose responses. Treatment of INS-1E cells with EX-527 also prevented resveratrol-induced up-regulation of Glut2, glucokinase, Pdx-1, and Tfam. Resveratrol markedly enhanced the glucose response of INS-1E cells and human islets, even after removal of the compound from the medium. These effects were mediated by and fully dependent on active SIRT1, defining a new role for SIRT1 in the regulation of insulin secretion.
Keywords: Glucose Metabolism, Insulin Secretion, Pancreatic Islet, Resveratrol, SIRT, Sirtuins
Introduction
Resveratrol (3,5,4′-trihydroxystilbene) is a polyphenolic compound found in grape skins that has been shown to increase lifespan in various organisms, promoting effects similar to those induced by caloric restriction (1–3). In rodents, a 20–40% reduction in calorie intake extends lifespan by ∼50% (4). Calorie restriction enhances insulin sensitivity and hepatic gluconeogenesis in mice (5) and increases muscle mitochondrial biogenesis in humans (6). These observations suggest a link between the molecular pathways activated by resveratrol and caloric restriction, sirtuins acting as common mediators (7, 8).
Sirtuins represent a conserved family of NAD+-dependent proteins homologous to the yeast deacetylase SIR2 (silent information regulator 2), found to be involved in aging processes. Overexpression of Sir2 increases lifespan of lower organisms, whereas deletion or mutations of Sir2 accelerates aging (9–11). Seven human homologs of Sir2 have been identified, named SIRT1 to SIRT7 (12, 13). They are ubiquitously expressed in human tissues but have diverse subcellular localizations, target substrates, and enzymatic activities (14, 15). Sirtuins can function as deacetylase or as mono(ADP-ribosyltransferase). Both reactions occur via cleavage of NAD+ to release nicotinamide, the latter being a sirtuin inhibitor. Because sirtuins hydrolyze NAD+ as a co-substrate, their activity depends on the NAD+/NADH ratio. Accordingly, sirtuins are sensitive to the cellular energy and redox states, conferring them a role as metabolic sensors.
The closest homolog to SIR2 is SIRT1, a protein deacetylase that is up-regulated and activated in adipose tissue, liver, muscle, and brain after fasting or calorie restriction in rodents (16). In these different organs, it has been shown that SIRT1 is implicated in glucose homeostasis and metabolism. In the liver, SIRT1 activates PGC-1α to increase gluconeogenic genes and to repress glycolysis (17), whereas in muscles, SIRT1 activates PGC-1α to increase mitochondrial biogenesis and function (18, 19). In white adipose tissue, SIRT1 decreases peroxisome proliferator-activated receptor γ activity and mobilizes free fatty acids (20).
Less clear is the role of SIRT1 on the endocrine pancreas, which controls glucose homeostasis via insulin secretion. In pancreatic β-cells, glucose entering through GLUT2 transporter is phosphorylated by glucokinase (21) and undergoes glycolysis leading to pyruvate formation that promotes mitochondrial activation and complete oxidation of glucose products (22). Mitochondrial metabolism generates ATP as well as additive factors. The resulting elevation of cytosolic ATP induces the closure of ATP-sensitive K+ channels and, as a consequence, depolarization of the plasma membrane (23). This induces the opening of the voltage-gated Ca2+ channels, resulting in Ca2+ influx within the cell (24) and stimulation of insulin exocytosis. Some reports have shown that SIRT1 positively regulates glucose-stimulated insulin secretion in pancreatic β-cells (25, 26). Mice with β-cell-specific Sirt1 overexpression exhibit improved glucose tolerance and enhanced insulin secretion when challenged with high glucose (26). In contrast, Sirt1 general knock-out animals have lower levels of circulating insulin, and their isolated islets show blunted insulin secretion (25). The authors proposed that SIRT1 would increase insulin secretion and ATP production through the transcriptional repression of the Ucp2 (uncoupling protein 2) gene expression (25).
Resveratrol is an allosteric activator of SIRT1 (2). Treatment of mice fed a high fat diet with resveratrol for >1 year was shown to improve insulin sensitivity and to increase lifespan (27). In another study, resveratrol treatment protected mice against high fat diet-induced obesity. These effects were associated with improved muscle function, explained by increased expression of genes involved in mitochondrial oxidative phosphorylation through SIRT1-mediated PGC-1α deacetylation (19).
Studied in insulin-secreting cells, resveratrol has been shown either to enhance exocytosis by one group (28) or to inhibit insulin secretion by another (29). Therefore, our comprehension of resveratrol effects on β-cell function is limited and contradictory. In view of the importance of SIRT1 in metabolic homeostasis and of the potential of resveratrol (or derivatives) for the treatment of metabolic disorders, it is striking how little is known about interactions between SIRT1, resveratrol, and β-cell function. In the present study, we investigated the effects of chronic resveratrol treatment on INS-1E β-cells and human islets. Because resveratrol markedly potentiated glucose-stimulated insulin secretion, we questioned the role of SIRT1 in resveratrol responses, showing that in β-cells, resveratrol effects were fully SIRT1-dependent.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
The clonal rat β-cell line INS-1E was cultured as described previously (30) in a humidified atmosphere containing 5% CO2 in RPMI 1640 medium supplemented with 5% (v/v) heat-inactivated fetal calf serum, 2 mm glutamine, 10 mm HEPES, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm sodium pyruvate, and 50 μm 2-mercaptoethanol. Freshly isolated human islets were obtained from five different donors with the family's consent. The use of human islets for research was approved by the institutional ethic committee. Donors had an average body mass index of 26 ± 2 (24–28) kg/m2 and age of 58 ± 7 (50–70) years. Islets were maintained in CMRL-1066 at 5.6 mm glucose supplemented with 10% FCS and antibiotics for 2–4 days before experiments. One extra batch of islets was used separately as it was obtained from a donor with a 22-year history of type 2 diabetes (44 years old, with a body mass index of 27.8 kg/m2, HbA1C of 6.7% at admission) under anti-diabetic medication (gliclazide, rosiglitazone, and exenatide). After 3–4 days of culture, cells were incubated in complete RPMI 1640 medium supplemented with the indicated concentrations of resveratrol (Sigma-Aldrich) prepared in ethanol. Control cells were incubated for the same culture period with 0.5% final (v/v) ethanol. Following chronic treatments, resveratrol was washed out at the end of the 24-h culture period and was not present during subsequent stimulations. The pharmacological inhibitor of SIRT1, EX-527 (31), was purchased from Tocris Bioscience (Ellisville, MO).
Adenoviral Treatments of INS-1E Cells
INS-1E cells were seeded in 24-well plates and cultured 3–4 days before adenovirus transduction with the respective adenoviruses, a kind gift from Joseph Rodgers and Pere Puigserver. For infection, cells were incubated for 2 h with adenoviral vectors encoding GFP (Ad-GFP,2 1.6 × 109 infectious units/ml), FLAG-tagged wild type mouse Sirt1 (Ad-FLAG-SIRT1, 2.1 × 1010 infectious units/ml), or inactive SIRT1 mutant that lacks deacetylase activity (Ad-SIRT1-H355A, 5.73 × 1010 infectious units/ml) (32). After transduction, cells were washed and cultured for another 2-day period before the 24-h resveratrol treatment.
Quantitative Real-time PCR
Total RNA from INS-1E cells and human islets was extracted using the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) and TRIzol reagent (Invitrogen), respectively. First-strand cDNA synthesis was performed with 1 or 2 μg RNA, reverse transcriptase (Super Script II, Invitrogen) and 1 μg random primers (Promega, Madison, WI). Primers for mitochondrial transcription factor A (Tfam), peroxisome proliferator-activated receptor coactivator-1α (Pgc-1α), hepatocyte nuclear factor-1 (Hnf-1α) and -4 (Hnf-4α), FoxO1, Ucp2 (uncoupling protein 2), Glut2 (glucose transporter 2), glucokinase, Pdx1 (pancreatic duodenal homeobox 1), Sirt1, NADH dehydrogenase 6 (ND6), cytochrome c oxidase I (CoxI), pyruvate carboxylase, and insulin were designed using the Primer Express Software (Applera Europe) and are listed in supplemental Table S1. Real-time PCR was performed using an ABI 7500 Sequence Detection System (Applera), and PCR products were quantified fluorometrically using the Power SYBR Green PCR Master Mix kit (Applied Biosystems). Values were normalized to transcription factor II B or cyclophilin genes.
Immunoblotting Analyses
Following the culture periods, cells and isolated mitochondria were harvested in radioimmune precipitation assay lysis buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 1 mm EDTA, 1% deoxycholic acid, 50 mm NaF, 0.2 mm Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 1 mm phenylmethylsulfonyl fluoride). For control of SIRT1 expression, INS-1E cells were transduced with respective adenoviruses, and 3 days later, cells were harvested in radioimmune precipitation assay lysis buffer. Protein extracts were separated by 10–12% SDS-PAGE before transfer onto Hybond-ECL nitrocellulose membrane (Amersham Biosciences). The membrane was blocked with 5% milk dissolved in TBS containing 0.1% Tween 20 and probed overnight at 4 °C with different antibodies: mouse anti-SIRT1 (1:1000, Chemicon-Millipore, Zug, Switzerland), rabbit anti-AMPK polyclonal antibody (Cell Signaling, Danvers, MA), mouse monoclonal antibody against the five subunits of oxidative phosphorylation complexes (1:250, catalog no. MS604, MitoSciences, Eugene, OR), goat anti-UCP2 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). After washing, the membranes were incubated 1 h at room temperature with secondary horseradish peroxidase-coupled anti-mouse IgG, anti-rabbit IgG, or anti-goat IgG (Amersham Biosciences), according to primary antibodies. The target proteins were visualized by chemiluminescence (ECL SuperSignal West Pico chemiluminescent substrate, Pierce) and analyzed by a ChemiDoc XRS System (Bio-Rad). Protein-related bands were quantified with Scion Image software (Scion Corporation, Frederick, MD).
Insulin Secretion Assay
Following the culture period and treatments as described, INS-1E cells were assayed for insulin secretion as detailed previously (30). Cells were maintained for 2 h in glucose-free medium. Then, cells were washed and preincubated in glucose-free KRBH buffer (Krebs-Ringer bicarbonate-HEPES buffer; 129 mm NaCl, 5 mm NaHCO3, 4.8 mm KCl, 1.2 mm KH2PO4, 1.2 mm MgSO4, 10 mm HEPES, and 1 mm CaCl2 at pH 7.4) containing 0.1% bovine serum albumin (KRBH/BSA). Next, cells were incubated for 30 min at 37 °C in KRBH/BSA containing different secretagogues. At the end of the incubation, supernatants were collected to measure insulin release, and cellular insulin contents were determined following acid-EtOH extraction (30) by radioimmunoassay (Linco, St. Charles, MO).
Cultured human islets were treated for 24 or 72 h with or without 25 μm resveratrol. On the day of the experiment, batches of 10 islets were hand-picked, starved for 1 h in KRBH/BSA 2.8 mm glucose, and then stimulated for 1 h at 37 °C in KRBH/BSA containing basal 2.8 mm or stimulatory 16.8 mm glucose concentrations.
Glucose Utilization and Oxidation
Glucose utilization, reflecting glycolytic rate, and oxidation were measured as described previously (33). Briefly, INS-1E cells seeded in six-well plates were preincubated for 2 h in glucose-free RPMI 1640 medium and then for 30 min in KRBH. Regarding glucose utilization, cells were then incubated in KRBH containing 2.5 or 15 mm glucose traced with d-[5-3H]glucose, and the reaction was stopped on ice after 30 min. Supernatants were collected, centrifuged to remove detached cells, and [3H]H2O was separated from d-[5-3H]glucose using a Dowex column. For glucose oxidation, cells were incubated for 1 h with KRBH containing 15 mm glucose traced with [U-14C]glucose. [3H]H2O and 14CO2 production was measured with an LKB-Wallak 1217 Rackbeta counter. Glucose metabolism was normalized to cellular protein contents (34).
Cellular ATP and Glutamate Measurements
Cellular ATP levels were monitored in INS-1E cells expressing the ATP-sensitive bioluminescent probe luciferase (35) following transduction with the viral construct AdCAG-luciferase the day before measurements (30). Cells were washed, incubated at 37 °C for 30 min at basal 2.5 mm glucose KRBH buffer containing 100 μm luciferin and then stimulated with 15 mm glucose for 20 min before adding 2 mm NaN3 as mitochondrial poison. Luminescence was monitored in a plate-reader luminometer (Fluostar Optima, BMG Labtechnologies, Offenburg, Germany). Total cellular ATP concentrations were determined in INS-1E cells after stimulation for 10 min with KRBH at 2.5 and 15 mm glucose, extraction in radioimmune precipitation assay lysis buffer and sonication. Cellular ATP concentrations were determined according to the manufacturer's instructions (ATP Bioluminescence Assay Kit HS II, Roche Diagnostics) and normalized to cellular protein contents. Glutamate concentrations were determined following a 60-min incubation at 2.5 and 15 mm glucose as described previously (36), normalized to cellular protein contents.
Oxygen Consumption
Oxygen consumption measurements were performed on isolated mitochondria as described previously (37). Cells were washed with PBS, scraped, lysed in mitochondria isolation buffer (250 mm sucrose, 20 mm Tris/HCl, 2 mm EGTA, pH 7.4) supplemented with 0.5% BSA, and pelleted by centrifugation (10 min at 500 × g). Pellets were resuspended in mitochondria isolation buffer and homogenized in a Teflon glass potter with a 500-rpm rotation rate for 20 up-and-down, followed by centrifugation for 8 min at 1500 × g to precipitate nuclei. Supernatants were further centrifuged for 15 min at 12,000 × g, and the resulting pellets were collected in respiration buffer (200 mm sucrose, 50 mm KCl, 20 mm Tris/HCl, 1 mm MgCl2, and 5 mm KH2PO4 at pH 7.0). Proteins were determined by Bradford assay, and 100 μg of mitochondria were used for measurements of oxygen consumption using a Clark-type electrode (Rank Brothers Ltd., Cambridge, UK). Oxygen consumption rates were measured on mitochondria directly after isolation and stimulated with succinate (5 mm) and ADP (100 μm).
Immunohistochemistry
INS-1E cells were seeded on polyornithine-treated glass coverslips 3 days prior to treatment with 25 μm resveratrol. Following the incubation time periods (2, 6, and 24 h), cells were fixed in 4% paraformaldehyde for 20 min, incubated with boiling 10 mm citrate, pH 6.0, for 10 min, and then permeabilized with 0.2% Triton PBS for 10 min. Slides were incubated overnight at 4 °C with primary rabbit antibodies against SIRT1 (1:250, Chemicon-Millipore, Zug, Switzerland) or FoxO1 (1:100). Next, after several washes with PBS, slides were treated with goat anti-rabbit biotinylated antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) and streptavidin-Alexa Fluor 488 (Invitrogen). Coverslips were then mounted on glass slides using fluorescent mounting medium (DakoCytomation AG, Untermüli, Switzerland), and samples were analyzed on a 63× objective Axiocam microscope (Zeiss, Zürich, Switzerland). Acquisition times were kept the same between groups according to channels to compare signal intensities.
Statistical Analysis
Statistics were done using the SPSS statistical package (version 15.0; Chicago, IL). Unless indicated, data are represented as the means ± S.E. of at least three independent experiments performed in triplicate. Differences between groups were assessed by the two-tailed unpaired t test for single comparison or by one-way analysis of variance, using a post hoc multiple comparison procedure (Fischer's least significant difference method). Results were considered statistically significant at p < 0.05.
RESULTS
Chronic Effects of Resveratrol on Insulin Secretion and Glucose Metabolism in INS-1E Cells
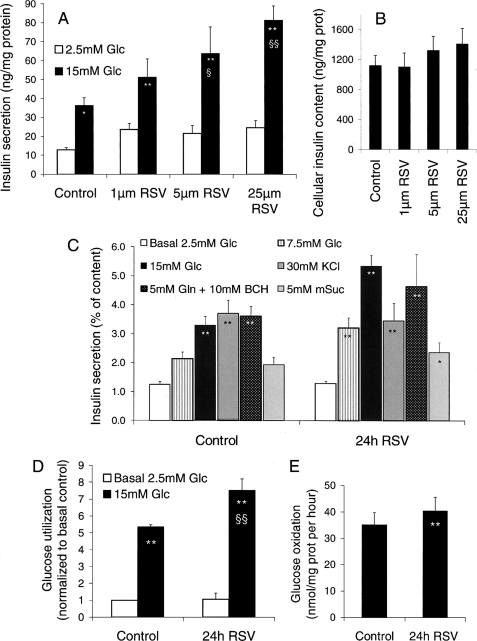
Cells were treated for the last 24 h of culture with 1, 5, and 25 μm resveratrol. Cells were then washed so that resveratrol was not present any longer during challenges. Insulin secretion tested in control cells showed a 2.8-fold increase upon 15 mm glucose stimulation compared with basal release at 2.5 mm glucose (p < 0.05). Incubating cells for 24 h with resveratrol did not change basal insulin release at any of the tested concentrations. However, the secretory response evoked by 15 mm glucose was potentiated in a dose-dependent manner (Fig. 1A). Compared with control INS-1E cells, those treated with 5 and 25 μm resveratrol exhibited a secretory response at 15 mm glucose that was enhanced 1.76-fold (p < 0.05) and 2.24-fold (p < 0.01), respectively (Fig. 1A). Cellular insulin contents were not statistically different following the 24 h treatment with the different concentrations of resveratrol (Fig. 1B), which did not induce noticeable side effects such as cell death or proliferative changes. The rest of the study was based on the effective and safe resveratrol concentration of 25 μm.
FIGURE 1.
Chronic effects of resveratrol on insulin secretion and glucose metabolism in INS-1E cells. INS-1E cells, cultured for 5 days after seeding, were exposed for 24 h to the indicated concentrations of resveratrol (RSV) in standard medium before experiments performed in the absence of resveratrol. A, chronic effects of resveratrol on insulin secretion. Insulin release was measured over a 30-min incubation period at basal 2.5 mm glucose (Glc) and stimulatory 15 mm glucose. B, cellular insulin contents following the 24-h treatment period described in A. C, the secretory responses of INS-1E cells treated for 24 h with 25 μm resveratrol (24h RSV) were assayed with different secretagogues: basal 2.5 mm glucose, intermediate 7.5 mm glucose, stimulatory 15 mm glucose, 30 mm KCl, 5 mm glutamine (Gln) with 10 mm 2-amino-bicycloheptane-2-carboxylic (BCH), and 5 mm methylsuccinate (mSuc). D, glucose utilization was measured at 2.5 and 15 mm glucose using d-[5-3H]-glucose in control and resveratrol (25 μm) treated INS-1E cells. E, glucose oxidation to 14CO2 was measured in control and resveratrol (25 μm) treated INS-1E cells over a 1-h stimulation period with 15 mm [U-14C]glucose. Values are means ± S.E. of 4–10 independent experiments, with each performed in triplicate. *, p < 0.05; **, p < 0.01 versus 2.5 mm glucose; §, p < 0.05; §§, p < 0.01 versus control group at 15 mm glucose.
Beside glucose, additional secretagogues were tested to nail down the target pathways of resveratrol. INS-1E cells were incubated for 24 h with 25 μm of resveratrol, washed, and then stimulated for 30 min with 7.5 and 15 mm glucose (Fig. 1C), showing that resveratrol was mostly effective at high glucose. Insulin secretion evoked by 30 mm KCl to directly induce plasma membrane depolarization and the accompanying calcium rise was not changed by the treatment. Similarly, resveratrol had no effects on glycolysis-independent mitochondrial activation using either 5 mm glutamine plus 10 mm 2-amino-bicycloheptane-2-carboxylic or 5 mm methyl-succinate (Fig. 1C). Data show that culturing cells in the presence of resveratrol potentiated the secretory response specifically to glucose.
These results prompted us to measure effects of resveratrol on glucose metabolism. Glucose utilization reflecting glycolytic rate was measured by [3H]H2O formation from d-[5-3H]glucose (Fig. 1D). In control cells, raising glucose concentration from 2.5 to 15 mm increased the glycolytic rate by a factor of 5.3 (p < 0.01). In cells previously incubated for 24 h with 25 μm resveratrol, there was a further 32% increase in 15 mm glucose utilization compared with control cells (p < 0.01), whereas basal glycolytic rate was unchanged. [U-14C]glucose complete oxidation to CO2 was measured in cells over a 60-min stimulation period following the 24-h treatment period (Fig. 1E). Incubation with 25 μm resveratrol enhanced 14CO2 formation by 15% (p < 0.01) upon 15 mm glucose stimulation (Fig. 1E), correlating with the observed increase in glucose utilization.
Mitochondrial Metabolism in INS-1E Cells after Resveratrol Treatment
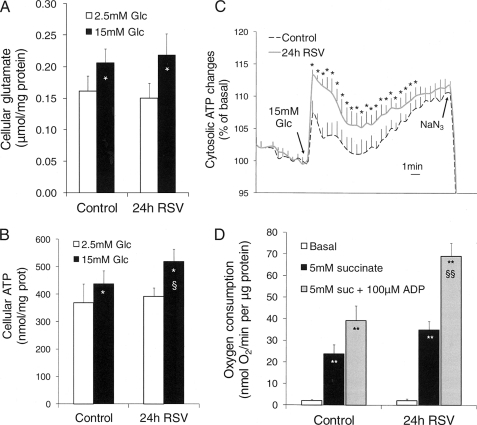
Resveratrol increased glucose metabolism, whereas CO2 production accounted only for half of the enhanced glucose utilization (Fig. 1, D and E). This suggested that glucose carbons underwent alternative cataplerotic pathways (38), such as the formation of glutamate that may contribute to the glucose response (39). Cellular glutamate levels were similarly elevated upon glucose stimulation in both resveratrol treated and nontreated cells (Fig. 2A).
FIGURE 2.
Effects of resveratrol on mitochondrial activation in INS-1E cells. INS-1E cells were exposed for 24 h to 25 μm resveratrol (RSV) in standard medium before glucose stimulation in the absence of resveratrol and measurements of cellular glutamate, ATP generation, and oxygen consumption. A, cellular glutamate concentrations measured at the end of the incubation period at 2.5 and 15 mm glucose (Glc). B, total cellular ATP concentrations were determined following a 10-min incubation period at 2.5 and 15 mm glucose. C, real-time cellular ATP changes were monitored in luciferase-expressing INS-1E cells as bioluminescence. ATP generation was induced by raising glucose from 2.5 to 15 mm (see arrow, 15 mm glucose) and collapsed by addition of the mitochondrial poison 2 mm NaN3 (see arrow, NaN3). D, O2 consumption measured on mitochondria isolated from INS-1E cells following the culture period and stimulated with 5 mm succinate, followed by further addition of 100 μm ADP. Values are means ± S.E. of five independent experiments. *, p < 0.05; **, p < 0.01 versus basal conditions; §, p < 0.05; §§, p < 0.01 versus control group at corresponding stimulatory conditions.
Next, ATP levels were measured in INS-1E cells, showing that in control cells, glucose stimulation increased cellular ATP concentrations by 19% (p = 0.017; see Fig. 2B). Following the 24-h resveratrol treatment, cellular ATP levels were further elevated at 15 mm glucose (+18%, p < 0.05) compared with control cells. We also monitored dynamic changes in cellular ATP, revealing elevated and sustained increases of ATP upon glucose stimulation in resveratrol-treated cells versus naïve cells (Fig. 2C).
Resveratrol-induced increases in ATP generation might be associated with changes in the electron transport chain efficiency. In this context, oxygen consumption was measured in mitochondria isolated from cells previously cultured for 24 h with resveratrol. Both state 4 and state 3 were tested, evoked respectively by 5 mm succinate and by succinate plus 100 μm ADP (Fig. 2D). In mitochondria isolated from control cells, induction of state 4 and state 3 resulted in 12- and 20-fold elevations, respectively, versus basal respiratory rates (p < 0.01 for both conditions). After 24 h of resveratrol (25 μm) treatment, state 3 was markedly enhanced (+73%, p < 0.01) compared with control mitochondria. The respiratory control ratio, defined as state 3 over state 4 ratio, was calculated to estimate coupling efficiency between oxygen consumption and phosphorylation of ADP to ATP. Respiratory control ratio was 24% higher following resveratrol treatment (2.09 versus 1.68 in control mitochondria, p < 0.05), indicating that resveratrol increased coupling of the mitochondria.
Effects of Resveratrol on Expression of Key Components of β-Cell
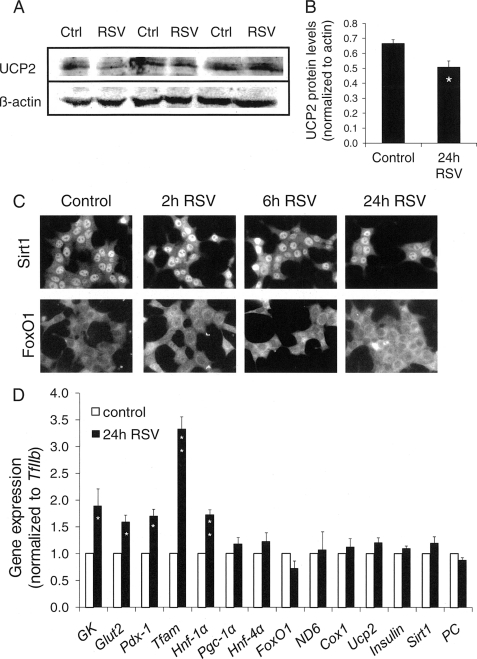
The increased efficiency of the respiratory chain observed following resveratrol treatment could be the consequence of changes in the machinery of the electron transport chain (37). Accordingly, the five respiratory chain complexes were analyzed by immunoblotting on mitochondria isolated from INS-1E cells after exposure to resveratrol for 24 h (supplemental Fig. S1), revealing similar expression levels in control and treated cells for all tested subunits, i.e. ND6 (complex I), FeS (complex II), COX I (complex IV), core 2 (complex III), and α (complex V).
Resveratrol is known to activate SIRT1, which, in turn, may repress Ucp2 expression (25). Although the uncoupling properties of UCP2 are debated, we checked expression of this protein following the 24-h resveratrol treatment (Fig. 3A). In mitochondria isolated from resveratrol-treated INS-1E cells, UCP2 expression was 24% lower versus controls (p < 0.05, Fig. 3B). This suggests that resveratrol mediated down-regulation of UCP2 through SIRT1-dependent activation.
FIGURE 3.
Effects of resveratrol on expression and location of key components of insulin-secreting cells. INS-1E cells were exposed for the indicated time period to 25 μm resveratrol (RSV) in standard medium before expression analyses and immune detection. A, INS-1E mitochondria were isolated to measure UCP2 protein levels by immunoblotting. B, immunoblotting for UCP2 (A) was normalized to β-actin and quantified by densitometry. Values are means ± S.E. of three independent experiments; *, p < 0.05 versus control. C, immunofluorescence images showing INS-1E cells stained for SIRT1 and FoxO1 following resveratrol treatment for 0, 2, 6, and 24 h. Images are representative of three independent preparations. D, after the 24-h treatment period with resveratrol, RNA was extracted from INS-1E cells, and expression of selected genes was measured by quantitative RT-PCR. Values were normalized to TFIIb expression. Results are means ± S.E. of three to five independent experiments, with each performed in triplicate. *, p < 0.05; **, p < 0.01 versus control (Ctrl).
SIRT1 can translocate between the cytoplasm and the nucleus, where expression of target genes may be regulated through deacetylation mechanism. To check whether resveratrol would affect SIRT1 subcellular localization, we performed immunohistochemistry on INS-1E cells following a time course treatment with 25 μm resveratrol. As shown in Fig. 3C, SIRT1 was mainly localized within the nucleus, independently of resveratrol treatment. As it has been demonstrated that SIRT1 can regulate the activity of the transcription factor FOXO, and in particular FoxO1, we also checked FoxO1 subcellular localization. Following resveratrol treatment, INS-1E cells displayed mostly cytosolic FoxO1 labeling over the whole 24-h period of resveratrol treatment (Fig. 3C). These data indicate that resveratrol effects are not mediated by translocation of the transcription factors SIRT1 and FoxO1.
Expression studies were complemented at the mRNA level by quantitative RT-PCR focusing on a panel of genes playing a key role in β-cell function and differentiation (Fig. 3D). Treatment for 24 h with 25 μm resveratrol increased expression of both the glucose transporter GLUT2 (+59%, p < 0.05) and the enzyme initiating glycolysis glucokinase (+89%, p < 0.05). The transcription factors HNF-1α and PDX-1, necessary for β-cell differentiation, were up-regulated (+73 and +70%, respectively, both with p < 0.05), as well as the nuclear-encoded transcription factor TFAM responsible for stability and transcriptional activity of the mitochondrial genome (+232%, p < 0.01). None of the tested genes participating to mitochondrial biogenesis were changed by resveratrol treatment; i.e. Pgc-1α, pyruvate carboxylase, ND6, and Cox1. Ucp2, which was found to be down-regulated at the protein level (Fig. 3A), was not modified at the mRNA level (Fig. 3D), an observation compatible with the know regulation of this mitochondrial protein at the translational level (40). Finally, expressions of both insulin and Sirt1 were unchanged by resveratrol, in agreement with cellular insulin contents (Fig. 1B) and SIRT1 protein levels analyzed by immunoblotting (supplemental Fig. S2).
Effects of Resveratrol on Insulin Secretion and Gene Expression in Human Islets
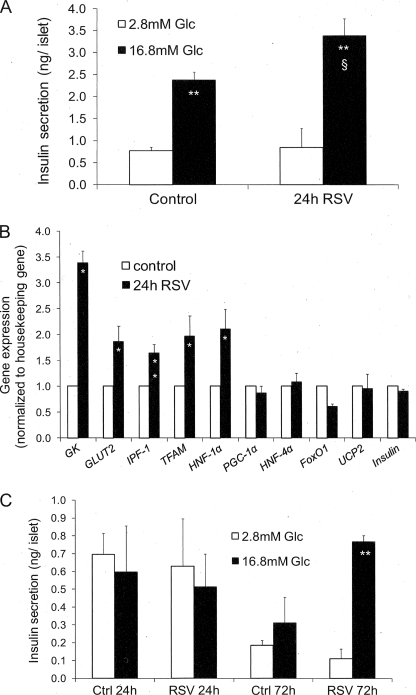
Given the strong effects of resveratrol observed in clonal INS-1E cells, we next evaluated selected key parameters in primary cells in the form of human islets. Human islets were treated for 24 h with 25 μm resveratrol and were tested the following day for glucose-stimulated insulin secretion in the absence of resveratrol (Fig. 4A). In control islets, 16.8 mm glucose stimulated insulin secretion 2.5-fold versus basal release (p < 0.01). Islets previously treated with resveratrol exhibited a secretory response enhanced by 42% compared with stimulated controls (p < 0.05), whereas basal insulin release was not changed.
FIGURE 4.
Effects of resveratrol on insulin secretion and gene expression in human islets. Islets obtained from human donors were exposed for 24 h to 25 μm resveratrol (RSV) in standard medium before measurements of the secretory response and expression of selected genes. A, following resveratrol treatment, human islets were hand-picked and washed, and insulin release was measured over a 60-min incubation period at basal 2.8 mm and stimulatory 16.8 mm glucose (Glc). Values are means ± S.E. of four independent experiments, with each performed in quintuplet. **, p < 0.01 versus basal 2.8 mm glucose; §, p < 0.05 versus control at 16.8 mm glucose. B, expression of selected genes measured by quantitative RT-PCR and normalized to tubulin and cyclophilin expression. Results are means ± S.E. of three to four independent experiments performed in triplicate. *, p < 0.05 versus control; **, p < 0.01 versus control (Ctrl). C, human islets obtained from a donor with type 2 diabetes were treated for periods of 24 and 72 h with 25 μm resveratrol and then tested for glucose-stimulated insulin secretion as described in A. Values are the means ± S.D. of one experiment performed in quintuplet. **, p < 0.01 versus 2.8 mm glucose of the corresponding group.
In human islets, exposure to resveratrol for 24 h significantly (p < 0.05) increased expression of glucokinase (+239%), GLUT2 (+87%), TFAM (+97%), HNF-1α (+111%), and IPF-1 (+64%), with the latter corresponding to Pdx-1 in rodents. Similarly to INS-1E cells, no significant changes of expression were observed for UCP2, PGC-1α, HNF-4α, FoxO1, and insulin. Altogether, secretion and expression data show good agreement between rat insulinoma and human islets regarding resveratrol effects.
Exceptionally, we had access to one batch of islets from a type 2 diabetic donor. In this case, we tested glucose-stimulated insulin secretion following periods of 24 and 72 h of treatment with 25 μm resveratrol (Fig. 4C). After 24 h, untreated type 2 islets did not respond to stimulatory glucose concentration, and extending the culture to 72 h only reduced basal insulin release without restoring the response to 16.8 mm glucose. Remarkably, type 2 islets treated with resveratrol for 72 h exhibited robust glucose-stimulated insulin secretion (7-fold versus basal, p < 0.05, Fig. 4C). Although extremely encouraging, it must be emphasized that these results are based on islets obtained from one type 2 diabetic patient, and it remains to be determined whether such beneficial effects could be extrapolated to others.
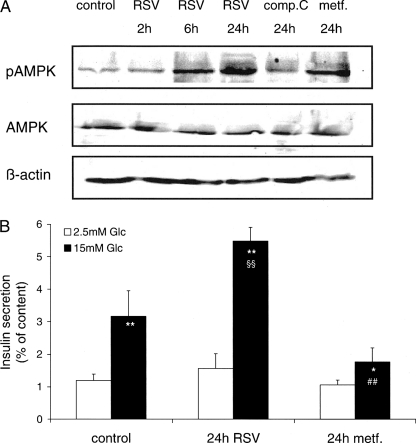
Effects of Resveratrol and Metformin on AMPK Activation in INS-1E Cells
Studies have shown that resveratrol could increase adenosine monophosphate kinase (AMPK) activity, i.e. phosphorylation, in hepatocytes (41), cardiac myocytes, and neurons (42, 43). This prompted us to test whether resveratrol could similarly activate AMPK in insulin secreting cells and potentially explain its potentiating effects. We used the AMPK inhibitor compound C (1 μm) as negative control and the AMPK activator metformin (5 mm) as positive control (Fig. 5A). Treatment of INS-1E cells with 25 μm resveratrol for 2, 6, and 24 h increased AMPK phosphorylation in a time-dependent manner.
FIGURE 5.
Effects of resveratrol and metformin on AMPK activation and insulin secretion in INS-1E cells. A, INS-1E cells were treated either with 25 μm resveratrol (RSV) for 2, 6, and 24 h; with 5 mm of the AMPK activator metformin (metf.) for 24 h; and with 1 μm of the AMPK inhibitor compound C (comp.C). Levels of phosphorylated AMPK protein (pAMPK) and total AMPK were assessed by immunoblotting of INS-1E cell extracts. Total AMPK and pAMPK protein were normalized to β-actin. Immunoblotting is representative of three independent experiments. B, the secretory responses of INS-1E cells treated with resveratrol and metformin for 24 h were assayed at basal 2.5 mm and stimulatory 15 mm glucose (Glc). Values are means ± S.D. of one of three representative experiments (n = 3). *, p < 0.05; **, p < 0.01 versus 2.5 mm glucose; §§, p < 0.01 versus control at 15 mm glucose; ##, p < 0.01 versus resveratrol at 15 mm glucose.
We then tested whether activation of AMPK could correlate with the secretory response. Treatment of INS-1E cells with 25 μm resveratrol for 24 h potentiated glucose-stimulated insulin secretion (+173%, p < 0.01) compared with control cells (Fig. 5B). On the contrary metformin (5 mm) treatment reduced the secretory response (−44%, p < 0.01) versus control group. The data show that resveratrol activated AMPK similarly to metformin. However, the reduction in glucose-stimulated insulin secretion associated with AMPK activation was overruled by the potentiating effects of resveratrol, pointing to alternative molecular targets.
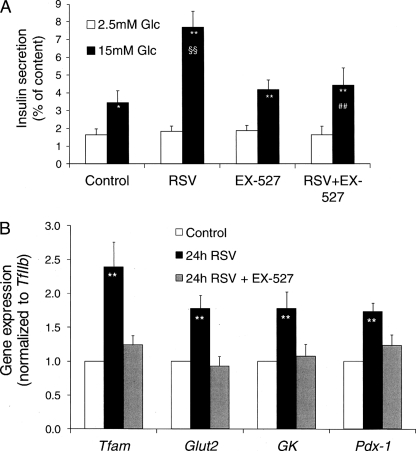
SIRT1 Inhibitor EX-527 Abrogates Resveratrol Effects in INS-1E Cells
Besides AMPK, SIRT1 is another candidate as molecular target for resveratrol effects, although resveratrol treatment modified neither expression nor translocation of SIRT1 (Fig. 3). Alternatively, resveratrol can promote SIRT1 deacetylase activity, an effect mediated by the lowering of the Michaelis constant of SIRT1 for acetylated substrates (2). In this context, we used the pharmacological SIRT1 inhibitor EX-527 (31) to further investigate the putative link between resveratrol and SIRT1 in insulin-secreting cells. Addition of EX-527 (1 μm) in the culture medium during the 24-h period preceding glucose-stimulated insulin secretion assay did not modify the secretory response. The 2.1-fold secretory response to 15 mm glucose observed in control INS-1E cells was enhanced by 122% (p < 0.01) following the 24-h resveratrol (25 μm) treatment (Fig. 6A). Culturing INS-1E cells with resveratrol in the presence of EX-527 totally abolished the potentiating effect induced by resveratrol (−42%, p < 0.01, see Fig. 6A). In human islets, addition of 1 μm EX-527 during the 24-h culture period in the presence of resveratrol reduced the secretory response at 16.8 mm glucose by 46% compared with resveratrol treatment alone (2.61 ± 0.55 versus 4.83 ± 0.39 ng/ml insulin per islet, respectively, p < 0.01, n = 4). When we used splitomicin, another inhibitor of SIRT1 catalytic activity (44), similar results were obtained (data not shown).
FIGURE 6.
Effects of the SIRT1 inhibitor EX-527 on insulin secretion and gene expression in INS-1E cells. INS-1E cells were exposed for 24 h to 25 μm resveratrol (RSV) and 1 μm EX-527 in standard medium before secretory responses and gene expression analyses. A, the secretory responses of INS-1E cells treated with 25 μm resveratrol with or without 1 μm EX-527 (EX-527) were assayed at basal 2.5 mm glucose (Glc) and stimulatory 15 mm glucose. Values are means ± S.E. of three independent experiments each performed in sextuplet. B, after the treatment period, RNA was extracted from INS-1E cells, and expression of selected genes was measured by quantitative RT-PCR. Values were normalized to TFIIb expression. Results are means ± S.E. of four to seven independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01 versus basal 2.5 mm glucose; §§, p < 0.01 versus control at 15 mm glucose; ##, p < 0.01 versus resveratrol at 15 mm glucose.
Effects of EX-527 were tested on the expression of those genes that were up-regulated by resveratrol treatment (Fig. 3D). As shown in Fig. 6B, addition of the SIRT1 inhibitor EX-527 in the culture medium abolished resveratrol-mediated up-regulation of Tfam, Glut2, glucokinase, and Pdx-1. These results indicate that SIRT1 enzymatic activity was responsible for resveratrol effects on INS-1E cells.
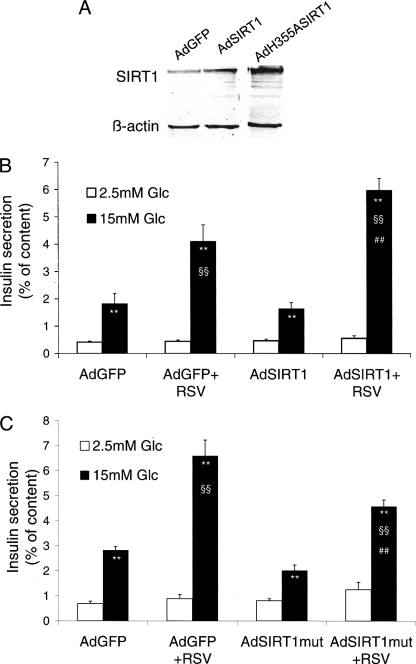
Effects of SIRT1 Overexpression and of an Inactive Mutant on Insulin Secretion
A pharmacological approach pointed to SIRT1 enzymatic activity as a mediator of resveratrol effects on insulin-secreting cells (Fig. 6). To further scrutinize this link, the presence of active Sirt1 in INS-1E cells was modulated by means of adenoviral gene transfer. Practically, we either overexpressed the wild type form (Ad-SIRT1) or expressed a mutant lacking deacetylase activity (Ad-H355A-SIRT1). Efficiency of the corresponding adenoviruses was assessed by quantitative RT-PCR and by immunoblotting 3 days after transduction of INS-1E cells. At the protein level, Ad-SIRT1 increased Sirt1 3.5-fold and Ad-H355A-SIRT1 4.3-fold over endogenous expression level (Fig. 7A).
FIGURE 7.
Effects of SIRT1 overexpression and inactive mutant expression on insulin secretion in INS-1E cells. INS-1E cells cultured for 3–4 days were transduced for 2 h with adenoviral vectors encoding GFP (AdGFP, control), wild type mouse SIRT1 (AdSIRT1, SIRT1 overexpression), and mutant H355A-SIRT1 lacking deacetylase activity (AdSIRT1mut, inactive mutant). Next, cells were washed and cultured for another 2-day period before the 24-h resveratrol (RSV, 25 μm) treatment followed by analyses. A, levels of SIRT1 protein were assessed by immunoblotting on INS-1E cell extracts 3 days after transduction. Immunoblotting is representative of three independent experiments. B, the secretory responses of INS-1E cells treated with resveratrol with or without SIRT1 overexpression (AdSIRT1) were assayed at basal 2.5 mm glucose (Glc) and stimulatory 15 mm glucose. C, the secretory responses of INS-1E cells treated with resveratrol with or without expression of an inactive SIRT1 mutant (AdSIRT1mut) were assayed at basal 2.5 mm glucose and stimulatory 15 mm glucose. Values are means ± S.E. of five independent experiments, with each performed in sextuplet. **, p < 0.01 versus 2.5 mm glucose; §§, p < 0.01 versus AdGFP at 15 mm glucose; ##, p < 0.01 versus AdGFP and resveratrol at 15 mm glucose.
Two days after transduction with the indicated adenoviruses, INS-1E cells were further cultured for the remaining 24 h in the presence of 25 μm resveratrol before glucose-stimulated insulin secretion assay. Cells transduced with control Ad-GFP and treated with resveratrol exhibited a glucose (15 mm) response enhanced 2.3-fold (p < 0.01) compared with untreated Ad-GFP controls (Fig. 7B). On its own, overexpression of SIRT1 did not modify the secretory pattern, neither at basal nor stimulatory glucose concentrations. In contrast, resveratrol treatment of INS-1E cells overexpressing SIRT1 further potentiated glucose-stimulated insulin release compared with resveratrol-treated Ad-GFP cells (+46%, p < 0.01).
Finally, we tested the effects of the inactive deacetylase mutant H355A-SIRT1 on resveratrol-treated INS-1E cells. Compared with nontreated Ad-GFP controls, the secretory response to 15 mm glucose was potentiated 2.3-fold (p < 0.01) following the 24-h culture period in the presence of resveratrol. Expression of the mutant H355A-SIRT1 partially abrogated such potentiation (−31%, p < 0.01) evoked by resveratrol (Fig. 7C), indicating that H355A-SIRT1 acted as a dominant negative.
These results show that resveratrol, although it is does not change SIRT1 expression, directly depends on cellular levels of active SIRT1 to exert its effects. However, in the absence of resveratrol, endogenous expression of SIRT1 seems to meet β-cell function requirement.
DISCUSSION
The present study shows that chronic treatment with the polyphenolic compound resveratrol rendered insulin-secreting cells highly responsive to glucose stimulation. This potentiating effect was specific to glucose and associated with enhanced metabolism of the sugar. Increased mitochondrial activation in resveratrol-treated cells was favored by up-regulation of GLUT2 and glucokinase, thereby accelerating the glycolytic rate at high glucose and funneling more substrates into the Krebs cycle. Increased expression of GLUT2 and glucokinase might be secondary to the observed up-regulation of the transcription factors PDX-1 and HNF-1α in resveratrol-treated cells. PDX-1 is a β-cell master gene (45) that has been shown to regulate GLUT2 transcription in β-cells (46), whereas effects on glucokinase expression remain conflicting (47–49). Regarding HNF-1α, its expression is required for GLUT2 transcription in differentiated insulin-producing cells (50, 51) and, consequently, necessary to maintain mitochondrial catabolism of the glycolytic product pyruvate (52). TFAM, another transcription factor controlling the mitochondrial genome (53) and which is under the control of PDX-1 (54), was also up-regulated upon resveratrol treatment. Overall, data indicate a sequence of events in which resveratrol activates SIRT1, inducing up-regulation of key transcription factors, in turn promoting expression of GLUT2 and metabolic enzymes, ultimately increasing metabolism-secretion coupling.
Similar effects were observed in human islets, even after removal of resveratrol from the medium. The resveratrol effects were fully dependent on the presence of active SIRT1 in the cells. Indeed, potentiation of the glucose responses was inhibited by both pharmacological and genetic inhibition of SIRT1 and, conversely, was further increased by overexpression of the sirtuin.
Resveratrol treatment enhanced efficiency of mitochondrial function, such as oxygen consumption and ATP generation. In particular, we observed an increased respiratory control ratio, reflecting tighter coupling between ADP to ATP phosphorylation and oxygen consumption. This correlated with up-regulation of TFAM, both in INS-1E cells and human islets, following resveratrol treatment. However, expression levels of the electron transport chain subunits were not modified, arguing against changes in mitochondrial biogenesis. At the protein levels, UCP2 was down-regulated, an effect possibly associated with mitochondrial coupling. These results indicate that resveratrol modified mitochondrial efficiency through qualitative rather than quantitative changes on this organelle.
It has been shown that resveratrol improves the metabolic state in animal models by increasing tissue responsiveness to insulin (19, 42) and exerts hypoglycemic effects in diabetic rats (55). Only few studies focused on resveratrol and insulin-secreting cells, still providing conflicting results. In MIN6 insulinoma cells, it was shown that resveratrol enhances insulin secretion by blocking KATP and KV channels (28). Conversely, resveratrol was reported to inhibit insulin secretion in rat pancreatic islets (29) and to counteract stimulus-induced depolarization of INS-1E cells (56). A close link between resveratrol and SIRT1 has been demonstrated in muscles and adipose tissues (19). Here, we provide the first evidence for SIRT1-dependent effects of resveratrol in β-cells, both in insulinoma cells and human islets. In agreement with positive effects of SIRT1 on insulin secretion (26), our results show that resveratrol exerts strong potentiating effects on glucose-stimulated insulin secretion, mediated by SIRT1.
Thus, present data show that a major target of resveratrol in β-cells is SIRT1. Although effects of resveratrol positively correlate with SIRT1 protein levels, the polyphenolic compound exerts its effects through SIRT1 activation without changing its expression. Downstream of SIRT1 activation, we identified molecular targets such as PDX-1, HNF-1α, TFAM, GLUT2, glucokinase, and UCP2 at the protein level. Two studies proposed that SIRT1 positively regulates glucose-stimulated insulin secretion in pancreatic β-cells through down-regulation of UCP2 and increased ATP production (25, 26). Here, upon SIRT1 activation, we similarly observed a correlation between increased glucose-stimulated insulin secretion and ATP generation, accompanied by down-regulation of UCP2 protein. Careful examination of mitochondrial activation revealed increased coupling when SIRT1 was activated by resveratrol. Although increased UCP2 expression has been associated with inhibition of glucose-stimulated insulin secretion (57, 58), the supposedly uncoupling properties of UCP2 have been disputed recently (59) or proposed to require additional activators (60, 61). In view of past and present data, the role of UCP2 in β-cell function remains a matter of debate.
Several papers have shown that resveratrol also stimulates AMPK in different tissues (41–43) and that phosphorylation of AMPK is associated with changes in SIRT1 expression and activity (62). Accordingly, activation of AMPK by resveratrol could account for some of the beneficial effects in mice on a high fat diet (19, 27). Here, in agreement with what was demonstrated in other tissues, we observed that resveratrol treatment increased AMPK phosphorylation in insulin-secreting cells. However, at least in β-cells, SIRT1 and AMPK seem to act in opposite directions because activation of the former by resveratrol increased insulin secretion, whereas metformin, used to selectively activate the latter, reduced the glucose response. This shows that stimulating actions of SIRT1 fully counteract inhibitory effects of AMPK activation (63, 64).
In conclusion, data show that resveratrol potentiates glucose-stimulated insulin secretion, glucose metabolism, and mitochondrial activity in insulin-secreting cells. These effects of resveratrol are dependent on the presence of active SIRT1, which mediates up-regulation of key genes for β-cell function. SIRT1 is not the only member of the sirtuin family to be involved in the regulation of insulin secretion. For instance, SIRT4, a mitochondrial ADP-ribosyl transferase, decreases glutamate dehydrogenase activity, thereby reducing the secretory response of β-cells (15, 65). SIRT3, another mitochondrial sirtuin, deacetylates the enzymes glutamate and isocitrate dehydrogenases (66) and maintains ATP levels through regulation of complex I in the electron transport chain (67). Thus, sirtuin modulators might represent novel therapeutic tools in the control of β-cell function. However, such promising drugs might fulfill drastic criteria such as sirtuin and tissue specificity.
Supplementary Material
Acknowledgments
We are grateful to Deborah Strebel, Clarissa Bartley, and Gaelle Chaffard for technical assistance; Françoise Assimacopoulos-Jeannet for helpful scientific advice; and to Joseph Rodgers and Pere Puigserver for SIRT1-related adenoviruses and accompanying precious instructions. Fresh human islets were obtained thanks to the European Consortium for Islet Transplantation “Islet for Research” distribution program sponsored by the Juvenile Diabetes Research Foundation. This study was part of the Geneva Programme for Metabolic Disorders (GeMet).
This work was supported by the Swiss National Science Foundation (to P. M.) and the State of Geneva.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- Ad
- adenovirus
- KRBH
- Krebs-Ringer bicarbonate-HEPES
- AMPK
- adenosine monophosphate kinase.
REFERENCES
- 1. Borra M. T., Smith B. C., Denu J. M. (2005) J. Biol. Chem. 280, 17187–17195 [DOI] [PubMed] [Google Scholar]
- 2. Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. (2003) Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 3. Woods T. M., Eichner S. F., Franks A. S. (2004) Ann. Pharmacother. 38, 887–891 [DOI] [PubMed] [Google Scholar]
- 4. Pugh T. D., Klopp R. G., Weindruch R. (1999) Neurobiol. Aging 20, 157–165 [DOI] [PubMed] [Google Scholar]
- 5. Dhahbi J. M., Mote P. L., Wingo J., Tillman J. B., Walford R. L., Spindler S. R. (1999) Am. J. Physiol. 277, E352–360 [DOI] [PubMed] [Google Scholar]
- 6. Civitarese A. E., Carling S., Heilbronn L. K., Hulver M. H., Ukropcova B., Deutsch W. A., Smith S. R., Ravussin E. (2007) PLoS Med. 4, e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guarente L., Picard F. (2005) Cell 120, 473–482 [DOI] [PubMed] [Google Scholar]
- 8. Guarente L. (2000) Genes Dev. 14, 1021–1026 [PubMed] [Google Scholar]
- 9. Kaeberlein M., McVey M., Guarente L. (1999) Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tissenbaum H. A., Guarente L. (2001) Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 11. Rogina B., Helfand S. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frye R. A. (1999) Biochem. Biophys. Res. Commun. 260, 273–279 [DOI] [PubMed] [Google Scholar]
- 13. Frye R. A. (2000) Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 14. Michan S., Sinclair D. (2007) Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 16. Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 17. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 18. Fulco M., Schiltz R. L., Iezzi S., King M. T., Zhao P., Kashiwaya Y., Hoffman E., Veech R. L., Sartorelli V. (2003) Mol. Cell 12, 51–62 [DOI] [PubMed] [Google Scholar]
- 19. Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 20. Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iynedjian P. B. (2009) Cell Mol. Life Sci. 66, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maechler P., Carobbio S., Rubi B. (2006) Int. J. Biochem. Cell Biol. 38, 696–709 [DOI] [PubMed] [Google Scholar]
- 23. Ashcroft F. M. (2005) J. Clin. Invest. 115, 2047–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S. N., Berggren P. O. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E16–28 [DOI] [PubMed] [Google Scholar]
- 25. Bordone L., Motta M. C., Picard F., Robinson A., Jhala U. S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., Easlon E. J., Lin S. J., Guarente L. (2006) PLoS Biol. 4, e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moynihan K. A., Grimm A. A., Plueger M. M., Bernal-Mizrachi E., Ford E., Cras-Méneur C., Permutt M. A., Imai S. (2005) Cell Metab. 2, 105–117 [DOI] [PubMed] [Google Scholar]
- 27. Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen W. P., Chi T. C., Chuang L. M., Su M. J. (2007) Eur. J. Pharmacol. 568, 269–277 [DOI] [PubMed] [Google Scholar]
- 29. Szkudelski T. (2006) Eur. J. Pharmacol. 552, 176–181 [DOI] [PubMed] [Google Scholar]
- 30. Merglen A., Theander S., Rubi B., Chaffard G., Wollheim C. B., Maechler P. (2004) Endocrinology 145, 667–678 [DOI] [PubMed] [Google Scholar]
- 31. Solomon J. M., Pasupuleti R., Xu L., McDonagh T., Curtis R., DiStefano P. S., Huber L. J. (2006) Mol. Cell. Biol. 26, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodgers J. T., Puigserver P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Andrade P. B., Rubi B., Frigerio F., van den Ouweland J. M., Maassen J. A., Maechler P. (2006) Diabetologia 49, 1816–1826 [DOI] [PubMed] [Google Scholar]
- 34. Casimir M., Rubi B., Frigerio F., Chaffard G., Maechler P. (2009) Biochem. J. 424, 459–466 [DOI] [PubMed] [Google Scholar]
- 35. Maechler P., Wang H., Wollheim C. B. (1998) FEBS Lett. 422, 328–332 [DOI] [PubMed] [Google Scholar]
- 36. Broca C., Brennan L., Petit P., Newsholme P., Maechler P. (2003) FEBS Letters 545, 167–172 [DOI] [PubMed] [Google Scholar]
- 37. Li N., Brun T., Cnop M., Cunha D. A., Eizirik D. L., Maechler P. (2009) J. Biol. Chem. 284, 23602–23612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Owen O. E., Kalhan S. C., Hanson R. W. (2002) J. Biol. Chem. 277, 30409–30412 [DOI] [PubMed] [Google Scholar]
- 39. Maechler P., Wollheim C. B. (1999) Nature 402, 685–689 [DOI] [PubMed] [Google Scholar]
- 40. Pecqueur C., Alves-Guerra M. C., Gelly C., Levi-Meyrueis C., Couplan E., Collins S., Ricquier D., Bouillaud F., Miroux B. (2001) J. Biol. Chem. 276, 8705–8712 [DOI] [PubMed] [Google Scholar]
- 41. Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan A. Y., Dolinsky V. W., Soltys C. L., Viollet B., Baksh S., Light P. E., Dyck J. R. (2008) J. Biol. Chem. 283, 24194–24201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dasgupta B., Milbrandt J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7217–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bedalov A., Gatbonton T., Irvine W. P., Gottschling D. E., Simon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15113–15118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oliver-Krasinski J. M., Stoffers D. A. (2008) Genes Dev. 22, 1998–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waeber G., Thompson N., Nicod P., Bonny C. (1996) Mol. Endocrinol. 10, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 47. Brissova M., Shiota M., Nicholson W. E., Gannon M., Knobel S. M., Piston D. W., Wright C. V., Powers A. C. (2002) J. Biol. Chem. 277 [DOI] [PubMed] [Google Scholar]
- 48. Wang H., Maechler P., Ritz-Laser B., Hagenfeldt K. A., Ishihara H., Philippe J., Wollheim C. B. (2001) J. Biol. Chem. 276, 25279–25286 [DOI] [PubMed] [Google Scholar]
- 49. Watada H., Kajimoto Y., Miyagawa J., Hanafusa T., Hamaguchi K., Matsuoka T., Yamamoto K., Matsuzawa Y., Kawamori R., Yamasaki Y. (1996) Diabetes 45, 1826–1831 [DOI] [PubMed] [Google Scholar]
- 50. Boj S. F., Parrizas M., Maestro M. A., Ferrer J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14481–14486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shih D. Q., Screenan S., Munoz K. N., Philipson L., Pontoglio M., Yaniv M., Polonsky K. S., Stoffel M. (2001) Diabetes 50, 2472–2480 [DOI] [PubMed] [Google Scholar]
- 52. Wang H., Antinozzi P. A., Hagenfeldt K. A., Maechler P., Wollheim C. B. (2000) EMBO J. 19, 4257–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 54. Gauthier B. R., Wiederkehr A., Baquié M., Dai C., Powers A. C., Kerr-Conte J., Pattou F., MacDonald R. J., Ferrer J., Wollheim C. B. (2009) Cell Metab. 10, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Su H. C., Hung L. M., Chen J. K. (2006) Am. J. Physiol. Endocrinol. Metab. 290, E1339–1346 [DOI] [PubMed] [Google Scholar]
- 56. Jakab M., Lach S., Bacová Z., Langelüddecke C., Strbák V., Schmidt S., Iglseder E., Paulmichl M., Geibel J., Ritter M. (2008) Cell Physiol Biochem. 22, 567–578 [DOI] [PubMed] [Google Scholar]
- 57. Chan C. B., De Leo D., Joseph J. W., McQuaid T. S., Ha X. F., Xu F., Tsushima R. G., Pennefather P. S., Salapatek A. M., Wheeler M. B. (2001) Diabetes 50, 1302–1310 [DOI] [PubMed] [Google Scholar]
- 58. Hong Y., Fink B. D., Dillon J. S., Sivitz W. I. (2001) Endocrinology 142, 249–256 [DOI] [PubMed] [Google Scholar]
- 59. Produit-Zengaffinen N., Davis-Lameloise N., Perreten H., Bécard D., Gjinovci A., Keller P. A., Wollheim C. B., Herrera P., Muzzin P., Assimacopoulos-Jeannet F. (2007) Diabetologia 50, 84–93 [DOI] [PubMed] [Google Scholar]
- 60. Brand M. D., Esteves T. C. (2005) Cell Metab. 2, 85–93 [DOI] [PubMed] [Google Scholar]
- 61. Nicholls D. G. (2006) Biochim. Biophys. Acta 1757, 459–466 [DOI] [PubMed] [Google Scholar]
- 62. Suchankova G., Nelson L. E., Gerhart-Hines Z., Kelly M., Gauthier M. S., Saha A. K., Ido Y., Puigserver P., Ruderman N. B. (2009) Biochem. Biophys. Res. Commun. 378, 836–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leclerc I., Woltersdorf W. W., da Silva Xavier G., Rowe R. L., Cross S. E., Korbutt G. S., Rajotte R. V., Smith R., Rutter G. A. (2004) Am. J. Physiol. Endocrinol. Metab. 286, E1023–1031 [DOI] [PubMed] [Google Scholar]
- 64. Salt I. P., Johnson G., Ashcroft S. J., Hardie D. G. (1998) Biochem. J. 335, 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ahuja N., Schwer B., Carobbio S., Waltregny D., North B. J., Castronovo V., Maechler P., Verdin E. (2007) J. Biol. Chem. 282, 33583–33592 [DOI] [PubMed] [Google Scholar]
- 66. Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 67. Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.