Abstract
Madin Darby canine kidney (MDCK) cells are a well characterized epithelial cell line used to study mechanisms of polarized delivery. As glycans on apically expressed proteins have been identified as targeting signals, and crosslinking by the abundant galectin-3 has been implicated in the mechanism of glycan-dependent sorting, we wanted to identify other members of the galectin (Gal) family expressed in MDCK cells. By analyzing intron-exon boundaries, we identified canine genes that were highly homologous to mammalian Gal-1, 2, 3, 4, 7, 8, 9, and 12, and galectin-related HSPC159 and GRIFIN. Transcripts for Gal-2 and -12 were not detected in MDCK cells, but we found transcript levels for Gal-3 > Gal-9 > Gal-8 > Gal-1 ⋙ Gal-4 > Gal-7. Canine Gal-1, -2, -3, -4, -7, -8, -9, and -12 were cloned and expressed in Escherichia coli as GST fusion proteins to characterize binding specificities on arrays of synthetic glycans on glass slides from Core H of the NIH Consortium for Functional Glycomics. Individual expression of the N-terminal (GST-Gal-9N) and C-terminal (GST-Gal-9C) carbohydrate recognition domains greatly improved protein yield and the ability to characterize Gal-9 binding on the array. Canine galectins differentially bound sulfated disaccharides as well as human blood groups A, B, and H on both N-glycans and linear glycan structures on the array. Analysis of GST-Gal-1, -3, -4, -7, -8, -9N, and -9C binding to immunopurified human MUC1 expressed in MDCK cells revealed a preference for binding GST-Gal-3 and -9, which interestingly reflects the two most abundant galectins expressed in MDCK cells.
Keywords: Carbohydrate-binding Protein, Carbohydrate Function, Glycoprotein, Oligosaccharide Receptors, Trafficking, MDCK Cells, MUC1, Galectin, Glycan Array, Mucin
Introduction
The functions of glycans on glycoproteins are quite varied, with immature N-linked glycans playing a key role in protein folding and quality control within the endoplasmic reticulum, and mature terminally processed N- and O-linked glycans playing key roles in glycoprotein interactions as diverse as subcellular targeting, signal transduction, cell-matrix interactions, cell-cell interactions, and cell-microbe interactions (1). We are specifically interested in the role of glycans in apical targeting in polarized epithelial cells as N-glycans on secreted model proteins, and both N-glycans and O-glycans on transmembrane model proteins, have been identified as apical targeting signals (for review, see Refs. 2, 3).
Recent studies on the mechanism of apical targeting in the model system of polarized Madin Darby canine kidney (MDCK)3 epithelial cells has revealed that proteins with unique types of apical sorting signals are differentially sorted into subpopulations of vesicles budding from the trans-Golgi network (TGN) (4–6). There is also evidence that some apical proteins traffic through unique endocytic compartments within the biosynthetic pathway such that proteins like influenza hemagglutinin and GPI-anchored proteins that exhibit a lipid raft-dependent targeting signal, transit the apical early endosome (AEE), while proteins like endolyn and p75 neurotrophin receptor that exhibit an N-glycan- and O-glycan-dependent targeting signal, respectively, transit the apical recycling endosomes (ARE) (7, 8). MUC1 is a transmembrane glycoprotein with a mucin-like ectodomain that is localized on the apical surface of epithelial cells; it provides protection from pathogens and likely plays a key role in epithelial development, repair and tumorigenesis (for review, see Ref. 9). Although the targeting signal for MUC1 apical delivery in polarized epithelial cells has not been defined, studies with MUC1 chimeras in MDCK cells did reveal that the signal is within the mucin-like ectodomain of MUC1 (10). We also observed that apical delivery of MUC1 in polarized MDCK cells was blocked by ablation of the ARE, but not the AEE, consistent with a glycan-dependent apical targeting signal for MUC1 (11). Interestingly, Delacour et al. (12, 13) reported that vesicles budding from the TGN carrying lipid-raft-independent cargo contain galectin-3, and that galectin-3 crosslinking of the glycoprotein cargo is required for apical sorting. As galectin-3 (Gal-3) constitutes only one member of the family of galectins and MUC1 interactions with Gal-3, and correlative expression with Gal-1 and -4 have been reported (14–17), we were interested in the identification, cloning, and characterization of all canine galectins expressed in MDCK cells.
The family of galectins is characterized by (i) their affinity for binding glycoconjugates containing β-galactose, (ii) their homologous sequence and structural elements, and (iii) their conserved exon/intron junctions (18–21). Galectins were previously named S-type lectins as many in this group are stabilized by the presence of thiols, and oxidation of the invariant Cys correlates with inactivation. There are more than a dozen galectins described in the literature and they are more or less numbered based on the order of their discovery although they fall into three subgroups. Prototypical galectins like Gal-1, with a single carbohydrate recognition domain (CRD) of about 130 residues, usually form dimers. Other members of this subgroup include Gal-2, -7, -10, -13, -14, -15, and galectin-related proteins PP13, PPL13, HSPC157, and GRIFIN (the latter four lack essential glycan-interacting residues and thereby the ability to bind glycoconjugates). Gal-3 comprises the only member of the chimeric subgroup with one CRD and a unique N-terminal domain involved in formation of multimers. Members of the third subgroup termed tandem-repeat, have two non-identical CRD joined by a linker domain, and include Gal-4, -6, -8, -9, and -12. Interestingly, a phylogenetic analysis of gene locations, exon-intron structures and sequences of chordate galectins revealed that Gal-1 is not the prototypical galectin (22). Instead, duplication of a mono-CRD galectin gene early in chordate evolution gave rise to a bi-CRD galectin gene, and independent evolution of the N-terminal CRD and C-terminal CRD gave rise to two distinct mono-CRD types including Gal-7, -10, -14, PP13, and PPL13 in the F4-group, and Gal-1, -2, -3, -5, HSPC159, and GRIFIN in the F3-group, respectively (22). Although canine Gal-3 and Gal-8 were included in this latter analysis, there have been no additional studies of canine galectin genes despite the availability of the dog genome and the widespread interest in the well-characterized model of MDCK cells. Friedrichs et al. (23) did estimate the levels of transcripts for some of the galectins in MDCK cells using real-time polymerase chain reaction (PCR). We now report the identification of eight canine galectin genes (and two galectin-like genes), of which six galectins are expressed in MDCK cells. We have cloned the canine galectin cDNAs and expressed them as GST-tagged proteins in order to assess their binding specificities on synthetic glycan arrays and their differential interactions with MUC1 synthesized in MDCK cells.
MATERIALS AND METHODS
Amplification of Canine Galectin RNA by RT-PCR
RNA from dog jejunum and heart (breed, Beagle) was purchased from Zyagen (San Diego, CA). MDCK type II cells (breed, Cocker Spaniel) were obtained from Gerard Apodaca (University of Pittsburgh), and RNA was isolated using Ambion RNAqueous 4PCR Kit (Austin, TX) as described by the manufacturer. First strand cDNA was synthesized from 1 μg RNA using RNA Superscript II reverse transcriptase and amplified using TaqDNA polymerase (Invitrogen, Carlsbad, CA). cDNAs were amplified using internal primers listed in supplemental Table S1 by heating at 95 °C for 30 s, annealing primers at 56 °C for 30 s and polymerization at 72 °C for 30 s. Amplified DNA (∼200 bp) for Gal-1, -3, -8, and -9 was visible on ethidium bromide-stained agarose gels after 28 cycles. Amplified cDNAs for Gal-2, -4, -7, and -12 were prepared with nested primers using the amplified product obtained with external primers after 28 cycles as substrate for a second amplification with the internal primers for 28 cycles.
Cloning of Canine Galectin cDNAs by RT-PCR
First strand cDNA was synthesized from heart, jejunum and MDCK RNA as already described and amplified using external primers listed in supplemental Table S1 by heating at 95 °C for 30 s, annealing primers at 56 °C for 30 s and polymerization at 72 °C for 60 s. Amplified cDNAs for Gal-1, -3, -4, -7, and -8 prepared from MDCK RNA, amplified cDNAs for Gal-2 and -9 from dog jejunum RNA, and Gal-12 from dog heart RNA were cloned into pCR2.1 TOPO vector from Invitrogen. Amplified cDNAs for Gal -7, -9, and -12 were prepared with nested primers using the amplified product obtained with UTR primers after 28 cycles as substrate for a second amplification with the external primers for 28 cycles. Multiple plasmid preps for each cloned cDNA were subjected to nucleotide sequencing using M13rev and T7 primers (GenBankTM accession numbers HQ637384-HQ637391 for eight galectins).
Expression of GST-galectins in Bacteria
Gal-1, -2, -3, -4, -7, -8, -9, and -12 cDNAs were subcloned into pGEX-6P-1 for expression in bacteria as N-terminal GST fusion proteins as previously described (24). As GST-Gal-9 repeatedly aggregated in the bacteria, we subcloned the N-terminal and C-terminal CRDs separately as GST-Gal-9N (residues 1–148) and GST-Gal-9C (residues 225–355), respectively, for further study. GST-Gal-12 also aggregated and was not further studied. Briefly, 2 liters of liquid culture were inoculated with E. coli strain BL21 transformed with pGEX-6P-1(GST-galectin). Isopropyl β-d-1-thiogalactopyranoside was added at mid-phase (OD ∼0.8), and bacteria were grown with shaking overnight at room temperature. The supernatant from the lysed bacteria was incubated at 4 °C overnight with 0.75 ml of a 50% slurry of glutathione-conjugated Sepharose (GE Healthcare, Piscataway, NJ) in a 15-ml conical tube with end-over-end mixing. GST-galectins were released from the washed beads with 1 ml of buffer (0.05 m Tris-HCl, pH 8, 150 mm NaCl) containing 14 mm β-mercaptoethanol and 15 mm glutathione. After centrifugation, the supernatant was incubated with 1 ml of a 50% slurry of lactose-conjugated Sepharose (Sigma) in a 15-ml conical tube at 4 °C for 30 min with end-over-end mixing. The slurry was poured into a 1-cm diameter column containing a sintered glass filter and the packed column bed was overlaid with 1 ml of a 50% slurry of Sepharose 6B (Sigma). The column bed was washed with 5 ml of buffer containing 14 mm β-mercaptoethanol (βME). 2 ml of buffer containing βME and 0.1 m α-lactose (Sigma) were added to the column, 0.5 ml collected, and the column capped for 30 min at 4 °C. Fractions of 0.5 ml were collected, and the absorbance at 280 nm was determined with a spectrophotometer. The peak fractions were pooled and lactose was removed on a PD-10 column as described by the manufacturer (GE Healthcare). Final protein concentrations were calculated using individual extinction coefficients determined from the amino acid content of each GST-Gal.
Glycan Array Screening of Canine GST-galectins
Freshly prepared GST-galectins in buffer containing βME were shipped overnight on ice to Core H of the Consortium for Functional Glycomics for analysis on glycan microarrays. GST-galectins were diluted to 0.1–100 μg/ml in standard binding buffer (Tris-buffered saline with MgCl2 and CaCl2, 1% BSA and 0.05% Tween 20). Seventy microliters were applied to the printed surface of the array before applying the coverslip and incubation at room temperature in a humidified chamber away from light for 1 h. The coverslip was removed and the slide was rinsed four times in standard TSM washing buffer containing 0.05% Tween and four times in TSM buffer without Tween. Seventy microliters of Alexa 488 conjugated anti-GST antibody (Invitrogen) was diluted in TSM buffer and applied for one hour in a humidified chamber away from light as described. Washes were performed as above, followed by four washes in distilled water before fluorescence of the spots was measured.
MUC1 Pull-down with Recombinant GST-galectins
Polarized MDCK type II cells growing on Costar permeable supports (Corning, NY) were cultured as previously described (7, 11, 25). Where indicated, MUC1 expression was achieved by infection with recombinant adenovirus (7, 11). A clonal MDCK cell line expressing MUC1 was previously described (7). Polarized MDCK cells expressing MUC1 were pulse labeled for 30 min with [35S]Met/Cys and chased for 90 min, and immunoprecipitates recovered with mouse monoclonal antibody B27.29 and protein G-conjugated Sepharose as previously described (7, 11). Proteins were eluted from protein G-conjugated Sepharose by incubation at 90 °C for 2 min in buffer A (10 mm HEPES, pH 7.4, 150 mm NaCl) containing 1% SDS. The eluted material was diluted 10-fold with buffer A containing 1% Triton X-100 (and adjusted to 14 mm βME). Identical samples from six 12 mm filters of polarized MDCK cells were combined and equal aliquots were incubated with either GST-Gal-1, -3, -4, -7, -8, -9N, or -9C prebound to glutathione-conjugated Sepharose (or no GST-Gal as a control), overnight on a rotating wheel at 4 °C. A fresh aliquot of each GST-galectin was prepared for every experiment by affinity purification (starting material ∼1 μg) on lactose-conjugated Sepharose (25 μl of a 50% slurry) and removal of lactose after subsequent binding to glutathione-conjugated Sepharose (25 μl of a 50% slurry) as described above. The following day, the beads were washed with buffer A and proteins eluted by successive incubations for 15 min each on a rotating wheel at 4 °C with buffer A containing (i) 0.1 m sucrose (as control), (ii) 0.1 m lactose (for specific binding) and (iii) 15 mm glutathione (to recover GST-galectins). The eluents (10 μl) were recovered with a Hamilton syringe after centrifugation (10,000 × g for 30 s) and mixed with 10 μl of Bio-Rad sample buffer for SDS-PAGE on a precast 4–15% gradient gel (Bio-Rad) and transfer to nitrocellulose (Millipore). Radiolabeled proteins were detected with a Bio-Rad Imager using a TR phosphorimager screen before cutting the nitrocellulose at the 100 kDa marker for immunoblotting of the large or small subunit with mouse monoclonal antibody B27.29 or Armenia hamster monoclonal antibody CT-2, respectively. Bands were detected using Perkin Elmer Western LightningTM Plus-ECL with a Bio-Rad Versadoc or Kodak BioMax MR film, and GST-galectins were detected by Coomassie staining (Bio-Rad Bio-safe Coomassie). The levels of radiolabeled proteins, immunoblotted subunits or Coomassie-stained GST-galectins were determined with Bio-Rad Quantity One software. The level of galectin binding of radiolabeled or immunoblotted subunits was normalized to the level of galectin recovered for the same sample based on Coomassie staining (divided by the molecular weight of the galectin as reported in Table 1). In each experiment, maximal binding to a single galectin was set as 1 and binding to other galectins was calculated as a fraction of maximal binding. Data were combined by averaging the values for four experiments (presented as mean and S.E.).
TABLE 1.
Properties of canine galectins
See supplemental figures and tables for details.
| Canine galectin | Chrom number | Number of coding exons | FW | FW GST-Gal | Class | Glycan-binding preference on synthetic array (see supplemental data) |
|---|---|---|---|---|---|---|
| Galectin-1 | 10 | 4 | 14,732 | 41,555 | Prototypical | Branched N-glycans with terminal Galβ4GlcNAc and 3-O-sulfated Galβ4GlcNAc-R |
| Galectin-2 | 10 | 4 | 14,476 | 41,251 | Prototypical | Lewisx (with Galα3 extension) on branched N-glycans |
| Galectin-3 | 8 | 5 | 29,506 | 56,329 | Chimeric | Blood group A or B on branched N-glycans or type 2 polylactosamine regardless of terminal sugar such as Neu5Acα2,6 |
| Galectin-4 | 1 | 9 | 35,170 | 61,993 | Tandem repeat | Blood group A on type 2 polylactosamine or biantennary N-glycans (less for blood group B) |
| Galectin-7 | 1 | 4 | 15,263 | 42,086 | Prototypical | H-antigen on type 2 polylactosamine; terminal Neu5Acα2,6 tolerated |
| Galectin-8 | 4 | 9 | 66,461 | 62,803 | Tandem repeat | Blood group A on biantennary N-glycans and type 2 polylactosamine chains but not with Neu5Acα2,6 |
| Galectin-9 | 9 | 10 | 39,639 | 66,461 | Tandem repeat | Not done (see Gal-9N and Gal-9C, below) |
| Galectin-9N (residues 1–148) | 43,333 | N-terminal CRD | Blood group A on biantennary N-glycans and 3-O-sulfated disaccharides | |||
| Galectin-9C (residues 149–355) | 41,670 | C-terminal CRD | 3-O-sulfated disaccharides and blood group B on biantennary N-glycans | |||
| Galectin-12 | 18 | 8 | 34,224 | 61,047 | Tandem repeat | Not done |
RESULTS
Identification of Canine Galectin Genes
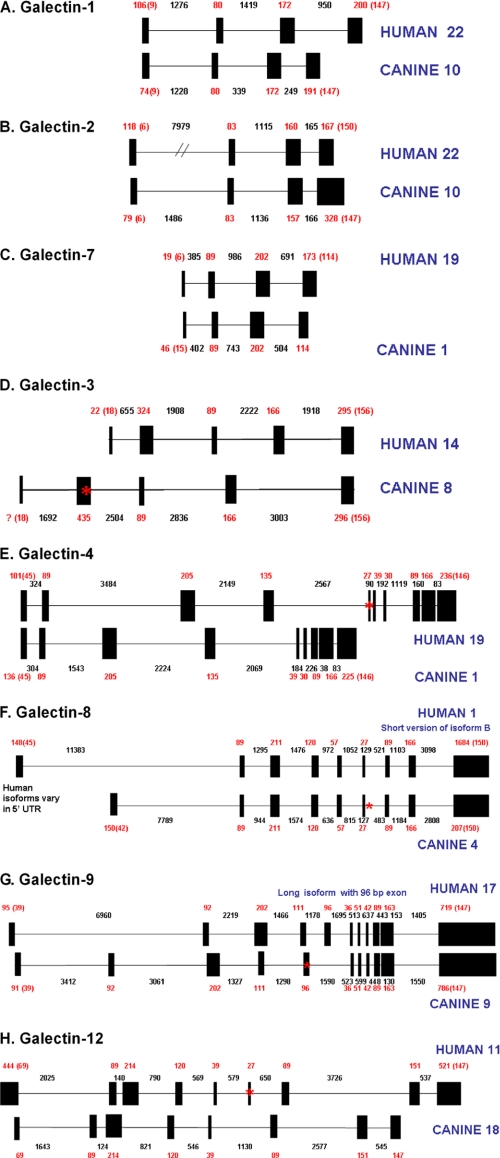
The genes for canine Gal-1, -2, -3, -4, -5, -8, -9, and -12 as well as galectin-related GRIFIN, were identified by a query of the dog genome for the term “galectin” (breed, Boxer). Identical references to canine genes were found with a query of the NCBI Nucleotide Database and searches using Nucleotide BLAST and TBLASTN. A pseudogene for Gal-1 containing a polyadenylation motif and no introns was also reported on the X chromosome and was not analyzed further. The predicted coding sequence of the Gal-4 gene included three CRDs but upon closer study, actually included the genes for both Gal-4 and Gal-7. A study of the gene attributed to Gal-5 revealed that it was actually homologous to mammalian galectin-related HSPC159. No canine genes homologous to rat Gal-5, mouse Gal-6, human Gal-10, human Gal-13, ovine Gal-14, ovine Gal-15, human PP13, or human PPL13 were identified. In general, genes for canine Gal-1, -2, -3, -4, -7, -8, -9, -12, HSPC159, and GRIFIN were highly homologous to the corresponding mammalian galectin genes with similar exon/intron structures and amino acid sequence (see Table 1, Fig. 1, and supplemental Figs. S1–S8). Canine GRIFIN and HSPC159 lack two and five essential residues, respectively, in the CRDs for sugar binding so they were not further investigated in this study. Primers for amplification of galectins from canine RNA by RT-PCR are described in supplemental Table S1. External primers overlapped the Met start codon and the stop codon in each case. UTR primers were designed within 5′ and 3′ UTR as needed. Internal primers were designed from exon sequences to produce a ∼200-bp product.
FIGURE 1.
Intron and exon structure of canine galectin genes. Exons and introns of the canine galectins are compared with the corresponding human genes with exons as black boxes and introns as black lines. The numbers represent base pairs for exons (red) and introns (black). Numbers in blue are the chromosome location. See text for further discussion of the canine genes. Alignments of amino acid sequences between canine and human galectins are presented in supplemental Figs. S1–S8.
Identification of Endogenous Galectins Expressed in MDCK Cells
RNA was prepared from MDCK cells and amplified by RT-PCR as described in Materials and Methods to yield ∼200-bp products. RNA from a commercial source of dog jejunum was used as a positive control. RNA for Gal-1, -3, -8, and -9 was most abundant in both MDCK and jejunum as the ∼200 bp products from RT-PCR were visible on agarose gels with ethidium bromide staining using only internal primers (Fig. 2A). Amplification of RNA from Gal-2, -4, -7, and -12 was apparent only with nested primers; the ∼200 bp product of amplification was produced only when the product from RT-PCR with external primers was subsequently used as template for PCR with internal primers. The data indicated that all the canine galectins could be amplified with the selected primers using jejunum RNA as a positive control, but only Gal-1, -3, -4, -7, -8, and -9 were amplified from MDCK cell RNA. The product of β-actin amplification by RT-PCR from jejunum and MDCK cell RNAs was similar.
FIGURE 2.
Cloning and expression of canine galectins. A, RNA was prepared from MDCK cells (M) and a commercial source of dog jejunum (J) and amplified by RT-PCR. Amplified DNA (∼200 bp) was visible by ethidium bromide staining for Gal-1, -3, -8, and -9 after 28 cycles, while amplified cDNAs for Gal-2, -4, -7, and -12 required nested primers. See supplemental Table S1 for primer sequences. B, cDNAs for Gal-1, -2, -3, -4, -7, -8, -9, and -12 were expressed in bacteria as GST fusion proteins and subjected to SDS-PAGE and Coomassie staining. The N-terminal and C-terminal carbohydrate recognition domains of Gal-9 were individually expressed as GST-Gal-9N and GST-Gal-9C, respectively.
Cloning of Canine Galectins Expressed in MDCK Cells
Amplified cDNAs for Gal-1, -3, -4, -7, and -8 prepared from MDCK cell RNA, amplified cDNAs for galectins-2 and -9 from dog jejunum RNA, and amplified cDNA for Gal-12 from dog heart RNA were cloned into pCR2.1 TOPO vector from Invitrogen and sequenced for comparison to the canine genes we identified above (see Table 1, Fig. 1, and supplemental Figs. S1–S8).
The cloned canine Gal-1 cDNA had the same nucleotide sequence as predicted by a query of the dog genome. The division of the coding sequence between the four exons of the canine gene was identical to the human Gal-1 gene and there was 82% identity in amino acid sequence.
The cloned canine Gal-2 cDNA had the same nucleotide sequence as predicted by a query of the dog genome (breed, Boxer) except that we found Val-124 is Leu-124 in our cloned cDNAs (breed, Beagle). Interestingly, the division of the coding sequence between exons 3 and 4 (four exons total) was different than reported for the human Gal-2 gene but the amino acids are identical (77% identity between human and canine Gal-2).
The cloned canine Gal-3 cDNA had the same nucleotide sequence as predicted by a query of the dog genome including six 9-mer repeats in coding exon 2. There are a variable number of 9-mer repeats (Q-A/G-P-P-G-G/A/PT-Y-P-G) reported for cow (four), mouse (three), pig (three), human (two), rat (two), and rabbit (one) Gal-3 (see asterisk in Fig. 1D). Otherwise, the division of the coding sequence between the five exons was identical to the human Gal-3 gene (with 73% identity of amino acids). The exon containing the 5′-UTR has not been identified for the canine Gal-3.
The cloned canine Gal-4 cDNA sequence was very different from that predicted by the canine genome data base where it is fused with a sense and antisense copy of canine Gal-7. The division of the coding sequence between the nine exons was identical to the human Gal-4 gene except that the canine Gal-4 is missing one exon of 27 bp (9 amino acids) between exons 4 and 5 (with 77% identity of amino acids) (see asterisk in Fig. 1E).
The cloned canine Gal-7 cDNA was very different from that predicted by the canine genome data base where its sequence is fused with canine Gal-4 and predicted to have an additional exon between exons 3 and 4. In our cloned sequence, the division of the coding sequence in exons 2, 3, and 4 was identical to the human Gal-7 gene with 76% identity of amino acids. There are a variable number of residues within the first coding exon of mouse (two), dog (five), cow (five), horse (six), and human (six).
The cloned canine Gal-8 cDNA had the same nucleotide sequence predicted by the canine genome database, except that it lacks the seventh coding exon (123 bp) within the linker region and thereby corresponds to the short isoform of Gal-8 (see asterisk in Fig. 1F). The division of the coding sequence between exons 2–9 of the canine gene was identical to the human Gal-8 gene, while the coding sequence in the first exon is one residue shorter. There is 86% identity between the human and canine Gal-8 short isoforms.
The cloned canine Gal-9 cDNA has the same nucleotide sequence as that predicted by the canine genome database, except that it includes exon 5 (96 bp) and thereby represents the long isoform (see asterisk in Fig. 1G). Our clone also had Thr-216 instead of Met-216, and Met-281 instead of Thr-281. The division of the coding sequence between the eleven exons of the canine gene was identical to the human Gal-9 gene and there was 74% identity of amino acids.
The cloned canine Gal-12 cDNA has the same nucleotide sequence as that predicted by the canine genome database, except that it lacks an exon (27 bp) between exons 5 and 6 and thereby represents the short isoform (see asterisk in Fig. 1H). Our clone also had Leu-300 instead of Val-300. The division of the coding sequence between the eight exons of the canine gene was identical to the human Gal-12 gene with 85% identity of amino acids. The exon containing the 5′-UTR has not been identified for the canine Gal-12.
Characterization of Glycan Binding for Canine Galectins
The cDNAs for canine Gal-1, -2, -3, -4, -7, -8, -9, and -12 were subcloned into the bacterial vector pGEX-6P-1 for expression in E. coli as N-terminal GST fusion proteins as previously described (24). The fusion proteins were affinity purified on glutathione-conjugated Sepharose and then lactose-conjugated Sepharose to obtain epitope-tagged galectins for characterization on the NIH Consortium Core H glycan array. GST-Gal-2, -9, and -12 consistently gave low yields, most likely due to aggregation in bacterial inclusion bodies (see Fig. 2B for a Coomassie-stained gel of all the GST-Gal). As Gal-9 is expressed in MDCK cells, which is the focus of this study, we attempted to improve the yield by sub-cloning the two carbohydrate recognition domains separately. Expression of GST-Gal-9N (residues 1–148) and GST-Gal-9C (residues 225–355) in bacteria gave yields similar to those obtained for other affinity purified GST-galectins (0.2–0.5 mg total from a 2-liter culture) (Fig. 2B).
Binding of Gal-1, -2, -3, -4, -7, -8, -9N, and -9C to glycan arrays on glass slides was carried out at multiple concentrations and analyzed with a fluorescently tagged anti-GST antibody. Greater specificity was consistently revealed at the lower concentrations and the preferred binding was determined by comparing the ranked order at multiple concentrations as previously described (26). Briefly, each GST-Gal was analyzed at multiple concentrations. At each concentration the Relative Fluorescence Units (RFU) corresponding to the glycan with the highest binding signal was set at 100 and the relative binding of the other glycans was calculated to provide a rank order of binding. The average of the three rankings for each glycan at three concentrations of GST-Gal was determined and the bound glycans were ordered from highest to lowest ranking as shown in supplemental Tables S2–S9. A representative profile at a single concentration for each GST-Gal is presented in Fig. 3. Structures of bound glycans are indicated and the average rank (in red numbering with 1 as the best binding) is provided in each case for comparison to the representative profile. A summary of the results are addressed below. Array data are also available online.
FIGURE 3.
Preferential binding of canine GST-tagged galectins on synthetic arrays. GST-tagged canine galectins were incubated at multiple concentrations (between 0.1–5.0 μg/ml) with synthetic glycan arrays printed on glass slides. The binding was analyzed with a fluorescently tagged anti-GST antibody. The results were ranked at each concentration and the average ranking from all concentrations is shown on the representative profiles in red with 1 being the best binding (see supplemental Tables for all data). Numbers in black represent the position of the specific glycan on the array. One profile is shown for GST-Gal-1 (A), GST-Gal-2 (B), GST-Gal-3 (C), GST-Gal-4 (D), GST-Gal-7 (E), GST-Gal-8 (F), GST-Gal-9N (G), and GST-Gal-9C (H). Yellow circle, Gal; yellow square, GalNAc; blue circle, Glc; blue square, GlcNAc; green circle, Man; red triangle, Fuc; purple diamond, Neu5Ac.
GST-Gal-1 binding was analyzed at three concentrations (1, 10 and 200 μg/ml) and preferentially bound to N-glycans with terminal Galβ4GlcNAc as well as linear 3-O-sulfated Galβ4GlcNAc-R (Galβ4GlcNAc is LacNAc) (see Fig. 3A). Interestingly, the highest ranking glycans for binding GST-Gal-1on the array were tetra-antennary (1) and tri-antennary (3) N-glycans with terminal Galβ4GlcNAc and a biantennary N-glycan with terminal Galα1,3Galβ4GlcNAc (2). Linear structures of both Galα1,3Galβ4GlcNAc-R and Galβ4GlcNAc-R exhibited 100-fold lower binding (data not shown), but linear Galβ4GlcNAc1,6Galβ4GlcNAc-R exhibited significant binding (10). The biantennary N-glycan terminating in Galβ1,4GlcNAc was similarly bound with (7) and without (6) terminal Neu5Acα2,3 while binding to tri- and tetra-antennary structures with terminal Galβ4GlcNAc was lost when sialylated. Slightly better binding was observed for 6-sulfated linear glycans [3-OSO3]Galβ1,4[6-OSO3]GlcNAc-R (4 and 5), when compared with [3-OSO3]Galβ1,3GlcNAc-R (9) as shown in supplemental Table S2.
GST-Gal-2 was analyzed at two concentrations (5 and 50 μg/ml) and showed notable preference for biantennary N-glycans with terminal Galα1,3 extended Lewisx, with or without core GlcNAc fucosylation (see supplemental Table S3 and Fig. 3B). Binding to other glycans was generally 100-fold lower.
GST-Gal-3 binding was determined at three concentrations (0.1, 1, and 10 μg/ml) and preferentially bound N-linked glycans with type 2 chain core (Galβ1,4GlcNAc) and terminal blood group A or B (1 and 2), as well as polylactosamine (PL) with various modifications including terminal blood group A (3 and 7), extended blood group A (8), Neu5Acα2,6 (4 and 9) or fucose (10). In general, longer PL with three Galβ1,4GlcNAcβ1,3 are recognized better than PL with two Galβ1,4GlcNAcβ1,3 (compare 5 and 12, 4 and 9, 3 and 7). Details are provided in supplemental Table S4 and Fig. 3C.
GST-Gal-4 binding was determined at three concentrations (0.5, 5, and 50 μg/ml) and preferentially bound linear glycans with terminal blood group A (3, 4, 6) or Galβ1,3 extended structures (1) as shown in supplemental Table S5 and Fig. 3D. Blood group A on a biantennary N-glycan (5) had reduced binding when compared with blood group A on lactose (4) or PL (3). Blood group B on lactose (2) or biantennary N-glycan (8) was also bound.
GST-Gal-7 binding was determined at three concentrations (0.1, 1, and 10 μg/ml) as shown in supplemental Table S6 and Fig. 3E and preferentially recognizes blood group H, but only on type 2 PL (1 and 2). Termination of type 2 PL chains with Neu5Acα2,6 (6 and 7) or blood group A (5 and 8), instead of fucose, decreased binding somewhat, whereas termination with Neu5Acα2,3 or blood group B was not tolerated.
GST-Gal-8 binding was determined at three concentrations (0.1, 1 and 10 μg/ml) as shown in supplemental Table S7 and Fig. 3F and preferentially bound poly-NAc-lactosamine (PL) with various modifications, as well as biantennary N-glycans with blood group A (2) (weaker for blood group B, 7). Binding to PL terminating with Gal (6) was enhanced by addition of terminal Neu5Acα2,3Galβ1,4 (1) but not GlcNAc alone (5), and was reduced substantially by addition of terminal Neu5Acα2,6Galβ1,4. Binding of PL terminating in blood group A was enhanced by longer PL chains (compare 3 and 9).
GST-Gal-9N binding was determined at three concentrations (0.1, 1, and 5 μg/ml) as shown in supplemental Table S8 and Fig. 3G and showed preferential binding to blood group A as well as 3-O-sulfated disaccharides. Bivalent blood group A found on biantennary N-glycans (1) was bound better than monovalent blood group A found on either lactose (8) or PL (4 and 7). Binding to blood group B on biantennary N-glycans (12) was 6-fold lower than the corresponding N-glycan with blood group A (1).
GST-Gal-9C binding was determined at three concentrations (0.1, 1, and 5 μg/ml) as shown in supplemental Table S9 and Fig. 3H and showed preferential binding to primarily sulfated disaccharides (eight of top ten). Binding to blood group B on a biantennary N-glycan (6) and PL with terminal Neu5Acα2,3 (8) was also observed.
As canine HSPC159 was initially listed as Gal-5 in the dog genome data base, we also cloned the corresponding cDNA and expressed it as “GST-Gal-5.” This GST-Gal-5 did bind and elute from the glutathione column but it did not subsequently bind to lactose conjugated to Sepharose during the purification step.4 This lack of binding was consistent with the lack of five essential residues within its carbohydrate recognition domain.
Differential Binding of MUC1 Subunits to GST-galectins
To evaluate the potential interaction of canine galectins with MUC1 in MDCK cells, we assessed the ability of the recombinant canine galectins to precipitate immunopurified MUC1 expressed in MDCK cells. Polarized cultures of MDCK cells expressing MUC1 were metabolically labeled and MUC1 was immunoprecipitated from cell extracts as described under “Materials and Methods.” The immunoprecipitates were recovered in buffer containing 1% SDS to dissociate the MUC1 large and small subunits and denature any bound endogenous galectins. MUC1 is synthesized as a single peptide but exhibits autocatalytic cleavage within the ectodomain yielding a small subunit containing the transmembrane domain and one N-linked glycan while the large subunit contains four N-linked glycans and the mucin-like tandem repeat domain with numerous sites for O-linked glycans. The samples were diluted into buffer containing 1% Triton X-100 and β-mercaptoethanol prior to mixing with the GST-galectins to stabilize recombinant galectin structure and activity. Fresh aliquots of each GST-galectin were affinity purified on lactose-conjugated beads immediately before binding to glutathione-conjugated beads and incubation with immunopurified MUC1. After overnight incubation at 4 °C, the washed beads were incubated sequentially with buffer containing 0.1 m sucrose (control), 0.1 m lactose (specific-binding), and glutathione (GST-Gal recovery). MUC1 released by sucrose and lactose washes were analyzed by SDS-PAGE and transferred to nitrocellulose before analyzing the radiolabeled bands with a phosphorimager (Fig. 4A). The nitrocellulose was subsequently cut in half and immunoblotted for either the large (Mr ∼250,000) or small (Mr ∼25,000 or a dimer at Mr ∼50,000) subunit of MUC1 (Fig. 4B). Eluted GST-galectins were subjected to SDS-PAGE, the gel stained with Coomassie, and the bands quantified after scanning. One representative experiment is shown in Fig. 4 and the combined results from four independent experiments are shown in Fig. 5.
FIGURE 4.
Screen of MDCK-expressed MUC1 binding to recombinant canine galectins. MDCK cells stably expressing MUC1 were pulse labeled for 30 min with [35S]Met/Cys and chased for 90 min prior to immunoprecipitation of MUC1 from cell extracts. Resuspended immunoprecipitates were incubated with individual GST-galectins and glutathione-conjugated beads and eluted with sucrose (S, control), then lactose (L, specific binding) and then glutathione (for Coomassie stain of GST-Gal) and subjected to SDS-PAGE and transferred to nitrocellulose. A representative profile of [35S]MUC1 (A), immunoblotted large or small subunits of MUC1 (B), and glutathione-eluted GST-Gal stained with Coomassie (C) are shown. Analysis of multiple experiments is shown in Fig. 5.
FIGURE 5.
Differential preference of galectin binding to the large and small subunits of MUC1. Polarized MDCK cells stably expressing MUC1 or MDCK cells infected with an adenovirus encoding MUC1 were metabolically labeled and immunopurified MUC1 was recovered and analyzed for binding to GST-galectins as described in legend to Fig. 4. Maximal binding to a single GST-galectin was set at 1 in each experiment and binding to other galectins was calculated as a fraction. Data are presented as mean and S.E. from four independent experiments.
In general, we observed >10-fold greater recovery of MUC1 from GST-galectins bound to glutathione-conjugated beads when the beads were incubated with lactose (L) than with sucrose (S), consistent with specific binding of MUC1 to all of the canine galectins (Fig. 4, A and B, compare lanes S with L in each case). We also observed similar levels of each GST-galectin released from the beads with glutathione when analyzed by Coomassie staining (Fig. 4C). The combined results from four independent experiments reported in Fig. 5A indicated that the large subunit preferentially bound both GST-Gal-3 and GST-Gal-9N. No significant difference was observed for the results obtained by analysis of either radiolabeled or immunoblotted large subunit. Binding of the large subunit to GST-Gal-4, -7, and -9C was 2–3-fold less, while binding to GST-Gal-1 and GST-Gal-8 was 5- and 10-fold less, respectively. We consistently observed that the small subunit preferentially bound to GST-Gal-3 in every experiment. The combined results in Fig. 5B indicate that binding of the small subunit to GST-Gal-7, -9N, and -9C was 3–4-fold lower while binding to GST-Gal-1, -4, and 8 was 5–10-fold lower. We did observe a trend toward greater binding of the small subunit to GST-galectins other than GST-Gal-3 when analyzed by metabolic labeling than by immunoblotting.
DISCUSSION
Our interest in the role that galectins might play in glycan-dependent apical targeting in polarized MDCK cells led us to search for the canine galectin genes. We identified canine genes corresponding to Gal-1, -2, -3, -4, -7, -8, -9, and -12 as well as the galectin-related genes (lacking functional CRD) HSPC159 and GRIFIN. Canine genes homologous to the species-specific rat Gal-5, mouse Gal-6, human Gal-10, human Gal-13, ovine Gal-14, ovine Gal-15, human PP13, and human PPL13 were not found.
Using RT-PCR we estimate that the levels of transcripts expressed in MDCK cells are Gal-3 > Gal-9 > Gal-8 > Gal-1 ⋙ Gal 4 > Gal-7, as evidence of Gal-4 and Gal-7 transcripts was obtained only with nested primers. Using real-time PCR and primers based on the canine exon sequences, Friedrichs et al. found that Gal-3 transcripts were 100-times more abundant than transcripts for Gal-1, -8, and -9, and 100,000 times more abundant than transcripts for Gal-12 which is consistent with our findings (23). However, the same group mistakenly identified transcripts for Gal-6, but not Gal-4, using erroneous sequences for the PCR primers (exon sequences for canine Gal-4 were used for the sense primer to amplify Gal-4 and the antisense primer for Gal-6, while the sense primer for Gal-6 was homologous to both mouse and canine Gal-4 exon sequences and the antisense primer for Gal-4 has no match in any data base) (23). Some of the confusion likely comes from the fact that the mouse-specific Gal-6 gene diverged from the more common Gal-4 gene found in a large variety of species (22). Our findings are most consistent with the existence of a canine Gal-4 gene rather than a Gal-6 gene based on gene intron/exon structure and amino acid homology of the coding sequence (see Fig. 1E and supplemental Fig. S4).
We expressed Gal-1, -2, -3, -4, -7, -8, -9N, and -9C as N-terminal tagged GST-fusion proteins in bacteria and glycan preferences were assessed on a synthetic glycan array using a fluorescently tagged anti-GST antibody. Glycan preferences for our canine GST-Gal-1 and -3 were similar to those previously determined for biotinylated human Gal-1 and Gal-3 on the array using galectins at varying concentrations to assess specificity (27): (i) GST-Gal-1 specifically bound sulfated Galβ4GlcNAc, while GST-Gal-3 did not; (ii) GST-Gal-1 preferred branched N-glycans with terminal Galβ4GlcNAc, while GST-Gal-3 bound branched N-glycans with terminal fucose-containing human blood groups A and B; and (iii) GST-Gal-3 binds type 2 polylactosamine regardless of the terminal sugar while GST-Gal-1 prefers terminal Galβ4GlcNAc. Representative results for GST-Gal-1 and GST-Gal-3 binding on the arrays are presented in Fig. 3. In contrast, we observed a notable difference in glycan preference for the canine GST-Gal-2 when compared with the reported preferences of the human biotinylated Gal-2 on the same array (27). Whereas the human Gal-2 showed significant binding to human blood group A on LacNAc and PL, the canine Gal-2 bound significantly to only biantennary N-glycans with terminal Galα1,3 extended Lewisx, with or without core GlcNAc fucosylation.
Glycan preferences for canine GST-Gal-4 on the array were notably different to those previously published for the mouse Gal-4 labeled with Alexa Fluor 488 on an array of biotinylated glycans on streptavidin-coated microtiter plates (28). The mouse Gal-4 showed preference for blood group A and B structures due to binding to the C-terminal CRD and N-terminal CRD, respectively, as well as 3-O-sulfated lactose (28). We found that canine GST-Gal-4 also had preference for blood group A and B although we have not prepared the canine C-terminal CRD and N-terminal CRD separately. Nevertheless, as shown in the supplemental data, GST-Gal-4 binding to sulfated glycans was limited to [3-OSO3]Galβ1,4[6-OSO3]Glcβ-R (ranked at 12) which was 10-fold lower than binding to fucosylated trisaccharides.
Human Gal-7 affinity for glycans containing LacNAc was previously studied using isothermal titration microcalorimetry, and was reportedly 6–11-fold weaker than that observed for bovine Gal-1 and murine Gal-3 (29). More recently, the GST-tagged human Gal-7 was analyzed on the Core H arrays but only at one concentration (200 μg/ml) which does not reveal the specificity usually observed at lower concentrations (data is online). Interestingly, we found that the canine GST-Gal-7 preferentially recognized blood group H.
Glycan preferences for canine GST-Gal-8 on the array were similar to those previously published for biotinylated human Gal-8 and the two separate CRDs (30). Whereas human Gal-8 and Gal-8N recognized the same sulfated and sialylated glycans, human Gal-8N did not bind polylactosamine or blood groups antigens; and human Gal-8 and Gal-8C did bind polylactosamine, but not sialylated or sulfated glycans (30). In contrast, we observed that the canine GST-Gal-8 prefers polylactosamine with terminal Neu5Acα2,3 (ranked at 1), but not Neu5Acα2,6, and it also prefers polylactosamine and branched N-glycans with terminal blood group A (weaker for B). GST-Gal-8 interaction with sulfated glycans was limited to [3-OSO3]Galβ1,4[6-OSO3]Glcβ-R (ranked at 31), which was significantly lower than maximal binding on the array.
Glycan preferences for human Gal-9 have not been studied at varying concentrations on the Core H array. Using frontal affinity chromatography, human Gal-9C and Gal-9N CRDs are both reported to bind branched N-glycans and polylactosamine, while human Gal-9N also binds glycolipid-type glycans such as the Forssman pentasaccharide (31). We found that the canine GST-Gal-9N binds preferentially branched N-glycans with terminal blood group A as well as sulfated disaccharides, while GST-Gal-9C prefers sulfated disaccharides but will bind to branched N-glycans with terminal blood group B.
Several galectins appear to have retained an exquisite specificity for human ABO(H) blood group antigens (27, 31, 33, 34). Blood group antigens represent polymorphic antigen structures (35). As a result, it is unlikely that the galectin-blood group antigen interactions regulate key processes in the trafficking and localization of cell surface glycoproteins and the initiation of key regulatory events. Indeed, the polymorphic nature of blood group antigens likely arose from unique selective pressures to reduce pathogen invasion (36, 37). However, while blood group modification may reduce pathogen invasion, their presence also eliminates the ability of an individual to develop antibodies against these antigens (35), exposing blood group positive individuals to pathogens that bear cognate antigens. Indeed, blood group positive pathogens are responsible for the induction of anti-blood group antibodies in blood group negative individuals (38, 39), suggesting that blood group positive individuals would be uniquely sensitive to infection by blood group positive pathogens. Given the ability of several galectin family members to recognize blood group antigens, recent studies examined whether galectins might uniquely fill this gap in adaptive immunity. Interestingly, Gal-4 and Gal-8 specifically recognize and kill blood group positive E. coli, while failing to recognize or alter the viability of other blood group negative bacteria (40). In this way, Gal-4 and Gal-8 provide protection against blood group positive bacteria regardless of the blood group status of an individual. The ability of several canine galectins to also recognize blood group antigens suggests that these innate immune functions may be conserved, as recently suggested for murine Gal-4 (40). Future studies will examine these intriguing possibilities.
The possibility still remains that galectins have evolved to play some role in membrane trafficking of transmembrane proteins. There are published data showing that Gal-3, -9, and -1 binding to cell surface proteins such as epidermal growth factor receptor, the glucose transporter Glut-2 and the Ca+2 channel TRPV5, respectively, stabilizes cell surface expression (32, 41, 42). Gal-3 and galectin-4 have also been implicated in glycan-dependent and raft-dependent sorting, respectively, of apically expressed proteins in polarized epithelial cells (12, 13). As MDCK cells are the best characterized model for studying membrane trafficking in polarized epithelial cells, we used the cloned canine galectins to begin to address the potential role of galectins in MUC1 apical targeting in these canine cells. Using the GST-tagged canine galectins in pull-down assays we determined that the small subunit of human MUC1 expressed in MDCK cells binds preferentially to Gal-3 while the large subunit binds preferentially to both Gal-3 and Gal-9N. We also observed that both subunits bound to Gal-1, -4, -7, -8, and -9C but with 2–10-fold lower levels. MUC1 binding to galectin-3 has been previously reported, but this is the first report that MUC1 exhibits significant binding to other galectins, including galectin-9. Yu et al. (17) observed co-immunoprecipitation of endogenous MUC1(large subunit) with galectin-3 from extracts of a human colon cancer cell line (HT29), as well as recombinant galectin-3 binding to MUC1 overexpressed in either MDCK or a human breast epithelial cell line. Argüeso et al. (14) also reported that MUC1 (large subunit) expressed in a human corneal cell line (HCLE) bound to a Gal-3 affinity column in a galactose-dependent manner. However, Ramasamy et al. (15) observed co-immunoprecipitation of galectin-3 and a truncated MUC1 (small subunit only) overexpressed in a ZR-75 breast tumor cell line; and pull-down of the truncated MUC1 with a recombinant GST-tagged human galectin-3 was blocked by mutation of the single N-glycosylation site or by MUC1 expression in glycosylation-defective CHO cells that lack normal N-glycan processing. Therefore, our findings are consistent with studies of MUC1 in other cell types but extend the possibility that MUC1 interaction with additional galectins in MDCK cells are significant and should be studied further.
Supplementary Material
Acknowledgments
We thank Sandra Gendler for the CT-2 antibody, and Ora Weisz and Rick Cummings for helpful discussions. Glycan microarray data were obtained through the Protein-Glycan Interaction Core (Core H) of the Consortium for Functional Glycomics supported by National Institutes of Health Grant GM62116.
This work was supported, in whole or in part, by National Institutes of Health Grants DK054787 (to R. P. H.), GM85448 (to D. F. S.), and Genzyme Renal Innovations Program (to R. P. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S9 and Figs. S1–S8.
P. A. Poland and R. P. Hughey, unpublished data.
- MDCK
- Madin Darby canine kidney
- CRD
- carbohydrate recognition domain
- Gal-
- galectin
- LacNAc
- N-acetyl-lactosamine
- PL
- poly-NAc-lactosamine.
REFERENCES
- 1. Varki A., Lowe J. B. (2009) Essentials of Glycobiology, Second Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Potter B. A., Hughey R. P., Weisz O. A. (2006) Am. J. Physiol. Cell Physiol. 290, C1–C10 [DOI] [PubMed] [Google Scholar]
- 3. Weisz O. A., Rodriguez-Boulan E. (2009) J. Cell Sci. 122, 4253–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jacob R., Naim H. Y. (2001) Curr. Biol. 11, 1444–1450 [DOI] [PubMed] [Google Scholar]
- 5. Polishchuk R., Di Pentima A., Lippincott-Schwartz J. (2004) Nat. Cell Biol. 6, 297–307 [DOI] [PubMed] [Google Scholar]
- 6. Guerriero C. J., Lai Y., Weisz O. A. (2008) J. Biol. Chem. 283, 18040–18047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cresawn K. O., Potter B. A., Oztan A., Guerriero C. J., Ihrke G., Goldenring J. R., Apodaca G., Weisz O. A. (2007) EMBO J. 26, 3737–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yeaman C., Le Gall A. H., Baldwin A. N., Monlauzeur L., Le Bivic A., Rodriguez-Boulan E. (1997) J. Cell Biol. 139, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hattrup C. L., Gendler S. J. (2008) Annu. Rev. Physiol. 70, 431–457 [DOI] [PubMed] [Google Scholar]
- 10. Pemberton L. F., Rughetti A., Taylor-Papadimitriou J., Gendler S. J. (1996) J. Biol. Chem. 271, 2332–2340 [DOI] [PubMed] [Google Scholar]
- 11. Mattila P. E., Kinlough C. L., Bruns J. R., Weisz O. A., Hughey R. P. (2009) Biol. Chem. 390, 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delacour D., Cramm-Behrens C. I., Drobecq H., Le Bivic A., Naim H. Y., Jacob R. (2006) Curr. Biol. 16, 408–414 [DOI] [PubMed] [Google Scholar]
- 13. Delacour D., Greb C., Koch A., Salomonsson E., Leffler H., Le Bivic A., Jacob R. (2007) Traffic. 8, 379–388 [DOI] [PubMed] [Google Scholar]
- 14. Argüeso P., Guzman-Aranguez A., Mantelli F., Cao Z., Ricciuto J., Panjwani N. (2009) J. Biol. Chem. 284, 23037–23045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramasamy S., Duraisamy S., Barbashov S., Kawano T., Kharbanda S., Kufe D. (2007) Mol. Cell. 27, 992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morelle W., Stechly L., André S., Van Seuningen I., Porchet N., Gabius H. J., Michalski J. C., Huet G. (2009) Biol. Chem. 390, 529–544 [DOI] [PubMed] [Google Scholar]
- 17. Yu L. G., Andrews N., Zhao Q., McKean D., Williams J. F., Connor L. J., Gerasimenko O. V., Hilkens J., Hirabayashi J., Kasai K., Rhodes J. M. (2007) J. Biol. Chem. 282, 773–781 [DOI] [PubMed] [Google Scholar]
- 18. Drickamer K., Taylor M. E. (1993) Annu. Rev. Cell Biol. 9, 237–264 [DOI] [PubMed] [Google Scholar]
- 19. Cooper D. N. (2002) Biochim. Biophys. Acta 1572, 209–231 [DOI] [PubMed] [Google Scholar]
- 20. Leffler H., Carlsson S., Hedlund M., Qian Y., Poirier F. (2004) Glycoconj. J. 19, 433–440 [DOI] [PubMed] [Google Scholar]
- 21. Cummings R. D., Liu F. T. (2009) Galectins, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 22. Houzelstein D., Gonçalves I. R., Fadden A. J., Sidhu S. S., Cooper D. N., Drickamer K., Leffler H., Poirier F. (2004) Mol. Biol. Evol. 21, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 23. Friedrichs J., Torkko J. M., Helenius J., Teräväinen T. P., Füllekrug J., Muller D. J., Simons K., Manninen A. (2007) J. Biol. Chem. 282, 29375–29383 [DOI] [PubMed] [Google Scholar]
- 24. Self A. J., Hall A. (1995) Methods Enzymol. 256, 3–10 [DOI] [PubMed] [Google Scholar]
- 25. Kinlough C. L., McMahan R. J., Poland P. A., Bruns J. B., Harkleroad K. L., Stremple R. J., Kashlan O. B., Weixel K. M., Weisz O. A., Hughey R. P. (2006) J. Biol. Chem. 281, 12112–12122 [DOI] [PubMed] [Google Scholar]
- 26. Smith D. F., Song X., Cummings R. D. (2010) Methods Enzymol. 480, 416–442 [DOI] [PubMed] [Google Scholar]
- 27. Stowell S. R., Arthur C. M., Mehta P., Slanina K. A., Blixt O., Leffler H., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marková V., Smetana K., Jr., Jeníková G., Láchová J., Krejciríková V., Poplstein M., Fábry M., Brynda J., Alvarez R. A., Cummings R. D., Maly P. (2006) Int. J. Mol. Med. 18, 65–76 [PubMed] [Google Scholar]
- 29. Brewer C. F. (2004) Glycoconj. J. 19, 459–465 [DOI] [PubMed] [Google Scholar]
- 30. Stowell S. R., Arthur C. M., Slanina K. A., Horton J. R., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 20547–20559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 32. Cha S. K., Ortega B., Kurosu H., Rosenblatt K. P., Kuro-O M., Huang C. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9805–9810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leffler H., Barondes S. H. (1986) J. Biol. Chem. 261, 10119–10126 [PubMed] [Google Scholar]
- 34. Sparrow C. P., Leffler H., Barondes S. H. (1987) J. Biol. Chem. 262, 7383–7390 [PubMed] [Google Scholar]
- 35. Yamamoto F., Clausen H., White T., Marken J., Hakomori S. (1990) Nature 345, 229–233 [DOI] [PubMed] [Google Scholar]
- 36. Neil S. J., McKnight A., Gustafsson K., Weiss R. A. (2005) Blood 105, 4693–4699 [DOI] [PubMed] [Google Scholar]
- 37. Rowe J. A., Handel I. G., Thera M. A., Deans A. M., Lyke K. E., Koné A., Diallo D. A., Raza A., Kai O., Marsh K., Plowe C. V., Doumbo O. K., Moulds J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17471–17476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Springer G. F., Horton R. E. (1969) J. Clin. Invest. 48, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Springer G. F., Williamson P., Brandes W. C. (1961) J. Exp. Med. 113, 1077–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stowell S. R., Arthur C. M., Dias-Baruffi M., Rodrigues L. C., Gourdine J. P., Heimburg-Molinaro J., Ju T., Molinaro R. J., Rivera-Marrero C., Xia B., Smith D. F., Cummings R. D. (2010) Nat. Med. 16, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. (2004) Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 42. Ohtsubo K., Takamatsu S., Minowa M. T., Yoshida A., Takeuchi M., Marth J. D. (2005) Cell. 123, 1307–1321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.