Abstract
Methylmercury (MeHg) toxicity is a continuous environmental problem to human health. The critical role of oxidative stress in the pathogenesis of MeHg cytotoxicity has been clarified, but the molecular mechanisms underlying MeHg-mediated oxidative stress remain to be elucidated. Here we demonstrate a post-transcriptional effect of MeHg on antioxidant selenoenzymes by using a MeHg-susceptible cell line. MeHg-induced selenium deficiency leads to failure of the recoding of a UGA codon for selenocysteine and results in degradation of the major antioxidant selenoenzyme glutathione peroxidase 1 (GPx1) mRNA by nonsense-mediated mRNA decay (NMD), a cellular mechanism that detects the premature termination codon (PTC) located 5′-upstream of the last exon-exon junction and degrades PTC-containing mRNAs. In contrast, thioredoxin reductase 1 (TrxR1), another antioxidant selenoenzyme of the thioredoxin system, was likely skipped by NMD because of a UGA codon in the last exon. However, TrxR1 activity was decreased despite mRNA up-regulation, which was probably due to the synthesis of aberrant TrxR1 protein without selenocysteine. Changes in selenoenzyme GPx1 and TrxR1 mRNAs were observed earlier than was the incidence of oxidative stress and up-regulation of other antioxidant enzyme mRNAs. Results indicated that the MeHg-induced relative selenium-deficient condition affects the major antioxidant selenoenzymes GPx1 and TrxR1 through a post-transcriptional effect, resulting in the disturbance of cellular redox systems and the incidence of oxidative stress. Treatment with ebselen, a seleno-organic compound, effectively suppressed oxidative stress and protected cells against MeHg-induced relative selenium deficiency and cytotoxicity.
Keywords: Antioxidant, Oxidative Stress, Selenium, Transfer RNA (tRNA), Translation, Antioxidant Selenoenzyme, Methylmercury, Nonsense-mediated mRNA Decay, Post-transcriptional Defect
Introduction
Methylmercury (MeHg)2 is a well- established toxicant to humans as recognized in some countries like Japan (Minamata Disease) (1), Iraq (2), and the United States (3). MeHg toxicity is a continuous environmental problem to human health, especially in susceptible populations who frequently eat plenty of fish or fish predators (4).3 The critical role of oxidative stress in the pathogenesis of MeHg cytotoxicity has been clarified both in vitro (5–10) and in vivo (11–13). Failure to protect cells against the early oxidative stress triggers subsequent ER stress and apoptosis (9). However, the molecular mechanism underlying the early incidence of oxidative stress by MeHg exposure remains to be elucidated.
The high affinity of MeHg for the selenohydryl group, sulfhydryl group, or selenide has been described (14). In addition, there has been some research on the interaction between MeHg and selenium since Ganther et al. (15) first reported it. Sodium selenite treatment can suppress MeHg-mediated fetotoxicity, neurotoxicity, or developmental toxicity (16–18). Further, MeHg toxicity was enhanced in selenium-deficient animals (19, 20). These reports suggest the possibility that the selenium component of naturally occurring selenium-containing proteins and tRNAs is the protecting factor against MeHg cytotoxicity. However, the role of selenium in protection against MeHg cytotoxicity has not been well elucidated.
The glutathione (GSH) and thioredoxin (Trx) systems, which are the central regulators of cellular redox status, include many antioxidant enzymes with the selenohydryl group or sulfhydryl group at the redox-active centers. Thus cellular antioxidant systems may be disturbed under MeHg exposure. In fact, exposure to MeHg mediates the decrease of glutathione peroxidase 1 (GPx1) activity in vitro and in vivo (12, 21–23) and the inhibition of recombinant TrxR activity in vitro (24).
GPx1, the most abundant selenoenzyme, plays a role in preventing the production of reactive oxygen species (ROS) by reducing hydrogen peroxide (H2O2) and free fatty acid hydroperoxides. In contrast, thioredoxin reductase 1 (TrxR1) and its substrate Trx are known to be involved in the redox regulation of cell signaling. Both GPx1 and TrxR1 contain 1 nonstandard amino acid, selenocysteine (Sec). Sec is encoded by the UGA codon, which is common for termination of protein synthesis. Under selenium deficiency, the UGA codon for Sec may be recognized as a nonsense codon, known as a premature termination codon (PTC), because of its unique specific biosynthesis (25). mRNAs harboring PTCs are known to be deleted by nonsense-mediated mRNA decay (NMD), an mRNA quality control mechanism that is active when PTC is located sufficiently upstream of the exon-exon junction (26). These findings prompted us to study the responses of antioxidant selenoenzymes to MeHg exposure.
Here we investigated the effect of MeHg on the cellular redox systems, especially on the dynamics of cytosolic selenoenzymes GPx1 and TrxR1, by using the MeHg-susceptible cell line to know the molecular mechanism underlying the incidence of oxidative stress after MeHg exposure. We clarified for the first time a post-transcriptional effect of MeHg on antioxidant selenoenzymes, which resulted in the disturbance of cellular redox systems and the incidence of oxidative stress.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Treatments
The C2C12 myogenic cell line expressing human mutant DMPK cDNA containing 160 CTG repeats (C2C12-DMPK160) was employed. The cells were cultured and exposed to MeHg as described previously (7, 8). For antioxidant treatment, cells were treated with 100 μm Trolox (SIGMA-Aldrich) as described previously (7–8). Ebselen (Sigma-Aldrich) stock was dissolved in dimethyl sulfoxide. Sodium selenite (Sigma-Aldrich) stock was prepared in Ca2+- and Mg2+-free phosphate buffered saline (PBS (−)) and added to the cells 16 h before the MeHg treatment.
Samples from MeHg-intoxicated Rats
Frozen soleus skeletal muscle samples were prepared from rats as previously reported (12). The samples were divided into 3 groups: 1) MeHg-treated, 2) (MeHg + Trolox 2.5 mg/kg)-treated, and 3) control without MeHg and Trolox. Each group included 5 rats. Pathological changes were detected in the cerebellum, dorsal root ganglia, dorsal root nerve, and skeletal muscles of MeHg-treated rats, whereas these changes were suppressed in rats treated with MeHg and Trolox (12).
Flow Cytometry Analysis
For ROS analysis, cells (1.3 × 105 cells/35-mm dish) were incubated with 500 nm 5-(and 6-)chloromethyl- 2′,7′-dichlorodihydrofluorescin diacetate, acetyl ester (CM-H2DCFDA) (Invitrogen) for 30 min, and washed twice with PBS (−). Cells were then exposed to 0.4 μm MeHg with or without 10 μm ebselen. Cells were harvested using 2 mm EDTA/PBS (−) that was prewarmed at 37 °C. The cell pellet was resuspended in 1 ml of PBS (−), and ROS was analyzed on a FACSCalibur flow cytometer (BD Biosciences) equipped with a 488 nm argon-ion laser and a 525 ± 10 nm bandpass emission filter. A total of 5,000 cells/sample were analyzed.
For analysis of apoptosis, cells (1.3 × 105 cells/35-mm dish) exposed to 0.4 μm MeHg with or without 10 μm ebselen for 16 h were harvested using 2 mm EDTA/PBS (−) that was pre-warmed at 37 °C. The cell pellet was then washed twice with PBS (−). An Annexin V-FITC apoptosis detection kit I (BD Biosciences) was used according to the manufacturer's directions. Cells were incubated with Annexin V-FITC in a buffer containing propidium iodide (PI) and analyzed. A total of 10, 000 cells/sample were analyzed on the FACSCalibur.
Quantitative Real-time PCR
Total RNA was extracted using a QuickGene-800 System and a QuickGene RNA Cultured Cell Kit S (FUJIFILM). First-strand cDNA was prepared with a QuantiTect Reverse Transcription kit (Qiagen). Quantitative real-time PCR was performed using a LightCycler DX 400 System (Roche). GPx1, manganese-superoxide dismutase (Mn-SOD), copper, zinc-superoxide dismutase (Cu, Zn-SOD), catalase, and TrxR1 mRNAs were amplified with a SYBR Green Master Mix (Roche) using the specific primer sets described in supplemental Table S1. Each mRNA transcript level was normalized to β-actin mRNA as described previously (9). Values are shown as means ± S.E. of more than four separate experiments.
siRNA Preparation and Cell Transfection
The following siRNA target sequences were used: mouse SMG-1, FlexiPlate siRNA SI00871857 (Qiagen); mouse SMG-7, FlexiPlate siRNA SI02765546 (Qiagen); and NS siRNA, All Star Negative Control siRNA (Qiagen). The transfections of synthetic siRNAs were carried out with HiPerFect Transfection Reagent (Qiagen) or LipofectamineTM RNAiMAX (Invitrogen). The cells were analyzed 52 h after transfection, and the results were confirmed in more than three independent experiments.
Antibodies and Western blot Analysis
Samples were prepared as described previously (27). Cell lysates were separated by 5.5% sodium dodecyl sulfate-polyacryamide gel electrophoresis for Upf1, phospho-Upf1, SMG-1, and SMG-7 or 7.5% for TrxR1 in the presence of DTT and transferred to nitrocellulose membranes (Bio-Rad). The membranes were then subjected to a monoclonal mouse anti-Upf1 antibody (5C3) (28), a monoclonal mouse anti-phospho-Upf1 antibody (7H1) (28), a polyclonal rabbit anti-SMG-1 antibody (Bethyl), a polyclonal rabbit anti-SMG-7 antibody (29), a monoclonal mouse anti-TrxR1 antibody (Santa Cruz Biotechnology), or a mouse monoclonal anti-α-tubulin antibody (Santa Cruz Biotechnology). Proteins were detected as described previously (27).
Measurement of TrxR1 Activity
Cells were harvested using a rubber policeman at the indicated time of MeHg exposure. Cell suspension was centrifuged at 150 × g for 5 min. The cell pellet was washed twice in PBS (−) and then homogenized in ice cold buffer (50 mm potassium phosphate, pH 7.4, containing 1 mm EDTA) by sonication and centrifuged at 12,000 rpm for 15 min at 4 °C. The sample was then concentrated using a MICROCON YM-30 with a molecular weight cut-off of 30,000 (Millipore). TrxR1 activity was measured by a Thioredoxin Reductase Assay kit (Cayman Chemical Company) according to the manufacturer's directions. Protein content was measured using a DC Protein Assay Kit II (Bio-Rad).
Statistical Analysis
Statistical significance was determined by a one-way Welch's t test. Data are expressed as mean ± S.E. A difference was considered statistically significant when p < 0.05.
RESULTS
Increase in Intracellular ROS 3–4 h after Exposure to MeHg
The C2C12-DMPK160 cell line was susceptible to MeHg-induced oxidative stress and showed apoptosis within 24 h after exposure to low levels of MeHg (7, 9). To clarify the onset of the oxidative burst after MeHg exposure, a time course study of flow cytometry analysis of C2C12-DMPK160 cells labeled with CM-H2DCFDA was performed. Results indicated that 0.4 μm MeHg exposure increased intracellular ROS levels 3–4 h after exposure compared with non-treated cells (Fig. 1).
FIGURE 1.
Effect of MeHg on intracellular ROS. Flow cytometry analysis of C2C12-DMPK160 cells labeled with CM-H2DCFDA for time course of intracellular ROS after 0.4 μm MeHg exposure. Data shown are representative of three separate experiments.
Up-regulation of mRNAs of Antioxidant Enzymes Except GPx1 after MeHg Exposure
MeHg-mediated increase in intracellular ROS was expected to promote ROS-dependent changes in antioxidant gene expression. Consistent with this notion, quantitative real-time PCR analysis showed up-regulation of mRNAs of anti-oxidant enzymes, Mn-SOD, Cu, Zn-SOD, catalase, and TrxR1 after MeHg exposure in C2C12-DMPK160 cells (Fig. 2, A–D). Treatment with 100 μm Trolox, a vitamin E-derived antioxidant, suppressed the increase in these mRNAs after 0.4 μm MeHg exposure, suggesting that the expression of these mRNAs was mediated by ROS. In contrast, the expression of selenium-dependent GPx1 mRNA was unexpectedly decreased 3–9 h after exposure to MeHg. In addition, treatment with Trolox did not alter MeHg-mediated decrease in GPx1 mRNA (Fig. 2E). Note that increase in TrxR1, another selenoenzyme, and decrease in GPx1 mRNAs were observed earlier than the response of other mRNAs.
FIGURE 2.
Effect of MeHg on the expression of Mn-SOD, Cu-Zn-SOD, catalase, TrxR1, and GPx1 mRNAs in C2C12-DMPK160 cells. Cells were exposed to 0.4 μm MeHg with or without 100 μm Trolox. Total RNAs prepared at the times indicated were analyzed by quantitative real-time PCR for quantified relative amount of Mn-SOD (A), Cu, Zn-SOD (B), catalase (C), TrxR1 (D), or GPx1 mRNA (E). The histogram depicts the indicated mRNA normalized to the β-actin mRNA value. Values shown are means ± S.E. of four separate experiments.
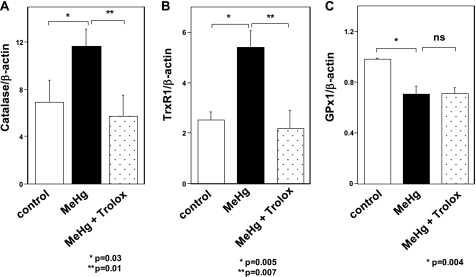
Down-regulation of GPx1 mRNA in MeHg-intoxicated Rats
To investigate the effect of MeHg on the expression of antioxidant enzyme mRNAs in vivo, we analyzed the expression of mRNAs extracted from soleus skeletal muscles of rats treated with MeHg for 4 weeks. The results showed that the catalase and TrxR1 mRNAs were up-regulated (Fig. 3, A and B), whereas GPx1 mRNA was downregulated (Fig. 3C). Co-addition of the antioxidant Trolox could suppress the change in catalase and TrxR1 mRNAs in soleus skeletal muscls of MeHg-exposed rats. However, down-regulation of GPx1 mRNA could not be rescued by treatment with Trolox. Results were consistent with those observed in C2C12-DMPK160 cells.
FIGURE 3.
Effect of MeHg on the expression of catalase, TrxR1, and GPx1 mRNAs in rats. Total RNAs from soleus muscles of rats treated with MeHg for 4 weeks were analyzed by quantitative real-time PCR for quantified relative amount of catalase (A), TrxR1 (B), or GPx1 mRNA (C). The histogram depicts the indicated mRNA normalized to the β-actin mRNA value. Values shown are means ± S.E. of four independent experiments.
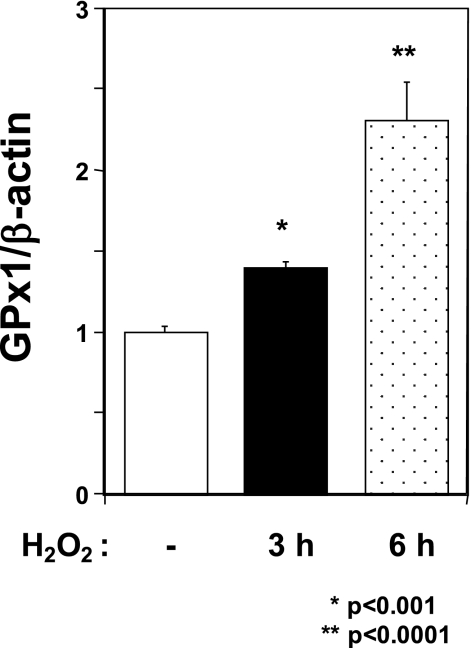
Up-regulation of GPx1 mRNA Was Induced by H2O2 Treatment
To investigate whether down-regulation of GPx1 mRNA is specific to MeHg exposure, we examined the change in GPx1 mRNA under oxidative stress by the general burden of H2O2. As shown in Fig. 4, treatment with H2O2 increased GPx1 mRNA in C2C12-DMPK160 cells. This result suggests that down-regulation of GPx1 mRNA after MeHg exposure is specific to MeHg.
FIGURE 4.
Effect of H2O2 on the expression of GPx1 mRNA in C2C12-DMPK160 cells. Total RNAs were prepared from cells treated with 0.5 mm H2O2 at the indicated time points, and then analyzed by quantitative real-time PCR for quantified relative amount of GPx1 mRNA. The histogram depicts GPx1 mRNA normalized to the β-actin mRNA value. Values shown are means ± S.E. of three independent experiments.
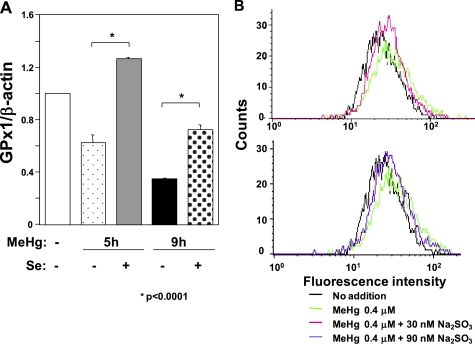
Pretreatment with Selenium Rescues Down-regulation of GPx1 mRNA and Diminishes the Increase in ROS after MeHg Exposure
Trolox-irresponsive decrease in selenium-dependent GPx1 mRNA abundance after MeHg exposure in this study and the high affinity of MeHg for the selenohydryl group and selenide (14) directed our attention to the notion that down-regulation of GPx1 mRNA might be caused by MeHg-induced selenium deficiency because COS medium, a C2C12-DMPK160 cell culture medium, contains only 8 nm selenium. To assess this possibility, we analyzed GPx1 mRNA abundance in the presence of additional 30 nm sodium selenite, a water-soluble selenium compound, under MeHg exposure. As shown in Fig. 5A, pretreatment with the additional 30 nm sodium selenite suppressed the MeHg-induced decrease in GPx1 mRNA. Together with the inability of Trolox to rescue the decrease in GPx1 mRNA abundance in the MeHg-exposed condition (Fig. 2E), these results suggest that the down-regulation of GPx1 mRNA after MeHg exposure was caused by the mechanism related to MeHg-induced selenium deficiency. To further support this notion, we analyzed the MeHg-mediated increase in ROS in the presence of sodium selenite. Flow cytometry analysis for ROS clarified that pretreatment with sodium selenite for 16 h suppressed the increase in intracellular ROS 7 h after MeHg exposure, although even 90 nm sodium selenite could not completely suppress the increase in ROS (Fig. 5B).
FIGURE 5.
Effect of sodium selenite on MeHg-induced down-regulation of GPx1 mRNA and intracellular ROS in C2C12-DMPK160 cells. A, cells were added to 30 nm sodium selenite 16 h before exposure to 0.4 μm MeHg. Total RNAs prepared at the indicated time points were analyzed by quantitative real-time PCR for quantified relative amount of GPx1 mRNA. The histogram depicts GPx1 mRNA normalized to the β-actin mRNA value. Values shown are means ± S.E. of four separate experiments. B, cells were added to 30 nm or 90 nm sodium selenite 16 h before exposure to 0.4 μm MeHg. Flow cytometry analysis of cells labeled with CM-H2DCFDA was performed for intracellular ROS 7 h after 0.4 μm MeHg exposure. Data shown are representative of three separate experiments.
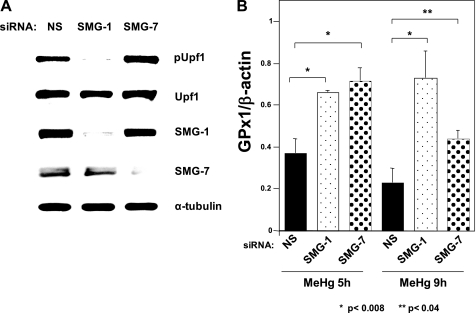
Down-regulation of GPx1 mRNA after MeHg Exposure Is Mediated by NMD
Under selenium-depleted conditions, the UGA codon for Sec on GPx1 mRNA is recognized as a PTC and GPx1 mRNA is degraded by NMD (30). If the MeHg-mediated decrease in GPx1 mRNA was caused by selenium deficiency, inhibition of NMD should rescue the MeHg-mediated GPx1 mRNA suppression. To assess this hypothesis, we performed synthetic siRNA-mediated knockdown of the mouse NMD components SMG-1 or SMG-7 and examined GPx1 mRNA expression under MeHg exposure. As shown in Fig. 6A, transfection of synthetic siRNA targeted to mouse SMG-1 or SMG-7 into C2C12-DMPK160 cells knocked down each protein. NMD inhibition was supported by the decrease (in the case of SMG-1 knockdown) or increase (in the case of SMG-7 knockdown) in Upf1 phosphorylation, a central component of NMD, since the Upf1 phosphorylation and dephosphorylation cycle is essential for NMD (31). These NMD-suppressed cells demonstrated increases in GPx1 mRNA accumulation under MeHg-exposed conditions (Fig. 6B). These results suggest that MeHg-induced decreases in GPx1 mRNA are mediated by NMD.
FIGURE 6.
Effect of NMD suppression on down-regulation of GPx1 mRNA after MeHg exposure. A, NMD suppression by synthetic siRNA-mediated knockdown of SMG-1 or SMG-7. Western blots of C2C12-DMPK160 transfected with the indicated synthetic siRNAs were analyzed with the indicated antibody probes. NS, non-silencing; pUpf1, anti-phospho-Upf1 antibody. B, synthetic siRNA-mediated knockdown of SMG-1 or SMG-7 results in the rescue of GPx1 mRNA down-regulation after MeHg exposure. C2C12-DMPK160 transfected with the indicated siRNAs was exposed to 0.4 μm MeHg, and total RNA was isolated 5 h or 9 h after exposure. Levels of GPx1 or β-actin mRNA were determined by quantitative real-time PCR. The histogram depicts the GPx1 mRNA normalized to the β-actin mRNA value represented as a fold-increase over NS siRNA- and non-MeHg-treated control. SMG-1- or SMG-7-knockdown cells revealed significantly higher values than NS-siRNA transfectants. Values shown are means ± S.E. of four separate experiments.
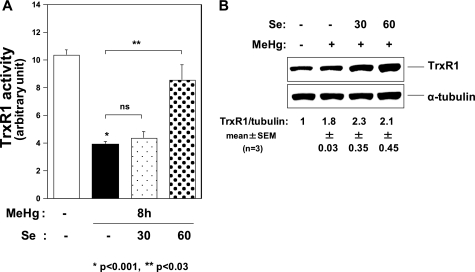
MeHg Suppresses TrxR1 Activity
The NMD-dependent decrease in GPx1 mRNA indicated failure of recoding events of the UGA codon for Sec under MeHg exposure. This probably occurs due to selenium depletion under MeHg exposure owing to the high affinity of MeHg for the selenohydryl group and selenide. In this case, TrxR1 activity should decrease because of the synthesis of the aberrant TrxR1 protein without Sec. To investigate this possibility, we analyzed the effect of MeHg on TrxR1 activity and found that it was significantly decreased 8 h after exposure to MeHg compared with non-treated cells (Fig. 7A) despite up-regulation of TrxR1 mRNA (Fig. 2D) and a slight increase in TrxR1 protein levels (Fig. 7B). The decrease in TrxR1 activity after MeHg exposure was rescued by pretreatment with 60 nm sodium selenite (Fig. 7A).
FIGURE 7.
Effect of MeHg on TrxR1. A, TrxR1 activity 8 h after exposure to MeHg. Pretreatment with 30 nm sodium selenite could not suppress the decrease in TrxR1 activity after 0.4 μm MeHg exposure. However, pretreatment with 60 nm sodium selenite rescued the MeHg-induced decrease in TrxR1 activity. Values shown are means ± S.E. of triplicate samples in 3 separate experiments. B, Western blots of C2C12-DMPK160 cells were analyzed with the indicated antibody probes. The densitometric quantification of TrxR1 protein normalized to the α-tubulin protein is represented as a fold-increase over the control. Representative images of three samples were shown with quantitative data (means ± S.E.).
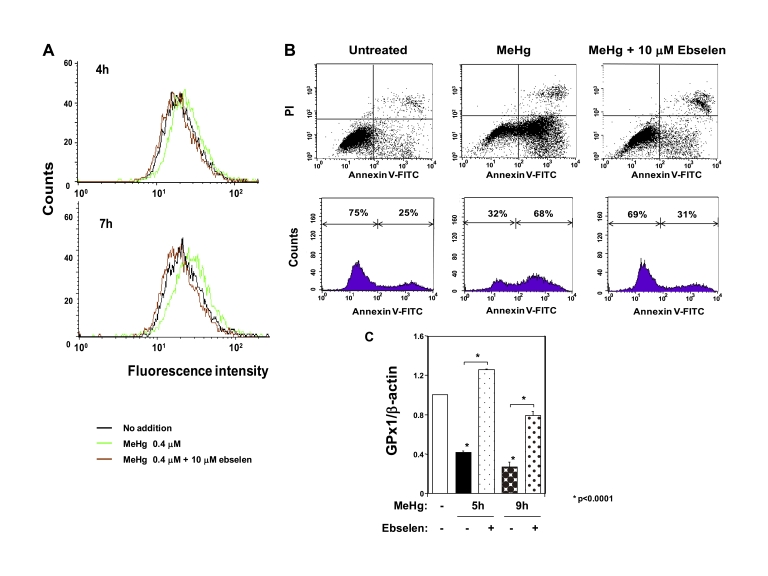
Ebselen Treatment Suppresses MeHg Cytotoxicity
To further assess the pathophysiological importance of MeHg- mediated suppression of GPx1 for MeHg cyototoxicity, we investigated the effect of ebselen, a seleno-organic compound that mimics GPx1, on MeHg cytotoxicity. Analysis of intracellular ROS by flow cytometry showed that treatment with 10 μm ebselen completely suppressed the ROS increase after exposure to MeHg in C2C12-DMPK160 cells (Fig. 8A). We next analyzed cell viability to further clarify the protective effect of ebselen against MeHg cytotoxicity. Flow cytometry analysis demonstrated that MeHg-induced apoptosis was clearly suppressed by treatment with ebselen (Fig. 8B). These results indicate that ebselen effectively protects against MeHg cytotoxicity. In addition, ebselen unexpectedly inhibited GPx1 mRNA down-regulation after MeHg exposure (Fig. 8C).
FIGURE 8.
Effect of ebselen on MeHg cytotoxicity. A, flow cytometry analysis of C2C12-DMPK160 cells for ROS after MeHg exposure for 7 h. B, upper panel shows flow cytometry analysis of C2C12-DMPK160 cells stained with PI and FITC-Annexin V. Vertical axis indicates PI fluorescence intensity and horizontal axis Annexin V fluorescence. Untreated C2C12-DMPK160 cells were primarily Annexin V-FITC and PI negative, indicating that they were viable. Exposure to 0.4 μm MeHg for 16 h increased cells undergoing apoptosis (Annexin V-FITC positive and PI negative). A minor population of cells were observed to be Annexin V-FITC and PI positive, indicating that they were in end stage apoptosis or already dead. The lower panel shows profile of frequency of viable cells (Annexin V-FITC and PI negative) and undergoing apoptosis cells (Annexin V-FITC positive and PI negative). Treatment with ebselen decreased frequency of undergoing apoptosis cells. C, quantitative real-time PCR analysis of GPx1 mRNA. The histogram depicts the indicated mRNA normalized to the β-actin mRNA value. Values shown are means ± S.E. of four independent experiments.
DISCUSSION
In this study we demonstrated for the first time to our knowledge that MeHg has a post-transcriptional effect on the main antioxidant selenoenzymes GPx1 and TrxR1, most likely through MeHg-induced selenium deficiency, resulting in the disturbance of cellular redox systems and the incidence of oxidative stress. The mechanism shown in this study is most likely universal in MeHg toxicity, because the same phenomenon was recognized in soleus muscle in a usual MeHg-intoxicated rat model (Fig. 3) and a wild-type C2C12-DMPK5 cell line (supplemental Fig. S1).
The down-regulation of the major antioxidant selenoenzyme GPx1 mRNA under MeHg exposure was specific to the burden of MeHg because up-regulation of GPx1 mRNA was observed under oxidative stress by the general burden of H2O2 (Fig. 4). This is in striking contrast with other antioxidant enzymes, including TrxR1, because their mRNAs are up-regulated under MeHg-induced ROS increase (Figs. 1, 2, A–D, and 3, A and B). Intriguingly, the antioxidant Trolox failed to rescue GPx1 mRNA decrease (Figs. 2E and 3C), even though it was able to suppress MeHg-induced increase in other antioxidant enzyme mRNAs (Figs. 2, A–D and 3, A and B) through suppression of intracellular ROS (9). These results suggest that the effect of MeHg on GPx1 mRNA was not mediated by intracellular ROS.
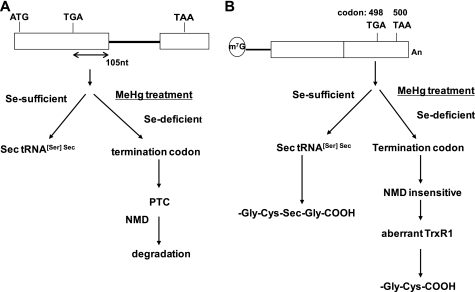
Our results indicated that the MeHg-induced decrease in GPx1 mRNA is a post-transcriptional event that is most likely mediated by cellular selenium deficiency. Several lines of evidence support this conclusion. First, the MeHg-mediated decrease in GPx1 mRNA was rescued by pretreatment with sodium selenite (Fig. 5A). This is consistent with the idea that MeHg treatment induces a relative intracellular selenium-deficient condition because MeHg has high affinity for the selenohydryl group and selenide (14). Secondly, NMD inactivation impaired GPx1 mRNA suppression by MeHg exposure. NMD is a post-transcriptional gene expression mechanism that recognizes a UGA codon for Sec on GPx1 mRNA that resides 105 nucleotides upstream of the sole exon-exon junction (Fig. 9A) as a PTC and degrades GPx1 mRNA under active selenium-deficient conditions (30).
FIGURE 9.
A model for the post-transcriptional effect of MeHg for antioxidant selenoenzymes. A, GPx1 cDNA. The UGA codon encoded for Sec resides 105 nucleotides upstream of the sole exon-exon junction. When a UGA codon is recognized as a nonsense codon under selenium deficiency, GPx1 mRNA should be a natural substrate for NMD. B, TrxR1 cDNA. The Sec codon UGA-498 resides in the last exon on TrxR1 mRNA, so TrxR1 mRNA cannot be a substrate for NMD even when a UGA codon is recognized as a nonsense codon under selenium deficiency.
In contrast to GPx1, mRNA of another antioxidant selenoenzyme, TrxR1 was not down-regulated by MeHg exposure. Sec codon UGA-498 on TrxR1 resides in the last exon (Fig. 9B) cannot be a substrate for NMD because downstream intron is required to trigger NMD (32). In theory, TrxR1 protein synthesized by NMD-skipped TrxR1 mRNA should be truncated because the Sec codon was recognized as a nonsense codon under selenium-deficient conditions. However, a low activity form of TrxR with a cysteine substituted for Sec codon UGA-498 in place of truncated TrxR was detected in in vivo study using rat liver selenium-starvation condition in unknown mechanism(s) (33). Further study will be needed to investigate a novel mechanism for decoding the UGA codon under selenium deficiency. Our study showed that TrxR1 activity was significantly decreased after MeHg exposure despite up-regulation of TrxR1 mRNA and a slight increase in TrxR1 protein levels (Figs. 2D and 7, A and B). The rescue of the decrease in TrxR1 activity by pretreatment with sodium selenite suggested that the decrease in TrxR1 activity was caused by the synthesis of aberrant TrxR1 protein (Fig. 7A).
Taken together with the above results, a MeHg-induced relative selenium-deficient condition disturbs intracellular redox systems through a post-transcriptional effect on the major antioxidant selenoenzymes GPx1 and TrxR1, in addition to the direct attack of cysteine and/or selenocysteine residues of antioxidant enzymes by MeHg (24). GPx1 is the most abundant selenoprotein and plays a critical role in reducing cellular H2O2. In contrast, TrxR1 transfers electrons from NADPH to thioredoxin, which in turn reduces thioredoxin peroxidase and other redox proteins (34). In addition, TrxR1 is able to reduce a number of substrates other than thioredoxin, including selenite (35), lipid hydroperoxides (36), and H2O2 (37). Therefore, MeHg disturbs the glutathione and thioredoxin cellular redox systems at the early stage of cytotoxicity, which is followed by cellular stress responses. In fact, changes in the selenoenzyme GPx1 and TrxR1 mRNAs together with the increase in intracellular ROS were recognized 3 h earlier after MeHg exposure than those of other antioxidant mRNAs (Figs. 1 and 2, A–E). Furthermore, ROS-mediated activation of the signal transduction system, namely, dissociation of Trx-ASK1 (apoptosis signal-regulating kinase 1) complex, phosphorylation of ASK1, and activation of the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) pathways were also recognized around the same time after the ROS increase in C2C12-DMPK160 cells (9).
The incidence of oxidative stress should depend on the capacity of the intracellular redox systems to deal with GPx1 and TrxR1 impairment under MeHg-induced relative selenium deficiency. Pretreatment with sodium selenite for 16 h could not completely suppress the increase in intracellular ROS after MeHg exposure in C2C12-DMPK160 cells (Fig. 5B), indicating that more H2O2 or other ROS might have been produced than could have been eliminated by the antioxidant selenoenzymes in this cell line. Therefore, we needed to use antioxidants such as Trolox (9) or ebselen (Fig. 8, A and B) to completely suppress the ROS increase and protect cells against MeHg-induced cytotoxicity. Ebselen, a seleno-organic compound, exhibits the GPx1 mimic activity (38) whose major substrate is H2O2. In addition, ebselen can also quench free radicals and singlet oxygen (38). Furthermore, ebselen is an excellent direct substrate for mammalian TrxR and thioredoxin (39, 40). We demonstrated here that ebselen restores down-regulation of GPx1 mRNA after MeHg exposure (Fig. 8C). Ebselen treatment does not increase the amount of bioavailable selenium (38), but generates the selenol form of the compound (39). MeHg may react with the ebselen-transformed reducing intermediate selenol in this context. This finding raises the possibility that ebselen may solve the problem due to MeHg-induced relative selenium deficiency. The results indicate that the GPx1-homologous antioxidant ebselen protects cells against MeHg-induced relative selenium deficiency and cytotoxicity.
Supplementary Material
This study was supported in part by KAKENHI (20591015) (to F. U.). This work was also supported in part by grants from the Japan Society for the Promotion of Science (to A. Y.), from the Japan Science and Technology Corporation (to A. Y.), and from the Yokohama Foundation for Advancement of Medical Science (to A. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
United Nations Environment Programme, Chemicals, Global Mercury Assessment. 2002: iii. (http://www.chem.unep.ch).
- MeHg
- methylmercury
- GPx1
- glutathione peroxidase 1
- NMD
- nonsense-mediated mRNA decay
- PTC
- premature termination codon
- TrxR1
- thioredoxin reductase 1
- Trx
- thioredoxin
- ROS
- reactive oxygen species
- H2O2
- hydrogen peroxidase
- Sec
- selenocysteine
- PI
- propidium iodide
- Mn-SOD
- manganese-superoxide dismutase
- Cu,Zn-SOD
- copper, zinc-superoxide dismutase
- ASK1
- apoptosis signal-regulating kinase 1.
REFERENCES
- 1. Hamada R., Osame M. (1996) Toxicology of Metals (Chang L. W. ed) pp. 337–351, CRC Press, London [Google Scholar]
- 2. Bakir F., Damluji S. F., Amin-Zaki L., Murtadha M., Khalidi A., al-Rawi N. Y., Tikriti S., Dahahir H. I., Clarkson T. W., Smith J. C., Doherty R. A. (1973) Science 181, 230–241 [DOI] [PubMed] [Google Scholar]
- 3. Davis L. E., Kornfeld M., Mooney H. S., Fiedler K. J., Haaland K. Y., Orrison W. W., Cernichiari E., Clarkson T. W. (1994) Ann. Neurol. 35, 680–688 [DOI] [PubMed] [Google Scholar]
- 4. Deleted in proof.
- 5. Park S. T., Lim K. T., Chung Y. T., Kim S. U. (1996) Neurotoxicology 17, 37–45 [PubMed] [Google Scholar]
- 6. Yee S., Choi B. H. (1996) Neurotoxicology 17, 17–26 [PubMed] [Google Scholar]
- 7. Usuki F., Ishiura S. (1998) NeuroReport 9, 2291–2296 [DOI] [PubMed] [Google Scholar]
- 8. Usuki F., Takahashi N., Sasagawa N., Ishiura S. (2000) Biochem. Biophys. Res. Commun. 267, 739–743 [DOI] [PubMed] [Google Scholar]
- 9. Usuki F., Fujita E., Sasagawa N. (2008) Neurotoxicology 29, 22–30 [DOI] [PubMed] [Google Scholar]
- 10. Shanker G., Aschner M. (2003) Brain Res. Mol. Brain Res. 110, 85–91 [DOI] [PubMed] [Google Scholar]
- 11. Yee S., Choi B. H. (1994) Exp. Mol. Pathol. 60, 188–196 [DOI] [PubMed] [Google Scholar]
- 12. Usuki F., Yasutake A., Umehara F., Tokunaga H., Matsumoto M., Eto K., Ishiura S., Higuchi I. (2001) Neurosci. Lett. 304, 199–203 [DOI] [PubMed] [Google Scholar]
- 13. Usuki F., Yasutake A., Umehara F., Higuchi I. (2004) Acta Neuropathol. 108, 1–9 [DOI] [PubMed] [Google Scholar]
- 14. Sugiura Y., Tamai Y., Tanaka H. (1978) Bioinorg. Chem. 9, 167–180 [DOI] [PubMed] [Google Scholar]
- 15. Ganther H. E., Goudie C., Sunde M. L., Kopecky M. J., Wagner P., Oh S-H., Hoekstra W. G. (1972) Science 175, 1122–1124 [DOI] [PubMed] [Google Scholar]
- 16. Nobunaga T., Satoh H., Suzuki T. (1979) Toxicol. Appl. Pharmacol. 47, 79–88 [DOI] [PubMed] [Google Scholar]
- 17. Satoh H., Yasuda N., Shimai S. (1985) Toxicol. Lett. 25, 199–203 [DOI] [PubMed] [Google Scholar]
- 18. Imura N. (1986) Dev. Toxicol. Environ. Sci. 12, 115–123 [PubMed] [Google Scholar]
- 19. Nishikido N., Furuyashiki K., Naganuma A., Suzuki T., Imura N. (1987) Toxicol. Appl. Pharmacol. 88, 322–328 [DOI] [PubMed] [Google Scholar]
- 20. Watanabe C., Yin K., Kasanuma Y., Satoh H. (1999) Neurotoxicol. Teratol. 21, 83–88 [DOI] [PubMed] [Google Scholar]
- 21. Hirota Y., Yamaguchi S., Shimojoh N., Sano K. (1980) Toxicol. App. Pharmacol. 53, 174–176 [DOI] [PubMed] [Google Scholar]
- 22. Usuki F., Yasutake A., Matsumoto M., Higuchi I. (2001) J. Health Science 47, 162–167 [Google Scholar]
- 23. Farina M., Soares F. A., Zeni G., Souza D. O., Rocha J. B. (2004) Toxicol. Lett. 146, 227–235 [DOI] [PubMed] [Google Scholar]
- 24. Carvalho C. M., Chew E. H., Hashemy S. I., Lu J., Holmgren A. (2008) J. Biol. Chem. 283, 11913–11923 [DOI] [PubMed] [Google Scholar]
- 25. Hatfield D. L., Gladyshev V. N. (2002) Mol. Cell. Biol. 22, 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamashita A., Kashima I., Ohno S. (2005) Biochim. Biophys. Acta 1754, 305–315 [DOI] [PubMed] [Google Scholar]
- 27. Usuki F., Yamashita A., Kashima I., Higuchi I., Osame M., Ohno S. (2006) Mol. Ther. 14, 351–360 [DOI] [PubMed] [Google Scholar]
- 28. Yamashita A., Izumi N., Kashima I., Ohnishi T., Saari B., Katsuhata Y., Muramatsu R., Morita T., Iwamatsu A., Hachiya T., Kurata R., Hirano H., Anderson P., Ohno S. (2009) Genes Dev. 23, 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohnishi T., Yamashita A., Kashima I., Schell T., Anders K. R., Grimson A., Hachiya T., Hentze M. W., Anderson P., Ohno S. (2003) Mol. Cell 12, 1187–1200 [DOI] [PubMed] [Google Scholar]
- 30. Moriarty P. M., Reddy C. C., Maquat L. E. (1998) Mol. Cell. Biol. 18, 2932–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S. (2006) Genes Dev. 20, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagy E., Maquat L. E. (1998) Trends Biochem. Sci. 23, 198–199 [DOI] [PubMed] [Google Scholar]
- 33. Lu J., Zhong L., Lönn M. E., Burk R. F., Hill K. E., Holmgren A. (2009) FASEB J. 23, 2394–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mustacich D., Powis G. (2000) Biochem. J. 346, 1–8 [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar S., Björnstedt M., Holmgren A. (1992) Eur. J. Biochem. 207, 435–439 [DOI] [PubMed] [Google Scholar]
- 36. Björnstedt M., Hamberg M., Kumar S., Xue J., Holmgren A. (1995) J. Biol. Chem. 270, 11761–11764 [DOI] [PubMed] [Google Scholar]
- 37. Björnstedt M., Xue J., Huang W., Akesson B., Holmgren A. (1994) J. Biol. Chem. 269, 29382–29384 [PubMed] [Google Scholar]
- 38. Sies H. (1993) Free Rad. Biol. Med. 14, 313–323 [DOI] [PubMed] [Google Scholar]
- 39. Zhao R., Masayasu H., Holmgren A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8579–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao R., Holmgren A. (2002) J. Biol. Chem. 277, 39456–39462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.