Abstract
Diabetic tissues are enriched in an “activated” form of human aldose reductase (hAR), a NADPH-dependent oxidoreductase involved in sugar metabolism. Activated hAR has reduced sensitivity to potential anti-diabetes drugs. The C298S mutant of hAR reproduces many characteristics of activated hAR, although it differs from wild-type hAR only by the replacement of a single sulfur atom with oxygen. Isothermal titration calorimetry measurements revealed that the binding constant of NADPH to the C298S mutant is decreased by a factor of two, whereas that of NADP+ remains the same. Similarly, the heat capacity change for the binding of NADPH to the C298S mutant is twice increased; however, there is almost no difference in the heat capacity change for binding of the NADP+ to the C298S. X-ray crystal structures of wild-type and C298S hAR reveal that the side chain of residue 298 forms a gate to the nicotinamide pocket and is more flexible for cysteine compared with serine. Unlike Cys-298, Ser-298 forms a hydrogen bond with Tyr-209 across the nicotinamide ring, which inhibits movements of the nicotinamide. We hypothesize that the increased polarity of the oxidized nicotinamide weakens the hydrogen bond potentially formed by Ser-298, thus, accounting for the relatively smaller effect of the mutation on NADP+ binding. The effects of the mutant on catalytic rate constants and binding constants for various substrates are the same as for activated hAR. It is, thus, further substantiated that activated hAR arises from oxidative modification of Cys-298, a residue near the nicotinamide binding pocket.
Keywords: Calorimetry, Crystal Structure, Diabetes, Enzyme Catalysis, Enzyme Structure, Enzyme Kinetics, Enzyme Mutation, Enzymes, Protein Structure, X-ray Crystallography
Introduction
The aldo-keto reductases constitute a superfamily of NADPH-dependent oxidoreductases (1) that catalyze the reduction of a wide variety of aldehydes and ketones to their corresponding alcohols. The aldo-keto reductase family member aldose reductase (AR)2 is implicated in the pathogenesis of a variety of diabetic complications including retinopathy, neuropathy, nephropathy, and cardiovascular diseases (2–7). Hence, AR has long been studied as a target for the design of potent active site inhibitors for the treatment of diabetes (8).
Human AR (hAR) exists in both a native and an activated form, the later of which poses a potential obstacle to the development of diabetes drugs. Kinetic studies of activated hAR revealed differences (typically increases) in Km and Vmax for aldehyde substrates and, more importantly from a physiological standpoint, a marked reduction in sensitivity to hAR inhibitors (e.g. 1000-fold increase in Ki for sorbinil, an otherwise potent active site inhibitor) (9–13). Because activated enzyme forms are generally less liable to inhibition by hAR inhibitors, the therapeutic effectiveness of such drugs would be compromised if a large fraction of the enzyme underwent conversion to this activated form as has been discovered in diabetic tissues (9).
The nature of the activated hAR form is not clearly understood but may result from oxidation of one of the six cysteines in the enzyme. Of the six cysteines found in hAR, only Cys-298 is located in sufficient proximity to the active site to directly perturb both enzyme activity and inhibitor sensitivity. Also, Cys-298 is the predominant site of thiolation on hAR, suggesting that its oxidation could be a mechanism for triggering conversion of hAR to the activated form (14). Indeed, when complexed with NADP+, hAR is less susceptible to modification by many agents, including glutathione, nitric oxide, and 4-hydroxy-2-nonenal (HNE). That is, of all the cysteine residues in hAR, only Cys-298 is sufficiently close to NADP+ to be shielded from modification.
Chemical modification and mutagenesis studies further distinguish Cys-298 as an important regulatory site on hAR. Either activation or inactivation of hAR can be effected by chemical modification of Cys-298. For example, the addition of oxidized glutathione results in glutathiolation of Cys-298 (14, 15) and renders the enzyme catalytically inactive (14), whereas dithiodiethanol also targets Cys-298 but leads to hAR activation (16). Furthermore, as a consequence of abolishing the potential for thiolation at residue 298, C298S and C298A mutants are rendered resistant to modification with reagents that are known to cause functional changes in the enzyme activity of the wild-type hAR (16, 17). Such an effect would be expected only if Cys-298 were directly involved in activating hAR.
Last, modification of Cys-298 by thiolation recapitulates another key characteristic of the activated enzyme, the reduction of hAR sensitivity to inhibitors. Such is the case for the HNE, which targets the modification of Cys-298 (18).
Presuming that thiolation of Cys-298 is indeed the trigger for conversion of hAR to the activated form and that its potential for thiolation is blocked by NADP(H), it follows that the enzyme could be converted to the activated form only during a catalytic cycle when nucleotide exchange occurs. In the reduction of aldehyde (forward reaction), AR follows a sequential ordered kinetic mechanism in which the co-factor NADPH binds before the aldehyde substrate and NADP+ is discharged after release of the alcohol product (19, 20). Due to tight binding affinity, most endogenous hAR is anticipated to exist as a complex with NADP(H) in vivo. Under steady state conditions, 90–95% of the enzyme will be present as an enzyme-nucleotide binary complex (21). Consequently, endogenous regulators that act through Cys-298 should be most effective when substrate levels rise to a sufficient level to stimulate catalytic turnover.
Previous studies have reported changes in relative specificity of hAR substrates caused by the C298S mutation (22). In the current study we report the effects of this mutation on additional substrates, including a thermodynamic and structural characterization of the C298S mutant. We suggest how structural changes caused by the mutation may explain the characteristics of activated hAR.
EXPERIMENTAL PROCEDURES
Production of Recombinant Wild-type and C298S hAR
His-tagged recombinant wild-type and C298S mutant hAR were expressed in Escherichia coli BL21 cells that were grown in Luria-Bertani broth containing 50 mg/liter ampicillin with constant shaking in rotary shaker at 240 rpm to reach the A600 between 0.6 and 0.8 at 37 °C. The protein expression was induced by supplementing 1 mm isopropyl-1-thio-galactopyranoside in the culture medium. The cells were harvested after 3–4 h by centrifugation (6000 × g, 10 min) and resuspended in 50 mm sodium phosphate buffer (pH 7.0) containing 300 mm NaCl and 1 mm 2-mercaptoethanol and lysed by ultrasonication. The hAR was isolated from the lysate separated by centrifugation at 10,000 × g for 1.0 h at 4 °C. The supernatant containing hexa-His-hAR was incubated for 1–2 h by constant gentle mixing with Talon metal affinity matrix (Clontech, Mountain View, CA), and later matrix slurry was passed through column and washed with 50 mm sodium phosphate buffer (pH 7.0) having 300 mm NaCl and 1.0 mm 2-mercaptoethanol. The protein was eluted with 150 mm imidazole in 50 mm sodium phosphate buffer (pH 7.0) containing 300 mm NaCl and 1 mm 2-mercaptoethanol and dialyzed in the 50 mm sodium phosphate buffer (pH 7.0) containing 1 mm 2-mercaptoethanol. The His tag was removed by thrombin cleavage (Novagen) as per the manufacturer's instructions, and the hAR protein was further purified by anion exchange on a DEAE Sephadex A25 column by binding with DEAE Sephadex A 25 matrix. The concentration of hAR proteins was determined by the Bradford assay (Bio-Rad) (23), the purity was assessed by SDS-PAGE, and the enzyme activity was determined by using 10 mm dl-glyceraldehyde and 0.15 mm NADPH as the substrate and cofactor, respectively.
Aldehyde Reduction Kinetics of Wild-type and C298S Mutant hAR
Aldehyde reduction activity of wild-type and C298S mutant hAR at 25 °C were monitored by UV spectrophotometry by a measuring decrease in the absorbance of the cofactor NADPH at 340 nm (24, 25). The assay was carried out in 10 mm potassium phosphate buffer (pH 6.2) using 0.15 mm NADPH, 0.5 μm hAR, and varied concentrations of dl-glyceraldehyde, d-glucose, and HNE. The unit of enzyme activity is defined as μmol of NADPH oxidized/min.
Alcohol Oxidation Kinetics of Wild-type and C298S Mutant hAR
The alcohol oxidation by hAR wild-type and C298S mutant proteins has been studied with benzyl alcohol as the substrate and 3-acetylpyridine adenine dinucleotide phosphate (3-APADP+) as the cofactor. The oxidation of benzyl alcohol was monitored spectrophotometrically at 25 °C as the increase in the absorbance of 3-APADP+ at 363 nm (25, 26). The assay was carried out in 0.125 m MES-Tris-HEPES (0.04 m each component) triple buffer (pH 8.5) with 0.5 μm of hAR, 0.10 mm 3-APADP+ and at varying concentrations of benzyl alcohol. The unit of enzyme activity is defined as μmol of 3-APADP+ reduced/min.
Data Analysis of the Enzyme Kinetics
The aldehyde reduction and alcohol oxidation rates for wild-type and C298S mutant hAR were analyzed according to the Michaelis-Menten model for enzyme kinetics. The Michaelis-Menten constant Km, the maximum velocity, Vmax, for the substrates for oxidation, and reduction reactions catalyzed by AR were determined by plotting rate of carbonyl reduction, v, versus substrate concentration, S, using the Equation 1.
The catalytic turnover number, kcat, was determined from the ratio of Vmax/[E], where [E] is the molar concentration of enzyme (from molecular weight and the weight/volume concentration).
Isothermal Titration Calorimetry
The thermodynamic binding parameters consisting of the binding constant, the binding enthalpy, and the binding entropy were determined from analysis of ITC experiments on wild-type and C298S-mutated hAR interacting with the co-factors NADPH and NADP+. For all the ITC measurements, the solution vessel of the ITC instrument was filled up with 1.43 ml of the hAR solution at concentrations ∼10–20× lower than the co-factor concentrations in the stirrer syringe. Before loading in the ITC cell the protein solution was dialyzed against the dialysis buffer consisting of 0.1 m potassium phosphate buffer (pH 7.0) with 1 mm EDTA and 1 mm DTT. The dialysis buffer was used in the reference vessel of the ITC as well as for rinsing of the vessels during the experiment. The co-factor solution was first titrated into the dialysate to determine any heats of dilution of the co-factor solution. Then the solution vessel in the ITC was rinsed thoroughly several times with the buffer and filled with the hAR solution. 5–10-μl aliquots of the co-factor solution were added to the hAR solution until well past saturation of the hAR binding sites as evident by the appearance of titration peaks the same as those of the dilution titration peaks. The analysis of the titration of the co-factor solution into the hAR solution consisted of first subtracting any average heat of dilution of the co-factor solution from the binding isotherm and then fitting the resulting binding isotherm to a single site binding model (27). Values of the binding free energy change (ΔbGo) and the binding entropy (ΔbSo) were then determined from the fundamental Equation 2 of thermodynamics,
The heat capacity change for the binding reaction, ΔbCp, is determined from the slope of the assumed linear dependence of the binding enthalpy on temperature. Each ITC reaction was performed at least twice, and thus, the uncertainties are reported as the S.D. between the Kb and ΔbH values. ITC experiments for binding of cofactors to C298S mutant hAR were performed following the procedures described above with the use of purified recombinant C298S mutant hAR instead of wild-type hAR.
Crystal Structure Determination
Purified wild-type as well as the C298S mutant hAR were concentrated by ultrafiltration (Amicon YM-10 membrane) to ∼12 mg/ml. Protein solutions were mixed with NADP+ solution to achieve a molar ratio of 1:3 for the protein to cofactor. Protein was further concentrated to ∼40 mg/ml (as determined using the molar extinction coefficient and absorbance readings at 280 nm wavelength) in 25 mm citrate (pH 5.3) and 1 mm β-mercaptoethanol. Crystals were produced using the hanging drop vapor diffusion method. Two μl of protein solution were pipetted onto a siliconized glass coverslip, and an equal volume of reservoir solution was pipetted on top of the protein drop. The reservoir solution contained 2.04 m ammonium sulfate, 7% (w/v) polyethylene glycol 400, and 0.1 m HEPES (pH 7.5). Crystals were briefly transferred to a solution containing the reservoir solution supplemented with 35% glycerol and flash-cooled by plunging them into liquid nitrogen. The preliminary diffraction data set from 20 to 1.8 Å resolution for the wild-type and C298S mutant hAR were collected on a Rigaku R-Axis-IV++ area detector mounted on a rotating anode generator equipped with a graphite monochromator and operated at 50 kV and 90 mA. The indexing of this data set indicated the crystals belong to P212121 space group containing two enzyme molecules in the asymmetric unit. For the wild-type enzyme, 2.1 Å data were collected at 100 K using beamline 9-1 at the Stanford Synchrotron Radiation Laboratory with an exposure time of 45 s per 1° frame, a 200-mm crystal-to-detector distance, and wavelength of 0.9795 Å. For the C298S mutant, 1.55 Å data were collected at 100 K using beamline 8.2.2 at the Advanced Light Source with an exposure time of 8 s per 1° frame, a 200-mm crystal-to-detector distance, and wavelength of 1.000 Å. Both beamlines utilized are equipped with a ADSC Quantum 315 CCD detector. The data were processed and scaled with Denzo/Scalepack (28) to an Rmerge of 11.1% and 6.2% for wild-type and C298S mutant hAR, respectively. Initial phases of the binary complex of wild-type hAR were obtained by molecular replacement method using the program EPMR (29) and the refined 1.8 Å holoenzyme (1AZ1) (excluding solvent molecules and ligands) as a search model. Initial rigid-body refinement of the protein atoms followed by repeated cycles of conjugate gradient energy minimization, simulated annealing, and B-factor refinement were performed using the CNS program package (30). For model building, electron density maps with 2Fo − Fc and Fo − Fc coefficients were visualized with the program COOT (31). The presence of clear electron density allowed the positioning of the cofactor as well as the solvent molecules. The program REFMAC (32) was employed at the last stages of the refinement of the final structure containing the cofactor, protein, and solvent molecules. The refined structure of wild-type protein without solvent molecules was used as the starting model for the refinement of the isomorphous C298S mutant hAR crystal structure. After initial rigid-body and simulated annealing refinements, electron density maps with 2 Fo − Fc and Fo − Fc coefficients were employed to rebuild the mutant protein structure and to locate the cofactor and solvent molecules following the procedures described above. Both the final models were validated with the following structure validation tools: PROCHECK (33), ERRAT (34), and VERIFY3D (35). For both the wild-type and the C298S models, 91% of the residues lie in the most favored regions of the Ramachandran plot, and 9% of the residues lie in the additionally favored regions. Both wild-type and C298S models scored 96.5% in Errat. Details of the data collections and the refinement statistics are shown in Table 4.
TABLE 4.
Crystallographic statistics of wild-type and C298S mutant hAR structures
| Structures | Wild-type-NADP+ | C298S-NADP+ |
|---|---|---|
| Data collection, processing, and structure refinement | ||
| Wavelength (Å) | 0.9795 | 1.000 |
| Space group | P212121 | P212121 |
| Unit cell parameters | ||
| a, b, c (Å) | 83.9, 86.1, 105.1 | 83.7, 86.2, 104.4 |
| α, β, γ (°) | α = β = γ = 90 | α = β = γ = 90° |
| Diffraction data | ||
| Resolution range (Å) | 100-2.1 | 90-1.55 |
| Unique reflections | 45,182 (4,373) | 105,836 (10,039) |
| Rmerge (square) (%) | 11.1 (31.9) | 6.2 (48.2) |
| Completeness (%) | 99.3 (98.2) | 96.6 |
| Redundancy | 6.2 (5.1) | 5.3 (5.2) |
| I/σ(I) | 10.4 (1.9) | 18.0 (2.9) |
| Refinement | ||
| Resolution range used in refinement (Å) | 66-2.1 | 1.5 |
| Reflections used in refinement (work/free) | 42,785 (3,019) | 100,472 (7,134) |
| Final R values (work/free) (%) | 21.0 (31.6)/26.3 (37.6) | 17.0 (24.6)/18.9 (30.0) |
| Protein atoms | 5,064 | 5,079 |
| Cofactor atoms | 96 | 96 |
| Water molecules | 341 | 719 |
| Sulfate atoms | 45 | 40 |
| r.m.s. deviation values | ||
| Bonds (Å) | 0.010 | 0.010 |
| Angles (°) | 1.3 | 1.4 |
RESULTS
Comparison of the Wild-type and C298S Mutant hAR Kinetic Parameters
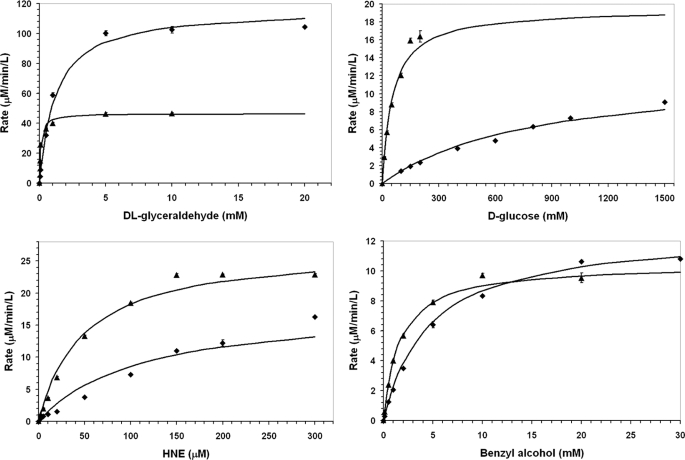
Typical results from enzyme assays on the reduction kinetics of wild-type and C298S mutant hAR for various aldehyde substrates and benzyl alcohol were performed two-three times. Values for Km and kcat were determined from the results in the initial linear increase in the rate of product formation using Equation 1 to determine Km and the ratio of Vmax/[hAR] to calculate kcat and are shown in Fig. 1 and summarized in Table 1. The comparison of the kinetic parameters of the wild-type and the C298S mutant hAR indicates that the substitution of serine for cysteine at position 298 reduces the affinity of the protein (1/Km) for dl-glyceraldehyde, d-glucose, and HNE by about 12, 15, and 2-fold, respectively. Overall this mutation doubled the catalytic constant (kcat) for dl-glyceraldehyde but reduced by 34% for HNE and d-glucose. This mutation also produces about a 3-fold decrease in the affinity of reverse reaction substrate benzyl alcohol and slightly increased the oxidation activity compared with wild-type enzyme.
FIGURE 1.
Michaelis-Menten kinetics forward and reverse reactions catalyzed by hAR. Comparison of wild-type (•) and C298S mutant (▴) hAR reduction kinetics for various aldehyde substrates; dl-glyceraldehyde, d-glucose, and HNE and oxidation kinetics for benzyl alcohol, respectively.
TABLE 1.
Comparison of catalytic activity of wild-type and C298S mutant hAR for carbonyl reduction and alcohol oxidation reactions at 25 °C
The uncertainties are S.D. of the average value.
| hAR Protein | Substrate | Km | kcat | kcat/Km |
|---|---|---|---|---|
| s−1 | ||||
| Wild-type | dl-Glyceraldehyde | 0.1 ± 0.01 mm | 1.55 ± 0.01 | 14.5 mm−1s−1 |
| C298S | 1.2 ± 0.05 mm | 3.72 ± 0.3 | 3.10 mm−1s−1 | |
| Wild-type | d-Glucose | 55.0 ± 6.0 mm | 0.64 ± 0.05 | 0.011 mm−1s−1 |
| C298S | 800 ± 50.0 mm | 0.42 ± 0.04 | 0.0005 mm−1s−1 | |
| Wild-type | HNE | 48.0 ± 4.0 μm | 0.90 ± 0.06 | 0.019 μm−1 s−1 |
| C298S | 110.0 ± 8.0 μm | 0.60 ± 0.02 | 0.006 μm−1 s−1 | |
| Wild-type | Benzyl alcohol | 1.62 ± 0.1 mm | 0.35 ± .025 | 0.215 mm−1s−1 |
| C298S | 4.55 ± 0.3 mm | 0.42 ± .02 | 0.092 mm−1s−1 |
Thermodynamics of Cofactor Binding to Wild-type and C298S Mutant hAR
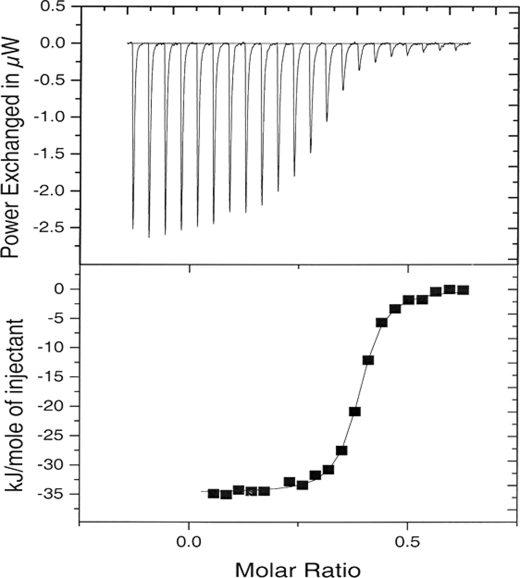
A typical binding scan of a co-factor solution titrated into a hAR solution is shown in Fig. 2. The titration consisted of 5-μl aliquots of a 0.292 mm NADP+ solution titrated into a solution of 0.051 ± 0.005 mm of wild-type hAR (pH 6.5) at 25 °C. The thermodynamic parameters from a fit of a single site binding model to the data are presented as part of the average under these conditions in Table 2. As shown in Table 2, the binding reactions are all enthalpy-driven, and the enthalpies decrease with increase in temperature. The binding constants of the NADPH-wild-type hAR complexes are higher than those of the NADPH-C298S mutant hAR complexes, whereas the binding constants of the NADP+ complexes are almost the same at all temperatures regardless of the enzyme form. The binding heat capacity changes were determined as the slope of a linear plot of −ΔbH versus T and are presented in Table 3. The binding heat capacity change for formation of the NADPH-C298S hAR complex is twice as large as that of the NADPH-wild-type hAR complex. However, the heat capacity change for binding of NADP+ to the C298S hAR mutant is the same as that of the NADP+ binding to the wild-type hAR and NADPH binding to the wild-type hAR (Table 3).
FIGURE 2.
ITC titration of 5-μl aliquots of a 0.29 mm NADP+ solution into a solution of 0.051 ± 0.005 mm of wild-type hAR (pH 6.5) and 25 °C, shown in top panel. The binding isotherm for the titration (■) along with a least squares fit of a one to one binding model to the binding isotherm (—). μW, microwatts.
TABLE 2.
The thermodynamics of cofactors binding to wild-type and to its C298S mutant hAR (pH 6.5) as a function of temperature
ITC experiments conducted for the NADPH binding at 15 °C revealed weak binding, which is supported by the close to zero binding enthalpy, i.e. −38.9 to −4.00 (15–25) = 0, produced by extrapolating the binding enthalpy of −38.9 kJ/mol for NADPH at 25 °C down to 15 °C. The uncertainties are S.D. of the average value.
| Protein | Cofactor | Kb | −ΔbG | −ΔbH | TΔbS |
|---|---|---|---|---|---|
| m−1 | kJ mol−1 | kJ mol−1 | kJ mol−1 | ||
| At 15 °C | |||||
| Wild-type | NADP+ | 16.7 ± 2.1 × 106 | 39.8 ± 0.3 | 17.7 ± 1.1 | 22.1 ± 1.1 |
| C298S | NADP+ | 11.6 ± 1.1 × 106 | 34.0 ± 0.3 | 21.0 ± 2.9 | 13.0 ± 2.9 |
| At 25 °C | |||||
| Wild-type | NADP+ | 9.2 ± 2.1 × 106 | 39.7 ± 0.6 | 36.3 ± 1.4 | 3.4 ± 1.5 |
| C298S | NADP+ | 10.4 ± 1.2 × 106 | 40.1 ± 0.3 | 40.2 ± 0.7 | 0 |
| Wild-type | NADPH | 6.6 ± 1.6 × 106 | 38.9 ± 0.6 | 32.8 ± 3.2 | 6.1 ± 3.3 |
| C298S | NADPH | 3.0 ± 1.3 × 106 | 37.0 ± 1.1 | 36.7 ± 3.8 | 0 |
| At 35 °C | |||||
| Wild-type | NADP+ | 7.6 ± 1.0 × 106 | 40.6 ± 0.3 | 68.2 ± 5.7 | −27.6 ± 5.7 |
| C298S | NADP+ | 8.0 ± 0.7 × 106 | 40.7 ± 0.2 | 64.6 ± 2.5 | −23.9 ± 2.5 |
| Wild-type | NADPH | 7.8 ± 0.2 × 106 | 40.7 ± 0.1 | 51.2 ± 5.4 | −10.5 ± 5.4 |
| C298S | NADPH | 1.1 ± 0.3 × 106 | 35.7 ± 0.4 | 79.5 ± 9.8 | −43.8 ± 9.8 |
TABLE 3.
Heat capacity changes for the binding of the co-factors to wild-type and its C298S mutant hAR
The uncertainties are S.D. of the average value.
| System | −ΔCP |
|---|---|
| kJ mol−1K−1 | |
| Wild-type hAR + NADP+ | 2.52 ± 0.38 |
| Wild-type hAR + NADPH | 1.84 ± 0.26 |
| C298S hAR + NADP+ | 2.18 ± 0.15 |
| C298S hAR + NADPH | 4.00 ± 0.54 |
Structure of Wild-type hAR
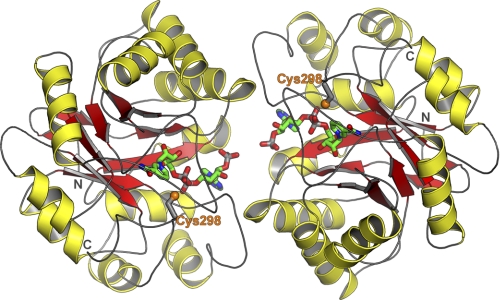
The binary complex structure of the wild-type hAR with cofactor NADP+ has been determined in a new crystal form with two molecules in the asymmetric unit (Fig. 3). Crystallographic parameters for the hAR structures reported here are shown in Table 4. Protein molecules, namely molA and molB, are related by a non-crystallographic rotation of 180° in the wild-type binary complex structure. The dimeric form of the hAR-NADP+ binary complex structure was refined to 2.1 Å resolution with the final R factor of 21.0% and Rfree of 26.3%. The refined final structure contains a total of 630 protein residues corresponding to 315 amino acids for each protein molecule, 2 cofactors NADP+ (Fig. 4), and 341 water sites. The r.m.s. deviation between the 315 CA atoms of molA and molB is 0.15 Å in the wild-type hAR binary complex structure. All dihedral angles of the protein backbone fall in the most favored (91%) and additionally allowed (9%) regions of the Ramachandran plot as defined in the program PROCHECK (33). The mean B values for the protein, cofactor/sulfate, and water atoms are 23, 31, and 28 Å2, respectively.
FIGURE 3.
Structure of hAR binary complex. Shown is the crystal structure of hAR complexed with NADP+ in the new crystal form with two molecules in the asymmetric unit.
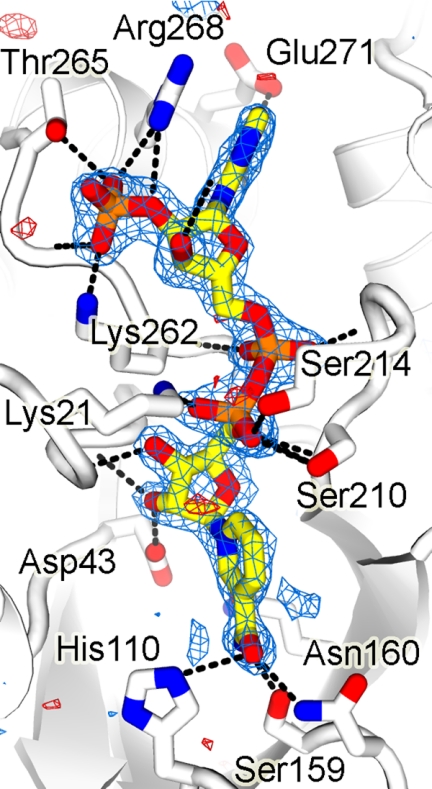
FIGURE 4.
Wild-type omit map corresponding to NADP+. In the refined structure, cofactor is bound to wild-type hAR in the extended conformation through a network of polar interactions, OP1···OG of Ser-263, OP1···G1 of Thr-265, OP2···NZ of Lys-262, OP3···HN of W128, OP3···HN of W49, O2*···HN of W128, O3*···OE1 of Gln-26, N7···ND2 of Asn-272, N1···HN of W-35, N6···OD1 of Asn-272, N6···OE2 of Glu-272, O1···N of Ser-214, O1···N of Leu-212, O2···N of Lys-262, O1···NZ of Lys-21, NO2···OG of Ser-210, O2*···OD2 of Asp-43, O3*···N of Trp-20, N7···OE1 of Gln-183, N7···OG of Ser-159, and O7···ND2 of Asn-160 and a number of van der Waals contacts. Labels OP1, OP2, and OP3 denote oxygen atoms of phosphate group in NADP+; O2* and O3* indicate hydroxyl group oxygen atoms O2' and O3' in NADP+; HN, N, and CA are main chain and OG, ND2, OD1, OD2, OE1, OE2, and NZ represent the side chain gamma oxygen, delta nitrogen, delta oxygen, epsilon oxygen, and zeta nitrogen atom(s) of the corresponding amino acid residues.
Structural Comparisons of Wild-type and C298S Mutant hAR Complexes with NADP+
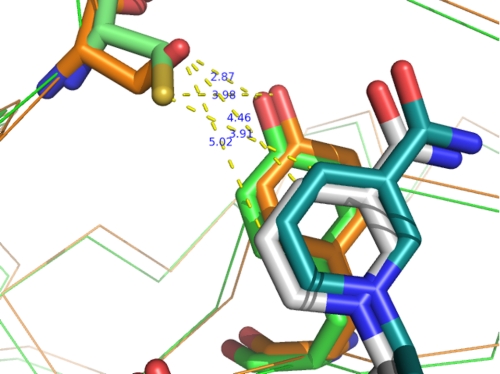
The binary complex structure of C298S mutant hAR with NADP+ was determined to 1.55 Å resolution with the final R factor of 17.0% and Rfree of 18.9%. It is isomorphous with the wild-type crystal form reported in this study. There are a total of 630 protein residues, 2 NADP+ (Fig. 5) molecules, and 719 water molecules in the dimer of the refined mutant structure. Monomers, molA and molB, are related by a rotation of 180° in the C298S mutant hAR binary complex structure. The r.m.s. deviation between the 315 CA atoms of molA and molB is 0.07 Å in the C298S mutant hAR binary complex structure. The r.m.s. deviation between the CA atoms of the dimer (630) and the monomers (315) of the wild-type with the dimer and the monomers of C298S mutant hAR are 0.29 and 0.16 Å, respectively. Superimposition of the wild-type and C298S mutant hAR structures indicate that there are a few regions with a noticeable difference between their conformations. One such region corresponds to movements of 0.5 Å in the main-chain conformation of the loop between residues 124 and 130 in molA and molB. In another region, residue Cys-298 of the wild-type structure sits on the edge of the active site with its side chain oriented to form van der Waals interactions with the C4 and C5 atoms of the nicotinamide group (about 3.9 Å distance). In the C298S mutant, the Ser-298 side chain adopts a different conformation compared with the Cys-298 side chain in the wild-type hAR structure, allowing the OH group of Ser to form hydrogen bonds with the OH group of Tyr-209 (Fig. 6) and main chain O of Ser-298. Also, the OH group of Ser-298 is 4.5 and 5.0 Å away from the C4 and C5 atoms of the nicotinamide ring in the C298S mutant structure. Most interactions found across the dimer interface of the wild-type (Table 5) binary complex are maintained in the C298S binary complex with minor differences in their distances between the atoms.
FIGURE 5.
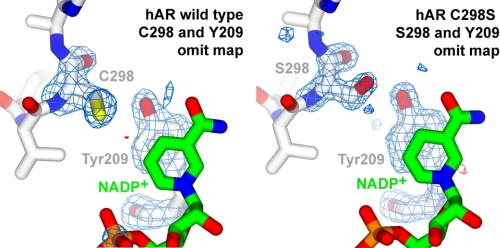
Omit map corresponding to Tyr-209 in wild-type and C298S mutant hAR. Omit maps of wild-type and C298S mutant hAR corresponding to the omitted regions of the residues 298 and 209 are shown. The maps were calculated with CNS simulated annealing algorithm of the coordinates without the residues 298 and 209, and Fo − Fc omit maps are contoured at ±4σ.
FIGURE 6.
Superposition of wild-type (brown) and C298S (green) hAR. The side chain for residues Cys-298 (green), Ser (brown), and their interactions with residue Tyr-209 and nicotinamide ring of NADP+ in the wild-type and C298S mutant hAR is shown.
TABLE 5.
Dimer contacts in wild-type and C298S mutant hAR
| molA |
molB |
Distance | ||
|---|---|---|---|---|
| Residue | Atom | Residue | Atom | |
| Å | ||||
| Pro-3 | CB | Asp-216 | O | 3.55 |
| CG | Asp-216 | O | 3.69 | |
| Pro-4 | CG | Pro-218 | O | 3.50 |
| CD | Pro-218 | O | 3.74 | |
| CB | Ala-220 | O | 3.50 | |
| Gly-5 | C | Pro-215 | O | 3.85 |
| CA | Pro-215 | O | 3.86 | |
| CA | Ala-220 | CB | 3.79 | |
| N | Ala-220 | CB | 3.75 | |
| Gln-6 | NE2 | Asp-216 | CB | 3.31 |
| OE1 | NADP318 | AO3* | 3.16 | |
| CG | Pro-215 | O | 3.45 | |
| Glu-9 | OE1 | Glu-229 | OE2 | 3.93 |
| Asn-2 | ND2 | Pro-222 | CG | 3.96 |
| Glu-3 | OE2 | Pro-222 | CD | 3.69 |
| OE2 | Pro-222 | CG | 3.63 | |
| Pro-15 | O | Gln-26 | CG | 3.62 |
| CG | Gln-26 | OE1 | 3.95 | |
| Asp-16 | CB | Gln-26 | CD | 3.98 |
| O | Pro-23 | CB | 3.54 | |
| O | Pro-23 | CG | 3.66 | |
| N | Gln-26 | OE1 | 4.04 | |
| CB | Gln-26 | NE2 | 3.43 | |
| CB | Pro-23 | CG | 3.87 | |
| CA | Pro-23 | CG | 3.90 | |
| CA | Gln-26 | CG | 4.00 | |
| Ala-220 | CB | Gly-25 | CA | 3.78 |
| Pro-222 | CG | Asn-52 | ND2 | 3.74 |
| Glu-229 | OE2 | Glu-29 | OE1 | 4.06 |
| Thr-265 | CG2 | Arg-268 | NH2 | 3.62 |
| Arg-268 | NH2 | Thr-265 | CG2 | 3.80 |
| NADP318 | AO3* | GLN26 | OE1 | 3.27 |
Cofactor Binding and Conformation
There are two molecules of NADP+ bound to each molecule of the dimer of the wild-type as well as to the C298S mutant hAR with the r.m.s. deviations between 48 cofactor atoms in molA and 48 atoms in molB being 0.18 and 0.03 Å, respectively. The extended conformation of NADP+ found in the wild-type dimer is very similar to that seen in the C298S mutant hAR with the r.m.s. deviation of 0.13 Å. The detailed interactions between the cofactor and the protein atoms are shown in Fig. 4 and Table 6. There is a water (W49) molecule that forms hydrogen bonding interactions between the APO3 atoms of molA (2.73 Å) and molB (2.54 Å) in wild-type and C298 structures.
TABLE 6.
Cofactor interactions with the atoms of wild-type, C298S mutant hAR, and solvent
B, second molecule (molB) of the dimer; W, water molecule.
| NADP atoms | Dimeric hAR |
Distance | |
|---|---|---|---|
| Atom | Residue | ||
| Å | |||
| AOP1 | OG | Ser-263 | 2.67 |
| OG1 | Thr-265 | 2.64 | |
| AOP2 | NZ | Lys-262 | 2.63 |
| AOP3 | H | W132 | 2.79 |
| H | W49 | 2.54 | |
| AO2* | H | W132 | 2.9 |
| AO2* | NH1 | Arg-268 | 3.18 |
| AO3* | OE1 | BGln-26 | 3.16 |
| AO3* | OD2 | Asp-216 | 3.12 |
| AN7 | ND2 | Asn-272 | 3.07 |
| AN1 | H | W35 | 2.55 |
| AN6 | OD1 | Asn-272 | 2.93 |
| OE2 | Glu-272 | 2.81 | |
| AO1 | N | Ser-214 | 3.12 |
| N | Leu-212 | 2.86 | |
| AO2 | N | Lys-262 | 2.97 |
| NO1 | NZ | Lys-21 | 2.83 |
| NO2 | OG | Ser-210 | 2.76 |
| NO5* | N | Ser-210 | 3.07 |
| NO2* | OD2 | Asp-43 | 2.58 |
| NO3* | N | Trp-20 | 2.88 |
| NO3* | N | Thr-19 | 3.11 |
| NN7 | OE1 | Gln-183 | 3.09 |
| OG | Ser-159 | 2.80 | |
| NO7 | ND2 | Asn-160 | 2.79 |
Comparison with Other AR Structures
Previously binary complex structure of hAR in orthorhombic crystal form with only one molecule in the asymmetric unit was reported (36). The r.m.s. deviation between the hAR structure (PDB code 1ADS) in the monomeric orthorhombic form and the molA or molB of the dimeric form of the wild-type hAR in the current crystal form is 0.5 Å over 315 CA atoms. Corresponding r.m.s. deviation between PDB code 1ADS and the molA or molB of the current dimer form of the C298S mutant hAR is 0.70 Å over 315 CA atoms.
DISCUSSION
hAR follows a classic sequential kinetic mechanism with the cofactor NADPH binding to the enzyme first followed by binding of the aldehyde substrate to the hAR-NADPH binary complex; a redox reaction then occurs,
and the product is released and followed by the ejection of the oxidized co-factor, NADP+. Most endogenous hAR is already bound with NADPH, and this complexed form is the precursor of the catalytic step. Binding of NADPH induces a re-orientation of a surface loop consisting of residues 213–217, and this re-orientation appears to be the rate-limiting step in the enzyme kinetics. An important question centers on the nature of an activated form of hAR that typically increases Km for binding of the substrate and exhibits a reduction in sensitivity to drug (inhibitors) binding to the substrate site. A likely explanation for the altered kinetics of the activated form involves modification of Cys-298. Cys-298 is implicated by its proximity to the substrate binding site and the observation that its conversion to Ser resulted in an activated form that is resistant to modification by reagents known to cause functional changes in enzyme activity of the wild-type hAR (16, 17). This study reports the differences caused by mutation of Cys-298 to Ser-298 in the kinetic parameters of substrates, glucose, glyceraldehyde, HNE and benzyl alcohol. The ITC results show that mutation of Cys-298 to Ser reduces the binding constant of NADPH while not significantly affecting the binding affinity of NADP+ and indicates that Cys-298 plays a role in the catalytic process of the reduction of the physiological metabolites tested.
The decreased binding constants observed by ITC measurements are consistent with increased Km values for the corresponding complexes reported in this study. In the Michaelis-Menten enzyme kinetic model, values for 1/Km are considered to reflect the binding constants of the substrate to the catalytic form, and these values are lower for all the substrates binding to the C298S mutant of hAR, including benzyl alcohol, which is a substrate for reverse reaction. This feature could be explained by the lower binding strength of NADPH to the Cys-298 mutant as this is the first step in the sequential enzyme mechanism of hAR catalysis. This trend indicates that the strength of the binary complexes for the forward and reverse reactions are weaker for the C298S mutant compared with that for the wild-type enzyme. Consequently the weaker binary complex binds the substrate less tightly in the Cys-298 mutant than the wild-type in the forward and reverse reactions. Notably, the decreases in 1/Km are dependent on the nature of the substrate and are manifested in their reduction potential from 12-, 14.5-, 2.3-, and 2.8-fold for dl-glyceraldehyde, d-glucose, HNE, and benzyl alcohol, respectively. Similarly, a reduction in the binding affinities of drug (inhibitors) to the substrate binding site would then also be expected for the C298S hAR mutant as the drug (inhibitors) mimic the structures of the substrates and or aim to bind at the same binding site.
Values of kcat increased by a factor of 2.4 for dl-glyceraldehyde, reduced to 66% for d-glucose and HNE, and increased for benzyl alcohol by 1.2-fold due to mutation. The variation in kcat does not compensate for the larger changes in Km, but the catalytic efficiencies defined by kcat/Km are reduced with mutation of Cys-298 to Ser. These trends are consistent with those observed in the activated form of hAR. That is, hAR is distinguished from the native enzyme by increased Km and Vmax values for aldehyde reduction. The similarity in trends supports the hypothesis that Cys-298 is key to the conversion of native enzyme to the activated form. Furthermore, when this residue is modified under physiological conditions as in the present mutagenesis study, values of Km increase for aldehyde substrates and presumably the binding of drugs (inhibitors) designed to mimic the structures of the substrates would be less potent. Such an expectation is also consistent with the characteristics of the activated form of hAR; namely, a marked reduction in sensitivity to hAR inhibitors (e.g. 1000-fold increase in Ki for sorbinil) (9–13).
The larger negative heat capacity change observed for binding of the NADPH to the C298S mutant (relative to wild-type) would include 1) contributions from hydrophobic effects where the ligand leaves the bulk water surroundings for a hydrophobic environment in the hAR binding site, 2) from dehydration of the serine, apolar amino acid residue at the binding site, and 3) from differences in the vibrational modes between the NADPH-wild-type and NADPH-C298S mutant hAR complexes. Overall differences in the heat capacity change between NADPH binding to the wild-type and C298S mutant hAR are attributed to the dissimilarity between the hydrophobic side chain of cysteine and the polar side chain of serine. However, dehydration of a hydrophobic side chain would increase a negative heat capacity change, whereas dehydration of the polar side chain of serine would increase a positive heat capacity change (37, 38). Therefore, the increase in the negative heat capacity of the NADPH-C298S mutant hAR binding reaction would more likely result from changes in the vibrational mode of the complex and not from differences between the hydrophobicity of the sites. As a result, differences in the changes in the vibrational modes of the NADPH-hAR complex seen would have arisen from re-positioning of the amino acid residues around the catalytic site by substitution of Ser for Cys-298. This may imply the positional rearrangements caused by binding of NADPH are greater in magnitude in the wild-type compared with the C298S mutant enzyme.
Evidence to support this hypothesis arises from comparison of wild-type and mutant crystal structures (Figs. 5 and 6). Structural differences due to the C298S mutation are evident near the nicotinamide binding pocket. The side chain of Cys-298 is located in a cavity formed by hydrophobic residues Trp-20, Trp-209, Trp-219, and the nicotinamide ring and is about 5 Å from these residues, but the side chain of Ser-298 has moved away from these residues. A hydrogen bond between Ser-298 and Tyr-209 is apparent in the mutant form of the enzyme but not the wild-type. Although this hydrogen bond appears compatible with binding of the planar oxidized nicotinamide ring in the crystal structure, additional conformational maneuvering might be required to accommodate the non-planar reduced nicotinamide ring. The energetic penalty for the maneuvering would be more severe in the C298S mutant because hydrogen bonding between the Tyr-209 and side chain OG of C298S makes this residue less flexible compared with the wild-type enzyme, which lacks the hydrogen bond.
In accordance with the mutation's more pronounced effect on NADPH binding relative to NADP+, the polarity of the mutated Ser-298 side chain is likely to be less compatible with the NADPH complex compared with the NADP+. In the case of the oxidized cofactor, the nicotinamide group is aromatic and primarily stabilized by π-stacking interactions with the aromatic ring of Tyr-209. However, in the case of the reduced cofactor, the nicotinamide ring is not in aromatic form and is uncharged. The increased polarity of the nicotinamide binding pocket imparted by the cysteine to serine mutation would destabilize the uncharged NADPH binding but not that of NADP+. The two-fold decrease in NADPH binding constants to C298S relative to wild-type might be due to the increased polarity of the nicotinamide binding pocket as well as the additional hydrogen bonding interaction accompanying mutation of the hydrophobic cysteine side chain to a polar hydroxyl group of the serine side chain.
The component of ΔCp that is associated with the hinge-like movement of the surface loop (213–219) that is expected to take place after NADPH binding and before NADP+ release in the wild-type may be comparable, but there may be a considerable discrepancy in these steps when C298S mutant enzyme is considered. This is because O of Val-297, OG of Ser-298, and NE1 of Trp-111 are at 2.84, 3.43, and 3.98 Å away from NE1 of Trp-219, CZ2 of Trp-111, and NO7 of NAP318, respectively, in C298S mutant, whereas the corresponding distances in the wild-type enzyme are significantly different; NE1 of Trp-111 and O of Val-297 are at 3.50 and 3.08 Å away from NO7 of NAP318 and NE1 of Trp-219. Therefore, synchronized movements of the loop 213–219 may be hindered by the modifications of residue 298 as residues 213–219, which form part of the phosphate binding region, and residue 298 are located on the cofactor binding site of the active site.
The difference between a cysteine and a serine residue might appear small with the substitution of the sulfur atom with an oxygen atom. However, sulfur has a larger radius and a more diffuse electron cloud than oxygen. These differences impart the cysteine sulfhydryl side chain with structural and chemical properties distinct from the serine hydroxyl side chain. In the current wild-type and C298S hAR structures, the Cβ-Sγ and Cβ-Oγ bond lengths differ by nearly half an angstrom, respectively, 1.80 and 1.41 Å. Discrepancies in the electronic properties of the sulfur and oxygen lead to a large difference in the pKa values of the cysteine and serine amino acids, respectively, 8.37 and 13.0. In the context of the enzyme active site, these values will be strongly affected by the local environment. Indeed, the pKa for the serine OH is depressed from 13.0 to 8.5 in the C298S hAR mutant.3 Incidentally, the experimentally determined pKa for the wild-type hAR is 8.4 ± 0.1 (21, 25). Analysis of high resolution structures in the Protein Data Bank reveal that serine predominantly displays gauche+, gauche−, and trans stereochemistry, but the gauche+ orientation is preferred in cysteine (39). Differences also exist in the observed hydrogen bond lengths and donor/acceptor angles. The bond lengths and angles are 3.5 (±0.1) Å and 104 (±27)° for cysteine and 3.0 (±0.2) Å and 115 (±19)° for serine (39). The differences in kinetic, thermodynamic, and structural properties described for the wild-type and C298S mutant hAR ultimately result from the deceptively simple replacement of a sulfur atom with an oxygen atom. Therefore, oxidative modifications surrounding the sulfur atom of Cys-298 caused by activation are anticipated to alter cofactor binding, conformational rearrangements, catalysis, and inhibition of hAR and related enzymes.
Acknowledgments
We thank David Eisenberg and Todd Yeates for use of their facilities, Peter Kasvinsky and the Youngstown State University Graduate School, for encouragement and support the staff at Synchrotron Radiation Laboratory beamline 9-1 and Advanced Light Source beamline 8.2.2 for expert assistance, and Dino Moras and Alberto Podjarny for the gift of the His-tag recombinant hAR overexpression cells. Data collection facilities at the Stanford Synchrotron Radiation Laboratory are funded by The Office of Biological and Environmental Research, United States Department of Energy, the National Institutes of Health, National Center for Research Resources, Biomedical, and the National Institute of General Medical Sciences.
This study was supported by Grant DK085496 from the National Institute of Diabetes and Digestive and Kidney Diseases.
The atomic coordinates and structure factors (codes 3Q65 and 3Q67) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
G. K. Balendiran, G. Ponniah, and M. Verma, unpublished results.
- AR
- aldose reductase
- hAR
- human AR
- HNE
- - hydroxyl-2-nonenal
- ITC
- isothermal titration calorimetry
- 3-APADP+
- 3-acetylpyridine adenine dinucleotide phosphate
- r.m.s.
- root mean square.
REFERENCES
- 1. Flynn T. G. (1982) Biochem. Pharmacol. 31, 2705–2712 [DOI] [PubMed] [Google Scholar]
- 2. Gabbay K. H. (1973) N. Engl. J. Med. 288, 831–836 [DOI] [PubMed] [Google Scholar]
- 3. Jaspan J. (1986) Drugs 32, 23–29 [DOI] [PubMed] [Google Scholar]
- 4. Greene D. A., Lattimer S. A., Sima A. A. (1987) N. Engl. J. Med. 316, 599–606 [DOI] [PubMed] [Google Scholar]
- 5. Kinoshita J. H., Nishimura C. (1988) Diabetes Metab. Rev. 4, 323–337 [DOI] [PubMed] [Google Scholar]
- 6. Newfield R. S., Polak M., Marchase R., Czernichow P. (1997) Diabetologia 40, B62–B64 [DOI] [PubMed] [Google Scholar]
- 7. Caprio S., Wong S., Alberti K. G., King G. (1997) Diabetologia 40, B78–B82 [DOI] [PubMed] [Google Scholar]
- 8. Lorenzi M. (2007) Exp. Diabetes Res. 2007, 61038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das B., Srivastava S. K. (1985) Diabetes 34, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 10. Grimshaw C. E., Lai C. J. (1996) Arch. Biochem. Biophys. 327, 89–97 [DOI] [PubMed] [Google Scholar]
- 11. Grimshaw C. E., Shahbaz M., Jahangiri G., Putney C. G., McKercher S. R., Mathur E. J. (1989) Biochemistry 28, 5343–5353 [DOI] [PubMed] [Google Scholar]
- 12. Srivastava S. K., Ansari N. H., Hair G. A., Awasthi S., Das B. (1986) Metabolism 35, 114–118 [DOI] [PubMed] [Google Scholar]
- 13. Srivastava S. K., Hair G. A., Das B. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 7222–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cappiello M., Voltarelli M., Cecconi I., Vilardo P. G., Dal Monte M., Marini I., Del Corso A., Wilson D. K., Quiocho F. A., Petrash J. M., Mura U. (1996) J. Biol. Chem. 271, 33539–33544 [DOI] [PubMed] [Google Scholar]
- 15. Dixit B. L., Balendiran G. K., Watowich S. J., Srivastava S., Ramana K. V., Petrash J. M., Bhatnagar A., Srivastava S. K. (2000) J. Biol. Chem. 275, 21587–21595 [DOI] [PubMed] [Google Scholar]
- 16. Bohren K. M., Gabbay K. H. (1993) Adv. Exp. Med. Biol. 328, 267–277 [DOI] [PubMed] [Google Scholar]
- 17. Petrash J. M., Harter T. M., Devine C. S., Olins P. O., Bhatnagar A., Liu S., Srivastava S. K. (1992) J. Biol. Chem. 267, 24833–24840 [PubMed] [Google Scholar]
- 18. Srivastava S., Chandra A., Ansari N. H., Srivastava S. K., Bhatnagar A. (1998) Biochem. J. 329, 469–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grimshaw C. E., Shahbaz M., Putney C. G. (1990) Biochemistry 29, 9947–9955 [DOI] [PubMed] [Google Scholar]
- 20. Kubiseski T. J., Hyndman D. J., Morjana N. A., Flynn T. G. (1992) J. Biol. Chem. 267, 6510–6517 [PubMed] [Google Scholar]
- 21. Grimshaw C. E., Bohren K. M., Lai C. J., Gabbay K. H. (1995) Biochemistry 34, 14356–14365 [DOI] [PubMed] [Google Scholar]
- 22. Del Corso A., Camici M., Mura U. (1987) Biochem. Biophys. Res. Comm. 148, 369–375 [DOI] [PubMed] [Google Scholar]
- 23. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 24. Nishimura C., Yamaoka T., Mizutani M., Yamashita K., Akera T., Tanimoto T. (1991) Biochim. Biophys. Acta 1078, 171–178 [DOI] [PubMed] [Google Scholar]
- 25. Balendiran G. K., Rajkumar B. (2005) Biochem. Pharmacol. 70, 1653–1663 [DOI] [PubMed] [Google Scholar]
- 26. Liu S. Q., Bhatnagar A., Srivastava S. K. (1992) Biochem. Pharmacol. 44, 2427–2429 [DOI] [PubMed] [Google Scholar]
- 27. Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. Kissinger C. R., Gehlhaar D. K., Smith B. A., Bouzida D. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1474–1479 [DOI] [PubMed] [Google Scholar]
- 30. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33. Laskowski R. A., McArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 34. Colovos C., Yeates T. O. (1993) Protein Sci. 2, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lüthy R., Bowie J. U., Eisenberg D. (1992) Nature 356, 83–85 [DOI] [PubMed] [Google Scholar]
- 36. Wilson D. K., Bohren K. M., Gabbay K. H., Quiocho F. A. (1992) Science 257, 81–84 [DOI] [PubMed] [Google Scholar]
- 37. Privalov P. L., Makhatadze G. I. (1992) J. Mol. Biol. 224, 715–723 [DOI] [PubMed] [Google Scholar]
- 38. Spolar R. S., Livingstone J. R., Record M. T., Jr. (1992) Biochemistry 31, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 39. Ippolito J. A., Alexander R. S., Christianson D. W. (1990) J. Mol. Biol. 215, 457–471 [DOI] [PubMed] [Google Scholar]