Abstract
The extracellular domain of the epithelial Na+ channel (ENaC) is exposed to a wide range of anion concentrations in the kidney. We have previously demonstrated that extracellular Cl− inhibits ENaC activity. To identify sites involved in Cl− inhibition, we mutated residues in the extracellular domain of α-, β-, and γENaC that are homologous to the Cl− binding site in acid-sensing ion channel 1a and tested the effect of Cl− on the activity of ENaC expressed in Xenopus oocytes. We identified two Cl− inhibitory sites in ENaC. One is formed by residues in the thumb domain of αENaC and the palm domain of βENaC. Mutation of residues at this interface decreased Cl− inhibition and decreased Na+ self-inhibition. The second site is formed by residues at the interface of the thumb domain of βENaC and the palm domain of γENaC. Mutation of these residues also decreased Cl− inhibition yet had no effect on Na+ self-inhibition. In contrast, mutations in the thumb domain of γENaC and palm of αENaC had little or no effect on Cl− inhibition or Na+ self-inhibition. The data demonstrate that Cl− inhibits ENaC activity by two distinct Na+-dependent and Na+-independent mechanisms that correspond to the two functional Cl− inhibitory sites. Furthermore, based on the effects of mutagenesis on Cl− inhibition, the additive nature of mutations, and on differences in the mechanisms of Cl− inhibition, the data support a model in which ENaC subunits assemble in an αγβ orientation (listed clockwise when viewed from the top).
Keywords: Acid-sensing Ion Channel (ASIC), Epithelial Cell, Ion Channels, Kidney, Sodium Channels, Amiloride, Chloride, ENaC
Introduction
The epithelial Na+ channel, composed of three homologous subunits (α-, β-, and γENaC),3 functions as a pathway for Na+ reabsorption across epithelia in the kidney collecting duct, lung, distal colon, and sweat duct (1, 2). In this role, the channel is critical for the maintenance of Na+ homeostasis and control of the composition and quantity of fluid on the apical membrane of these epithelia. ENaC mutations, and defects in its regulation, cause inherited forms of hypertension and hypotension (3) and may contribute to the pathogenesis of lung disease in cystic fibrosis (4).
At the apical membrane of epithelia, ENaC is exposed to highly varied extracellular environments. Through its large extracellular domain, ENaC detects concentration changes in a number of molecules, producing changes in ENaC gating. This mechanism may fine tune ENaC activity to respond to diverse challenges. For example, the urine Na+ concentration in the kidney collecting duct ranges between ∼1 and 100 mm under conditions of volume depletion and volume excess. Binding of Na+ to ENaC extracellular domains inhibits channel gating through a process known as Na+ self-inhibition (5–7). pH also fluctuates widely (e.g. 4.5–8 in kidney collecting duct) (8); by modulating Na+ self-inhibition, acidic pH increases and alkaline pH reduces ENaC activity (9).
In previous work, we found that extracellular Cl− also modulates ENaC activity (10). As the predominant anion in the extracellular fluid, the concentration of Cl− varies over a wide range (1–100 mm in kidney collecting duct), in parallel with changes in Na+ (8). Within this concentration range, Cl− inhibits ENaC gating through two distinct mechanisms (10). First, Cl− enhances Na+ self-inhibition of the channel. Second, Cl− inhibits ENaC through a direct effect that is independent of extracellular Na+.
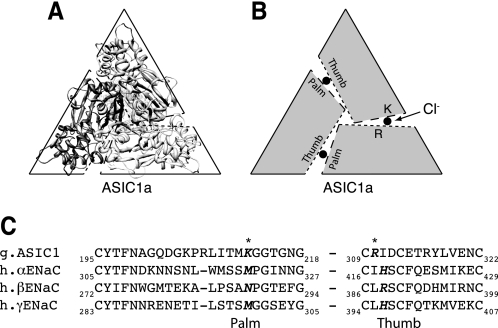
Crystallization of ASIC1a has begun to provide structural insights into the DEG/ENaC family of ion channels. ASIC1a assembles as a homotrimer (11, 12), suggesting that other members of the family are also trimers. Surprisingly, the ASIC1a structure revealed three Cl− ions bound to the extracellular domains (11). The Cl− binding sites are located at each of the three interfaces between the subunits, formed by residues at the base of the thumb domain of one subunit (Arg-310, Glu-314) and the palm domain of the opposing subunit (Fig. 1, A and B, shows ASIC1a looking down from the top). Recent work found that Cl− binding slowed the rate of ASIC1a desensitization when the channel was activated by protons (13).
FIGURE 1.
Sequence alignment of ASIC1a with α-, β-, and γENaC. A, ribbon structure of chicken ASIC1a (2QTS) shown perpendicular to the plane of the membrane from an extracellular perspective. B, model of ASIC1a subunit arrangement. Dotted lines represent the thumb domain, dashed lines represent the palm domain. Approximate location of Cl− is depicted by a filled circle located between the thumb and palm domains of two adjacent subunits. C, partial sequence alignment of the palm and thumb domains of chicken ASIC1a with human α-, β-, and γENaC using the ClustalW method. ASIC1a Cl− coordinating residues are indicated with an asterisk. Potential ENaC Cl−-coordinating residues identified by sequence alignment are in bold.
Because ASIC1a and ENaC share a high degree of sequence similarity, including a number of conserved cysteine residues that form disulfide bonds, the ASIC1a crystal structure has been used as a starting point to investigate structure-function relationships in the extracellular domain of ENaC (10, 14). Using this approach, we identified homologous residues in the thumb domains of αENaC (His-418) and βENaC (Arg-388) that contribute to ENaC inhibition by Cl− (10). Importantly, the equivalent residue in γENaC (His-396) did not contribute. These findings support the possibility that Cl− regulates ENaC through sites that are analogous to the Cl− binding sites identified in ASIC1a. In the current work, we test whether ENaC palm domain residues also contribute to Cl− inhibition. Furthermore, because the residues that form the Cl− inhibitory sites are predicted to lie at the interface between two neighboring subunits, our approach provides insight into the subunit arrangement of the ENaC trimeric channel complex.
EXPERIMENTAL PROCEDURES
DNA Constructs
cDNAs for human α-, β-, and γENaC in pMT3 were cloned as described previously (15, 16). Mutations were generated by site-directed mutagenesis (QuikChange II; Stratagene) and sequenced in the University of Iowa DNA Core.
Expression and Whole Cell Electrophysiology in Xenopus Oocytes
Oocytes were harvested from albino Xenopus laevis females and manually defolliculated following a 1-h treatment with 0.75 mg/ml Type IV collagenase (Sigma) in Ca2+-free ND-96 (96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 5 mm HEPES, pH adjusted to 7.4 with NaOH). Following nuclear injection of cDNAs encoding α-, β-, and γENaC (0.02 μg/μl each), cells were incubated at 18 °C in modified Barth's saline (88 mm NaCl, 1 mm KCl, 0.33 mm Ca(NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 2.4 mm NaHCO3, 10 mm HEPES, 50 μg/ml gentamycin sulfate, 10 μg/ml sodium penicillin, 10 μg/ml streptomycin sulfate, pH adjusted to 7.4 with NaOH) for 20–24 h prior to the study. Oocytes were voltage clamped (two-electrode voltage clamp) and currents were amplified with an Oocyte Clamp OC-725C (Warner Instruments), digitized with a MacLab/200 interface (ADInstruments), and recorded and analyzed with Chart software (ADInstruments). Unless otherwise noted, recordings were done at −60 mV in a 116 mm NaCl solution (116 mm NaCl, 2 mm KCl, 0.4 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH adjusted to 7.4 with NaOH). Low Na+ solutions (1 mm NaCl, 115 mm N-methyl-d-glucamine Cl, pH adjusted with HCl) were used as indicated in the figure legends. Low Cl− solutions were prepared by replacing the anion of the sodium salt with SO42− and d-mannitol (to balance osmolarity), SCN−, I−, or Br−; the composition of the low Cl− solutions was otherwise identical to that of 116 NaCl Ringer's. Amiloride-sensitive current was determined by adding 10 μm amiloride to the bathing solution (supplemental Table 1). The Cl−-induced changes in amiloride-sensitive current were calculated as the -fold increase/decrease relative to the base-line current in 116 mm NaCl just prior to each test solution application. This was done to reduce the effect of time-dependent current run-down. Na+ self-inhibition was measured by rapidly changing the bathing solution from low sodium (1 mm NaCl) to high sodium (116 mm NaCl) and quantitated as ((peak current − steady state current)/peak current). To account for day-to-day variation, values were normalized to values recorded from cells expressing wild-type ENaC on the same day. All values are reported as averages ± S.E. Statistical significance was calculated using a two-tailed Student's t test (p < 0.05).
RESULTS
ENaC Palm Domain Residues Participate in Cl− Inhibition
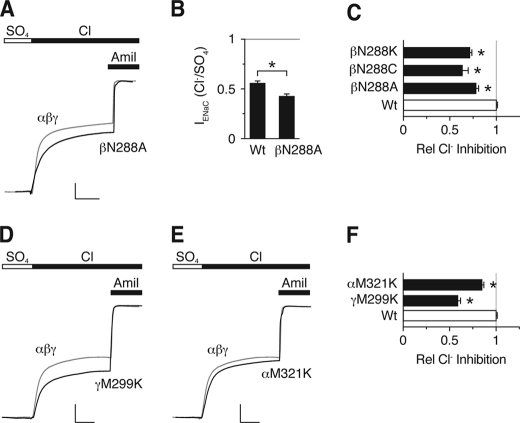
To identify ENaC palm domain residues that contribute to inhibition by Cl−, we did a sequence alignment between ASIC1a and α-, β-, and γENaC in the region of ASIC1a Lys-212 (Fig. 1C). The alignment was facilitated by adjacent conserved Gly residues as well as a highly conserved upstream Cys-containing motif. Lys-212 is not conserved in ENaC. The corresponding residues are Asn-288 in βENaC, Met-321 in αENaC, and Met-299 in γENaC. To test their potential role in ENaC regulation by extracellular Cl−, we mutated these residues and quantitated ENaC inhibition by Cl− in Xenopus oocytes. We first focused on βENaC. Fig. 2A shows representative traces from cells expressing human α-, β-, and γENaC. For wild-type ENaC (gray trace), addition of Cl− to the extracellular bathing solution (replacing SO42−, which does not inhibit ENaC) (10) decreased the amiloride-sensitive current 55.4%, consistent with our previous work (10). Mutation of Asn-288 to Ala in βENaC (co-expressed with wild-type αENaC and γENaC) reduced ENaC inhibition by Cl− (Fig. 2, A–C); Fig. 2B shows the amiloride-sensitive ENaC current with cells bathed in Cl− (relative to SO42−), and Fig. 2C shows the Cl−-inhibited current. Mutation of Asn-288 to Cys or Lys decreased Cl− inhibition to a similar extent (Fig. 2C). These findings support two conclusions. First, Asn-288 in βENaC contributes to inhibition by extracellular Cl−. Second, at this position, lysine (the amino acid at the equivalent position in ASIC1a) is not a functionally conservative substitution.
FIGURE 2.
Effect of ENaC palm domain mutations on Cl− inhibition. A, D, and E, representative recordings of current versus time from Xenopus oocytes expressing αβγENaC (A, C, and D gray trace; scale bars: vertical, 1 μA; horizontal,10 s), αβN288Aγ (A, black trace; scale bars: vertical, 1.5 μA; horizontal, 15 s), αβγM299K (D, black trace; scale bars: vertical, 1.7 μA; horizontal, 15 s), and αM321Kβγ (E, black trace; scale bars: vertical, 1.7 μA; horizontal, 14 s). Cl− inhibition was measured by exchanging SO42− for Cl− in extracellular bathing solution. 10 μm amiloride was added at the end of the recording to determine the amount of ENaC current. All traces are normalized to amiloride-sensitive current in the presence of Na2SO4. B, summary data of amiloride-sensitive current with cells bathed in Cl− (relative to SO42−) for wild-type (Wt) ENaC or αβN288Aγ (n = 6, *, p < 0.001). C, summary data of fraction of amiloride-sensitive current inhibited by Cl− (relative to SO42−) for the indicated βENaC palm domain mutations relative to wild-type ENaC (mean ± S.E. (error bars), n = 6–11, *, p < 0.001). F, summary data of Cl− inhibition for αM321K and γM299K (mean ± S.E., n = 13, *, p < 0.001).
To test the role of the equivalent palm domain residues in αENaC and γENaC, we mutated them to Lys (αM321K and γM299K). In γENaC, mutation of Met-299 to Lys resulted in a significant decrease in Cl− inhibition (Fig. 2, D and F). In contrast, mutation of αENaC Met-321 to Lys produced only a small decrease in Cl− inhibition (Fig. 2, E and F).
These results indicate that residues located in the palm domains of βENaC and γENaC contribute to Cl− inhibition, whereas the palm domain of αENaC plays a lesser role. In previous work, we identified thumb domain residues in α- and β-, but not γENaC, that contribute to Cl− inhibition (10). Together, these findings suggest that Cl− inhibits ENaC predominantly through two Cl− inhibitory sites, in contrast to the three sites identified in the ASIC1a crystal structure.
Mutations of γENaC Met-299 Alter Anion Inhibition Selectivity
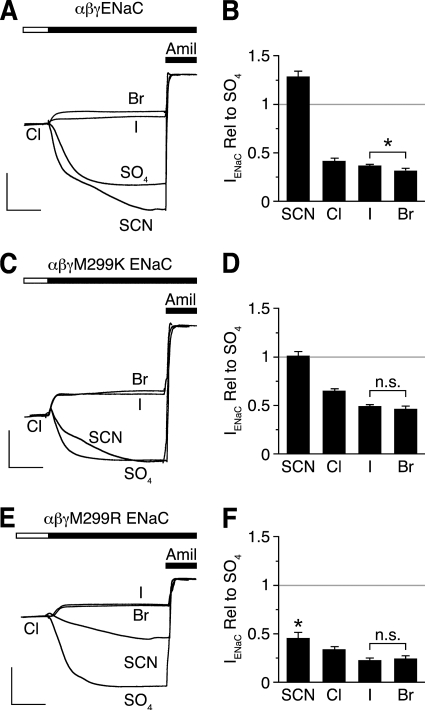
If palm domain residues contribute to anion binding, analogous to the equivalent residues in ASIC1a, then mutation of these residues might alter the anion selectivity of ENaC inhibition. To test this possibility, we focused on Met-299 in γENaC. Fig. 3, A and B, shows the effects of anion substitutions on wild-type ENaC current. Br− inhibited ENaC to the greatest extent (compared with SO42−), followed by I− and Cl−, whereas SCN− did not inhibit ENaC (10). Thus, the selectivity sequence for inhibition of wild-type ENaC is Br− > I− > Cl− ≫ SCN−. Mutation of Met-299 to Lys reduced ENaC inhibition by Cl−, I−, and Br− (Fig. 3, B and D). In addition, this mutation altered anion selectivity of inhibition, such that Br− and I− decreased ENaC current to an equivalent degree (Br− = I− > Cl− ≫ SCN−) (Fig. 3, C and D). Mutation of Met-299 to another basic residue, Arg, had the same effect as Lys on relative Br−/I− selectivity (Fig. 3, E and F). However, this mutation had two additional effects. First, it allowed SCN− to inhibit ENaC (ISCN− < ISO42−, p < 0.001), albeit less than Cl− (Br− = I− > Cl− > SCN−). Second, it did not reduce, but slightly enhanced, ENaC inhibition by Cl−, Br−, and I− (Fig. 3, D and F). Together, these data suggest that both electrostatic and steric factors at residue 299 influence selectivity of anion inhibition, which is consistent with a role for this residue in anion binding.
FIGURE 3.
γENaC Met-299 mutations alter selectivity of anion inhibition. A, C, and E, representative recordings of current versus time from Xenopus oocytes expressing αβγENaC (A), αβγM299K (C), or αβγM299R (E). Anion selectivity was measured by replacing extracellular Cl− with SCN−, SO42−, I−, or Br−. Traces are scaled to amiloride-sensitive current for wild-type ENaC in the presence of Cl−. B, D, and F, summary data of -fold change in amiloride-sensitive current for the indicated anion relative to SO42− for cells expressing αβγENaC (B), αβγM299K (D), or αβγM299R (F). Data are mean ± S.E. (error bars; n = 6–9, *, p < 0.001; n.s. indicates p not significant).
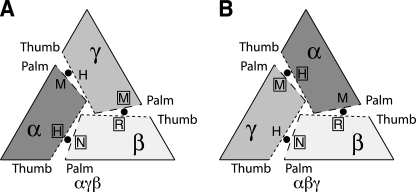
ENaC Subunits Are Arranged in an αγβ Orientation
Based on our findings and the ASIC1a crystal structure, we hypothesized that the Cl− inhibitory sites are positioned at the interfaces between ENaC subunits, with palm domain residues from one subunit paired with thumb domain residues from the adjacent subunit. A key question is which ENaC subunit provides the palm domain and which subunit provides the thumb domain at each Cl− site? There are two possible structural arrangements for the three ENaC subunits. When viewed from the top, they could adopt an αγβ (Fig. 4A) or an αβγ (Fig. 4B) orientation (listed clockwise). To determine the subunit orientation, we made two assumptions. First, we assumed that the palm domain residues that contribute to Cl− inhibition would be paired with thumb domain residues that also contribute to Cl− inhibition. Second, we assumed that the palm domain and thumb domain that had little or no effect on Cl− regulation would be paired together.
FIGURE 4.
Potential ENaC subunit arrangements. A trimeric assembly could result in an αγβ (A) or αβγ (B) channels (listed clockwise, viewing channel from the top). Dotted lines represent the thumb domain interface. Dashed lines represent the palm domain interface. Approximate locations of putative Cl− inhibitory residues are labeled accordingly: αH, His-418; αM, Met-321; γH, His-396; γM, Met-299; βR, Arg-388; βN, Asn-288. Approximate locations of chloride ions are identified with filled circles. Residues that had large effects on Cl− inhibition are identified with an open box.
Using these assumptions, we examined the pairing of palm and thumb domain residues predicted by the two models. We started with the βENaC palm domain residue Asn-288 which, when mutated, reduced Cl− inhibition (Fig. 2, A and B). In the αγβ model, this residue is paired with the αENaC thumb domain residue His-418 (Fig. 4A). We reported previously that mutation of this residue reduced Cl− inhibition (10). Conversely, in the αβγ model, Asn-288 pairs with the γENaC thumb domain residue His-396 (Fig. 4B), which had no effect on ENaC Cl− inhibition (10). Thus, these data support the αγβ orientation.
At a second interface in the αγβ model, Met-299 in γENaC is paired with Arg-388 in βENaC (Fig. 4A). Consistent with this model, mutation of both residues reduced Cl− inhibition (Fig. 2) (10). The αγβ model also places the residues that had little or no effect on Cl− inhibition (α Met-321 and γ His-396) together at the third interface (Fig. 4A). Contrary to the data, the αβγ model would pair these “nonfunctional” residues together with residues that contribute to Cl− inhibition (Fig. 4B). Taken together, the data suggest that ENaC assembles in an αγβ orientation.
Mutations of Residues in Separate Cl− Sites Are Additive
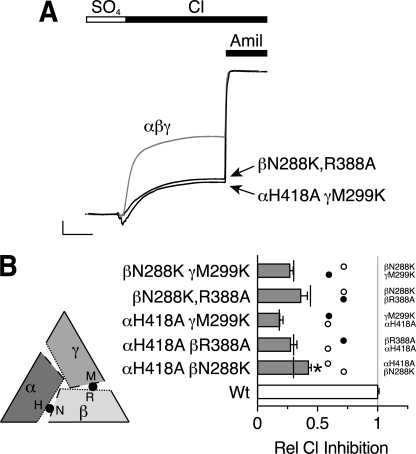
To test further whether the ENaC subunits form an αγβ heterotrimer, we combined mutations at the two Cl− inhibitory sites. If each site has an independent effect on ENaC activity, we predict that combining a mutation from the α-β interface with a mutation in the β-γ interface will produce an additive decrease in Cl− inhibition. Fig. 5A shows two examples. In βN288K,R388A, we simultaneously mutated the β subunit palm domain (predicted location at the α-β interface) and thumb domain (predicted location at the β-γ interface). When co-expressed with wild-type α- and γENaC, this double mutation greatly decreased the effect of extracellular Cl− on ENaC current (Fig. 5). A similar decrease in Cl− inhibition was observed when we combined mutations in the thumb domain of the α-β interface (αH418A) with the palm domain of the β-γ interface (γM299K) (Fig. 5). In Fig. 5B, we quantitated the results for each of the single and double mutations at the α-β and β-γ interfaces; single mutations are indicated by white circles (α-β interface) and black circles (β-γ interface), the double mutations are indicated by gray bars, and predicted additive values (sum of the individual mutations) are indicated by vertical black lines. When mutations from the α-β and β-γ interfaces were combined, there was an additive reduction in Cl− inhibition relative to the corresponding individual mutations (Fig. 5B, compare gray bars with vertical black lines). In contrast, when we mutated two residues predicted to lie at the same interface (α-β interface), αH418A and βN288K, the effects were less than additive (Fig. 5B). These data indicate that the two Cl− inhibitory sites have independent effects on ENaC regulation and provide support for the αγβ subunit model.
FIGURE 5.
Additivity of Cl− inhibitory site mutations. A, representative recordings of current versus time from Xenopus oocytes expressing αβγENaC (gray trace, scale bars: vertical, 1 μA; horizontal, 10 s), αβN288K,R388Aγ (black trace, scale bars: vertical, 1.6 μA; horizontal, 13 s) or αH418AβγM299K (black trace, scale bars: vertical, 1.75 μA; horizontal, 13 s). Cl− inhibition was measured by replacing SO42− in the bathing solution with Cl−. 10 μm amiloride was added at the end of the recording to quantitate ENaC current. All traces are normalized to amiloride-sensitive current in the presence of SO42−. B, summary data showing relative Cl− inhibition for each combination of double mutations (gray bars) at the α-β and β-γ interfaces. Individual α-β and β-γ interface mutations shown as white or black circles, respectively. Predicted values for additive double mutations (sum of individual mutations) are indicated with vertical black lines. Data are mean ± S.E. (error bars; n = 4–14). All values for single or double mutations are significantly different from wild-type (Wt) ENaC (p < 0.001). The αH418AβN288K double mutation is significantly less than the predicted additive value (*, p = 0.006). The other double mutants are not statistically different from the predicted additive values.
Different Cl− Sites Alter ENaC Activity through Distinct Mechanisms
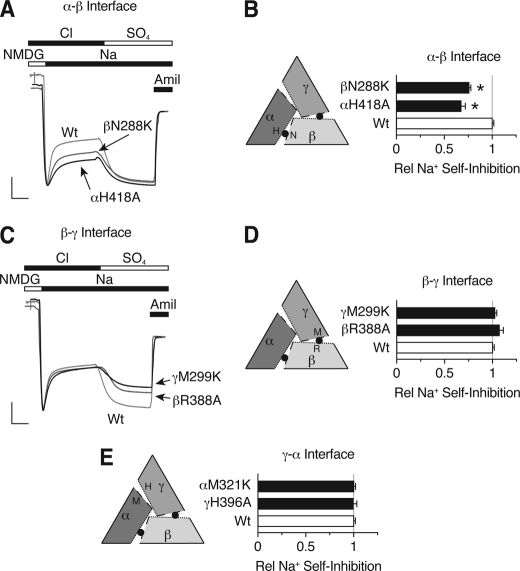
As a third test of the αγβ model, we examined the mechanisms by which Cl− inhibits ENaC. In previous work, we found that inhibition occurs through two different mechanisms. One is dependent on extracellular Na+ (Cl− enhanced Na+ self-inhibition) whereas the second is independent of Na+ (10). We hypothesized that these two mechanisms might correspond to the two Cl− inhibitory sites, one site modulating Na+ self-inhibition and the other site regulating ENaC in a Na+-independent manner. To test this hypothesis, we investigated the effect of Cl− site mutations on Na+ self-inhibition.
Fig. 6A shows representative current traces for mutations predicted to be in the α-β interface. We measured ENaC Na+ self-inhibition and Cl− inhibition in the same cells. At the start of the trace, addition of Na+ to the extracellular bathing solution generated a peak current that rapidly decreased to a lower steady-state level, as a result of Na+ self-inhibition. In Fig. 6B, we plotted the fraction of current inhibited by Na+ (relative to wild-type ENaC). αH418A and βN288K each reduced Na+ self-inhibition (0.683 ± 0.027 and 0.766 ± 0.023 relative to wild-type, respectively). In the second part of the trace, Cl− in the bathing solution was replaced with SO42− to measure Cl− inhibition (Fig. 6A). Both mutations decreased ENaC Cl− inhibition, similar to our data in Fig. 2. Because mutation of αH418A and βN288K had congruent effects on Na+ self-inhibition and Cl− inhibition, the data suggest that they are located at the same interface.
FIGURE 6.
Differential effects of Cl− site mutations on Na+ self-inhibition. A and C, representative recordings of current versus time from Xenopus oocytes expressing αβγENaC (A and C, light gray traces, scale bars: vertical, 1 μA; horizontal, 10 s), αβN288Kγ (A, dark gray trace, scale bars: vertical, 0.3 μA; horizontal, 9 s), αH418Aβγ (A, black trace, scale bars: vertical, 1 μA; horizontal, 10 s), αβR388Aγ (C, dark gray trace, scale bars: vertical, 1.22 μA; horizontal, 8 s), or αβγM299K (C, black trace, scale bars: vertical, 1 μA; horizontal, 9 s). Na+ self-inhibition was measured in the presence of extracellular Cl− by rapidly changing from low (1 mm Na+, 115 mm NMDG) to high Na+ (116 mm Na+) extracellular solutions. Cl− inhibition was measured by replacing extracellular Cl− with SO42−. Traces are scaled relative to amiloride-sensitive peak current. B, D, and E, summary data showing Na+ self-inhibition relative to wild-type (Wt) ENaC for single mutations at the predicted α-β interface (B), β-γ interface (D), and γ-α interface (E). Data are mean ± S.E. (error bars; n = 3–10, *, p < 0.001).
In Fig. 6, C and D, we tested mutations predicted to be located at the β-γ interface. Although βR388A and γM299K each reduced ENaC inhibition by Cl−, they had no effect on Na+ self-inhibition (1.075 ± 0.038 and 1.028 ± 0.015, respectively). This supports the idea that βR388A and γM299K are located at a common interface. Likewise, mutations at the γ-α interface, αM321K and γH396A (mutations that had minimal effect on Cl− inhibition), did not alter Na+ self-inhibition (1.007 ± 0.044 and 0.998 ± 0.018, respectively; Fig. 6E). Together, the data indicate that the two Cl− sites inhibit ENaC through different mechanisms. At the α-β interface, Cl− enhances Na+ self-inhibition of ENaC, whereas at the β-γ interface, Cl− inhibits ENaC through a mechanism that is independent of Na+ self-inhibition.
In addition to providing mechanistic insights, these results also confirm our hypothesis that ENaC adopts an αγβ channel architecture. This is the only model that pairs the residues that reduced Na+ self-inhibition (αH418A with βN288K) and the residues that had no effect on Na+ self-inhibition (βR388A and γM299K). In the alternative αβγ structure, αH396, a residue that functions through Na+ self-inhibition, would be paired with γM299, a residue that is Na+-independent (Fig. 4B and Fig. 6), a possibility not supported by the data.
DISCUSSION
In this work, we identified palm domain residues that participate in ENaC inhibition by extracellular Cl−. One residue is located in βENaC (Asn-288), and the second is at the equivalent position in γENaC (Met-299). In previous work, we identified thumb domain residues in αENaC (His-418) and βENaC (Arg-R388) that also contribute to Cl− inhibition. Together, the data suggest that ENaC has two predominant Cl− inhibitory sites that are positioned at the interfaces between ENaC subunits.
We began with the assumption that ENaC shares overall structural similarity with ASIC1a, including a trimeric stoichiometry. A key feature is the location of the ASIC1a Cl− inhibitory sites at the interfaces between adjacent subunits (11). Our findings suggest that the Cl− inhibitory sites are conserved between ASIC1a and ENaC, supporting the idea that these channels share structural similarity in their extracellular domains. We exploited this observation to deduce the subunit orientation of the ENaC channel structure. Based on the effects of mutagenesis on Cl− inhibition, the additivity of mutations, and on differences in the mechanisms of Cl− inhibition, the data support an αγβ orientation (listed clockwise when viewed from the top). Moreover, the data suggest that assembly of the ENaC subunits is not stochastic, but occurs in a favored configuration. Although unlikely, we cannot completely exclude the existence of a population of αβγ channels that are not modulated by changes in extracellular Cl− or by the mutations we tested.
In the ASIC1a crystal structure, Cl− is coordinated in part through its interaction with the positively charged side chain of Lys-212 in the palm domains (11, 13). It is interesting that the amino acids at the equivalent positions in ENaC (βN288 and γM299) do not carry a positive charge. Although it is clear from our mutagenesis data that these residues are critical for ENaC inhibition by Cl−, their specific role in this regulation is unclear. They may participate directly in Cl− binding. Direct interactions could occur on the backbone, as demonstrated by structural data from other halide binding sites (17, 18) or, analogous to their role in ASIC1a, direct interactions could occur with the residue side chain. Consistent with this possibility, we found that γM299 mutations altered the selectivity sequence for anion inhibition. In particular, mutation to Arg increased Cl− inhibition and created a binding site for SCN−. This is analogous to previous work on deoxyhemoglobin, where mutation of Trp-37 to Arg created a novel anion binding site (19). Our data, in combination with structural data from the literature, suggest that palm domain residues directly participate in Cl− binding. However, in the absence of an ENaC crystal structure, we cannot completely exclude the possibility that the residues we identified influence anion binding indirectly.
In previous work, we reported that Cl− inhibited ENaC through two different mechanisms (10). First, Cl− reduced ENaC current by facilitating Na+ self-inhibition of the channel. Second, Cl− also reduced ENaC current through a “direct” mechanism that was independent of extracellular Na+. Our current studies provide a structural basis for these two mechanisms. We found that Cl− modulation of Na+ self-inhibition occurred at the α-β interface, whereas Cl− inhibited ENaC through a direct effect at the β-γ interface. It is interesting that residues in βENaC contribute to both Cl− inhibitory sites. This raises the possibility that Cl− induces conformational changes restricted to the β subunit. However, we think it is more likely that conformational changes are distributed throughout all three subunits. This notion is supported by our findings that Cl− regulates ENaC through two distinct mechanisms, coupled with the additive effect of mutations at the two sites.
Although the three ENaC subunits share sequence similarity, there is a distinct lack of symmetry in the contribution of each subunit to ENaC regulation by Cl−. Of the three interfaces between ENaC subunits, two play a predominant role in this regulation. Additionally, at these two interfaces, the mechanisms by which Cl− regulates ENaC differ. There are several other examples of asymmetry in the regulation of ENaC gating. For example, ENaC is activated by proteolytic cleavage of αENaC and γENaC, but not βENaC (20–22). Also, mutation of an extracellular His in γENaC decreases Na+ self-inhibition, but mutation of the equivalent His in αENaC has the opposite effect (7). Mutations at the DEG position also have varying effects when introduced in each ENaC subunit (23, 24). Such asymmetry might expand the range of conformational changes available to fine tune channel gating.
Even though ENaC is a constitutively active channel, there is a growing list of molecules that modulate its function, including, but not limited to, Na+, Cl−, H+, Zn2+, and Ni2+ (5, 9, 10, 25). In this regard, it is clear that the extracellular domain functions as a sensor to detect the concentrations of these molecules in a variety of fluids, including urine, airway surface liquid, and sweat. This may allow ENaC to respond rapidly to a variety of challenges that threaten to disrupt Na+ homeostasis. Our current findings explain in part the mechanisms by which ENaC gating is modulated by extracellular Cl−. Moreover, they provide a mechanistic basis to explain functional interactions between ENaC regulation by Cl− and Na+, the two principal components of extracellular fluids. We speculate that defects in this regulation may contribute to the pathogenesis of hypertension and other disorders of Na+ transport.
Supplementary Material
Acknowledgments
We thank Diane Olson, Zeru Peterson, Caitlin Digman, and Abigail Hamilton for assistance, and we acknowledge the University of Iowa DNA Core Facility for reagents and DNA sequencing.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- ENaC
- epithelial Na+ channel
- ASIC1a
- acid-sensing ion channel 1a.
REFERENCES
- 1. Schild L. (2004) Rev. Physiol. Biochem. Pharmacol. 151, 93–107 [DOI] [PubMed] [Google Scholar]
- 2. Snyder P. M. (2005) Endocrinology 146, 5079–5085 [DOI] [PubMed] [Google Scholar]
- 3. Lifton R. P. (1996) Science 272, 676–680 [DOI] [PubMed] [Google Scholar]
- 4. Boucher R. C., Stutts M. J., Knowles M. R., Cantley L., Gatzy J. T. (1986) J. Clin. Invest. 78, 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garty H., Palmer L. G. (1997) Physiol. Rev. 77, 359–396 [DOI] [PubMed] [Google Scholar]
- 6. Chraïbi A., Horisberger J. D. (2002) J. Gen. Physiol. 120, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheng S., Bruns J. B., Kleyman T. R. (2004) J. Biol. Chem. 279, 9743–9749 [DOI] [PubMed] [Google Scholar]
- 8. Rose B. D. (1984) Clinical Physiology of Acid-Base and Electrolyte Disorders, 2nd Ed., pp. 274–275, McGraw-Hill, New York [Google Scholar]
- 9. Collier D. M., Snyder P. M. (2009) J. Biol. Chem. 284, 792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collier D. M., Snyder P. M. (2009) J. Biol. Chem. 284, 29320–29325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 12. Gonzales E. B., Kawate T., Gouaux E. (2009) Nature 460, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kusama N., Harding A. M., Benson C. J. (2010) J. Biol. Chem. 285, 17425–17431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kashlan O. B., Adelman J. L., Okumura S., Blobner B. M., Zuzek Z., Hughey R. P., Kleyman T. R., Grabe M. (2011) J. Biol. Chem. 286, 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McDonald F. J., Price M. P., Snyder P. M., Welsh M. J. (1995) Am. J. Physiol. Cell Physiol. 268, C1157–C1163 [DOI] [PubMed] [Google Scholar]
- 16. McDonald F. J., Snyder P. M., McCray P. B., Jr., Welsh M. J. (1994) Am. J. Physiol. Lung Cell. Mol. Physiol. 266, L728–L734 [DOI] [PubMed] [Google Scholar]
- 17. Burkhard P., Tai C. H., Jansonius J. N., Cook P. F. (2000) J. Mol. Biol. 303, 279–286 [DOI] [PubMed] [Google Scholar]
- 18. Fiedler T. J., Davey C. A., Fenna R. E. (2000) J. Biol. Chem. 275, 11964–11971 [DOI] [PubMed] [Google Scholar]
- 19. Kavanaugh J. S., Rogers P. H., Case D. A., Arnone A. (1992) Biochemistry 31, 4111–4121 [DOI] [PubMed] [Google Scholar]
- 20. Vallet V., Chraibi A., Gaeggeler H. P., Horisberger J. D., Rossier B. C. (1997) Nature 389, 607–610 [DOI] [PubMed] [Google Scholar]
- 21. Carattino M. D., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 25290–25295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 23. Snyder P. M., Olson D. R., Bucher D. B. (1999) J. Biol. Chem. 274, 28484–28490 [DOI] [PubMed] [Google Scholar]
- 24. Snyder P. M., Bucher D. B., Olson D. R. (2000) J. Gen. Physiol. 116, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheng S., Perry C. J., Kleyman T. R. (2004) J. Biol. Chem. 279, 31687–31696 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.