Abstract
GluA1 (formerly GluR1) AMPA receptor subunit phosphorylation at Ser-831 is an early biochemical marker for long-term potentiation and learning. This site is a substrate for Ca2+/calmodulin (CaM)-dependent protein kinase II (CaMKII) and protein kinase C (PKC). By directing PKC to GluA1, A-kinase anchoring protein 79 (AKAP79) facilitates Ser-831 phosphorylation and makes PKC a more potent regulator of GluA1 than CaMKII. PKC and CaM bind to residues 31–52 of AKAP79 in a competitive manner. Here, we demonstrate that common CaMKII inhibitors alter PKC and CaM interactions with AKAP79(31–52). Most notably, the classical CaMKII inhibitors KN-93 and KN-62 potently enhanced the association of CaM to AKAP79(31–52) in the absence (apoCaM) but not the presence of Ca2+. In contrast, apoCaM association to AKAP79(31–52) was unaffected by the control compound KN-92 or a mechanistically distinct CaMKII inhibitor (CaMKIINtide). In vitro studies demonstrated that KN-62 and KN-93, but not the other compounds, led to apoCaM-dependent displacement of PKC from AKAP79(31–52). In the absence of CaMKII activation, complementary cellular studies revealed that KN-62 and KN-93, but not KN-92 or CaMKIINtide, inhibited PKC-mediated phosphorylation of GluA1 in hippocampal neurons as well as AKAP79-dependent PKC-mediated augmentation of recombinant GluA1 currents. Buffering cellular CaM attenuated the ability of KN-62 and KN-93 to inhibit AKAP79-anchored PKC regulation of GluA1. Therefore, by favoring apoCaM binding to AKAP79, KN-62 and KN-93 derail the ability of AKAP79 to efficiently recruit PKC for regulation of GluA1. Thus, AKAP79 endows PKC with a pharmacological profile that overlaps with CaMKII.
Keywords: Calmodulin; Calcium/Calmodulin-dependent Protein Kinase (CaMK); Drug Action; Glutamate Receptors, Ionotropic (AMPA, NMDA); Protein Kinase C (PKC); Protein Phosphorylation; AKAP; LTP; Synaptic Plasticity
Introduction
Rapidly acting, cell-permeable kinase inhibitors represent a principal tool to dissect signaling pathways (1). However, inhibitor cross-reactivity, particularly to the common ATP binding site of various kinases, may confound delineation of signaling pathways and identification of physiologically relevant substrates (1). The classical CaMKII2 inhibitors KN-62 and KN-93 appear to be relatively specific for this kinase (and related members of this family including CaMKI and CaMKIV) as they bind to the Ca2+/CaM binding site on the kinase, thus preventing kinase activation via competition with Ca2+/CaM (1–3). Although KN-62 and KN-93 do not substantially inhibit other kinases including PKC, non-kinase targets of these compounds have been described (4–11). However, among the non-kinase targets of KN-62 and KN-93, only one appears to be sensitive to these compounds but insensitive to the inactive analog KN-92 (11). Thus, sensitivity to KN-62 and/or KN-93 combined with insensitivity to KN-92 has routinely been used as diagnostic criteria for implicating CaMKII and related kinases in physiological events.
One proposed physiological substrate of CaMKII is Ser-831 of the GluA1 AMPA receptor subunit (12, 13). Phosphorylation of GluA1 at Ser-831 is linked with learning-induced enhancement of synaptic strength and long-term potentiation (LTP) (14–19). However, this site is also a substrate for PKC (12, 13). Thus, the relative contribution of each kinase toward regulation of this site during LTP and other plasticity-producing paradigms has largely relied on the putative pharmacological specificity of inhibitors for each kinase.
Confinement of kinases and other signaling molecules with their physiological targets by scaffold and anchoring proteins imparts spatiotemporal specificity to signaling events (20). AKAP79 (AKAP150 in rodents) is a multivalent signaling scaffold that binds the cAMP-dependent protein kinase (PKA), calcineurin (CaN), and PKC and is targeted to GluA1 via the scaffold protein SAP97 (21–24). By recruiting PKA and CaN to GluA1, AKAP79 coordinates bidirectional regulation of GluA1 at Ser-845 (25), a PKA site linked to modulation of AMPA receptor function and trafficking during synaptic plasticity (15, 16, 26–30). Likewise, by concentrating PKC near GluA1, AKAP79 facilitates PKC phosphorylation of GluA1 at Ser-831 in response to PKC-activating signals, thereby making PKC a more potent regulator of GluA1 than CaMKII (31).
Residues 31–52 of AKAP79 bind the catalytic core of the PKC in a pseudosubstrate-like manner (32). However, this PKC binding domain is also a high affinity site for association with Ca2+/CaM (33). Given that both AKAP79 and CaMKII bind Ca2+/CaM and that classical CaMKII inhibitors act by binding to the Ca2+/CaM site on CaMKII to prevent activation of the kinase, we hypothesized that CaMKII inhibitors may alter the ability of AKAP79 to associate with either PKC or CaM. Here, we demonstrate that KN-62 and KN-93 target AKAP79 with high affinity to enhance CaM binding, thereby disrupting AKAP79-anchored PKC signaling to GluA1. In contrast, KN-92 does not disrupt AKAP79-dependent signaling to GluA1. Thus, AKAP79 endows PKC with a pharmacological profile that is indistinguishable from that commonly associated with CaMKII.
EXPERIMENTAL PROCEDURES
PKC and CaM Binding Assays
Neutravidin beads (20 μl; Pierce) were incubated for 2 h at 4 °C with 5 μg of biotinylated AKAP79(31–52) or biotinylated control peptides (all synthesized by Biomolecules Midwest Inc.) in 500 μl of PKC binding solution containing 150 mm NaCl, 10 mm HEPES, 5 mm EGTA, 5 mm EDTA, 0.1% Tween 20, 0.1% BSA, and protease inhibitors (Sigma) (pH 7.40). Following incubation, beads were washed four times with the binding solution to remove unbound peptide. For CaM binding experiments, AKAP79(31–52) or control peptides and neutravidin beads were incubated in a Ca2+-free solution containing 150 mm NaCl, 10 mm HEPES, 1 mm EGTA, 0.1% Tween 20, and protease inhibitors (pH 7.40). For CaM experiments, the unbound peptide was removed by washing the beads either four times with the Ca2+-free buffer or two times in this solution followed by twice in buffer in which EGTA was replaced with 100 μm CaCl2. All CaMKII reagents (KN-62, LC Laboratories or EMD Biosciences; KN-93, KN-92, and CaMKIINtide, EMD Biosciences) were prepared initially as 10 mm stocks in DMSO representing a 1000× stock for the highest concentration of each compound we tested. These stocks were subsequently serially diluted in DMSO to maintain a constant final 0.1% DMSO concentration in all experiments. DMSO was included to 0.1% for control (i.e. beads alone and/or no drug) experiments as well. Peptide-bound beads were then incubated overnight at 4 °C with either PKC isoforms (200 ng (∼5 nm); Biomol or EMD Biosciences) or CaM (8.5 μg (∼1 μm); EMD Biosciences). Following overnight incubation, beads were washed four times with the respective buffer in the presence or absence of the drug. Protein was eluted by boiling in 2× Laemmli sample buffer for 5 min and resolved via SDS-PAGE. Competition assays between CaM and PKC for binding to AKAP79(31–52) were performed as above for the Ca2+-independent CaM binding assay using 85 μg of CaM to approximate cellular concentrations (∼10 μm) of free CaM.
DNA Constructs and Recombinant Proteins
GluR1 in pRK5 and AKAP79 in pEGFP were used as described previously (31). A His-tagged C-terminal fusion of the CaM binding domain (CaMBD; residues 412–480) from the rat small conductance calcium-activated potassium channel (rSK2) in pET33b was kindly provided by John Adelman (Vollum Institute, Oregon Health and Science University). This CaMBD was expressed in BL-21(DE3) cells (Invitrogen) and purified on a nickel column (Qiagen) as described previously (34, 35). The ability of the CaMBD to bind CaM was confirmed by first incubating His-CaMBD (2.5 μg) with nickel-nitrilotriacetic acid-agarose beads (20 μl) in Ca2+-free buffer as described above for interactions between CaM and AKAP79(31–52). Following washing, CaMBD-bound beads were incubated overnight with CaM (85 μg) in the absence or presence of KN-62 or KN-93 (1 μm each). After overnight incubation, the beads were washed four times in the buffer in the continued presence or absence of drug, eluted by boiling in 2× Laemmli sample buffer for 5 min, and resolved by SDS-PAGE.
Cell Culture
HEK 293 cells (ATCC) were obtained at passage 36 and used for a maximum of eight passages. Cell cultures were maintained in DMEM with 10% FBS and penicillin/streptomycin. Cells were plated at low density on 15-mm coverslips and transfected by the calcium phosphate method as described previously (31). 1 μg of each construct was used for each condition. Hippocampal neurons were prepared from 1–2-day-old rat pups and maintained in Neurobasal A supplemented with B27 and penicillin/streptomycin. Experiments were performed at 12–14 days in vitro.
Cell Treatments
Cells were washed three times in extracellular solution containing 150 mm NaCl, 5 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, and 10 mm HEPES (pH 7.40). The CaN inhibitor cyclosporin A (1 μm; LC Laboratories) was included in this solution for all conditions to ensure against CaN-dependent dephosphorylation of GluA1 (25). Cells were pretreated in this solution for 15 min with or without either CaMKII reagents or the PKC inhibitor bisindolylmaleimide I (BIS-I) at the indicated concentrations. Cells were treated with phorbol 12-myristate 13-acetate (PMA; 1 μm; LC Laboratories) or ionomycin (10 μm; EMD Biosciences) for 3 min in the continued presence of the reagents. Treatments were stopped by washing three times with ice-cold PBS. Cells were then lysed in 200 μl of lysis solution containing 150 mm NaCl, 10 mm HEPES, 5 mm EGTA, 5 mm EDTA, 1% Triton X-100, protease inhibitors, and phosphatase inhibitors (pH 7.40). Lysates were incubated on ice for 15 min and then centrifuged at 14,000 × g for 10 min at 4 °C. Supernatants were collected, 2× Laemmli sample buffer was added, and the samples were boiled for 5 min.
Immunoblotting
Samples were separated by SDS-PAGE on 4–12 or 4–20% gels and transferred to nitrocellulose. For the in vitro binding assays, blots were probed with mouse monoclonal antibodies directed against specific PKC isoforms α, β, γ, δ, and ϵ (1:200–1:1000; all from BD Biosciences) or with a rabbit polyclonal antibody directed against PKCζ (1:200; Santa Cruz Biotechnology) or a mouse monoclonal antibody to CaM (1:500; Millipore). Goat anti-rabbit or anti-mouse IgG horseradish peroxidase-conjugated antibodies (1:10,000; Millipore) were used as secondary antibodies. Signals were visualized using enhanced chemiluminescence (Pierce) and digitally acquired and analyzed using Quantity One software (Bio-Rad). For cell-based assays, blots were first probed with either a rabbit monoclonal antibody directed against phospho-GluA1(Ser-831) (1:1000; Millipore) or rabbit antibody directed against phospho-CaMKII(Thr-286) (1:1000; Millipore) followed by the goat anti-rabbit antibody as secondary antibody. Following detection as indicated above, blots were stripped and reprobed with a rabbit antibody directed against the C terminus of GluA1 (0.5 μg/ml; Millipore) or a mouse monoclonal antibody to CaMKII (1:200; Santa Cruz Biotechnology), respectively, to determine the Ser-831 phosphorylation/GluA1 or Thr-286 phosphorylation/CaMKII ratio for each treatment that was normalized to control condition for each experiment. Data were averaged and are expressed as mean ± S.E. and were subjected to one-way analysis of variance followed by a Bonferroni post hoc analysis or by Student's t test. Statistical significance is reported as p < 0.05 or p < 0.01.
Electrophysiology
Whole-cell recordings were made with an Axopatch 200B or a Multiclamp 700A amplifier (Molecular Devices). Patch pipettes (2–4 megaohms) contained 140 mm cesium methanesulfonate, 10 mm HEPES, 5 mm adenosine triphosphate (sodium salt), 5 mm MgCl2, 2 mm CaCl2, and 10 mm BAPTA (pH 7.4). Extracellular solution contained 150 mm NaCl, 5 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, 10 mm glucose, and 1 μm cyclosporin A (pH 7.4). Cells were pretreated in the extracellular solution with or without the indicated CaMKII reagents for 10–15 min and recorded in the continued presence of these reagents. The exception to this was for CaMKIINtide where a myristoylated version was used for pretreatment and a non-myristoylated version was included in the pipette solution because of the limited availability of this reagent. Glutamate (1 mm) was applied in the presence of cyclothiazide (100 μm; Ascent Scientific) to evoke nondesensitizing AMPA receptor currents. Solution exchanges were achieved via a series of flow pipes that were controlled by solenoid valves (Warner Instruments) and moved into position by a piezoelectric bimorph. Currents were digitized at 5 kHz and filtered at 1 kHz with a Digidata 1322A board (Molecular Devices) and Clampex 9 software (Molecular Devices). Series resistance (90–95%) and whole-cell capacitance compensation were used. Series resistance was monitored throughout the experiments by 10-mV hyperpolarizing jumps prior to each application of glutamate. Only cells with series resistance <6 megaohms and stable throughout the recording were included for analysis. All experiments were initiated within 1 min of establishing the whole-cell configuration and were performed at a holding potential of −60 mV at 20 °C. Currents were normalized to the amplitude of current from the initial agonist application for each experiment. Data are expressed as means ± S.E. and were subjected to analysis of variance followed by Student's t test.
RESULTS
CaMKII Inhibitor KN-62 Modifies PKC Binding to AKAP79(31–52) in Isoform-dependent Manner
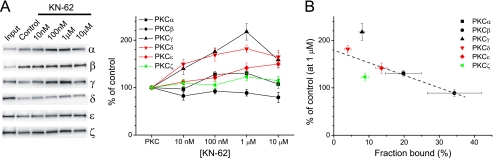
An in vitro binding assay was adopted to examine whether CaMKII inhibitors affect PKC interaction with AKAP79. These assays relied on the use of the AKAP79(31–52) peptide, which qualitatively replicates the essential properties of full-length AKAP79 interactions with PKC and CaM (32, 33). Biotinylated AKAP79(31–52) peptide was first coupled to neutravidin-coated beads and subsequently incubated with PKC in varying concentrations of KN-62. Control experiments indicated that PKC did not bind to the beads in the absence of AKAP79(31–52) or to other AKAP-related peptides including the CaN binding domain of AKAP79 or a prototypical AKAP-derived PKA binding domain (supplemental Fig. 1). Consistent with previous studies (32), all isoforms of PKC tested associated with AKAP79(31–52) (supplemental Fig. 1). However, each isoform associated with the peptide to varying degrees with the relative rank order of binding PKCβ > PKCα > PKCϵ > PKCζ ≈ PKCγ > PKCδ (Fig. 1, A and B). All PKC isoforms except for PKCβ and PKCζ exhibited a significant dose-dependent increase in binding to AKAP79(31–52) in response to KN-62 that was typically half-maximal at ∼10–100 nm (Fig. 1A), concentrations lower than that necessary to inhibit CaMKII in vitro (Ki ∼ 900 nm) and that do not cause PKC inhibition (2). However, PKC association to AKAP79(31–52) tended to decline at the highest concentration tested (10 μm). Interestingly, the degree to which the binding of each PKC isoform to AKAP79(31–52) was altered by KN-62 was inversely related to the relative affinity each isoform had for AKAP79(31–52) such that those isoforms that bound relatively weakly were more likely to be stabilized by the drug (Fig. 1, A and B). Collectively, these data suggest that AKAP79 may be a novel target for KN-62.
FIGURE 1.
KN-62 affects AKAP79-PKC interactions in isoform-dependent manner. AKAP79(31–52) was incubated with PKC isoforms (200 ng; Input lane, 25 ng) and KN-62 as indicated. A, left, representative blots showing the effect of KN-62 on PKC isoform binding AKAP79(31–52). Right, graphical summary of data normalized to their respective controls. Statistical significance for each group is as follows: PKCα: n = 6; 100 nm and 1 μm, p < 0.05; PKCβ: n = 4–5; not significant; PKCγ: n = 5–6; 100 nm and 1 μm, p < 0.01; 10 μm, p < 0.05; PKCδ: n = 4–5; 10 nm–10 μm, p < 0.01; PKCϵ: n = 7–8; 1–10 μm, p < 0.01; and PKCζ: n = 4–5; not significant. B, percentage of control binding to AKAP79(31–52) in response to KN-62 (1 μm) is plotted versus the fraction of input bound for each PKC isoform. Percentages are calculated as the amount of PKC recovered in control lanes normalized to the input lane. Black, red, and green symbols reflect typical, novel, and atypical PKC isoforms, respectively. The dashed line represents the best fit through the data determined by linear regression analysis (r = 0.85). All graphs depict mean ± S.E.
PKCα and PKCγ Binding to AKAP79 are Differentially Sensitive to CaMKII Reagents
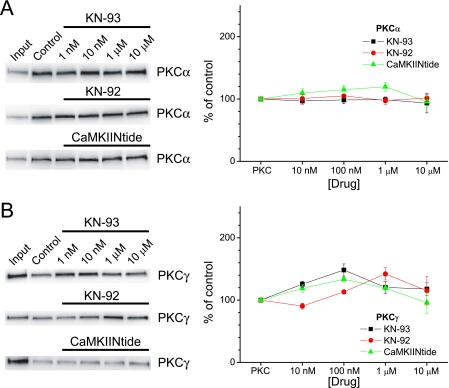
Although KN-62 is still widely used as a pharmacological agent to suppress CaMKII activity, it is often supplanted by KN-93, which benefits from higher potency (Ki ∼ 370 nm), lower molecular weight (which should enhance cell permeability), and a close structural analog (KN-92) that does not affect CaMKII and can thus be used to control for nonspecific effects (3). More recently, a high affinity peptide (CaMKIINtide; Ki ∼ 50 nm) derived from an endogenous CaMKII inhibitory protein has been used as a pharmacological tool to inhibit CaMKII activity (36). CaMKIINtide differs in that it inhibits Ca2+-dependent and Ca2+-independent (i.e. autophosphorylated) CaMKII activity, whereas KN-62 and KN-93 only inhibit Ca2+-dependent CaMKII activity (2, 3, 36). Given that KN-62 generally enhanced PKC binding to AKAP79, we tested whether other CaMKII reagents could likewise enhance association of PKC to AKAP79 using PKCα as a prototypical isoform. Neither KN-93 nor KN-92 affected PKCα binding to AKAP79(31–52), whereas CaMKIINtide only modestly, but not significantly, affected binding with a maximal enhancement of ∼20% (Fig. 2A). These data initially suggested that these compounds may be more selective for CaMKII compared with AKAP79/PKC interactions. However, further examination using PKCγ, which was the isoform most sensitive to KN-62, revealed that all CaMKII reagents could enhance association of PKCγ to AKAP79(31–52) to a similar maximal extent (∼35–45%) (Fig. 2B). As was observed for KN-62 (Fig. 1A), at the highest concentrations tested, all compounds exhibited noticeable declines in their ability to enhance PKCγ association to AKAP79(31–52) (Fig. 2B). These results were unexpected given that CaMKIINtide operates by a distinct mechanism from that of KN-62 and KN-93 to inhibit CaMKII and the lack of effect of these reagents toward PKCα. Whether this reflected the high sensitivity of our assay to detect quantitatively small changes in binding, which would be magnified when compared with the low affinity of PKCγ for AKAP79, is not clear. However, as these compounds do not directly affect PKC activity and thus are unlikely to bind PKC, these results further suggest that AKAP79 represents a novel high affinity target for CaMKII reagents.
FIGURE 2.
Differential effects of CaMKII inhibitors on PKCα and PKCγ. AKAP79(31–52) was incubated with PKCα and KN-93 (n = 3–4), KN-92 (n = 3–4), or CaMKIINtide (n = 8) as indicated (A) or PKCγ and KN-93 (n = 4–5; 100 nm, p < 0.01), KN-92 (n = 3; 1 μm, p < 0.05), or CaMKIINtide (n = 3–4; 100 nm, p < 0.05) (B). Left panels, representative blots for each condition. Right panels, summary data normalized to respective controls. All graphs depict mean ± S.E.
KN-62 and KN-93 Enhance Ca2+-independent Binding of CaM to AKAP79(31–52)
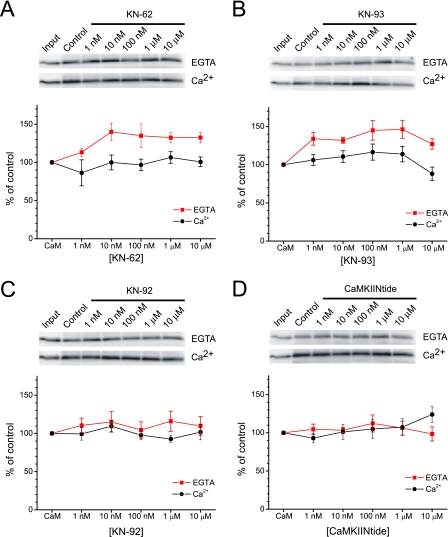
Because AKAP79(31–52) also serves as a binding site for CaM, we next tested whether the CaMKII reagents also affected CaM association with AKAP79(31–52). Control experiments indicated that CaM did not associate with neutravidin beads, in either the absence or presence of Ca2+, unless AKAP79(31–52) was present (supplemental Fig. 2). Previous studies suggested that CaM only associates with AKAP79 in the presence of Ca2+ (33). However, we found that apoCaM associated with AKAP79(31–52), although ∼33% less effectively than Ca2+/CaM (Fig. 3), as assessed by the ratio of control binding to input in the absence and presence of Ca2+. Importantly, both classical CaMKII inhibitors, KN-62 and KN-93, potently (EC50 ∼ 1–10 nm) enhanced apoCaM binding to AKAP79(31–52) by ∼ 40% without affecting the Ca2+-dependent association of CaM to the peptide (Fig. 3, A and B). Thus, in the presence of KN-62 and KN-93, apoCaM bound AKAP79(31–52) to nearly the same extent as Ca2+/CaM. The enhancement of apoCaM association to AKAP79 by KN-62 and KN-93 occurred at concentrations that were ∼10–100-fold lower than their previously measured Ki toward CaMKII. In contrast, the control analog KN-92 did not substantially alter CaM association with AKAP79(31–52) regardless of the presence of Ca2+ (Fig. 3C). Thus, apoCaM binding to AKAP79(31–52) possesses a pharmacological profile matching that of CaMKII yet with over an order of magnitude greater potency. The mechanistically distinct CaMKIINtide did not affect CaM association with AKAP79(31–52) (Fig. 3D). Because KN-62 and KN-93 do not appear to bind CaM or PKC (2, 3), these data further support the idea that AKAP79(31–52) is a high affinity target for common small molecule inhibitors of CaMKII.
FIGURE 3.
Classical CaMKII inhibitors enhance Ca2+-independent binding of CaM to AKAP79. AKAP79(31–52) was incubated with CaM (1 μm; Input lane, 50 ng) and KN-62 at the indicated concentrations in the absence or presence of Ca2+. A, top, representative blots of CaM binding to AKAP79(31–52) in 1 mm EGTA (upper) or 100 μm Ca2+ (lower). Bottom, summary data normalized to respective controls (n = 3–6; for EGTA: 10 nm–10 μm KN-62, p < 0.05). B, same as A but with KN-93 (n = 5–7; for EGTA: 1 nm, 100 nm, and 1 μm, p < 0.01; 10 nm, p < 0.05). C and D, same as above except with KN-92 (n = 6) and CaMKIINtide (n = 5–6), respectively. All graphs depict mean ± S.E.
KN-62 and KN-93 Favor ApoCaM-mediated Displacement of PKC from AKAP(31–52)
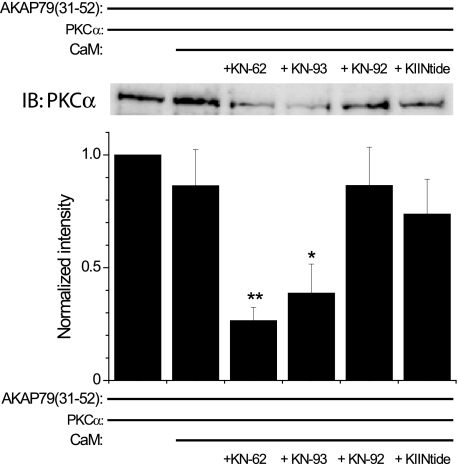
Given that Ca2+/CaM displaces PKC from AKAP79(31–52) and that KN-62 and KN-93 enhance apoCaM binding to AKAP79(31–52) to a similar extent as Ca2+/CaM, we hypothesized that these reagents could cause apoCaM, when present at relevant cellular concentrations (10 μm), to displace PKC from the AKAP. Given that our data suggested that the ability of the various CaMKII reagents to maximally enhance PKC binding to AKAP79 typically occurred at ∼1 μm (Figs. 1 and 2), we chose this concentration to stringently test our hypothesis. As shown in Fig. 4, apoCaM alone does not significantly affect PKCα binding to AKAP79(31–52). However, in the presence of KN-62 or KN-93 (1 μm each), apoCaM effectively displaced PKCα from AKAP79(31–52) by ∼69 and 55%, respectively, compared with that observed in the absence of the drugs (Fig. 4). Moreover, KN-92 and CaMKIINtide (1 μm each) failed to drive apoCaM-mediated displacement of PKCα from AKAP79(31–52) (Fig. 4). These data suggest that KN-62 and KN-93 may primarily act to displace PKC from AKAP79(31–52) particularly in cellular environments where CaM is abundant.
FIGURE 4.
Classical CaMKII inhibitors induce apoCaM-mediated displacement of PKC from AKAP79. AKAP(31–52) was incubated with PKCα (200 ng) in the absence or presence of apoCaM (10 μm) and various CaMKII reagents (each 1 μm). Top, representative Western blot of PKCα binding to AKAP79(31–52) in the absence or presence of apoCaM and the indicated CaMKII reagents. Bottom, graph summarizing the data (mean ± S.E.) from multiple experiments (n = 6). For each experiment, the data were normalized to the amount of PKC bound to AKAP79(31–52) alone. *, p < 0.05; **, p < 0.01 compared with PKCα + AKAP79(31–52) + apoCaM. KIINtide, CaMKIINtide; IB, immunoblot.
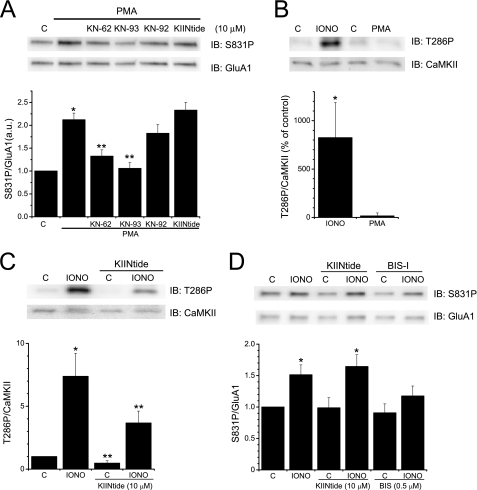
KN-62 and KN-93 Prevent PKC Phosphorylation of GluR1 in Hippocampal Neurons
Previous work suggests that AKAP79/150 contributes to PKC-mediated phosphorylation of GluA1 in hippocampal neurons (31). Moreover, the ability of AKAP79/150 to facilitate PKC signaling is critically dependent on its ability to associate with PKC (31, 37). If, as suggested by our data, KN-62 and KN-93 displace PKC from AKAP79, then these reagents should disrupt endogenous AKAP79/150-dependent PKC signaling within a cellular context. Thus, we tested whether CaMKII reagents affect PKC-mediated phosphorylation of GluA1 in cultured hippocampal neurons. Application of the phorbol ester PMA (1 μm; 3 min) to activate PKC led to approximately a doubling of GluA1 phosphorylation at Ser-831 (Fig. 5A). Under conditions commonly used to assess CaMKII involvement in cellular studies, pretreating neurons with either KN-62 or KN-93 (10 μm; 15 min) significantly (p < 0.01) inhibited the PMA-induced increase in Ser-831 phosphorylation (Fig. 5A). In contrast, similar pretreatment of neurons with the control compound KN-92 or CaMKIINtide did not affect PMA-induced Ser-831 phosphorylation (Fig. 5A.). CaMKII was not activated to a significant extent by the PMA treatment as assessed by its phosphorylation at Thr-286 (Fig. 5B); albeit treatment of cells with a Ca2+ ionophore, ionomycin (10 μm; 3 min), revealed that CaMKII could be substantially activated when appropriately stimulated (Fig. 5B). Thus, the ability of KN-62 and KN-93 (and the inability of KN-92) to inhibit PMA-induced phosphorylation of Ser-831 is consistent with their ability to interfere with AKAP79/150-dependent PKC signaling. Moreover, treatment of cells with CaMKIINtide attenuated both basal and Ca2+-induced phosphorylation of CaMKII at Thr-286 by ∼52% (Fig. 5C), consistent with the noncompetitive action of this reagent (Ref. 36 but see Ref. 38). This suggests that the inability of CaMKIINtide to attenuate the PMA-induced increase in Ser-831 phosphorylation was not due to a failure of this compound to enter cells and inhibit CaMKII but rather as a consequence of its inability to impair AKAP79/150-dependent PKC signaling. In contrast to the ability of CaMKIINtide to suppress Ca2+-induced CaMKII autophosphorylation, CaMKIINtide did not affect GluA1 phosphorylation at Ser-831 in response to ionomycin treatment (Fig. 5D). Instead, ionomycin-induced phosphorylation of GluA1 at this site was reduced by the PKC inhibitor BIS-I (500 nm; Fig. 5D), suggesting that PKC may be primarily responsible for rapid phosphorylation of GluA1 at Ser-831 in neurons, consistent with the targeting of PKC to GluA1 via AKAP79/150. Collectively, these data indicate that both KN-62 and KN-93 effectively block a PKC-mediated event in the absence of CaMKII activation. Moreover, these data indicate that the pharmacological profile of the CaMKII reagents toward Ser-831 phosphorylation more closely matches the ability of these reagents to enhance apoCaM association to AKAP79(31–52) and consequently displace PKC from AKAP79(31–52) than to inhibit CaMKII. Thus, our combined results suggest that KN-62 and KN-93 favor CaM binding over PKC binding to AKAP79/150 and therefore prevent the ability of AKAP79/150 to efficiently recruit PKC to GluA1 for phosphorylation of Ser-831.
FIGURE 5.
Classical CaMKII inhibitors block PKC phosphorylation of GluA1 in neurons. A, hippocampal neurons were untreated (i.e. control (C)) or pretreated with CaMKII reagents (10 μm each) as indicated prior to PMA application (1 μm; 3 min) to activate PKC. Top, representative blots of Ser-831 phosphorylation (S831P) visualized using a phosphospecific antibody (upper panel) or to total GluA1 (lower panel). Bottom, summary graph of the ratio of Ser-831 phosphorylation/GluR1 signals normalized to control (n = 8–13; *, p < 0.01 compared with control; **, p < 0.01 compared with PMA). B, neurons were untreated (C) or stimulated with ionomycin (IONO; 10 μm; 3 min) or PMA (1 μm; 3 min). Top, representative blots demonstrating increase in CaMKII phosphorylation at Thr-286 (T286P) in response to ionomycin but not to PMA (upper panel). Blots were reprobed with an antibody directed against CaMKII (lower panel). Bottom, summary graph of the ratio of phosphorylation at Thr-286/CaMKII signals as percentage of control (n = 3; *, p < 0.05 compared with control). C, neurons were untreated (C) or stimulated with ionomycin (10 μm; 3 min) with or without CaMKIINtide (KIINtide; 10 μm; 15 min) pretreatment. Top, representative blots demonstrating CaMKIINtide suppression of basal and ionomycin-induced phosphorylation of CaMKII at Thr-286 (T286P) (upper panel). Blots were reprobed with an antibody directed against CaMKII (lower panel). Bottom, summary graph of the ratio of phosphorylation of CaMKII at Thr-286/CaMKII signals normalized to control (n = 7; *, p < 0.01 compared with control; **, p < 0.05 compared with the corresponding condition in the absence of CaMKIINtide). D, neurons were untreated (C) or stimulated with ionomycin (10 μm; 3 min) with or without CaMKIINtide (KIINtide; 10 μm; 15 min) or BIS-I (0.5 μm; 15 min) pretreatment. Top, representative blots demonstrating that the PKC inhibition by BIS-I but not CaMKII inhibition by CaMKIINtide suppressed the ionomycin-induced phosphorylation of GluA1 at Ser-831 (S831P) (upper panel). Blots were reprobed with an antibody directed against GluR1 (lower panel). Bottom, summary graph of the ratio of phosphorylation of GluA1 at Ser-831/GluA1 signals normalized to control (n = 6; *, p < 0.01 compared with their respective controls). IB, immunoblot. All graphs depict mean ± S.E.
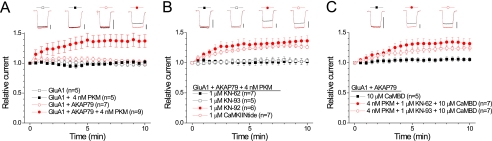
KN-62 and KN-93 Prevent AKAP79-anchored PKC Regulation of GluR1 Receptor Currents
To more directly test this hypothesis, we examined whether the CaMKII reagents could disrupt the ability of AKAP79 to facilitate PKC-mediated GluA1 regulation using a simplified preparation. As demonstrated previously (31), infusion of the constitutively active catalytic fragment of PKC (PKM; 4 nm) into HEK 293 cells expressing GluA1 leads to ∼35–40% augmentation of current only in cells co-expressing full-length AKAP79 (Fig. 6A). This facilitation of PKC regulation of GluA1 by AKAP79 requires the PKC binding domain on AKAP79 and is mediated via Ser-831 (31). Pretreating cells with either KN-62 or KN-93 (1 μm each) ablated this AKAP79-dependent PKC-mediated augmentation of GluA1 receptor currents (Fig. 6B). It is important to note that the high intracellular concentration of the Ca2+ chelator BAPTA used in these experiments prevented activation of endogenous CaMKII. In contrast, pretreating cells with the control compound, KN-92 (1 μm), did not alter the ability of AKAP79 to augment GluA1 receptor currents in response to PKM infusion (Fig. 6B). Similarly, pretreatment with myristoylated CaMKIINtide (1 μm) followed by co-infusion of CaMKIINtide (1 μm) did not prevent the PKM-mediated enhancement of GluA1 receptor currents in the presence of AKAP79 (Fig. 6B). If, as proposed above, KN-62 and KN-93 disrupt AKAP79-anchored PKC signaling to GluA1 by favoring CaM binding over PKC binding, then buffering intracellular CaM should restore the ability of PKM to augment GluA1 receptor currents in cells expressing AKAP79 even when these compounds are present. To test this idea, we infused the CaMBD derived from the rat SK2 small conductance Ca2+-activated potassium channel into cells expressing AKAP79 and GluA1 (34, 35). This CaMBD binds CaM with high affinity regardless of the presence of Ca2+ (34, 35). Consistent with our prediction, PKM enhanced GluA1 receptor currents in cells expressing AKAP79 upon co-infusion of the CaMBD (10 μm) despite the presence of KN-62 or KN-93 (Fig. 6C). This augmentation was not due to a nonspecific action of the CaMBD as control experiments revealed that infusion of the CaMBD (10 μm) alone did not modify GluA1 receptor currents in cells expressing AKAP79 (Fig. 6C). Further control experiments indicated that the ability of the CaMBD to bind CaM was not altered by either KN-62 or KN-93 (supplemental Fig. 3). This suggests that the CaMBD does not bind KN-62 or KN-93 and that it is unlikely to attenuate their action by acting as a buffer for these compounds. Collectively, these results demonstrate a novel mechanism of action for KN-62 and KN-93 that renders AKAP79-anchored PKC and CaMKII indistinguishable from each other based on sensitivity to these reagents and a common substrate.
FIGURE 6.
Classical CaMKII inhibitors prevent AKAP79-anchored PKC regulation of GluA1. A, HEK cells were transfected with GluA1 ± AKAP79. A summary time course of GluA1 receptor currents is shown, demonstrating that AKAP79 facilitates PKC regulation of GluA1 (GluA1 AKAP79 versus GluA1 + AKAP79 + PKM, p < 0.05). PKM (4 nm) was included in the patch pipette as indicated. All data are expressed as mean ± S.E. The number of observations for each condition is indicated. Insets, representative glutamate-evoked (500-ms) current traces from the first (black) and final (red) sweep (10 min) for each condition. Vertical scale bars equal 500 pA. B, KN-62 and KN-93, but not KN-92 or CaMKIINtide, inhibit AKAP79-anchored PKC regulation of GluR1 (KN-62 and KN-93 both p < 0.05 compared with GluA1 + AKAP79 + PKM from A). Cells were transfected with GluA1 + AKAP79. Patch pipettes contained PKM. Cells were pretreated with KN-62, KN-93, KN-92, or CaMKIINtide and recorded in the continued presence of these reagents. Data are depicted as in A except that vertical scale bars equal 1 nA. C, buffering CaM by infusion of the CaMBD (10 μm) restores AKAP79-anchored PKC-mediated up-regulation of GluA1 receptor currents in the presence of KN-62 and KN-93. Cells were transfected with GluA1 + AKAP79. Cells were infused with the CaMBD alone or the CaMBD + PKM in the presence of KN-62 or KN-93. Data are depicted as in A except that vertical scale bars equal 1 nA. The CaMBD alone did not modify the stability of GluR1 receptor currents (compare with GluA1 + AKAP79 in A). However, infusion of the CaMBD prevented KN-62- and KN-93-mediated inhibition of AKAP79-anchored PKC regulation of GluA1 (both p < 0.05 compared with the corresponding treatments in B).
DISCUSSION
Prior studies undeniably support a critical role for CaMKII in controlling synaptic structure, plasticity, learning, and memory (for a review, see Ref. 39). However, identification of bona fide CaMKII substrates contributing to and/or mediating enhancement of AMPA receptor responses during synaptic plasticity remains less well defined (40, 41). This stems from the fact that LTP likely arises from phosphorylation of multiple substrates, each participating in phosphorylation-dependent processes enacted sequentially and/or in parallel. Failure of one or more of these processes may be sufficient to impair LTP. Thus, kinase inhibitors might directly or indirectly reduce the phosphorylation of any specific LTP-related substrate.
The best characterized CaMKII substrate reliably phosphorylated during LTP and contributing to LTP, learning, and memory is CaMKII itself at Thr-286 (14, 42, 43). Aside from this site, Ser-831 of GluA1 is one of the best characterized sites that directly impacts AMPA receptor function linked to LTP and learning (14–19). Because this site is also a substrate for PKC, both kinases may have overlapping roles in synaptic plasticity. In fact, selective elevation of either kinase, by direct infusion of constitutively active forms of these enzymes into neurons, potentiates AMPA receptor-mediated synaptic transmission and occludes LTP, suggesting that common sites and/or processes underlie these phenomena (44, 45). However, assessing the role of endogenous kinases toward LTP generation typically necessitates a reliance on pharmacological tools. Although early studies found that peptide inhibitors of either kinase prevent LTP induction (46, 47), subsequent work found that CaMKII-“specific” peptide inhibitors were also potent PKC inhibitors, and the inhibitory action of these reagents toward LTP correlated well with PKC inhibition (48). Thus, sensitivity to small molecule inhibitors of CaMKII, such as KN-62 and KN-93, with negligible activity toward PKC has been used as key evidence for CaMKII mediation of LTP (14, 49–52).
Our data indicate that the classical CaMKII inhibitors KN-62 and KN-93 influence the association of PKC and CaM with AKAP79(31–52). ApoCaM association to AKAP79(31–52) was more potently enhanced by these compounds compared with PKC association to the peptide. This enhancement persisted throughout the concentration range tested. Quantitatively, these reagents enhanced apoCaM binding to AKAP79(31–52) to nearly the same extent observed for CaM in the presence of Ca2+. Neither the “control” reagent, KN-92, nor the novel peptide reagent, CaMKIINtide, affected association of CaM to AKAP79(31–52). Although these reagents enhanced binding of some PKC isoforms to AKAP79(31–52) to a similar degree as observed for apoCaM binding to the peptide, it must be emphasized that the cellular concentration of CaM is likely to be several orders of magnitude more abundant than PKC. Thus, under cellular conditions, it would be expected that these reagents would much more readily drive CaM association to AKAP79/150 at the expense of PKC as observed in our in vitro apoCaM/PKC competition assay (Fig. 4). Indeed, the in vitro pharmacological profile for apoCaM binding to AKAP79(31–52) and displacement of PKC was matched in two cellular assays that are reflective of AKAP79/150-anchored PKC signaling. Thus, the Ca2+-independent association of CaM to AKAP79 is likely the primary means by which these reagents attenuate AKAP79/150-anchored PKC signaling to GluA1. This idea was supported by experiments in which buffering of CaM restored AKAP79-anchored PKC regulation of GluR1 in the presence of these common CaMKII inhibitors.
At present, it is unclear at the structural level how the CaMKII reagents modify PKC and apoCaM association to AKAP79(31–52). However, it is interesting to note that although CaMKII and AKAP79 preferentially associate with the Ca2+-bound form of CaM these proteins likely utilize different CaM binding motifs (53). As CaM binding domains are known to exhibit a high degree of conformational flexibility (53, 54), KN-62 and KN-93 may stabilize distinct conformations of the CaMBDs in AKAP79 and CaMKII. Therefore, these reagents may induce conformations whereby the apoCaM association to AKAP79 is stabilized but Ca2+/CaM association to CaMKII is destabilized. It is also important to note that CaM also exhibits a high degree of conformational flexibility (53, 54). Thus, despite the apparent inability of these compounds to directly interact with CaM, it is possible that the remarkably high apparent affinity of these reagents toward the AKAP79/apoCaM interaction (relative to that of CaM/CaMKII) is achieved, in part, via reciprocal conformational adaptations in apoCaM that recognize the KN-62- and KN-93-induced state of AKAP79. Presumably, the catalytic core of PKC, which binds AKAP79(31–52), exhibits less conformational flexibility than CaM and consequently is more restrictive toward the conformations adopted by pseudosubstrates like AKAP79. This may contribute not only to the lower potency of these reagents toward stabilizing AKAP79/PKC interactions but also to the tendency for AKAP79/PKC interaction to decline at the highest concentration tested. Interestingly, neither reagent affected CaM association to the SK2-derived CaMBD. In light of our data and a previous study (11), it may be more appropriate to describe these reagents as modifiers of CaM binding. Given the prevalence of CaM-binding proteins (54), it will be important to determine whether other CaM-binding proteins also exhibit sensitivity to KN-62 and KN-93.
Our data complement and extend a recent study that found that AKAP79 modifies the cellular pharmacology of PKC by conferring resistance to some ATP-competitive PKC inhibitors including BIS-I (55). Although, we have found that BIS-I is effective in abrogating both PMA- and ionomycin-stimulated PKC phosphorylation of hippocampal GluA1 receptors (31), this does not necessarily contradict the aforementioned study as AKAP79-induced PKC resistance to BIS-I is time- and Ca2+-dependent (55). Thus, our pretreatment protocol was likely long enough to suppress the activity of AKAP150-localized pools of PKC. Additionally, ongoing synaptic activity and neuronal firing in our neuronal cultures may have provided sufficient levels of intracellular Ca2+ to enhance BIS-I access to PKC associated with AKAP150. AKAP79/150 thus not only restricts PKC sensitivity to some PKC-selective reagents but as illustrated here also endows PKC with pharmacological sensitivity to classical CaMKII inhibitors that do not normally target PKC.
Enhancement of GluA1 phosphorylation at Ser-831 is linked with LTP and is also a downstream consequence of other manipulations that alter synaptic function (56–72). KN-62 or KN-93 sensitivity and/or CaMKII autophosphorylation at Thr-286 has been used as central criteria for CaMKII involvement in these phenomena (57, 61, 62, 68, 70–72). However, PKC, like CaMKII, is a Ca2+-dependent enzyme, and thus, both would be expected to be rapidly activated in response to similar stimuli. Indeed, PKC inhibitors have been shown to be effective at preventing increases in GluA1 Ser-831 phosphorylation in several of these studies (58, 60, 63–65, 67). Given our data demonstrating that KN-62 and KN-93 disrupt AKAP79-dependent PKC regulation of GluA1 (mediated via Ser-831 phosphorylation), the combination of these inhibitors and CaMKII autophosphorylation is unlikely to be sufficient to discriminate between CaMKII and AKAP79-anchored PKC activity.
Although it is tempting to speculate that because of its precise targeting to GluA1 AKAP79/150-anchored PKC is principally responsible for Ser-831 phosphorylation, there are cases in which phosphorylation of this site appears to be selectively sensitive to PKC inhibitors (63, 73). Whether these instances reflect the tissue- and/or development-specific constraints on AKAP79/150-anchored PKC signaling or are due to differences in stimulus intensity that bypass the need for AKAP79/150 remains unknown. It is also important to point out that although AKAP79-anchored PKC shares a common substrate and overlapping pharmacology with CaMKII the presence of AKAP79 does not preclude CaMKII regulation of GluA1 at Ser-831 (31). Thus, it is plausible that both proteins are important for phosphorylation at this site but that they do so to discrete populations of GluA1-containing receptors and/or with distinct temporal profiles and/or by distinct mechanisms. Indeed, recent work indicates that CaMKII, by phosphorylation of SAP97, can abrogate AKAP79/150-directed signaling to GluA1 and thus indirectly influence GluA1 phosphorylation (74). Because the AKAP79/150 signaling complex is targeted to other ion channels (21, 37, 75–80), future studies will need to address whether AKAP79/150 similarly endows these potential targets with KN-62 and KN-93 sensitivity.
Supplementary Material
Acknowledgments
We are grateful to John Adelman for providing the CaMBD construct and Maria Schumacher for advice regarding bacterial expression and purification of the CaMBD.
This work was supported, in whole or in part, by National Institutes of Health Grant NS46661 (to S. J. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- CaMK
- Ca2+/calmodulin-dependent protein kinase
- AKAP
- A-kinase anchoring protein
- BIS-I
- bisindolylmaleimide I
- CaM
- calmodulin
- CaMBD
- CaM binding domain
- CaN
- calcineurin (also known as protein phosphatase 2B)
- KN-62
- 1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine
- KN-92
- 2-[N-(4′-methoxybenzenesulfonyl)]amino-N-(4′-chlorophenyl)-2-propenyl-N-methylbenzylamine phosphate
- KN-93
- 2-[N-(2-hydroxyethyl)-N-(4-methoxybenzenesulfonyl)]amino-N-(4-chlorocinnamyl)-N-methylbenzylamine
- LTP
- long-term potentiation
- PKA
- cAMP-dependent protein kinase (also known as protein kinase A or A-kinase)
- PMA
- phorbol 12-myristate 13-acetate
- SAP97
- synapse-associated protein 97
- BAPTA
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- PKM
- catalytic fragment of PKC.
REFERENCES
- 1. Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tokumitsu H., Chijiwa T., Hagiwara M., Mizutani A., Terasawa M., Hidaka H. (1990) J. Biol. Chem. 265, 4315–4320 [PubMed] [Google Scholar]
- 3. Sumi M., Kiuchi K., Ishikawa T., Ishii A., Hagiwara M., Nagatsu T., Hidaka H. (1991) Biochem. Biophys. Res. Commun. 181, 968–975 [DOI] [PubMed] [Google Scholar]
- 4. Gargett C. E., Wiley J. S. (1997) Br. J. Pharmacol. 120, 1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Humphreys B. D., Virginio C., Surprenant A., Rice J., Dubyak G. R. (1998) Mol. Pharmacol. 54, 22–32 [DOI] [PubMed] [Google Scholar]
- 6. Ledoux J., Chartier D., Leblanc N. (1999) J. Pharmacol. Exp. Ther. 290, 1165–1174 [PubMed] [Google Scholar]
- 7. Gao L., Blair L. A., Marshall J. (2006) Biochem. Biophys. Res. Commun. 345, 1606–1610 [DOI] [PubMed] [Google Scholar]
- 8. Rezazadeh S., Claydon T. W., Fedida D. (2006) J. Pharmacol. Exp. Ther. 317, 292–299 [DOI] [PubMed] [Google Scholar]
- 9. Marley P. D., Thomson K. A. (1996) Biochem. Biophys. Res. Commun. 221, 15–18 [DOI] [PubMed] [Google Scholar]
- 10. Yue C., Sanborn B. M. (2001) Mol. Cell. Endocrinol. 175, 149–156 [DOI] [PubMed] [Google Scholar]
- 11. Smyth J. T., Abbott A. L., Lee B., Sienaert I., Kasri N. N., De Smedt H., Ducibella T., Missiaen L., Parys J. B., Fissore R. A. (2002) J. Biol. Chem. 277, 35061–35070 [DOI] [PubMed] [Google Scholar]
- 12. Barria A., Derkach V., Soderling T. (1997) J. Biol. Chem. 272, 32727–32730 [DOI] [PubMed] [Google Scholar]
- 13. Mammen A. L., Kameyama K., Roche K. W., Huganir R. L. (1997) J. Biol. Chem. 272, 32528–32533 [DOI] [PubMed] [Google Scholar]
- 14. Barria A., Muller D., Derkach V., Griffith L. C., Soderling T. R. (1997) Science 276, 2042–2045 [DOI] [PubMed] [Google Scholar]
- 15. Lee H. K., Barbarosie M., Kameyama K., Bear M. F., Huganir R. L. (2000) Nature 405, 955–959 [DOI] [PubMed] [Google Scholar]
- 16. Lee H. K., Takamiya K., Han J. S., Man H., Kim C. H., Rumbaugh G., Yu S., Ding L., He C., Petralia R. S., Wenthold R. J., Gallagher M., Huganir R. L. (2003) Cell 112, 631–643 [DOI] [PubMed] [Google Scholar]
- 17. Whitlock J. R., Heynen A. J., Shuler M. G., Bear M. F. (2006) Science 313, 1093–1097 [DOI] [PubMed] [Google Scholar]
- 18. Shukla K., Kim J., Blundell J., Powell C. M. (2007) J. Biol. Chem. 282, 18100–18107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu H., Real E., Takamiya K., Kang M. G., Ledoux J., Huganir R. L., Malinow R. (2007) Cell 131, 160–173 [DOI] [PubMed] [Google Scholar]
- 20. Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 21. Colledge M., Dean R. A., Scott G. K., Langeberg L. K., Huganir R. L., Scott J. D. (2000) Neuron 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 22. Klauck T. M., Faux M. C., Labudda K., Langeberg L. K., Jaken S., Scott J. D. (1996) Science 271, 1589–1592 [DOI] [PubMed] [Google Scholar]
- 23. Coghlan V. M., Perrino B. A., Howard M., Langeberg L. K., Hicks J. B., Gallatin W. M., Scott J. D. (1995) Science 267, 108–111 [DOI] [PubMed] [Google Scholar]
- 24. Dell'Acqua M. L., Dodge K. L., Tavalin S. J., Scott J. D. (2002) J. Biol. Chem. 277, 48796–48802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tavalin S. J., Colledge M., Hell J. W., Langeberg L. K., Huganir R. L., Scott J. D. (2002) J. Neurosci. 22, 3044–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roche K. W., O'Brien R. J., Mammen A. L., Bernhardt J., Huganir R. L. (1996) Neuron 16, 1179–1188 [DOI] [PubMed] [Google Scholar]
- 27. Kameyama K., Lee H. K., Bear M. F., Huganir R. L. (1998) Neuron 21, 1163–1175 [DOI] [PubMed] [Google Scholar]
- 28. Lee H. K., Kameyama K., Huganir R. L., Bear M. F. (1998) Neuron 21, 1151–1162 [DOI] [PubMed] [Google Scholar]
- 29. Banke T. G., Bowie D., Lee H., Huganir R. L., Schousboe A., Traynelis S. F. (2000) J. Neurosci. 20, 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Esteban J. A., Shi S. H., Wilson C., Nuriya M., Huganir R. L., Malinow R. (2003) Nat. Neurosci. 6, 136–143 [DOI] [PubMed] [Google Scholar]
- 31. Tavalin S. J. (2008) J. Biol. Chem. 283, 11445–11452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Faux M. C., Rollins E. N., Edwards A. S., Langeberg L. K., Newton A. C., Scott J. D. (1999) Biochem. J. 343, 443–452 [PMC free article] [PubMed] [Google Scholar]
- 33. Faux M. C., Scott J. D. (1997) J. Biol. Chem. 272, 17038–17044 [DOI] [PubMed] [Google Scholar]
- 34. Schumacher M. A., Crum M., Miller M. C. (2004) Structure 12, 849–860 [DOI] [PubMed] [Google Scholar]
- 35. Schumacher M. A., Rivard A. F., Bächinger H. P., Adelman J. P. (2001) Nature 410, 1120–1124 [DOI] [PubMed] [Google Scholar]
- 36. Chang B. H., Mukherji S., Soderling T. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10890–10895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoshi N., Zhang J. S., Omaki M., Takeuchi T., Yokoyama S., Wanaverbecq N., Langeberg L. K., Yoneda Y., Scott J. D., Brown D. A., Higashida H. (2003) Nat. Neurosci. 6, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vest R. S., Davies K. D., O'Leary H., Port J. D., Bayer K. U. (2007) Mol. Biol. Cell 18, 5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Colbran R. J., Brown A. M. (2004) Curr. Opin. Neurobiol. 14, 318–327 [DOI] [PubMed] [Google Scholar]
- 40. Malenka R. C., Bear M. F. (2004) Neuron 44, 5–21 [DOI] [PubMed] [Google Scholar]
- 41. Lee H. K. (2006) Pharmacol. Ther. 112, 810–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fukunaga K., Muller D., Miyamoto E. (1995) J. Biol. Chem. 270, 6119–6124 [DOI] [PubMed] [Google Scholar]
- 43. Giese K. P., Fedorov N. B., Filipkowski R. K., Silva A. J. (1998) Science 279, 870–873 [DOI] [PubMed] [Google Scholar]
- 44. Lledo P. M., Hjelmstad G. O., Mukherji S., Soderling T. R., Malenka R. C., Nicoll R. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11175–11179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ling D. S., Benardo L. S., Serrano P. A., Blace N., Kelly M. T., Crary J. F., Sacktor T. C. (2002) Nat. Neurosci. 5, 295–296 [DOI] [PubMed] [Google Scholar]
- 46. Malenka R. C., Kauer J. A., Perkel D. J., Mauk M. D., Kelly P. T., Nicoll R. A., Waxham M. N. (1989) Nature 340, 554–557 [DOI] [PubMed] [Google Scholar]
- 47. Malinow R., Schulman H., Tsien R. W. (1989) Science 245, 862–866 [DOI] [PubMed] [Google Scholar]
- 48. Hvalby O., Hemmings H. C., Jr., Paulsen O., Czernik A. J., Nairn A. C., Godfraind J. M., Jensen V., Raastad M., Storm J. F., Andersen P., et al. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4761–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stanton P. K., Gage A. T. (1996) J. Neurophysiol. 76, 2097–2101 [DOI] [PubMed] [Google Scholar]
- 50. Bortolotto Z. A., Collingridge G. L. (1998) Neuropharmacology 37, 535–544 [DOI] [PubMed] [Google Scholar]
- 51. Huber K. M., Mauk M. D., Kelly P. T. (1995) Neuroreport 6, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 52. Ito I., Hidaka H., Sugiyama H. (1991) Neurosci. Lett. 121, 119–121 [DOI] [PubMed] [Google Scholar]
- 53. Yamauchi E., Nakatsu T., Matsubara M., Kato H., Taniguchi H. (2003) Nat. Struct. Biol. 10, 226–231 [DOI] [PubMed] [Google Scholar]
- 54. Ikura M., Ames J. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoshi N., Langeberg L. K., Gould C. M., Newton A. C., Scott J. D. (2010) Mol. Cell 37, 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takagi Y., Takagi N., Besshoh S., Miyake-Takagi K., Takeo S. (2003) Neurosci. Lett. 341, 33–36 [DOI] [PubMed] [Google Scholar]
- 57. Fu X. Z., Zhang Q. G., Meng F. J., Zhang G. Y. (2004) Neurosci. Res. 48, 85–91 [DOI] [PubMed] [Google Scholar]
- 58. Fang L., Wu J., Lin Q., Willis W. D. (2003) Brain Res. Mol. Brain Res. 118, 160–165 [DOI] [PubMed] [Google Scholar]
- 59. Lecrux C., Nicole O., Chazalviel L., Catone C., Chuquet J., MacKenzie E. T., Touzani O. (2007) Stroke 38, 3007–3015 [DOI] [PubMed] [Google Scholar]
- 60. Caldeira M. V., Melo C. V., Pereira D. B., Carvalho R., Correia S. S., Backos D. S., Carvalho A. L., Esteban J. A., Duarte C. B. (2007) J. Biol. Chem. 282, 12619–12628 [DOI] [PubMed] [Google Scholar]
- 61. Ba M., Kong M., Yang H., Ma G., Lu G., Chen S., Liu Z. (2006) Neurochem. Res. 31, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 62. Bevilaqua L. R., Medina J. H., Izquierdo I., Cammarota M. (2005) Neuroscience 136, 397–403 [DOI] [PubMed] [Google Scholar]
- 63. Jones T. L., Sorkin L. S. (2005) Pain 117, 259–270 [DOI] [PubMed] [Google Scholar]
- 64. Ménard C., Patenaude C., Massicotte G. (2005) Neurosci. Lett. 389, 51–56 [DOI] [PubMed] [Google Scholar]
- 65. Guan Y., Guo W., Robbins M. T., Dubner R., Ren K. (2004) Neurosci. Lett. 366, 201–205 [DOI] [PubMed] [Google Scholar]
- 66. Sharp J. W., Ross C. M., Koehnle T. J., Gietzen D. W. (2004) Neuroscience 126, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 67. Oh J. D., Geller A. I., Zhang G., Chase T. N. (2003) Brain Res. 971, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kramár E. A., Bernard J. A., Gall C. M., Lynch G. (2003) J. Biol. Chem. 278, 10722–10730 [DOI] [PubMed] [Google Scholar]
- 69. Svenningsson P., Tzavara E. T., Witkin J. M., Fienberg A. A., Nomikos G. G., Greengard P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3182–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao D., Watson J. B., Xie C. W. (2004) J. Neurophysiol. 92, 2853–2858 [DOI] [PubMed] [Google Scholar]
- 71. Yuen E. Y., Liu W., Yan Z. (2007) J. Biol. Chem. 282, 16434–16440 [DOI] [PubMed] [Google Scholar]
- 72. Anderson S. M., Famous K. R., Sadri-Vakili G., Kumaresan V., Schmidt H. D., Bass C. E., Terwilliger E. F., Cha J. H., Pierce R. C. (2008) Nat. Neurosci. 11, 344–353 [DOI] [PubMed] [Google Scholar]
- 73. Tsui J., Malenka R. C. (2006) J. Biol. Chem. 281, 13794–13804 [DOI] [PubMed] [Google Scholar]
- 74. Nikandrova Y. A., Jiao Y., Baucum A. J., Tavalin S. J., Colbran R. J. (2010) J. Biol. Chem. 285, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu G., Shi J., Yang L., Cao L., Park S. M., Cui J., Marx S. O. (2004) EMBO J. 23, 2196–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sandoz G., Thümmler S., Duprat F., Feliciangeli S., Vinh J., Escoubas P., Guy N., Lazdunski M., Lesage F. (2006) EMBO J. 25, 5864–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chai S., Li M., Lan J., Xiong Z. G., Saugstad J. A., Simon R. P. (2007) J. Biol. Chem. 282, 22668–22677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oliveria S. F., Dell'Acqua M. L., Sather W. A. (2007) Neuron 55, 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang X., Li L., McNaughton P. A. (2008) Neuron 59, 450–461 [DOI] [PubMed] [Google Scholar]
- 80. Jeske N. A., Patwardhan A. M., Ruparel N. B., Akopian A. N., Shapiro M. S., Henry M. A. (2009) Pain 146, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.