Abstract
Cellular stress induced by nutrient deprivation, hypoxia, and exposure to many chemotherapeutic agents activates an evolutionarily conserved cell survival pathway termed autophagy. This pathway enables cancer cells to undergo self-digestion to generate ATP and other essential biosynthetic molecules to temporarily avoid cell death. Therefore, disruption of autophagy may sensitize cancer cells to cell death and augment chemotherapy-induced apoptosis. Chloroquine and its analog hydroxychloroquine are the only clinically relevant autophagy inhibitors. Because both of these agents induce ocular toxicity, novel inhibitors of autophagy with a better therapeutic index are needed. Here we demonstrate that the small molecule lucanthone inhibits autophagy, induces lysosomal membrane permeabilization, and possesses significantly more potent activity in breast cancer models compared with chloroquine. Exposure to lucanthone resulted in processing and recruitment of microtubule-associated protein 1 light chain 3 (LC3) to autophagosomes, but impaired autophagic degradation as revealed by transmission electron microscopy and the accumulation of p62/SQSTM1. Microarray analysis, qRT-PCR, and immunoblotting determined that lucanthone stimulated a large induction in cathepsin D, which correlated with cell death. Accordingly, knockdown of cathepsin D reduced lucanthone-mediated apoptosis. Subsequent studies using p53+/+ and p53−/− HCT116 cells established that lucanthone induced cathepsin D expression and reduced cancer cell viability independently of p53 status. In addition, lucanthone enhanced the anticancer activity of the histone deacetylase inhibitor vorinostat. Collectively, our results demonstrate that lucanthone is a novel autophagic inhibitor that induces apoptosis via cathepsin D accumulation and enhances vorinostat-mediated cell death in breast cancer models.
Keywords: Apoptosis, Autophagy, Breast Cancer, Cancer Therapy, Caspase
Introduction
Autophagy is an evolutionarily conserved degradation pathway that eliminates certain proteins, defective organelles, and protein aggregates (1). It has been suggested that autophagy may function as a tumor suppressor pathway by degrading cellular components that are damaged or no longer needed (2, 3). Consistent with this idea, mice haploinsufficient for beclin 1 are tumor-prone, and loss of at least one beclin 1 allele has been reported in some primary tumors (4, 5). However, once cancer has formed activation of the autophagy pathway may be utilized to generate ATP via protein recycling to overcome metabolic/hypoxic stress and maintain survival (6, 7). Because nutrient and oxygen deprivation are hallmark characteristics of the tumor microenvironment, disrupting of autophagic degradation may be a promising approach to selectively target cancer cells.
Previous investigations have proposed that persistent autophagy may lead to a caspase-independent form of cell death following depletion of the minimal amount of organelles and proteins needed for survival (8). However, the process of autophagic breakdown of cellular components to maintain bioenergetics also inhibits cell death induced by growth factor withdrawal, insufficient angiogenesis, and various chemotherapeutic agents (9). The induction of autophagy has been observed in malignant cells following treatment with many cancer therapeutics including arsenic trioxide, etoposide, rapamycin, histone deacetylase (HDAC) inhibitors (sodium butyrate and suberoylanilide hydroxamic acid, SAHA, vorinostat), tamoxifen, temozolomide, imatinib, bortezomib, and ionizing radiation (10–16). Several studies have reported that inhibition of autophagy enhances the pro-apoptotic effects of anticancer agents, including vorinostat, cyclophosphamide, and the tyrosine kinase inhibitors imatinib, nilotinib, and dasatinib (17–21). Considering the dissimilar mechanisms of action of these agents, inhibition of autophagy may be a promising strategy to enhance the activity of a broad range of cancer therapeutics.
Disruption of autophagy is commonly achieved by targeted knockdown of genes essential for autophagy such as ATG5 and ATG7 or by using the phosphatidylinositol 3-kinase inhibitor 3-methyladenine (3-MA) or the lysosomotropic drug chloroquine (CQ)2 (17, 19, 22). CQ is a weak base that inhibits lysosomal acidification, which prevents the fusion of autophagosomes with lysosomes and subsequent autophagic degradation (23). CQ and its analog hydroxychloroquine (HCQ) have been used for many years for the treatment of rheumatoid arthritis and human immunodeficiency virus (HIV) and the prophylaxis and treatment of malaria and is now being investigated in clinical trails in combination with conventional cancer therapy (9, 24–27). Currently, CQ and HCQ are the only clinically relevant autophagic inhibitors being used in cancer therapy, but their ability to cause ocular toxicity, especially irreversible retinopathy, underscores the need for additional inhibitors of autophagy (28).
Lucanthone (Miracil D) has been extensively used as an anti-schistome agent. The drug also blocks topoisomerase II activity and has been reported to inhibit AP endonuclease (APE1), an important enzyme in DNA base excision repair (29, 30). Based on these properties, lucanthone is currently being investigated as a sensitizer to chemotherapy and radiation. Here we report a novel mechanism of action for lucanthone that is characterized by the disruption of lysosomal function, inhibition of autophagy, and induction of apoptosis. These effects enable lucanthone to potentiate the anticancer activity of vorinostat. Lucanthone-induced apoptosis occurred through a p53-independent mechanism and was, in part, mediated by an increase in cathepsin D levels. Importantly, lucanthone displayed more potent activity against a panel of breast cancer cell lines compared with CQ. Considering that previous clinical investigations of lucanthone did not note any signs of drug-related ocular toxicity, it may have a better safety profile than CQ or HCQ (31). Collectively, this study provides evidence that lucanthone inhibits autophagy and provides a rationale for its use in combination with anticancer agents that induce this survival pathway.
EXPERIMENTAL PROCEDURES
Cell Lines
MDA-MB-231, HCC1954, BT-474, SKBR-3, MDA-MB-435, HCC1937, and BT-20 breast cancer cell lines were obtained from the American Type Culture Collection (Rockville, MD). HCT116 53+/+ and p53−/− isogenic cell lines were kindly provided by Dr. Bert Vogelstein (John Hopkins, Baltimore, MD). Cell lines were maintained in RPMI supplemented with 10% fetal bovine serum under conditions of 5% CO2 at 37 °C.
Antibodies and Chemicals
Antibodies were obtained from the following commercial sources: anti-tubulin and p53 (Sigma), anti-LC3-II and anti-cleaved caspase-3 (Cell Signaling, Beverly, MA), anti-cathepsin D (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-SQSTM1 (Abnova, Walnut, CA). Horseradish peroxidase-conjugated secondary antibodies for immunoblotting were obtained from Amersham Biosciences (Piscataway, NJ). Alexa Fluor 488 goat anti-mouse and goat anti-rabbit were obtained from Molecular Probes (Eugene, OR). Bortezomib, etoposide, and vorinostat were purchased from the CTRC pharmacy. Chloroquine, giemsa, propidium iodide, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), bafilomycin A1, pepstatin A, Z-VAD-fmk, and acridine orange were obtained from Sigma. Lucanthone was kindly provided by Spectrum Pharmaceuticals.
Quantification of Drug-induced Cytotoxicity
Cell viability was assessed by MTT assay. Cells were seeded into 96-well microculture plates at 10,000 cells per well and allowed to attach for 24 h. Cells were then treated with lucanthone, chloroquine, vorinostat, or combinations for 72 h. Following drug treatment, MTT was added and cell viability was quantified using a BioTek (Winooski, VT) microplate reader. Effects on cell viability were also determined by measuring ATP levels using the ATPlite assay system (PerkinElmer, Waltham, MA) and by trypan blue exclusion. Pro-apoptotic effects following in vitro drug exposure were quantified by propidium iodide (PI) staining and fluorescence-activated cell sorting (FACS) analysis of sub-G0/G1 DNA content as previously described (32).
Immunocytochemistry
Cells were plated on chamber slides and allowed to attach overnight. Cells were then treated for 48 h with lucanthone or CQ. Following drug treatment, cells were fixed with 4% paraformaldehyde, permeabilized using 0.2% Triton X-100, and incubated overnight with indicated primary antibodies. Alexa Fluor 488 conjugated fluorescent secondary antibodies were used to visualize protein localization. Images were captured using an Olympus fluorescent microscope (Center Valley, PA) with a DP71 camera and a 60X objective. Image-Pro Plus software Version 6.2.1 (MediaCybernetics, Bethesda, MD) was used for image acquisition. Quantification of fluorescence was determined in 5 random fields using ImageJ software.
Giemsa Staining
Cells were plated in chamber slides and treated with lucanthone or CQ for 48 h. After drug treatment, cells were washed with PBS and fixed in methanol for 5 min. Cells were then incubated for 1 h in Giemsa stain diluted 1:20 with deionized water. Cells were rinsed with water and imaged using an Olympus fluorescent microscope. Image-Pro Plus software version 6.2.1 was used for image acquisition.
Acridine Orange Staining
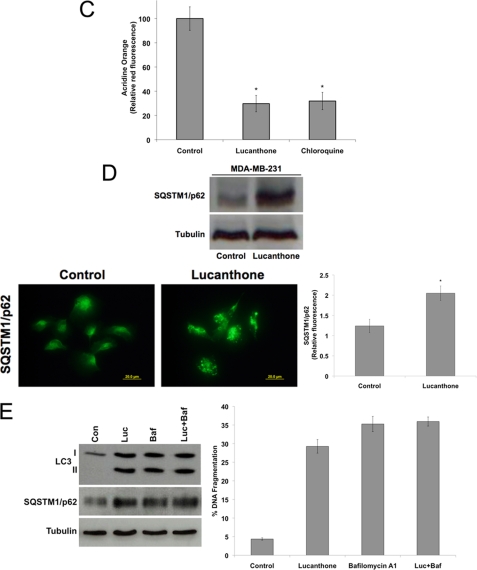
Acidic lysosomes were visualized by acridine orange staining. After treatment with lucanthone or CQ for 48 h, cells were stained with 1 μm acridine orange for 15 min at 37 °C. Cells were washed with PBS and images were captured using an Olympus fluorescent microscope. Based on the acidity, lysosomes appeared as orange fluorescent cytoplasmic vesicles. Quantification of 5 random fields of acridine orange intensity and image acquisition were performed using Image-Pro Plus software Version 6.2.1.
Transmission Electron Microscopy
Transmission electron microscopy of cells was performed as previously described (19). Sections were cut in an LKB Ultracut microtome (Leica, Deerfield, IL), stained with uranyl acetate and lead citrate, and examined in a JEM 1230 transmission electron microscope (JEOL, USA, Inc., Peabody, MA). Images were captured using the AMT Imaging System (Advanced Microscopy Techniques Corp, Danvers, MA).
Immunoblotting
Colon cancer cells were incubated with 10 μm lucanthone, 50 μm CQ, or 2.5 μm vorinostat for 48 h. Cells were harvested and were then lysed as previously described (33). Approximately 50 μg of total cellular protein from each sample were subjected to SDS-PAGE, proteins were transferred to nitrocellulose membranes, and the membranes were blocked with 5% nonfat milk in a Tris-buffered saline solution containing 0.1% Tween-20 for 1 h. The blots were then probed overnight at 4 °C with primary antibodies, washed, and probed with species-specific secondary antibodies coupled to horseradish peroxidase. Immunoreactive material was detected by enhanced chemiluminescence (West Pico, Pierce).
Preparation and Transfection of siRNA and shRNA
Cathepsin D and non-target SMARTpool siRNA were obtained from Dharmacon (Lafayette, CO). Cells were transfected with 100 nm of each siRNA using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Transfected cells were incubated at 37 °C for 24 h and then treated with drugs for 48 h. Efficiency of RNAi was measured at 48 h by immunoblotting. Lentiviral particles with ATG7 or empty vector shRNA were obtained from Dharmacon. MDA-MB-231 cells were infected for 24 h, and cells were selected with puromycin. ATG7 knockdown was determined by immunoblotting.
Expression Microarrays
Breast cancer cells were treated with 10 μm lucanthone for 48 h. Total RNAs were isolated using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD) and treated with TURBO DNA-freeTM kit (Applied Biosystems, Foster City, CA). 300 ng of total RNA per sample was amplified and hybridized to GeneChip® Human Gene 1.0 ST arrays (Affymetrix, Inc., Santa Clara, CA) according to the manufacturer's instructions. These arrays assay for the expression of about 28,869 well-annotated genes with 764,885 distinct probes. Affymetrix CEL files were imported into Partek® Genomics SuiteTM 6.4 (Partek Inc.) using the default Partek normalization parameters and the robust multi-array average (RMA) analysis adjusted for probe sequence and GC content (GC-RMA). Data normalization was performed across all arrays using quantile normalization. Significantly up-regulated genes (p < 0.05 and > 4-fold increase) were identified.
Quantitative Real Time Polymerase Chain Reaction
cDNA from lucanthone-treated cells were used for relative quantification by RT-PCR analyses. First-strand cDNA synthesis was performed from 1 μg of RNA in a 20 μl reaction mixture using the high-capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Cathepsin D (CTSD) and GAPDH transcripts were amplified using commercially available TaqMan® Gene expression assays (Applied Biosystems, Foster City, CA). Relative gene expression was calculated with the 2−ΔCt method using GAPDH as a housekeeping gene (34).
Statistical Analyses
Statistical significance of differences observed in samples was determined using the Tukey-Kramer Comparison Test or the Student's t test. Differences were considered significant in all experiments at p < 0.05.
RESULTS
Lucanthone Induces Lysosomal Membrane Permeabilization (LMP)
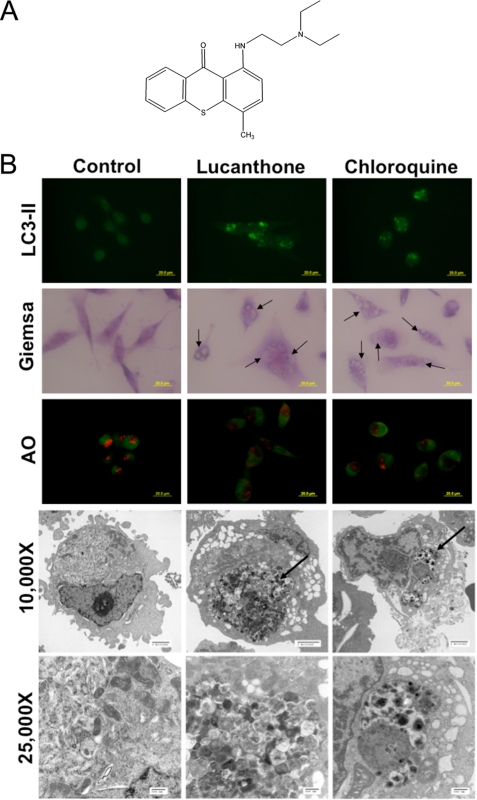
Autophagy promotes cell survival and leads to drug resistance by enabling cancer cells to recycle cellular components to generate ATP. In accordance with this, inhibition of autophagy genetically or using compounds such as 3-MA and CQ enhances the activity of many anticancer agents (9). Despite the promising potential of targeting this pathway for cancer therapy, CQ and its analog HCQ are currently the only FDA-approved drugs that inhibit autophagy. Lucanthone is an anti-schistome agent that is currently being evaluated for cancer therapy by Spectrum Pharmaceuticals (Fig. 1A). A previous report suggested that lucanthone induced autophagy in schistomes, which was characterized by the formation of autophagic vacuoles (35). However, based on its chemical structure, we hypothesized that lucanthone would disrupt lysosomal function and inhibit the final step in autophagic degradation. Consistent with this hypothesis, treatment of MDA-MB-231 breast cancer cells with lucanthone induced lipid modification of LC3-I into LC3-II, which is characterized by the punctate localization of LC3 to autophagosomes (Fig. 1B). LC3-I is recruited to autophagosomes, where LC3-II is generated by site-specific proteolysis and lipidation and is considered an early marker of autophagy (9). Lucanthone also induced cytoplasmic vacuolization, which is characteristic of LMP and autophagy (Fig. 1B). Furthermore, lucanthone decreased the red staining intensity of lysosomes with acridine orange indicating a loss of lysosomal acidity following treatment with either CQ or lucanthone (Fig. 1, B and C).
FIGURE 1.
Lucanthone inhibits autophagy. A, chemical structure of lucanthone. B, lucanthone induces LC3-II formation, vacuolization, and LMP. MDA-MB-231 cells were treated for 48 h with 10 μm lucanthone or 50 μm chloroquine. LC3-II was visualized by immunocytochemistry, vacuolization by Giemsa staining, and LMP by loss of acridine orange fluorescence. Electron microscopy demonstrates vacuolization and electron dense particle accumulation. Arrows indicate undegraded protein accumulation. C, quantification of LMP. The degree of red acridine orange staining was measured in MDA-MB-231 cells by immunofluorescence and quantified using ImageJ software. D, lucanthone stimulates SQSTM1/p62 accumulation and aggregation. Cells were treated with 10 μm lucanthone for 48 h. Levels and aggregation of p62 were determined by immunoblotting and immunocytochemistry, respectively. Fluorescence was quantified using ImageJ software. Mean ± S.D., n = 5. * indicates a significant difference from the controls. p < 0.05. E, Bafilomycin A1 does not augment lucanthone-mediated p62 or LC3-II accumulation or apoptosis. MDA-MB-231 cells were treated with 10 μm lucanthone, 100 nm bafilomycin A1, or both agents for 48 h.
Lucanthone Inhibits Autophagic Degradation
Inhibition of autophagy results in an accumulation of proteins normally degraded by this pathway. 48-hour exposure to lucanthone or CQ induced an accumulation of electron dense particles when visualized by transmission electron microscopy, suggesting protein aggregation (Fig. 1B). The polyubiquitin-binding protein p62 or sequestosome 1 (SQSTM1) is degraded by autophagy, localized to cellular inclusion bodies, and has been proposed to play a role in facilitating protein aggregate clearance by autophagy (36, 37). Consistent the other markers of autophagy inhibition we observed following treatment with lucanthone, p62 levels were strongly increased following drug treatment (Fig. 1D). Immunocytochemistry revealed that p62 displayed a diffuse staining pattern under basal conditions, but aggregated in response to lucanthone. This effect is in accord with its reported interaction with ubiquitinated proteins (37) (Fig. 1D). To further evaluate that lucanthone inhibits autophagy, we treated cells with lucanthone in the presence of bafilomycin A1, an inhibitor of vacuolar type H(+)-ATPase (V-ATPase) that prevents the acidification of lysosomes and disrupts autophagic degradation. Consistent with both agents acting as autophagy inhibitors, co-treatment did not induce a further increase in p62 expression or apoptosis (Fig. 1E). Therefore, both lucanthone and CQ induce many of the hallmark features of autophagy (LC3-II formation and vacuolization), but ultimately inhibit autophagy through deacidification of lysosomes.
Lucanthone Is Cytotoxic to Breast Cancer Cells
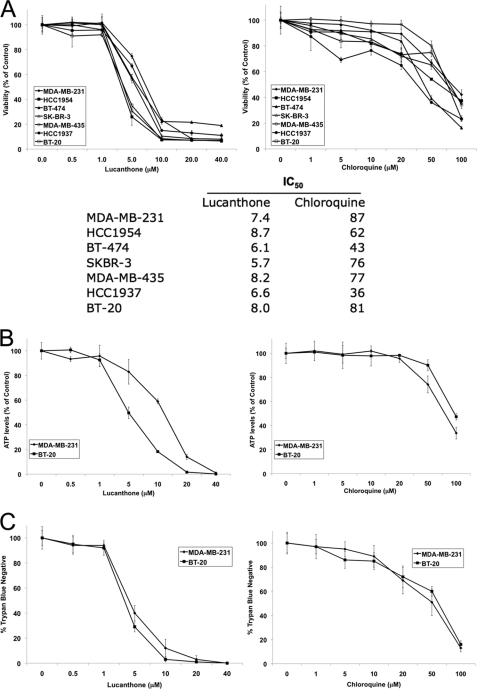
LMP and subsequent inhibition of autophagy had been reported to induce cell death in cancer cells (38, 39). To investigate the anticancer activity of lucanthone, cell viability was measured by MTT assay. Lucanthone reduced cell viability to a similar extent in a panel of seven breast cancer cell lines (Fig. 2A). In addition, a direct comparison revealed that lucanthone was significantly more potent than CQ at reducing breast cancer cell viability with a mean IC50 of 7.2 μm versus 66 μm for CQ (Fig. 2A). Measurement of cell viability in two representative cell lines (MDA-MB-231 and BT-20) by ATPlite assay (Fig. 2B) and trypan blue exclusion (Fig. 2C) revealed comparable results.
FIGURE 2.
Lucanthone is significantly more effective at reducing the viability of breast cancer cells than CQ. A, lucanthone and CQ decrease breast cancer cell line viability. Seven breast cancer cell lines were treated with varying concentrations of lucanthone or CQ for 72 h. Cell viability was measured using the MTT assay. Mean ± S.D., n = 3. IC50 values were calculated from the results of the MTT assays. B, lucanthone reduces ATP levels in the MDA-MB-231 and BT-20 cell lines. Cells were treated for 72 h, and ATP levels were measured using the ATPlite assay. Mean ± S.D., n = 3. C, lucanthone reduces the viability of breast cancer cells. Cells were treated for 72 h, and viability was determined by trypan blue exclusion. Mean ± S.D., n = 3.
Lucanthone Induces Cathepsin D Expression
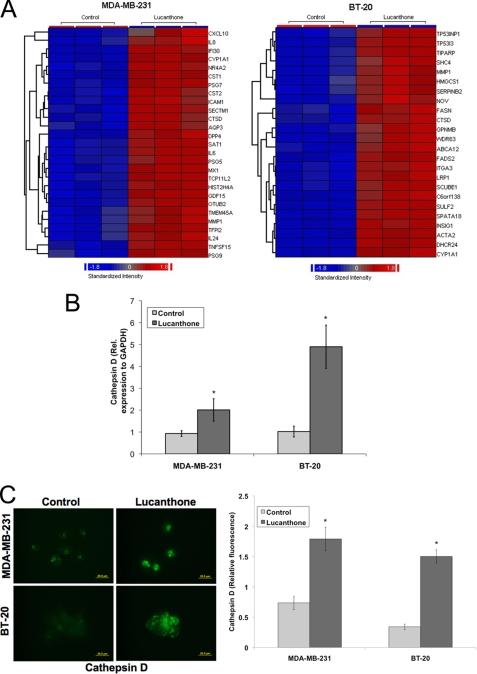
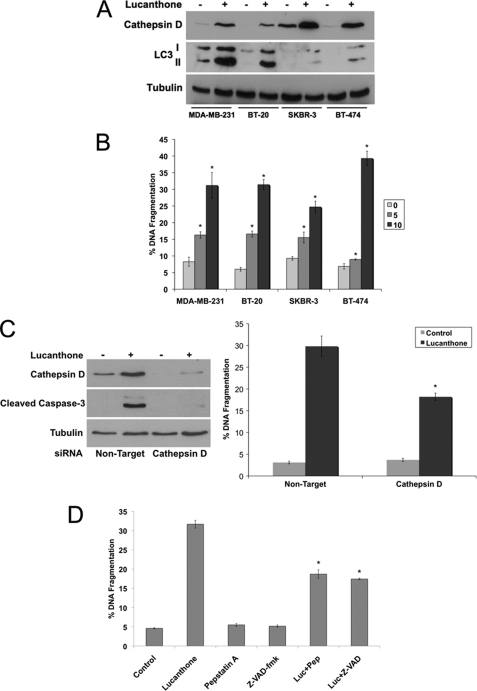
To further characterize the effects of lucanthone on breast cancer cells, expression profiling was performed on the MDA-MB-231 and BT-20 cell lines (supplemental Table S1). Of the genes induced by lucanthone, cathepsin D (CTSD), matrix metalloproteinase-1 (MMP1), and cytochrome P450, family 1, member A1 (CYP1A1) were increased in both cell lines (Fig. 3A). The lysosomal protease cathepsin D is a key mediator of apoptosis and its release into the cytosol has been reported to promote cell death (40–42). Considering this, we further evaluated the role of cathepsin D during lucanthone-mediated cell death. Quantitative real-time PCR (qRT-PCR) confirmed that lucanthone induced a significant increase in cathepsin D levels in both cell lines (Fig. 3B). Furthermore, lucanthone also increased cathepsin D protein levels 2–3-fold and promoted its cytosolic aggregation as measured by immunocytochemistry (Fig. 3C).
FIGURE 3.
Cathepsin D expression is highly elevated following lucanthone treatment. A, affymetrix expression arrays identify cathepsin D (CTSD) as a strongly up-regulated gene in both MDA-MB-231 and BT-20 cells. Cells were treated with 10 μm lucanthone for 48 h. RNA isolation and expression arrays were performed as described under “Experimental Procedures.” Data represent genes up-regulated by at least 4-fold following lucanthone treatment. B, quantitative real-time PCR analysis of cathepsin D expression in MDA-MB-231 and BT-20 breast cancer cells. Cells were treated with 10 μm lucanthone for 48 h and then harvested for analysis. Levels of mRNAs were standardized to the expression of GAPDH. Mean ± S.D., n = 4. *, indicates a significant difference from the controls. p < 0.05. C, lucanthone increases cathepsin D levels and promotes its aggregation. Breast cancer cells were treated for 48 h with 10 μm lucanthone. Cathepsin D was detected by immunofluorescence and quantified using ImageJ software. Mean ± S.D., n = 5. * indicates a significant difference from the controls. p < 0.05.
Cathepsin D Is an Essential Mediator of Lucanthone-induced Apoptosis
We next measured the expression of cathepsin D in four breast cancer cell lines by immunoblotting to investigate its contribution to lucanthone-mediated apoptosis. As expected, cathepsin D levels were strongly increased following lucanthone treatment (Fig. 4A) and correlated with apoptosis (Fig. 4B). To further establish the mechanistic role of cathepsin D in lucanthone-induced apoptosis, siRNA was used to knockdown its expression (Fig. 4C). Cells with reduced cathepsin D levels were significantly less sensitive to lucanthone-mediated apoptosis (Fig. 4C). We next evaluated the role of the aspartyl protease inhibitor pepstatin A, which inhibits cathepsin D and the broad-spectrum caspase inhibitor Z-VAD-fmk on lucanthone-mediated apoptosis. Both inhibitors blunted apoptosis induced by lucanthone (Fig. 4D).
FIGURE 4.
Induction of cathepsin D contributes to lucanthone-mediated apoptosis. A, lucanthone induces cathepsin D and LC3-II expression. Breast cancer cell lines were treated with 10 μm lucanthone for 48 h. Protein levels were determined by immunoblotting. B, lucanthone induces apoptosis in breast cancer cell lines. Cells were treated with 5 or 10 μm lucanthone for 48 h. Apoptosis was determined by PI staining and flow cytometry. Mean ± S.D., n = 3. * indicates a significant difference from controls. p < 0.05. C, cathepsin D knockdown diminishes lucanthone-induced apoptosis. RNAi was performed as described under “Experimental Procedures.” MDA-MB-231 cells transfected with non-target or cathepsin D siRNA were treated with lucanthone for 48 h. Immunoblotting confirmed knockdown of cathepsin D, which was associated with reduced cleavage of caspase-3 following lucanthone treatment. Apoptosis was determined by PI staining and flow cytometry. Mean ± S.D., n = 3. * indicates significant difference from non-target siRNA transfected cells treated with lucanthone. p < 0.05. D, pepstatin A and Z-VAD-fmk blunt lucanthone-mediated apoptosis. Cells were treated with 10 μm lucanthone in the presence or absence of 100 μm pepstatin A or 20 μm Z-VAD-fmk for 48 h, and apoptosis was measured by PI-FACS analysis. Mean ± S.D., n = 3. * indicates a significant difference from lucanthone-treated cells. p < 0.05.
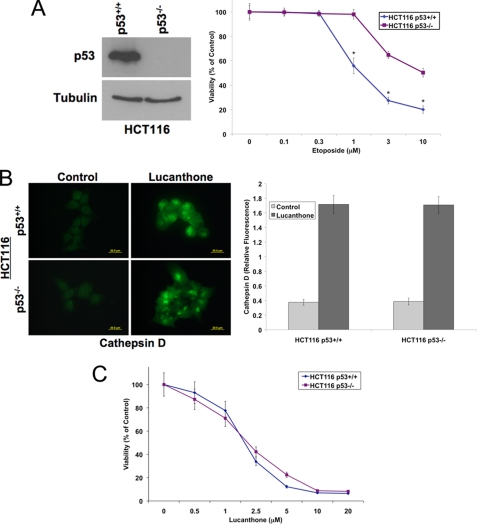
Loss of p53 Does Not Diminish Cathepsin D Accumulation or Activity of Lucanthone
Loss of function of p53 is a frequent event in human cancer that is associated with tumorigenesis and drug resistance (43). Therefore, agents that possess efficacy independent of p53 status are highly desirable. We investigated the role of p53 in lucanthone-mediated cell death using HCT116 53+/+ and p53−/− isogenic cell lines (Fig. 5A). As expected, p53-deficient cells were less sensitive to etoposide, a topoisomerase inhibitor that stimulates p53-mediated apoptosis (44). Lucanthone induced cathepsin D accumulation equally regardless of p53 status (Fig. 5B). Consistent with equipotent induction of cathepsin D, lucanthone reduced viability to a similar extent in both HCT116 p53+/+ and p53−/− cell lines (Fig. 5C).
FIGURE 5.
Lucanthone induces cathepsin D expression and decreases cell viability independently of p53. A, HCT116 p53+/+ and p53−/− cells were used to evaluate whether p53 was required for lucanthone-mediated apoptosis. Immunoblotting confirmed the lack of p53 in the HCT116 p53−/− cell line. 72 h MTT assay determined that HCT116 p53−/− cells were less sensitive to etoposide. Mean ± S.D., n = 3. * indicates a significant difference compared with HCT116 p53+/+-treated cells. B, lucanthone increases cathepsin D levels in HCT116 cells independent of p53 status. Cells were treated with 5 μm lucanthone for 48 h. Cathepsin D was visualized by immunocytochemistry and quantified using ImageJ software. Mean ± S.D., n = 5. C, HCT116 p53+/+ and p53−/− cells are equally sensitive to lucanthone. Cells were treated with varying concentrations of lucanthone for 72 h. Cell viability was determined by MTT assay. Mean ± S.D., n = 3.
Inhibition of Autophagy Enhances the Activity of Vorinostat
Inhibitors of autophagy such as CQ and 3-MA have previously been shown to augment the efficacy of many chemotherapeutic agents and radiation therapy (9). The HDAC inhibitor vorinostat and the proteasome inhibitor bortezomib have been shown to induce both apoptosis and autophagy and disruption of autophagy strongly potentiates their pro-apoptotic activity (14, 16, 19, 21). To inhibit autophagy, we used shRNA against the essential autophagy gene ATG7 to interfere with this pathway (Fig. 6A). Cells with silenced ATG7 levels displayed increased sensitivity to vorinostat, but surprisingly, not bortezomib. Consistent with our previous data with bafilomycin A1 (Fig. 1E), knockdown of ATG7 did not augment lucanthone-mediated apoptosis (Fig. 6A). Thus, we hypothesized that lucanthone would enhance vorinostat-mediated cell death due to its ability to inhibit autophagy. The combination of lucanthone and vorinostat led to increased induction of cathepsin D in MDA-MB-231 cells over what was achieved by either single agent treatment (Fig. 6B), which was associated with decreased cell viability and increased apoptosis (Fig. 6C). Consistent with bortezomib not showing increased efficacy in ATG7 knockdown cells, lucanthone did not augment bortezomib-mediated apoptosis (Fig. 6D). Collectively, these data provide evidence lucanthone inhibits autophagy and can successfully enhance the anticancer activity of vorinostat.
FIGURE 6.
Lucanthone enhances the anticancer activity of vorinostat. A, knockdown of ATG7 sensitizes MDA-MB-231 cells to vorinostat-induced apoptosis. Cells were transfected with non-target or ATG7 shRNA. Reduced expression of ATG7 was confirmed by immunoblotting at 48 h (left). Cells were treated for 48 h with 2.5 μm vorinostat, 10 nm bortezomib, or 10 μm lucanthone. Apoptosis was measured by PI-FACS analysis (right). Mean ± S.D., n = 3. * denotes a significant difference from vector control-treated cells. p < 0.05. The combination of lucanthone and vorinostat increases cathepsin D levels. MDA-MB-231 cells were treated with 10 μm lucanthone, 2.5 μm vorinostat, or the combination for 48 h. Following treatment, cells were harvested and cathepsin D levels were determined by immunoblotting (left). Relative intensity of the bands was quantified using ImageJ software (right). B, effects of lucanthone and vorinostat on breast cancer cell viability and apoptosis. Cells were treated with 5 μm lucanthone, 2.5 μm vorinostat, or the combination for 72 h. Cell viability was measured by MTT assay. Mean ± S.D., n = 3. Lucanthone enhances vorinostat-mediated apoptosis. Cells were treated with 10 μm lucanthone, 2.5 μm vorinostat, or the combination for 48 h. Apoptosis was measured by PI-FACS analysis. Mean ± standard deviation, n = 3. *, indicates a significant difference compared with controls. ** indicates a significant difference compared with single agent groups. p < 0.05. D, lucanthone does not augment bortezomib-induced apoptosis. Cells were treated with 10 μm lucanthone, 10 nm BZ, or both agents for 48 h. Apoptosis was determined by PI-FACS analysis. Mean ± S.D., n = 3.
DISCUSSION
Autophagy is the primary mechanism that stimulates the turnover of cellular components under nutrient-deficient conditions. The degradation of these components enables cells to produce the metabolic building blocks necessary to maintain survival. Cancer cells can utilize this pathway to survive in hypoxic conditions and resist apoptosis in the presence of chemotherapy (3, 45). Therefore, inhibition of autophagy is a viable therapeutic strategy to sensitize cancer cells to chemotherapy-mediated apoptosis. CQ and its analog HCQ are the only autophagic inhibitors currently being evaluated in clinical trials for cancer therapy. These agents are lysosomotropic agents that disrupt lysosomal function and prevent the acid-dependent degradation of proteins within the autophagosome (46). Although these agents are under clinical investigation for cancer therapy, CQ and HCQ induce ocular toxicities, such as retinopathy, which may limit their utility.
Lucanthone (Miracil D) has been used for years as an anti-schistome agent and has been previously reported to disrupt topoisomerase II activity and inhibit APE1, an important enzyme in DNA base excision repair (29, 30). Our data demonstrate that lucanthone induced vacuolization, interfered with lysosomal function, and led to the accumulation of undegraded proteins. Furthermore, lucanthone-treated cells displayed increased expression and aggregation of p62, a protein that was recently identified to be degraded by autophagy (36). Taken together, our data demonstrates that lucanthone inhibits autophagy via a similar mechanism to CQ by disrupting lysosomal function.
The efficacy of lucanthone was evaluated in a panel of seven breast cancer cell lines. Lucanthone exhibited similar activity across all cell lines tested and importantly was on average 10 times more potent than CQ at reducing cell viability. To further investigate the mechanism of action of lucanthone, microarray analyses were conducted on the MDA-MB-231 and BT-20 cell lines to quantify drug-induced changes in gene expression. These assays revealed a strong induction in the protease cathepsin D, which is a hallmark feature of altered lysosome function (42). Several studies have shown that cathepsin D possesses activity at physiologic pH and thus, promotes apoptosis upon its translocation to the cytosol (40–42, 47). Consistent with these results, siRNA-mediated knockdown of cathepsin D significantly reduced lucanthone-mediated apoptosis demonstrating that its expression contributed to the anticancer activity of this agent. These results are in agreement with our previous data showing that cathepsin D accumulation also plays a significant role during CQ-induced cell death (21).
Agents that induce LMP have been shown to trigger apoptosis independent of functional p53 status (48). To investigate if this was also true of lucanthone, we evaluated this possibility in HCT116 p53+/+ and p53−/− cells. As expected, lucanthone induced cathepsin D expression and reduced cell viability equivalently in both cell lines. Because p53 defects are a major contributor to drug resistance, the ability of lucanthone to kill cells independently of p53 status may have important clinical implications.
Lucanthone is currently being investigated as a sensitizer to chemotherapy and radiation due to its ability to interfere with DNA repair (30). However, since lucanthone is equally effective at reducing cancer cell viability independent of p53 status, inhibition of autophagy may be a more significant contributor to the lucanthone mechanism of action that effects on DNA repair. Because lucanthone inhibits autophagy, it may also be able to enhance the activity of chemotherapeutic agents that induce this pathway. Vorinostat and bortezomib have been reported to induce both apoptosis and autophagy and inhibition of autophagy augments its activity (16, 19, 21). We demonstrate that lucanthone enhances the activity of vorinostat in a panel of breast cancer cell lines. Consistent with this result, prior reports have demonstrated improved efficacy of vorinostat when autophagy is inhibited with CQ, 3-MA, or the knockdown of essential autophagy genes, such as ATG7 (19, 21). However, the ability of autophagic inhibitors to enhance bortezomib-induced apoptosis may be cell-type specific. While we report no benefit of bortezomib-induced apoptosis when autophagy is inhibited with lucanthone or with ATG7 shRNA, other investigators have observed conflicting results (16, 49–51). Several studies have shown that proteasome inhibitors activate autophagy and that inhibition of this pathway enhances the activity of bortezomib (16, 49). However, another report in multiple myeloma demonstrated that inhibition of autophagy blunted bortezomib antimyeloma activity (50). Further studies are warranted to better evaluate the role that autophagy plays during bortezomib-induced cell death.
In addition to vorinostat, many other cancer treatments including arsenic trioxide, etoposide, rapamycin, tamoxifen, temozolomide, imatinib, and ionizing radiation induce autophagy (2, 9). Therefore, disrupting this pathway may become an important strategy to improve conventional chemotherapy. While inhibition of autophagy is a relatively new approach, a small study in glioblastoma found that the median survival was increased 2-fold for patients receiving CQ with conventional therapy compared with controls (52, 53). Similar to CQ, lucanthone also readily crosses the blood brain barrier suggesting that it may have therapeutic activity against brain tumors.
CQ is an FDA-approved antimalarial agent that is currently being evaluated in clinical trials in combination with conventional chemotherapy. Our findings demonstrate that lucanthone disrupts lysosomal function and accordingly blocks autophagy. Furthermore, lucanthone is significantly more potent in a panel of breast cancer cell lines compared with CQ and appears to lack ocular toxicity that is characteristic of CQ derivatives. Taken together, our work provides a platform for future studies with lucanthone in combination with modalities that induce autophagy, such as certain chemotherapeutic agents and radiation therapy.
Supplementary Material
This work was supported by a grant from the Martha and Frank Herberth Memorial Fund and Beta and Melvin Leazar Memorial Fund and Robert F. and Anna M. Harper Memorial Fund of the San Antonio Area Foundation and funds provided by Spectrum Pharmaceuticals. G. R. is employed at Spectrum Pharmaceuticals. This study was funded in part by Spectrum Pharmaceuticals.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- CQ
- chloroquine
- LMP
- lysosomal membrane permeabilization
- HCQ
- hydroxychloroquine
- PI
- propidium iodide.
REFERENCES
- 1. Yorimitsu T., Klionsky D. J. (2005) Cell Death Differ. 12, Suppl. 2, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Apel A., Zentgraf H., Büchler M. W., Herr I. (2009) Int. J. Cancer 125, 991–995 [DOI] [PubMed] [Google Scholar]
- 3. Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 5. Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kondo Y., Kanzawa T., Sawaya R., Kondo S. (2005) Nat. Rev. Cancer 5, 726–734 [DOI] [PubMed] [Google Scholar]
- 7. Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., Thompson C. B. (2005) Cell 120, 237–248 [DOI] [PubMed] [Google Scholar]
- 8. Hippert M. M., O'Toole P. S., Thorburn A. (2006) Cancer Res. 66, 9349–9351 [DOI] [PubMed] [Google Scholar]
- 9. Carew J. S., Nawrocki S. T., Cleveland J. L. (2007) Autophagy 3, 464–467 [DOI] [PubMed] [Google Scholar]
- 10. Bursch W., Ellinger A., Kienzl H., Török L., Pandey S., Sikorska M., Walker R., Hermann R. S. (1996) Carcinogenesis 17, 1595–1607 [DOI] [PubMed] [Google Scholar]
- 11. Kanzawa T., Germano I. M., Komata T., Ito H., Kondo Y., Kondo S. (2004) Cell Death Differ. 11, 448–457 [DOI] [PubMed] [Google Scholar]
- 12. Kanzawa T., Kondo Y., Ito H., Kondo S., Germano I. (2003) Cancer Res. 63, 2103–2108 [PubMed] [Google Scholar]
- 13. Paglin S., Hollister T., Delohery T., Hackett N., McMahill M., Sphicas E., Domingo D., Yahalom J. (2001) Cancer Res. 61, 439–444 [PubMed] [Google Scholar]
- 14. Shao Y., Gao Z., Marks P. A., Jiang X. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 18030–18035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takeuchi H., Kondo Y., Fujiwara K., Kanzawa T., Aoki H., Mills G. B., Kondo S. (2005) Cancer Res. 65, 3336–3346 [DOI] [PubMed] [Google Scholar]
- 16. Zhu K., Dunner K., Jr., McConkey D. J. (2010) Oncogene. 29, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amaravadi R. K., Yu D., Lum J. J., Bui T., Christophorou M. A., Evan G. I., Thomas-Tikhonenko A., Thompson C. B. (2007) J. Clin. Invest. 117, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bellodi C., Lidonnici M. R., Hamilton A., Helgason G. V., Soliera A. R., Ronchetti M., Galavotti S., Young K. W., Selmi T., Yacobi R., Van Etten R. A., Donato N., Hunter A., Dinsdale D., Tirrò E., Vigneri P., Nicotera P., Dyer M. J., Holyoake T., Salomoni P., Calabretta B. (2009) J. Clin. Invest. 119, 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carew J. S., Medina E. C., Esquivel J. A., 2nd, Mahalingam D., Swords R., Kelly K., Zhang H., Huang P., Mita A. C., Mita M. M., Giles F. J., Nawrocki S. T. (2009) J Cell Mol. Med. 14, 2448–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carew J. S., Nawrocki S. T., Giles F. J., Cleveland J. L. (2008) Biologics 2, 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carew J. S., Nawrocki S. T., Kahue C. N., Zhang H., Yang C., Chung L., Houghton J. A., Huang P., Giles F. J., Cleveland J. L. (2007) Blood 110, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poole B., Ohkuma S. (1981) J. Cell Biol. 90, 665–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khraishi M. M., Singh G. (1996) Lupus 5, Suppl. 1, S41–S44 [PubMed] [Google Scholar]
- 25. Myint H. Y., Tipmanee P., Nosten F., Day N. P., Pukrittayakamee S., Looareesuwan S., White N. J. (2004) Trans. R Soc. Trop. Med. Hyg. 98, 73–81 [DOI] [PubMed] [Google Scholar]
- 26. Romanelli F., Smith K. M., Hoven A. D. (2004) Curr. Pharm. Des 10, 2643–2648 [DOI] [PubMed] [Google Scholar]
- 27. Solomon V. R., Lee H. (2009) Eur. J. Pharmacol. 625, 220–233 [DOI] [PubMed] [Google Scholar]
- 28. Tehrani R., Ostrowski R. A., Hariman R., Jay W. M. (2008) Semin. Ophthalmol. 23, 201–209 [DOI] [PubMed] [Google Scholar]
- 29. Dassonneville L., Bailly C. (1999) Biochem. Pharmacol 58, 1307–1312 [DOI] [PubMed] [Google Scholar]
- 30. Luo M., Kelley M. R. (2004) Anticancer Res. 24, 2127–2134 [PubMed] [Google Scholar]
- 31. Milligan A. J., Katz H. R., Leeper D. B. (1978) J Natl. Cancer Inst. 60, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 32. Mahalingam D., Medina E. C., Esquivel J. A., 2nd, Espitia C. M., Smith S., Oberheu K., Swords R., Kelly K. R., Mita M. M., Mita A. C., Carew J. S., Giles F. J., Nawrocki S. T. (2010) Clin. Cancer Res. 16, 141–153 [DOI] [PubMed] [Google Scholar]
- 33. Nawrocki S. T., Carew J. S., Maclean K. H., Courage J. F., Huang P., Houghton J. A., Cleveland J. L., Giles F. J., McConkey D. J. (2008) Blood 112, 2917–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pfaffl M. W. (2001) Nucleic. Acids Res. 29, e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clarkson J., Erasmus D. A. (1984) J Helminthol. 58, 59–68 [DOI] [PubMed] [Google Scholar]
- 36. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Ǿvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 38. Boya P., Andreau K., Poncet D., Zamzami N., Perfettini J. L., Metivier D., Ojcius D. M., Jäättelä M., Kroemer G. (2003) J. Exp. Med. 197, 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boya P., Kroemer G. (2008) Oncogene. 27, 6434–6451 [DOI] [PubMed] [Google Scholar]
- 40. Emert-Sedlak L., Shangary S., Rabinovitz A., Miranda M. B., Delach S. M., Johnson D. E. (2005) Mol. Cancer Ther. 4, 733–742 [DOI] [PubMed] [Google Scholar]
- 41. Fehrenbacher N., Jäättelä M. (2005) Cancer Res. 65, 2993–2995 [DOI] [PubMed] [Google Scholar]
- 42. Liaudet-Coopman E., Beaujouin M., Derocq D., Garcia M., Glondu-Lassis M., Laurent-Matha V., Prébois C., Rochefort H., Vignon F. (2006) Cancer Lett. 237, 167–179 [DOI] [PubMed] [Google Scholar]
- 43. Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 44. Lin X., Ramamurthi K., Mishima M., Kondo A., Christen R. D., Howell S. B. (2001) Cancer Res. 61, 1508–1516 [PubMed] [Google Scholar]
- 45. Amaravadi R. K., Thompson C. B. (2007) Clin. Cancer Res. 13, 7271–7279 [DOI] [PubMed] [Google Scholar]
- 46. Glaumann H., Ahlberg J. (1987) Exp. Mol. Pathol. 47, 346–362 [DOI] [PubMed] [Google Scholar]
- 47. Beaujouin M., Baghdiguian S., Glondu-Lassis M., Berchem G., Liaudet-Coopman E. (2006) Oncogene 25, 1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erdal H., Berndtsson M., Castro J., Brunk U., Shoshan M. C., Linder S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ding W. X., Ni H. M., Gao W., Chen X., Kang J. H., Stolz D. B., Liu J., Yin X. M. (2009) Mol. Cancer Ther. 8, 2036–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoang B., Benavides A., Shi Y., Frost P., Lichtenstein A. (2009) Mol. Cancer Ther. 8, 1974–1984 [DOI] [PubMed] [Google Scholar]
- 51. Milani M., Rzymski T., Mellor H. R., Pike L., Bottini A., Generali D., Harris A. L. (2009) Cancer Res. 69, 4415–4423 [DOI] [PubMed] [Google Scholar]
- 52. Savarino A., Lucia M. B., Giordano F., Cauda R. (2006) Lancet Oncol. 7, 792–793 [DOI] [PubMed] [Google Scholar]
- 53. Sotelo J., Briceño E., Lopez-González M. A. (2006) Ann. Intern. Med. 144, 337–343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.