Abstract
l-Fucose is a common constituent of Asn-linked glycans in vertebrates, invertebrates, and plants, but in fungal glycoproteins, fucose has not been found so far. However, by mass spectrometry we detected N-glycans and O-glycans containing one to six deoxyhexose residues in fruit bodies of several basidiomycetes. The N-glycans of chanterelles (Cantharellus cibarius) contained a deoxyhexose chromatographically identical to fucose and sensitive to α-l-fucosidase. Analysis of individual glycan species by tandem MS, glycosidase digestion, and finally 1H NMR revealed the presence of l-fucose in α1,6-linkage to an α1,6-mannose of oligomannosidic N-glycans. The substitution by α1,6-mannose of α1,2-mannosyl residues of the canonical precursor structure was yet another hitherto unknown modification. No indication for the occurrence of yet other modifications, e.g. bisecting N-acetylglucosamine, was seen. Besides fucosylated N-glycans, short O-linked mannan chains substituted with fucose were present on chanterelle proteins. Although undiscovered so far, l-fucose appears to represent a prominent feature of protein-linked glycans in the fungal kingdom.
Keywords: Fungi, Glycoprotein, Glycoprotein Structure, NMR, Post-translational Modification, Chanterelle, Fucose, N-Glycan
Introduction
The glycosylation capacity of fermentable yeasts and filamentous fungi has been well studied because of their potential biotechnological importance. In contrast to the members of other multicellular, eukaryotic organisms, i.e. green plants and metazoa, these fungi formed no complex-type N-glycans but rather oligomannosidic N-glycans often with characteristic extensions such as α1,3-mannose chains, phosphomannose or α-galactofuranose residues. This difference comfortably explains that fucose has so far not been found in fungal glycoproteins, as in animals and plants fucose occurs attached to N-acetylglucosamine (GlcNAc) or galactose residues in complex-type glycans only (1, 2). In fungi, fucose has only been detected in fungal polysaccharides (3–5) and in activated form as GDP-fucose (6) but not on protein-linked glycans.
Fungi are classified into basal fungi and dikarya, which are further divided into ascomycota and basidiomycota. Agaricomycotina is one of the three subphyla of the basidiomycetes (7). With the exception of morels and truffles belonging to the ascomycetes, most of the fungal species with macroscopic fruiting bodies well known as - often edible - mushrooms are found in the phylum basidiomycota. Glycoproteins of three species of this group, i.e. Agaricus bisporus (8), Schizophyllum commune (9) and Coprinopsis cinerea (10) have been studied and found to contain exclusively regular oligomannosidic N-glycans. C. cinerea additionally contained glycans with a bisecting α1,4-GlcNAc residue (10).
In this work, we studied the N-glycans of naturally growing mushrooms, in particular chanterelle (Cantharellus cibarius) by matrix assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Surprisingly, many species exhibited oligomannosidic N-glycans, with a deoxyhexose, which, for chanterelles, was shown to be fucose α1,6-linked to mannose by tandem MS (MS/MS),3 glycosidase digestion, and last but most importantly, high field 1H NMR. In addition, fucosylated O-glycans, more exactly O-mannans, were detected in chanterelles and penny buns (porcini, Boletus edulis).
EXPERIMENTAL PROCEDURES
Preparation of N-Glycans for NMR Analysis
Chanterelles (5 kg fresh weight; purchased from a local supermarket) were homogenized in distilled water and digested overnight at 37 °C with pepsin (0.1 mg/ml) in 5% (v/v) formic acid and 0.05% (v/v) 2-mercaptoethanol. After centrifugation for 10 min at 12,000 rpm and 4 °C, the supernatant was applied to Dowex 50W-X2 with 2% (v/v) acetic acid. (Glyco)peptides were eluted with 0.6 m ammonium acetate, pH 6. The orcinol-positive fractions were subjected to two different gel filtrations (Sephadex G-25 and G-50 in 1% acetic acid). Collected fractions were lyophilized, dissolved in 0.1 m citrate phosphate buffer, pH 5.0, and incubated at 95 °C for 10 min to inactivate pepsin. The glycopeptides were then treated with peptide N-glycosidase A (12 milliunits/50 mg of lyophilized glycopeptides = 100 g fresh chanterelles) at 37 °C overnight. The digest was purified by passage over C18 solid phase extraction cartridges (1 g, Bakerbond SPE; Fisher Scientific) equilibrated with 2% acetonitrile in water. Released glycans in the flow-through were desalted by size exclusion chromatography on Sephadex G15 (General Healthcare, Vienna). Orcinol-positive fractions were again lyophilized.
Preparation of Fluorescently Labeled N-Glycans
The dried glycans were subjected to pyridylamination by the modified method of the original overnight procedure (11). 2-Aminopyridine (5.86 mmol; Sigma-Aldrich) was dissolved in acetic acid and added to the dried glycans (2–6 mg). The sample was incubated at 90 °C for 1 h and allowed to cool, and acetone (water-free) was added. After vortexing for 30 s and centrifuging at 13,000 rpm for 5 min, the solvent was decanted. This procedure was repeated twice. After very brief drying under vacuum, reduction was accomplished by adding dimethylboran-complex solution and incubating at 90 °C for 20 min (11). After cooling to room temperature and dilution, excess reagent was removed by gel filtration.
Fractionation of Pyridylaminated Glycans
Pyridylaminated (PA-) oligosaccharides were fractionated by a “three-dimensional” technique comprising normal phase chromatography, reversed phase (RP), and porous, graphitic carbon (PGC) chromatography. Normal phase HPLC was performed on an Amide 80 column (250 × 4.6 mm, 5 μm; Tosoh Bioscience, Stuttgart, Germany) using 100 mm ammonium formate, pH 7.3, for PA-labeled sugars. After 1 min at 70% acetonitrile, a gradient from 70 to 58% acetonitrile was developed in 30 min at a flow rate of 1 ml/min. The excitation and emission wavelengths were 310 and 380 nm, respectively. RP-HPLC was performed on a Hypersil ODS column (4 × 250 mm, 5 μm; Thermo Scientific) at a flow rate of 1 ml/min. Glycans were eluted in 50 mm ammonium formate buffer, pH 4.4, by a gradient from 0 to 12% methanol in 40 min. Detection was done with the wavelengths 320 nm and 400 nm. PGC-HPLC was performed with a Hypercarb column (100 × 3 mm; Thermo Scientific) at a flow rate of 0.6 ml/min and a gradient from 15 to 30% of 95% acetonitrile in 0.1% ammonium formate buffer, pH 3.0, over 30 min. PA-glycans were detected with the wavelengths at 320 nm and 400 nm.
Preparation of N- and O-Glycans for MALDI-TOF MS
To determine the N-glycomes of various species of mushrooms, their N-glycans were isolated by using the “in-gel release method” (12). Briefly, 50 mg of fruit body was homogenized with a ball mill grinder under liquid nitrogen, dissolved in sample buffer, and subjected to SDS-PAGE, but only to the point where the unseparated protein zone had just entered the separation gel. N-Glycans were isolated directly from this band by N-glycosidase F digestion (12), desalted, and purified using a 10-mg HyperSep Hypercarb SPE cartridge (Thermo Scientific) (13). Analytes were eluted using 60% acetonitrile in 0.3% ammonium formate buffer, pH 9.0, dried in a Speed-Vac concentrator and analyzed by MALDI-TOF MS.
O-Glycans were released directly from SDS-PAGE bands by reductive β-elimination with 0.5 m sodium hydroxide in 1 m NaBH4 at 50 °C overnight (14, 15) and purified by Hypercarb SPE cartridge.
Monosaccharide Analysis
Monosaccharide composition of PA-labeled, purified oligomannosidic N-glycans from chanterelle was determined after hydrolysis with 4 m trifluoroacetic acid for 3 h at 100 °C. The released monosaccharides were analyzed by HPLC as anthranilic acid (16) and as 3-methyl-1-phenyl-2-pyrazolin-5-one derivatives (17). To identify the reducing sugar of O-glycans, labeling with anthranilic acid was done immediately after nonreductive release (18) and before hydrolysis with trifluoroacetic acid.
Glycosidase Digestions
Jack bean α-mannosidase (Sigma-Aldrich) was incubated with PA-labeled N-glycans in 0.1 m citrate phosphate buffer, pH 5.0. Bovine kidney α-l-fucosidase (Sigma) digestion was applied in 0.05 m sodium citrate buffer, pH 4.6. The dabsylated glycopeptide Ser-(GnGnF6-)Asn served as control substrate for α-l-fucosidase digestion. Both exoglycosidase digestions were performed at 37 °C overnight and analyzed by PGC-LC-ESI-MS or MALDI-TOF MS. The control glycopeptide was obtained from human IgG by digestion with Pronase and β-galactosidase (19).
Mass Spectrometry
Electrospray ionization mass spectrometry (ESI-MS) was performed on a Q-TOF Ultima Global instrument (Waters-Micromass, Manchester, UK). Analysis of O-glycans and borohydride-reduced N-glycans was performed by positive-ion LC-ESI-MS with a PGC column as described (20). For MS/MS experiments, fucosylated N-glycans were infused directly in a solution of 50% acetonitrile containing 0.1% formic acid and 0.5 mm sodium hydroxide (21). Free oligosaccharides of various mushrooms were analyzed by MALDI-TOF MS on an Ultraflex II instrument (Bruker, Bremen, Germany) using 2% 2,5-dihydroxybenzoic acid as matrix.
NMR Spectroscopy
Isolated oligosaccharides were lyophilized and dissolved in D2O (99.996% D%; Sigma-Aldrich) to concentrations of ∼120 μg 600 μl−1. Solutions were transferred into 5-mm high precision NMR sample tubes (Promochem, Wesel, Germany). All spectra were recorded on a Bruker DRX-600 AVANCE spectrometer (Bruker, Rheinstetten, Germany) at 600.13 MHz (1H) using the Bruker Topspin 1.3 software. Man6Fuc and Man5Fuc were measured at 298.1 K and 308.0 K, all other samples only at 298.1 K. Chemical shifts were referenced to external acetone (δH 2.225 ppm). The one-dimensional proton spectra were recorded with presaturation, acquisition of 32,000 data points, and a relaxation delay of 1.0 s. After zero filling to 64,000 data points and Fourier transformation, spectra were performed with a range of 7,200 Hz. To determine two-dimensional homonuclear COSY, TOCSY (100-ms mixing time), and ROESY (400-ms mixing time) spectra, 384 experiments with 2,048 data points and appropriate number of scans, each were recorded using standard Bruker programs. After linear forward prediction to 512 data points in the f2 dimension and sinusoidal multiplication in both dimensions, they were Fourier transformed to two-dimensional spectra with a range of 6,000 Hz in both dimensions (see also supplemental NMR Data).
RESULTS
Mass Profiles of Fungal N-Glycans
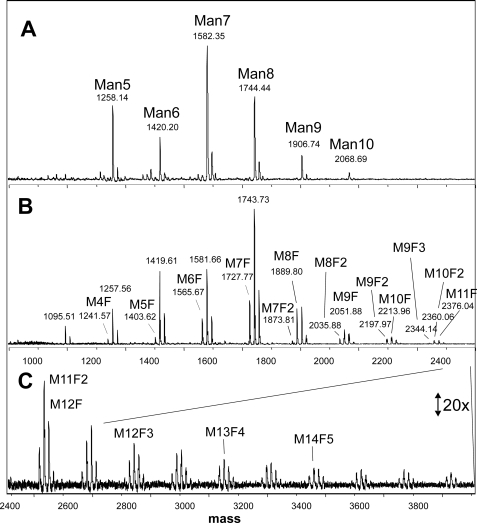
Pure research interest led us to subject chanterelles to our standard scheme of N-glycan preparation (22). Surprisingly, the MALDI-TOF MS profile deviated considerably from that of common mushroom (A. bisporus), which was recently shown to contain only oligomannosidic N-glycans (8). In chanterelles, additional glycans were seen, whose masses could be explained by the presence of one or more deoxyhexose residues (Fig. 1). The number of deoxyhexose residues, later shown to be fucose, varied from 1 to 6 and correlated with the number of hexose units (Fig. 1 and supplemental Fig. 1).
FIGURE 1.
N-Glycans from fruit bodies of A. bisporus (A) and C. cibarius (B and C). The masses refer to the sodium adduct peaks. Man5, Man6 etc. designate N-glycans of Man5GlcNAc2, Man6GlcNAc2 and so on in A. In B and C, due to lack of space, M stands for mannose, and F for fucose. Thus, M10F2 denotes an N-glycan of the composition Man10Fuc2GlcNAc2. The assignment of hexoses and deoxyhexoses as mannose and fucose is based on additional experiments (see “Results”).
Mass spectrometric N-glycan profiling of protein extracts of fruit bodies from 25 species of Agaricomycotina revealed either plain oligomannosidic N-glycans or such structures with one to several fucose residues (Table 1 and supplemental Figs. 2 and 3). Commercially cultivated fungi other than champignon such as oyster mushroom (Pleurotus ostreatus) and shitake (Lentinula edodes) also exhibited plain oligomannosidic N-glycans only. The same applied to the magic mushrooms Psilocybe semilanceata and Panaelus cyanescens and many other mushrooms of our adventitious collection. However, many other species contained fucosylated N-glycans, and notably most or all of these fungi live in symbiotic (ectomycorrhizal) or parasitic association with green plants. The host plants include angiosperm trees and conifers and tend to prefer acidic soil poor in nutrients (Table 1). The degree of fucosylation varied from 13 to 80% depending on the species. In all mushrooms, the dominant glycan structures comprised five to nine mannoses, but larger glycans were usually present indicative of elongation of the original Glc3Man9 precursor. The N-glycans of porcini (B. edulis), showed a fucosylation pattern similar to that of chanterelles but with a bias toward multiply fucosylated N-glycan structures (supplemental Fig. 2).
TABLE 1.
Fucosylation in diverse mushroom species
All fungi listed here belong to the phylum Basidiomycota and the subphylum Agaricomycotina (7). The degree of fucosylation is expressed as the fraction of N-glycans (in %) containing one or more fucose residues. The ecological type (M = mycorrhizal, p = parasitic, and S = saprophytic) as well as the soil pH preference (ac = acidic, alk = alkaline, neu = neutral or var = various habitats tolerated) are given (37). English names were taken from the home page of the British Mycological Society).
| Trivial name | Class/Order/Species | fucose (%) | ecology | soil |
|---|---|---|---|---|
| Tremellomycetes | ||||
| Tremellales | ||||
| Salmon salad | Tremiscus helvelloides | 0.0 | S | var |
| Dacrymycetes | ||||
| Dacrymycetales | ||||
| Yellow stagshorn | Calocera viscosa | 13.0 | S | var |
| Agaricomycetes | ||||
| Agaricales | ||||
| Liberty cab | Psilocybe semilanceata | 0.0 | S | var |
| Blue meanies | Panaeolus cyanescens | 0.0 | S | var |
| Deceiver | Laccaria laccata | 0.0 | M | var |
| Grisette | Amanita vaginata | 0.0 | M | var |
| Webcap | Cortinarius variicolor | 0.0 | M | alk |
| Common mushroom | Agaricus bisporus | 0.0 | S | var |
| Oyster mushroom | Pleurotus ostreatus | 0.0 | S | var |
| Shiitake | Lentinula edodes | 0.0 | S | var |
| Giant puffball | Calvatia gigantean | 0.0 | S | var |
| Fly agaric | Amanita muscaria | 0.0 | M | ac |
| Polyporales | ||||
| Birch polypore | Piptoporus betulinus | 0.0 | SP | var |
| Turkey tail | Trametes versicolor | 0.0 | SP | var |
| Boletales | ||||
| Scaly earthball | Scleroderma verrucosum | 80.6 | M | ac |
| Penny bun or Cep | Boletus edulis | 25.3 | M | ac |
| Bovine bolete | Suillus bovinus | 57.7 | M | ac |
| Cantharellales | ||||
| Chanterelle | Cantharellus cibarius | 29.2 | M | ac |
| Horn of plenty | Craterellus cornucopioides | 17.6 | M | neu |
| Russulales | ||||
| Saffron milkcap | Lactarius deliciosus | 0.0 | M | neu |
| Weeping milkcap | Lactarius volemus | 14.6 | M | ac |
| Sickener | Russula emetica | 19.0 | M | ac |
| Primrose brittlegill | Russula sardonia | 0.0 | M | ac |
| Root rot | Heterobasidion annosum | 17.3 | P | var |
| Hairy curtain crust | Stereum hirsutum | 44.0 | SP | var |
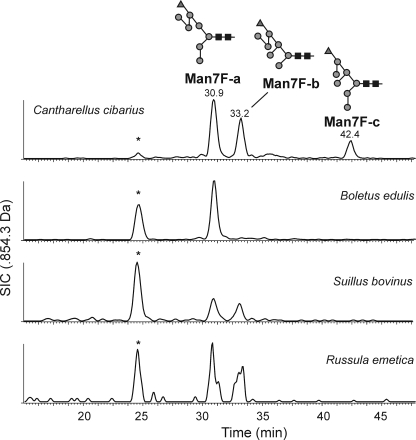
Comparative separation of reduced N-glycans from chanterelles, porcini (B. edulis) and others by PGC-LC-ESI-MS showed that these mushrooms contained the same isoforms albeit in different abundance as shown on the example of Man7Fuc (Fig. 2). Man7Fuc glycans of penny buns and chanterelles that had comparable elution times also had comparable collision-induced decay MS/MS spectra (data not shown).
FIGURE 2.
Chromatographic comparison of selected N-glycans from different mushroom species. Reduced N-glycans from chanterelles, penny buns, and other species were subjected to PGC-LC-ESI-MS. The selected ion chromatogram for Man7Fuc is shown. The peaks at 25 min labeled with an asterisk were caused by in-source fragmentation of Man9. The figures follow the Consortium for Functional Glycomics guidelines. Black squares represent GlcNAc; gray circles, mannose; and gray triangles, fucose.
Analysis of the Monosaccharide Composition
To elucidate the nature of the deoxyhexose, the monosaccharides of chanterelle N-glycans were at first analyzed as anthranilic acid derivatives. This revealed the presence of mannose, GlcNAc and fucose or rhamnose as these deoxyhexoses co-elute. To solve this ambiguity, the monosaccharides were labeled with 3-methyl-1-phenyl-2-pyrazolin-5-one (17), which resulted in a clear identification of fucose. This identification was strongly corroborated by digestion with α-l-fucosidase, which also demonstrated the l-configuration of this fucose (see later).
Preparation of Homogeneous Isoforms of Chanterelle N-Glycans
PA-labeled N-glycans were subjected to size separation by normal phase HPLC (supplemental Fig. 4). The monofucosylated species Man4Fuc, Man5Fuc, Man6Fuc each gave one dominant peak on reversed phase, whereas Man7Fuc was fractionated into three isomers (supplemental Fig. 4). The fractions were then subjected to carbon HPLC, which did not lead to further subfractionation.
Structural Characterization of Chanterelle N-Glycans by Mass Spectrometry and Glycosidase Digestions
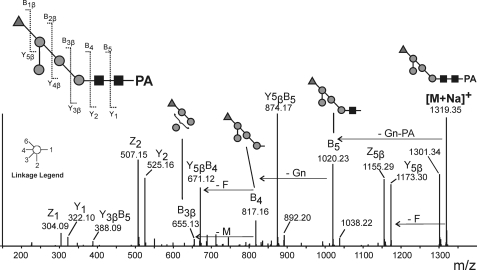
MS/MS experiments with Man4Fuc, the smallest fucosylated N-glycan, were at first conducted with the protonated ion. However, the fragments indicated that fucose was linked to the first GlcNAc as well as to one of the mannoses. Proton adducts of fucosylated N-glycans are prone to fucose rearrangement, which gives rise to misleading fragments and therefore, the sodium adduct was selected for analysis by collision-induced decay (23, 24). This yielded fragments unambiguously indicating the fucose to be linked to a mannose residue (Fig. 3).
FIGURE 3.
MS/MS spectrum of Man4Fuc from chanterelles. The [M+Na]+ ion of PA-labeled Man4Fuc (m/z 1319.35) was selected. Schemes label fragments that indicate the fucose to be linked to a mannose residue.
α-Mannosidase from jack bean could remove three mannoses from PA-labeled Man6Fuc, suggesting that the fucose terminated a chain of three mannose residues, although failure to remove further terminal mannoses due to steric hindrance could not be excluded at this point. The resulting artificial Man3Fuc was treated with a moderate dose of α-l-fucosidase, which led to partial defucosylation (supplemental Fig. 5). The rate of defucosylation was five times slower than that of human IgG glycopeptide with core α1,6-fucose, which is a particularly good substrate for bovine kidney α-l-fucosidase. Thus, fucose removal proceeded with decent swiftness as opposed to processes catalyzed by impurities of enzyme preparations applied at very high concentrations. We interpret this result as proof for the α-linkage as well as the l-configuration of fucose in chanterelle N-glycans.
An α-mannosidase digest was also performed with the whole N-glycan mixture. The tail of larger structures, in particular the nonfucosylated glycans with 10 and 11 hexose residues, disappeared. Instead Man3-Man6 glycans could be found, which insinuates that essentially all outer chain hexoses were α-mannoses.
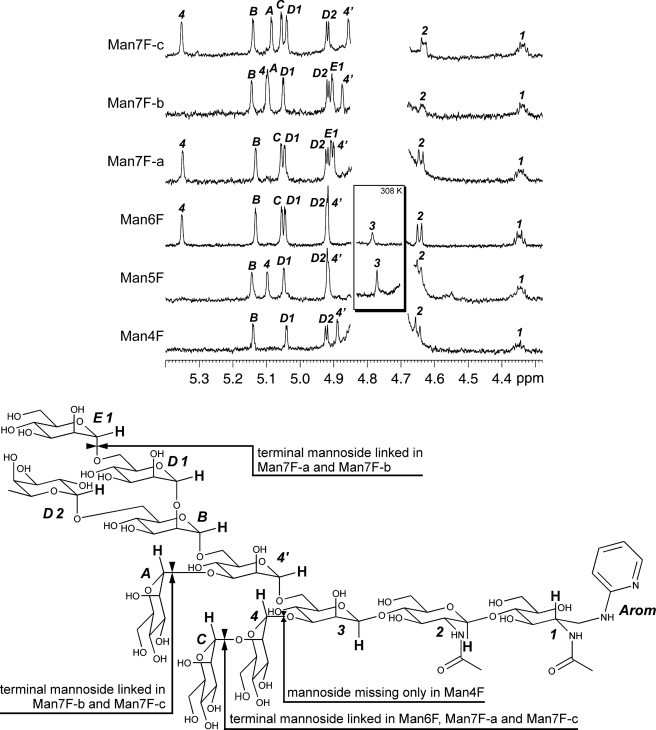
NMR Spectroscopy of N-Glycans
For the NMR-based structure determination only 1H NMR and homonuclear two-dimensional NMR spectra have been applied, as the small amounts of purified oligosaccharides did not allow measurements of 13C resonances with reasonable signal to noise ratio. However, based on data from mass spectrometric and wet chemical analyses, DQF-COSY and TOCSY spectra enabled the identification of all monosaccharide units. Information about spatial closeness of anomeric protons to connected units, determined from ROESY spectra, as well as 1H chemical shifts and coupling constants furthermore facilitated analysis of all anomeric configurations and interglycosidic linkages. All six investigated structures possess a conserved core oligosaccharide consisting of seven monosaccharide units. Parts of this structure are also present in similar oligomannosidic glycans from different sources, which have been analyzed earlier by NMR (25–28) and therefore allow comparisons with spectra of related structures.
The reduced and 2-aminopyridyl protected carbohydrate unit was identified as 2-acetylamino-2-deoxy-d-glucitol, which had originally been the d-GlcpNAc [1] forming the glycosidic linkage to the protein moiety in the natural source. Its proton H-2 causes an indicative 1H NMR signal at 4.35 ppm, optimal for NMR-based analysis. The second d-GlcpNAc [2] unit has a β-glycosidic linkage to position 4 of d-GlcNAc [1], indicated by a NOE between the characteristic H-1 signal of β-d-GlcpNAc [2] at 4.66 ppm and the H-4 at approximately 3.85 ppm. The consecutive saccharide unit is a β-d-Manp [3], linked to C-4 of β-d-GlcpNAc [2]. Its indicative proton signal is below the overwhelming HDO signal at 298 K (25). Therefore, the structure and interglycosidic linkages have been determined from Man6Fuc and Man5Fuc by two-dimensional NMR spectra recorded at 308 K (Fig. 4, inset). The β-d-Manp [3] carries on its position 6 the α-d-Manp [4′], which has the second α-d-Manp unit (B) bound to its position 6. Both units cause anomeric proton signals at 4.87 ppm and 5.14 ppm, respectively, which are usable for identification of the structure and of the interglycosidic bonds (25–27). α-d-Manp [B] carries two further saccharide units. A α-d-Manp [D1] is bound at position 2 causing the characteristic low field shift of H-1 in saccharide [B] (25, 29). On position 6 a α-l-Fucp [D2] is bound. Anomeric form and pyranose ring closure of the fucose was determined by 1H chemical shifts (30–32), and its absolute configuration was deduced from glycosidase digestion (see above).
FIGURE 4.
Proton NMR measurements of the isolated oligosaccharides. The frequency region with indicative signals of all saccharide units is shown, and these protons are highlighted in the structure scheme. Spectra have been recorded at 298.0 K. Additional measurements of Man5Fuc and Man6Fuc have been made at 308.0 K to cause a high field shift of the HDO signal. H-1 proton signals of β-Man [3] can hence be detected and are shown in the inset. Selected indicative 1H shifts of the isolated oligosaccharides are given in the supplemental NMR Data. The drawing at the bottom shows a fictive Man8Fuc, including all saccharide units occurring in the measured compounds. The distinctive units of each isolated oligosaccharide are specified. Proton atoms labeled in bold caused indicative 1H NMR signals. The numbering of saccharide units refers to Van Halbeek et al. (25).
These seven saccharide units represent our smallest isolated oligosaccharide Man4Fuc and build the conserved backbone of all further isolated oligosaccharides (Fig. 4). Man5Fuc further carries a α-d-Manp [4] linked to position 3 of the β-d-Manp [3]. This type of branching of the carbohydrate skeleton has been reported earlier for very similar oligosaccharides isolated from other sources (25–28). The indicative 1H NMR signal of the α-d-Manp [4] anomeric proton has a resonance at approximately 5.1 ppm. Man6Fuc carries an additional α-d-Manp [C] bound to α-d-Manp [4] in a 1→2 linkage. This unit causes an indicative low field shift of H-1 in unit [4] (Fig. 4), which has also been reported earlier (25, 29). The further three isolated Man7Fuc oligosaccharides had different substitution patterns of further mannosides. Man7Fuc-a has the Man6Fuc skeleton and additionally carries an α-d-Manp [E1] at position 6 of α-d-Manp [D1]. Man7Fuc-c also has the Man6Fuc skeleton with a further α-d-Manp [A] in 1→3 linkages to α-d-Manp [4′]. Man7Fuc-b carries both units [E1] and [A], but not the mannose [C] leading to a high field shift of H-1 signal to 5.09 ppm in α-d-Manp [4], comparable with those in Man5Fuc. Spectral regions with indicative signals of all six compounds are shown in Fig. 4, and the structural features are presented Fig. 4.
Determination of GDP-Fucose
Chanterelle extracts contained a compound isobaric with GDP-fucose and eluting at the same position on porous graphitic carbon. As the carbon column exerts a high selectivity with regard to the nature of the sugar and the linkage of phosphates (33), it is likely that chanterelles contain exactly the same GDP-fucose as mammals and plants. This agrees with the previous detection of GDP-fucose and the relevant enzymes for its synthesis in the filamentous fungus Mortierella alpine (6), which belongs to a phylum very distant to the Ascomycota investigated here. Common mushrooms (A. bisporus) also contained GDP-fucose despite the lack of fucosylated N-glycans.
O-Glycan Analysis of Chanterelle
O-Glycans were released from glycoproteins of chanterelles, champignons, and penny buns by reductive β-elimination from gel bands. Analysis of these glycans by PGC-ESI-MS revealed them to consist of hexoses plus one or two deoxyhexoses in chanterelle and porcini and only hexoses in champignons (Fig. 5). Fragmentation analysis of the different peaks showed the reducing sugar to be a hexose (data not shown). The first deoxyhexose is linked to the reducing sugar and the second deoxyhexose to the next hexose. Monosaccharide analysis with anthranilic acid identified the sugars as mannose and deoxyhexose, i.e. fucose as demonstrated by its sensitivity to α-fucosidase (data not shown). Anthranilic acid derivatization of the glycans obtained by nonreductive β-elimination (18) corroborated mannose as the reducing sugar (data not shown).
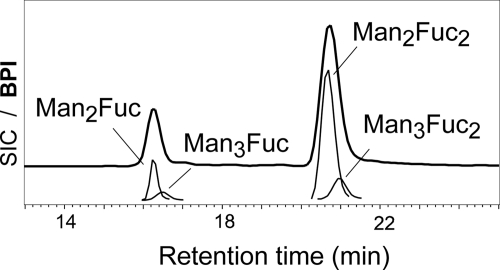
FIGURE 5.
O-Glycan structures of chanterelles. Reduced O-glycans were analyzed by carbon-LC-ESI-MS. Masses of 491.20, 653.25, and 799.31 Da were found, which indicated the glycan compositions given in the figure. The envelope line shows the base peak intensity chromatogram (BPI), and the thinner lines the selected ion chromatograms for the [M+H]+ ions (SIC).
DISCUSSION
Fucose has not yet been found as a constituent of fungal glycoproteins (34–36), even though not less than 10 of the 25 somewhat arbitrarily selected mushroom species contained obvious amounts of fucosylated N-glycans in their fruiting bodies. At least chanterelles and penny buns additionally contained fucosylated O-glycans. Some recent studies have dealt with mushroom N-glycans (8–10). Incidentally, all three species belong to the Agaricales order. Also, no other of the several investigated species of this order contained fucosylated glycoproteins.
The linkage of fucose to mannose has so far not been seen in eukaryotic glycans with the possible but enigmatic exception of the parasite glycans listed as entry 8486 in the glycome data base. The biosynthesis of this unusual structural element remains mysterious, as our efforts at demonstrating in vitro enzymatic activity remained unsuccessful. Possible reasons for this failure may be lack of the transferase in overground fungal organs, its rapid deterioration during storage, or the need for some labile cofactor or other unknown requirement. Apparently, fucose addition does not require the assistance of GlcNAc-transferase I, which initiates the generation of fucosylated complex-type N-glycans in animals or plants. This assumption is supported by the total absence of homologs to mammalian GlcNAc-transferase I among the so far known fungal proteins.
The data may point at a preferential occurrence of fucosylated glycoproteins in fungi that form mycorrhiza. More pronounced are the absence of fucose in cultivatable fungi and its presence in species preferring acidic habitats. These observations may point at a functional significance of this novel type of fucosylation.
Supplementary Material
Acknowledgments
We thank Karin Polacsek and Karin Hofbauer for helping with the purification of PA-labeled N-glycans and Dr. Ebrahim Razzazi-Fazeli (University of Veterinary Medicine, Vienna, Austria) for assistance with MALDI-TOF MS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and NMR data.
- MS/MS
- tandem mass spectrometry
- ESI
- electrospray mass ionization
- PA
- pyridylaminated
- PGC chromatography
- porous, graphitic carbon chromatography
- RP
- reversed phase.
REFERENCES
- 1. Staudacher E., Altmann F., Wilson I. B., März L. (1999) Biochim. Biophys. Acta 1473, 216–236 [DOI] [PubMed] [Google Scholar]
- 2. Ma B., Simala-Grant J. L., Taylor D. E. (2006) Glycobiology 16, 158R–184R [DOI] [PubMed] [Google Scholar]
- 3. Chakraborty I., Mondal S., Rout D., Chandra K., Islam S. S. (2007) Carbohydr. Res. 342, 982–987 [DOI] [PubMed] [Google Scholar]
- 4. Maiti D., Chandra K., Mondal S., Ojha A. K., Das D., Roy S. K., Ghosh K., Chakraborty I., Islam S. S. (2008) Carbohydr. Res. 343, 817–824 [DOI] [PubMed] [Google Scholar]
- 5. Mondal S., Chandra K., Maiti D., Ojha A. K., Das D., Roy S. K., Ghosh K., Chakarborty I., Islam S. S. (2008) Carbohydr. Res. 343, 1062–1070 [DOI] [PubMed] [Google Scholar]
- 6. Ren Y., Perepelov A. V., Wang H., Zhang H., Knirel Y. A., Wang L., Chen W. (2010) Biochem. Biophys. Res. Commun. 391, 1663–1669 [DOI] [PubMed] [Google Scholar]
- 7. Hibbett D. S., Binder M., Bischoff J. F., Blackwell M., Cannon P. F., Eriksson O. E., Huhndorf S., James T., Kirk P. M., Lücking R., Thorsten Lumbsch H., Lutzoni F., Matheny P. B., McLaughlin D. J., Powell M. J., Redhead S., Schoch C. L., Spatafora J. W., Stalpers J. A., Vilgalys R., Aime M. C., Aptroot A., Bauer R., Begerow D., Benny G. L., Castlebury L. A., Crous P. W., Dai Y. C., Gams W., Geiser D. M., Griffith G. W., Gueidan C., Hawksworth D. L., Hestmark G., Hosaka K., Humber R. A., Hyde K. D., Ironside J. E., Kõljalg U., Kurtzman C. P., Larsson K. H., Lichtwardt R., Longcore J., Miadlikowska J., Miller A., Moncalvo J. M., Mozley-Standridge S., Oberwinkler F., Parmasto E., Reeb V., Rogers J. D., Roux C., Ryvarden L., Sampaio J. P., Schüssler A., Sugiyama J., Thorn R. G., Tibell L., Untereiner W. A., Walker C., Wang Z., Weir A., Weiss M., White M. M., Winka K., Yao Y. J., Zhang N. (2007) Mycol. Res. 111, 509–547 [DOI] [PubMed] [Google Scholar]
- 8. Wilson I. B., Zeleny R., Kolarich D., Staudacher E., Stroop C. J., Kamerling J. P., Altmann F. (2001) Glycobiology 11, 261–274 [DOI] [PubMed] [Google Scholar]
- 9. Berends E., Ohm R. A., de Jong J. F., Rouwendal G., Wösten H. A., Lugones L. G., Bosch D. (2009) Appl. Environ. Microbiol. 75, 4648–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buser R., Lazar Z., Käser S., Künzler M., Aebi M. (2010) J. Biol. Chem. 285, 10715–10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kondo A., Suzuki J., Kuraya N., Hase S., Kato I., Ikenaka T. (1990) Agric. Biol. Chem. 54, 2169–2170 [PubMed] [Google Scholar]
- 12. Rendiæ D., Wilson I. B., Lubec G., Gutternigg M., Altmann F., Léonard R. (2007) Electrophoresis 28, 4484–4492 [DOI] [PubMed] [Google Scholar]
- 13. Packer N. H., Lawson M. A., Jardine D. R., Redmond J. W. (1998) Glycoconj. J. 15, 737–747 [DOI] [PubMed] [Google Scholar]
- 14. Pabst M., Wu S. Q., Grass J., Kolb A., Chiari C., Viernstein H., Unger F. M., Altmann F., Toegel S. (2010) Carbohydr. Res. 345, 1389–1393 [DOI] [PubMed] [Google Scholar]
- 15. Taylor A. M., Holst O., Thomas-Oates J. (2006) Proteomics 6, 2936–2946 [DOI] [PubMed] [Google Scholar]
- 16. Anumula K. R. (1994) Anal. Biochem. 220, 275–283 [DOI] [PubMed] [Google Scholar]
- 17. Zhang L., Xu J., Zhang L., Zhang W., Zhang Y. (2003) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 793, 159–165 [DOI] [PubMed] [Google Scholar]
- 18. Chai W., Feizi T., Yuen C. T., Lawson A. M. (1997) Glycobiology 7, 861–872 [DOI] [PubMed] [Google Scholar]
- 19. Roitinger A., Leiter H., Staudacher E., Altmann F. (1998) Glycoconj. J. 15, 89–91 [DOI] [PubMed] [Google Scholar]
- 20. Pabst M., Altmann F. (2008) Anal. Chem. 80, 7534–7542 [DOI] [PubMed] [Google Scholar]
- 21. Wuhrer M., Koeleman C. A., Hokke C. H., Deelder A. M. (2006) Rapid Commun. Mass Spectrom. 20, 1747–1754 [DOI] [PubMed] [Google Scholar]
- 22. Altmann F., Fabini G., Ahorn H., Wilson I. B. (2001) Biochimie 83, 703–712 [DOI] [PubMed] [Google Scholar]
- 23. Brüll L. P., Kovácik V., Thomas-Oates J. E., Heerma W., Haverkamp J. (1998) Rapid Commun. Mass Spectrom. 12, 1520–1532 [DOI] [PubMed] [Google Scholar]
- 24. Wuhrer M., Koeleman C. A., Deelder A. M. (2009) Anal. Chem. 81, 4422–4432 [DOI] [PubMed] [Google Scholar]
- 25. Van Halbeek H., Dorland L., Veldink G. A., Vliegenthart J. F., Michalski J. C., Montreuil J., Strecker G., Hull W. E. (1980) FEBS Lett. 121, 65–70 [DOI] [PubMed] [Google Scholar]
- 26. Petrescu A. J., Butters T. D., Reinkensmeier G., Petrescu S., Platt F. M., Dwek R. A., Wormald M. R. (1997) EMBO J. 16, 4302–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sato H., Fukae K., Kajihara Y. (2008) Carbohydr. Res. 343, 1333–1345 [DOI] [PubMed] [Google Scholar]
- 28. Bruch R. C., Bruch M. D. (1982) J. Biol. Chem. 257, 3409–3413 [PubMed] [Google Scholar]
- 29. Sandström C., Berteau O., Gemma E., Oscarson S., Kenne L., Gronenborn A. M. (2004) Biochemistry 43, 13926–13931 [DOI] [PubMed] [Google Scholar]
- 30. Hanniffy O. M., Shashkov A. S., Moran A. P., Prendergast M. M., Senchenkova S. N., Knirel Y. A., Savage A. V. (1999) Carbohydr. Res. 319, 124–132 [DOI] [PubMed] [Google Scholar]
- 31. Bilan M. I., Vinogradova E. V., Tsvetkova E. A., Grachev A. A., Shashkov A. S., Nifantiev N. E., Usov A. I. (2008) Carbohydr. Res. 343, 2605–2612 [DOI] [PubMed] [Google Scholar]
- 32. Ye L. B., Zhang J. S., Yang Y., Zhou S., Liu Y. F., Tang Q. J., Du X. J., Chen H., Pan Y. J. (2009) Food Chem. 112, 962–966 [Google Scholar]
- 33. Pabst M., Grass J., Fischl R., Léonard R., Jin C., Hinterkörner G., Borth N., Altmann F. (2010) Anal. Chem. 82, 9782–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deshpande N., Wilkins M. R., Packer N., Nevalainen H. (2008) Glycobiology 18, 626–637 [DOI] [PubMed] [Google Scholar]
- 35. Gemmill T. R., Trimble R. B. (1999) Biochim. Biophys. Acta 1426, 227–237 [DOI] [PubMed] [Google Scholar]
- 36. Goto M. (2007) Biosci. Biotechnol. Biochem. 71, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 37. Gminder A. (2008) Handbuch für Pilzsammler: 340 Arten Mitteleuropas sicher bestimmen, Kosmos, Stuttgart, Germany [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.