Abstract
The Rab6-binding kinesin, Rab6-KIFL, was identified in a two-hybrid screen for proteins that interact with Rab6, a small GTPase involved in membrane traffic through the Golgi apparatus. We find that Rab6-KIFL accumulates in mitotic cells where it localizes to the midzone of the spindle during anaphase, and to the cleavage furrow and midbody during telophase. Overexpression of Rab6-KIFL causes a cell division defect resulting in cell death. Microinjection of antibodies to Rab6-KIFL results in the cells becoming binucleate after one cell cycle, and time-lapse microscopy reveals that this is due to a defect in cleavage furrow formation and thus cytokinesis. These data show that endogenous Rab6-KIFL functions in cell division during cleavage furrow formation and cytokinesis, in addition to its previously described role in membrane traffic.
Keywords: cytokinesis/Golgi/kinesin/mitosis/Rab
Introduction
The Rab proteins are small ras-related GTPases that act as key regulators during the recognition events that occur between different cellular membranes to ensure the specificity of intracellular membrane traffic (Waters and Pfeffer, 1999). In the search for effector proteins of the Golgi-localized Rab protein, Rab6 (Martinez et al., 1994), a kinesin family motor protein and a GTPase activating protein (GAP), GAPCen-A, were identified using the yeast two-hybrid system (Echard et al., 1998; Cuif et al., 1999). Other studies investigating the function of the golgins, large coiled-coil proteins of the Golgi apparatus, have described the Rab6-binding GRIP domain shared by many of these proteins, including golgin-97 and golgin-245 in mammals, and Imh1 in budding yeast (Barr, 1999; Munro and Nichols, 1999). At the present time, the relative importance of GRIP-domain proteins and Rab6-KIFL as Rab6 effectors is unclear.
Kinesins are a large family of proteins that share a conserved motor domain, which binds to microtubules and couples ATP hydrolysis to mechanical force generation (Hirokawa et al., 1998). Most kinesins are microtubule plus end-directed motors, thus moving away from the microtubule organizing centre, although the NCD class possess minus end-directed movement (Hirokawa et al., 1998). They play a wide variety of roles in cellular function, being involved in processes as diverse as formation of the mitotic spindle and chromosome partitioning, and the intracellular movement of organelles and vesicles (Hirokawa et al., 1998; Allan and Schroer, 1999). The kinesin family protein identified as binding to Rab6, Rab6-KIFL, has two microtubule-binding sites, one within the N-terminal kinesin motor domain and the other at the C-terminus, with the Rab6-binding domain separating the two (Echard et al., 1998).
Rab6-KIFL is thought to function in membrane traffic within the Golgi apparatus, and from the Golgi apparatus to the endoplasmic reticulum, providing motility to transport vesicles (Echard et al., 1998; Allan and Schroer, 1999). At present the consequences of Rab6 binding to Rab6-KIFL remain unknown; it is possible that Rab6 acts either as a regulator of motor activity or to recruit Rab6-KIFL to Golgi apparatus membranes or vesicles. Another potential function of Rab6-KIFL is the partitioning of the Golgi apparatus during mitosis (Shima and Warren, 1998). Dividing mammalian cells must ensure that the two daughter cells receive their correct complement not only of chromosomes, but also of subcellular organelles such as the mitochondria and Golgi apparatus (reviewed in Shima and Warren, 1998). While the exact nature of the Golgi partitioning mechanism is not understood, it does appear to involve the microtubule cytoskeleton, and to be an active rather than a stochastic process and may thus require some form of motor protein, potentially Rab6-KIFL (Shima et al., 1998).
Here we report our findings on the localization and cellular function of the putative Rab6-interacting kinesin, Rab6-KIFL, in both interphase and mitotic cells.
Results
Characterization of Rab6-KIFL antibodies
In order to investigate the function of human Rab6-KIFL, an antibody was raised in sheep against the region previously shown to bind to the small GTPase Rab6 (Echard et al., 1998). This antibody was able to detect the antigen against which it was made, but did not detect a protein of the expected size in interphase HeLa cell extracts (Figure 1A). However, a 100 kDa protein matching the predicted size of Rab6-KIFL was detected when extracts of HeLa cells arrested in mitosis with the microtubule-depolymerizing drug nocodazole were western blotted (Figure 1A, left panel). Cyclin B2, a protein known to be present mostly in mitotic cells showed a similar behaviour (Jackman et al., 1995; Figure 1C, left panel), whereas α-tubulin was equally abundant in both interphase and mitotic samples as expected (Figure 1D). Pre-incubation of the affinity-purified Rab6-KIFL antibody with its antigen completely abolished the signal for Rab6-KIFL on blots, indicating that the antibody is indeed specific for its antigen (Figure 1A, right panel). Similar results were obtained with the published rabbit antiserum to Rab6-KIFL (generated by Echard et al., 1998); this also recognized a protein of 100 kDa in mitotic but not in interphase cells (Figure 1B, left panel), which was competed by the antigen (Figure 1B, right panel). In addition, this antibody displays a cross-reactivity to a protein of 200 kDa present in mitotic cells, again competed by the antigen (Figure 1B, right panel).
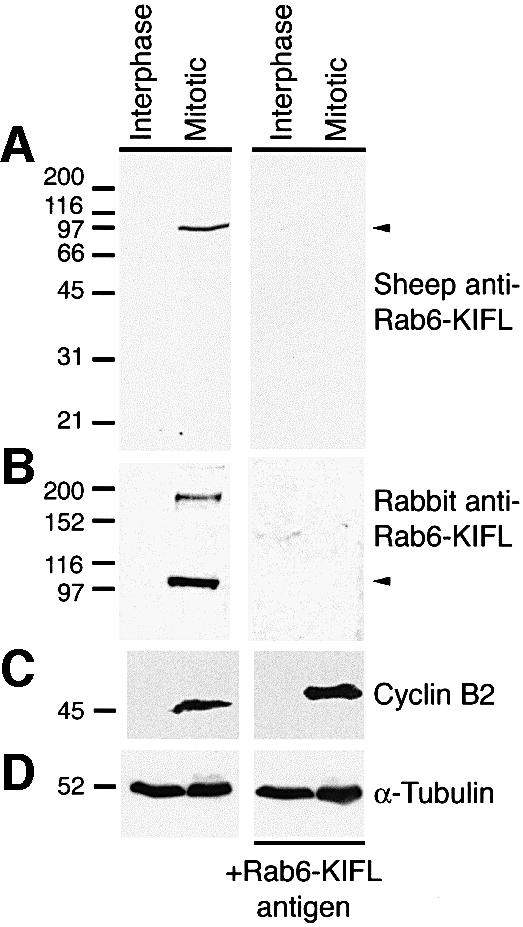
Fig. 1. Characterization of Rab6-KIFL antibodies. (A) Protein extracts, 100 µg, from either interphase or nocodazole-arrested mitotic HeLa cells, >90% in mitosis, were analysed by western blotting with an affinity-purified sheep polyclonal antibody raised against Rab6-KIFL. For control blots the antibody was pre-incubated with 50 µg of the recombinant protein antigen. (B) HeLa cell extracts were western blotted as above with the published rabbit polyclonal antibody to Rab6-KIFL again with a control in which the antibody was pre-incubated with its antigen. (C and D) The same samples were also probed for cyclin B2 as a marker for mitotic arrest, and α-tubulin as a loading control, respectively.
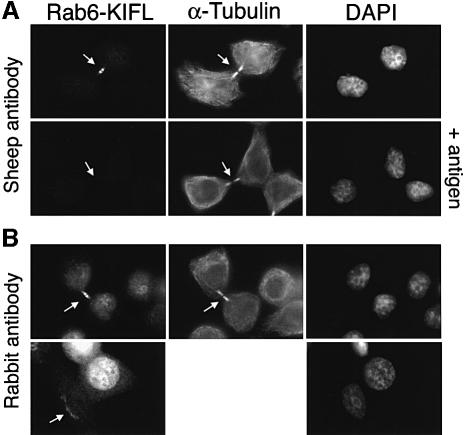
Immunofluorescence microscopy was then used to investigate the localization of the endogenous Rab6-KIFL (Figure 2). When stained with the affinity-purified sheep antibody to Rab6-KIFL, the majority of cells failed to display any specific staining. However, dividing cells undergoing cytokinesis showed a strong staining in the midbody region (Figure 2A, upper panels), which was competed by the antigen, thus demonstrating its specificity (Figure 2A, lower panels, +antigen). Essentially identical results were obtained with the published rabbit antiserum to Rab6-KIFL (Figure 2B, upper panels), and again this staining was competed by the antigen (not shown). This antibody also gave a weak perinuclear reticular staining pattern in some interphase cells, which is characteristic of the Golgi apparatus (Figure 2B, lower left panel; see also Figure 2B in Echard et al., 1998). The sheep antibody did not show this Golgi staining pattern, thus reflecting a difference in the two antisera.
Fig. 2. Rab6-KIFL localizes to the midbody in dividing cells. (A) HeLa cells were stained with the affinity-purified sheep polyclonal antibody to Rab6-KIFL, a monoclonal antibody to α-tubulin and DAPI to reveal the DNA. Control coverslips were stained as above, except that the antibody dilution was pre-incubated with 25 µg of the Rab6-KIFL antigen for 1 h on ice. (B) HeLa cells were stained as above except that the published rabbit antibody to Rab6-KIFL was used in place of the sheep antibody. Arrows denote areas of particular interest.
Periodic accumulation of Rab6-KIFL in mitotic cells
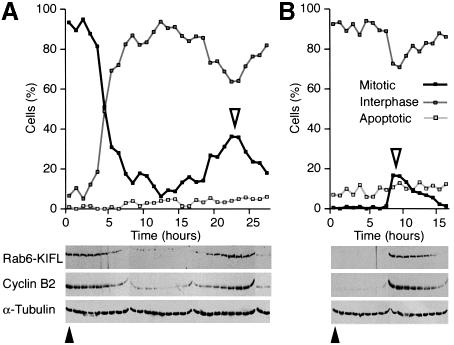
These data suggested that both the localization and the amount of Rab6-KIFL protein were cell cycle regulated. To investigate this further, HeLa cells were arrested either in M-phase with nocodazole or at the end of G1 with hydroxyurea. The drugs were then washed out, and samples taken at various times during the chase for analysis by western blotting and immunofluorescence microscopy to ascertain exactly what stage of the cell cycle the cells had reached (Figure 3). Consistent with the data shown above, Rab6-KIFL levels were high in nocodazole-arrested cells, but rapidly dropped once the drug was removed and the cells exited mitosis (Figure 3A). After 15–20 h, the cells were once again passing through mitosis as judged by quantitation of the mitotic index of the culture and the chromosomal and nuclear morphology (Figure 3A). In agreement with this the levels of Rab6-KIFL started to increase, peaked and then declined (Figure 3A). This cyclic behaviour was similar to that seen for the well-characterized cell cycle control protein, cyclin B2 (Figure 3A; Jackman et al., 1995). When cells were arrested in G1 with hydroxyurea, no Rab6-KIFL could be detected, consistent with the nocodazole arrest data (Figure 3B). Approximately 8 h after the drug was washed out the levels of both Rab6-KIFL and cyclin B2 showed a dramatic increase followed by a slow decline, which matched the profile of cells entering and subsequently exiting mitosis (Figure 3B). In both cases the level of α-tubulin was found to remain constant at all time points taken (Figure 3A and B, α-tubulin), indicating that the changes in the amount of Rab6-KIFL are specific. Rab6-KIFL therefore shows a cyclic behaviour with either of the two different drug arrests, and behaves in essentially the same fashion as cyclin B2. Together with the data in Figure 1, these results show that Rab6-KIFL is a kinesin found only in dividing cells during mitosis.
Fig. 3. Rab6-KIFL levels oscillate in a cell cycle controlled manner. (A) HeLa cells were arrested with nocodazole for 20 h, then released into fresh drug-free medium. Samples were collected every hour as shown, and the number of mitotic, interphase and apoptotic cells counted. Cell extracts were analysed by western blotting with antibodies to Rab6-KIFL, cyclin B2 and α-tubulin as a loading control. (B) HeLa cells were arrested with 2 mM hydroxyurea for 18 h, then released into fresh drug-free medium. The cells were analysed as before. Solid arrowheads indicate the time of drug washout, and open arrowheads the peak of mitotic cells.
Localization of Rab6-KIFL throughout the cell cycle
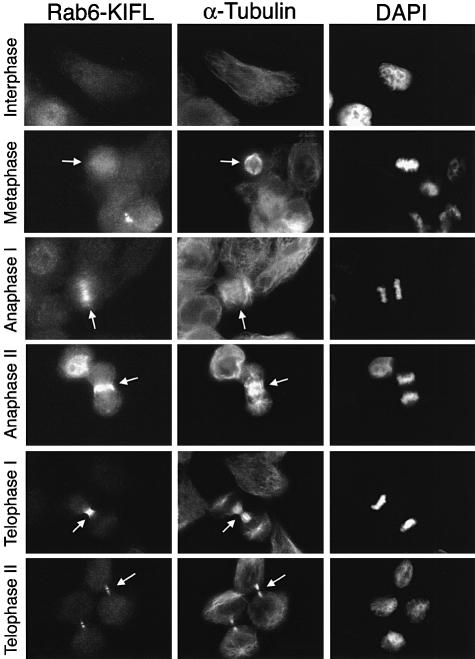
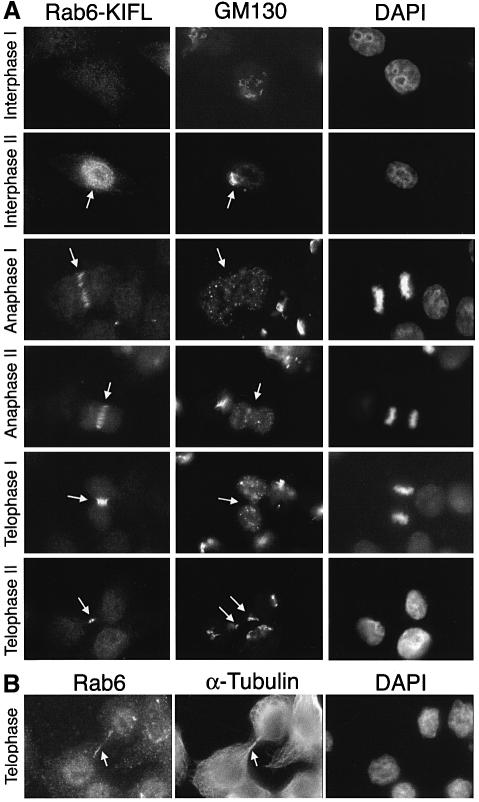
In order to obtain some insight as to the function of Rab6-KIFL, its localization was followed throughout the cell cycle by means of immunofluorescence microscopy. Rab6-KIFL staining was compared with both the microtubule cytoskeleton (Figure 4) because of its localization to the midbody (see Figure 2A), and the Golgi apparatus (Figure 5) because of a previous study implying a functional role in membrane traffic (Echard et al., 1998). Confirming the western blotting data, no signal above background was seen in the majority of interphase cells (Figure 4, Interphase; Figure 5, Interphase I), although some interphase cells showed a distinct nuclear staining (Figure 5A, Interphase II). The latter probably represent cells late in G2, or early in M-phase, since they often showed the early signs of Golgi disassembly (Figure 5, Interphase II arrow). This observation is in agreement with the localization predicted using a neural net algorithm, which gives a 68.7% certainty of nuclear localization (Reinhardt and Hubbard, 1998). Metaphase cells possessing a short spindle showed a staining pattern for Rab6-KIFL not localized to any discrete structures, indicating that the protein was mainly cytosolic at this point (Figure 4, Metaphase). In early anaphase cells when the spindle begins to elongate, Rab6-KIFL staining was observed as a series of short bars across the middle of the spindle, running parallel with its axis, together with some diffuse cytosolic staining (Figure 4, Anaphase I). In late anaphase cells when the cleavage furrow pinches in, the staining became entirely restricted to short filamentous structures, probably microtubules, in this region (Figure 4, Anaphase II). By telophase Rab6-KIFL staining was restricted to the midbody, and following cytokinesis this staining, by now associated with the midbody remnant, was gradually lost (Figure 4, Telophase I and II, arrows). At no point in the cell cycle was any significant co-localization between the Golgi marker GM130 and Rab6-KIFL observed (Figure 5A). For example, in anaphase when the Golgi is found as many small clusters of vesicles, Rab6-KIFL shows a clearly discrete staining across the spindle midzone, while GM130 is found in many punctate structures dispersed throughout both poles of the cell (Figure 5A, Anaphase I, arrow). During telophase when the Golgi apparatus starts to rebuild in the daughter cells there are two areas of Golgi staining. One is adjacent to the centrosomes, while the other minor pool is immediately adjacent to the bridge, possibly indicating a role for the Golgi in cytokinesis (Piel et al., 2000). However, even at this stage of the cell cycle, no Rab6-KIFL was observed on the fragments of the Golgi adjacent to the midbody (Figure 5A, Telophase II, arrows). Essentially the same staining pattern was observed when Rab6 was used as a Golgi marker, although cells late in telophase often showed additional staining along the midbody microtubules (Figure 5B, Telophase). The accumulation of Rab6-KIFL during mitosis, and the cell cycle control of its localization to the spindle midzone and cleavage furrow, indicate that it may function in the mechanism of cell division, while the lack of any significant Golgi localization during interphase or mitosis appears to rule out roles in membrane traffic during interphase or Golgi partitioning during mitosis.
Fig. 4. Rab6-KIFL localizes to the spindle midzone during anaphase and cleavage furrow during cytokinesis. HeLa cells were stained with the affinity-purified sheep antibody to Rab6-KIFL, a monoclonal antibody to α-tubulin, while the DNA was stained with DAPI. Representative images of cells at the indicated stages of the cell cycle are shown. Arrows denote areas of particular interest.
Fig. 5. Discrete localization of Rab6-KIFL and the Golgi apparatus throughout the cell cycle. (A) HeLa cells fixed with methanol were stained with the affinity-purified sheep antibody to Rab6-KIFL, a rabbit polyclonal antibody to the Golgi marker GM130, while the DNA was stained with DAPI. Representative images of cells at the indicated stages of the cell cycle are shown. (B) HeLa cells fixed with paraformaldehyde were stained with antibodies to Rab6 and α-tubulin; the DNA was stained with DAPI. A representative image of cells at the indicated stage of the cell cycle is shown. Arrows denote areas of particular interest.
Rab6-KIFL overexpression interferes with normal cell division
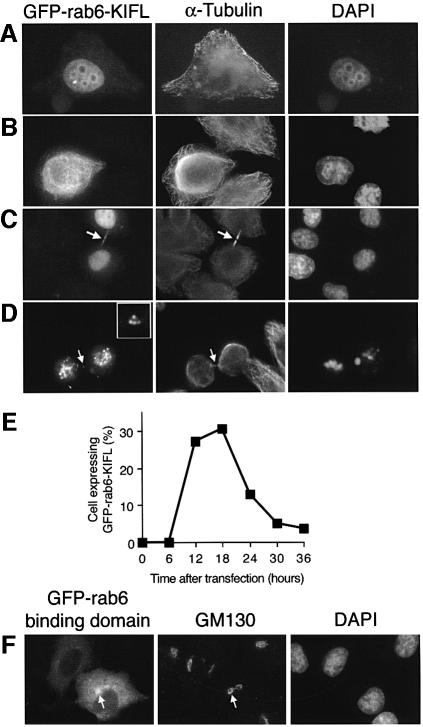
This localization of Rab6-KIFL to the spindle midzone and cleavage furrow is reminiscent of proteins known to function in cytokinesis, such as the Rho exchange factor ECT2, and Rho effector citron kinase (Madaule et al., 1998; Tatsumoto et al., 1999). In order to investigate the possibility that Rab6-KIFL functions in cytokinesis, two different types of experiment were performed, focusing on either exogenous or endogenous Rab6-KIFL. In the first type of experiment, HeLa cells were transfected with constructs corresponding to either the full-length protein or the Rab6-binding domain fused to green fluorescent protein (GFP). Initial experiments allowing cells to express Rab6-KIFL for 30 h according to our standard transfection protocol resulted in very few expressing cells, which we hypothesized might be due to a toxic effect of the protein. When early time points, between 12 and 18 h after transfection, were analysed, we found the expected number of expressing cells consistent with this hypothesis (see quantitation in Figure 6E). Full-length GFP-tagged Rab6-KIFL was found to localize to the nucleus at low levels of expression (Figure 6A), and at higher levels to associate with filamentous structures, probably microtubule bundles, in the cytoplasm, as well as to the nucleus (Figure 6B). Some binucleate cells were observed; in these cells Rab6-KIFL staining formed a meshwork around the nuclei (not shown). Due to the nature of transient transfection experiments, we cannot be certain that these cells were not binucleate before transfection. Many dividing cells, apparently late in telophase, displayed a filamentous cytoplasmic staining for Rab6-KIFL together with some staining in the midbody region (Figure 6C, arrow). After 24 h ∼60% of the transfected cells were found to be dead as judged by 4′,6′-diamidine- 2-phenylindole (DAPI) staining of their DNA (Figure 6E), being found in pairs joined by the remnants of the midbody (Figure 6D, arrow and enlarged in inset). When stained with a marker for the Golgi apparatus some cells showed changes to this organelle, as reported previously, in addition to the morphological abnormalities described above (data not shown; Echard et al., 1998). The GFP-tagged Rab6-binding domain, in contrast to the full-length protein, localized throughout the cytoplasm, in part to the Golgi apparatus, but never to microtubules (Figure 6F). However, little or no effect was seen on the Golgi apparatus (Figure 6F), and no effects were observed on the microtubule cytoskeleton or on the process of cell division, even at high levels of transfection (Figure 6F; data not shown). These data show that exogenously expressed full-length Rab6-KIFL can interfere with the process of cell division, and cause cell death rather than specifically perturbing the structure of the Golgi apparatus.
Fig. 6. Overexpression of Rab6-KIFL interferes with normal cell division. (A–D) HeLa cells were transfected with an expression plasmid for GFP-tagged Rab6-KIFL. Cells were fixed in methanol after 18 h and processed for immunofluorescence with antibodies to α-tubulin. Representative images demonstrating the various effects of Rab6-KIFL overexpression are shown. (E) Quantitation of the number of cells expressing GFP–Rab6-KIFL as a function of time after transfection. (F) HeLa cells were transfected with an expression plasmid for the GFP-tagged Rab6-binding domain of Rab6-KIFL. Cells were fixed in methanol after 18 h and processed for immunofluorescence with antibody to GM130. A representative image demonstrating the effect of overexpressing the Rab6-binding domain construct is shown.
Microinjection of antibodies to Rab6-KIFL blocks cytokinesis
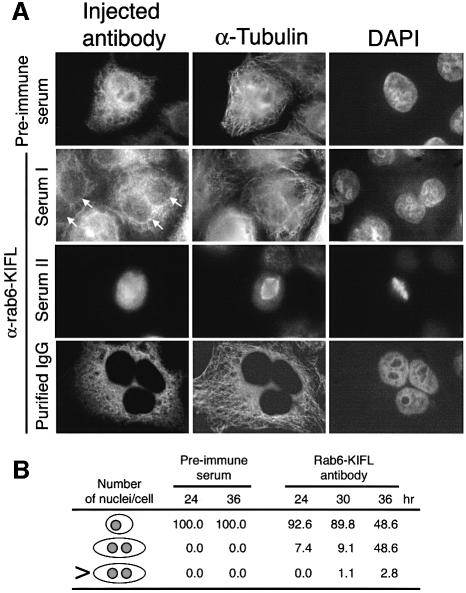
The second approach taken was aimed at providing evidence as to the function of endogenous Rab6-KIFL. In these experiments, mononucleate HeLa cells were microinjected with either the crude Rab6-KIFL antiserum, purified immunoglobulin to Rab6-KIFL or the corresponding pre-immune serum as a control, then left for 24, 30 or 36 h before being fixed and processed for immunofluorescence microscopy. At all the time points taken after microinjection an increase in binucleate or, rarely, in multinucleate cells was observed when either the Rab6-KIFL antiserum (Figure 7A, Serum I) or the purified immunoglobulin to Rab6-KIFL (Figure 7A, Purified IgG) was used. Very rarely, dividing cells could be seen; usually these had formed a metaphase plate, indicating that this process is not perturbed by the antibody microinjection (Figure 7A, Serum II). When the pre-immune serum corresponding to this antibody was microinjected, only mononucleate cells were observed at all the time points investigated (Figure 7A, Pre-immune serum). Quantitation of the effects of microinjection for the various conditions revealed that after 24–30 h, between 7 and 10% of the cells microinjected with the Rab6-KIFL antibody were found to be binucleate and that this rose to nearly 50% by 36 h (Figure 7B). These effects were specific for Rab6-KIFL since microinjection of the corresponding pre-immune serum resulted in no bi- or multinucleate cells at any of the time points taken (Figure 7B). These data indicate that microinjection of the Rab6-KIFL antibody causes a specific defect in cell division, preventing cytokinesis.
Fig. 7. Effects of antibody microinjection on cell division. (A) HeLa cells grown on coverslips were microinjected as described in Materials and methods with pre-immune serum, the Rab6-KIFL antiserum or purified sheep immunoglobulin (IgG) to Rab6-KIFL. Representative images of microinjected cells are shown for each condition, 36 h post-injection. (B) The number of normal, binucleate or multinucleate microinjected cells was counted at the times indicated, after microinjection of either the pre-immune serum or the affinity-purified antibody to Rab6-KIFL. The numbers shown are from a typical experiment in which 200 cells were microinjected for each condition. In each case similar results were obtained for three independent experiments performed with all the conditions shown.
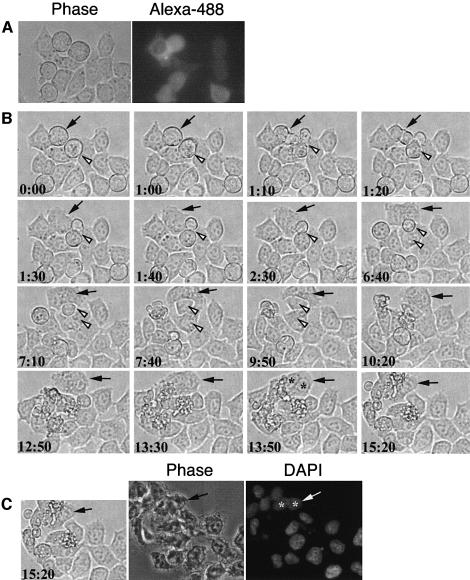
To identify the stage at which the Rab6-KIFL antibody interferes with cytokinesis, the microinjected cells were imaged using time-lapse microscopy (Figure 8). HeLa cells microinjected with the Rab6-KIFL antibody were allowed to recover for 6 h under normal culture conditions. Microinjected cells were then identified under the microscope by the presence of the fluorescent dye Alexa-488, which was co-injected with the Rab6-KIFL antibody (Figure 8A, right panel). In the example shown a microinjected cell that has already entered mitosis is seen to start forming a cleavage furrow, which then regresses before cytokinesis has taken place (Figure 8B, t = 0:00–1:40, filled arrow). The microinjected cell then flattens out completely and proceeds to migrate in a clockwise direction; the presence of two nuclei is clearly visible (Figure 8B, t = 13:50, asterisks). At the end of the imaging period the sample was fixed and stained with DAPI, which revealed that this cell does indeed contain two nuclei (Figure 8C, arrow and asterisks). In comparison, an adjacent non-injected cell is seen to form a cleavage furrow and then undergo cytokinesis, giving rise to two mononucleate daughter cells (Figure 8B, t = 0:00–7:40, open arrowhead). Cells microinjected with the corresponding pre-immune antibody divided normally, and displayed no defects in cleavage furrow formation (Figure 7B; E.Hill and F.A.Barr, unpublished observations). Together with the data localizing Rab6-KIFL to the spindle midzone and to the cleavage furrow, these observations suggest that Rab6-KIFL functions during cleavage furrow formation and cytokinesis.
Fig. 8. Time-lapse imaging of cells microinjected with antibodies to Rab6-KIFL. (A) HeLa cells grown on coverslips were microinjected as described in Materials and methods with the Rab6-KIFL antibody and purified donkey IgG conjugated to Alexa-488. Cells were then allowed to recover for 6 h before imaging. A phase contrast, and corresponding Alexa-488 fluorescence image are shown. (B) Time-lapse imaging of the cells shown in (A) above; time after commencement of imaging is indicated in hours and minutes. The filled arrow indicates a microinjected cell in mitosis that has started to divide. The open arrowhead indicates a non-injected cell in mitosis that divides normally. (C) After 15 h 20 min of imaging the cells were fixed and the DNA stained with DAPI. The same cells are shown as in (B) above. A movie corresponding to this figure is available as Supplementary data at The EMBO Journal Online.
Discussion
We have shown that mammalian Rab6-KIFL accumulates in mitotic cells, where it localizes to central spindle microtubules and to the midbody during cytokinesis. Unlike kinesins such as XKLP1 required for chromosome positioning, or CENP-E involved in kinetochore capture (Vernos et al., 1995; Wood et al., 1997), Rab6-KIFL is not observed at either the spindle poles or the kinetochores, and therefore we believe it plays no role in these processes. It is also unlikely that Rab6-KIFL is required for spindle formation, since it is only recruited to the spindle once the metaphase plate has formed. Furthermore, Rab6-KIFL staining is most intense at the early stages of cleavage furrow formation, after which point it gradually decreases in intensity. Microinjection of antibodies specific to Rab6-KIFL blocks cytokinesis and causes the cells to become binucleate after one cell cycle, but does not interfere with spindle formation in any obvious manner. Microinjected cells appear normal until metaphase, since they are able to align their chromosomes to form a metaphase plate, but then fail to undergo cytokinesis, indicative of a defect in cleavage furrow formation or of cytokinesis itself. Time-lapse imaging of microinjected cells reveals that they start to form a cleavage furrow but do not undergo cytokinesis; instead the cleavage furrow relaxes and the now binucleate cells flatten out. Consistent with these observations, cells overexpressing Rab6–kinesin have a disorganized microtubule cytoskeleton with many microtubule bundles, and although the majority of cells are able to form a cleavage furrow they appear to be unable to undergo cytokinesis.
Members of the kinesin family of motor proteins are required for many aspects of the formation, maintenance and elongation of the mitotic spindle, as well as for cleavage furrow formation and cytokinesis (Heck, 1999; Robinson and Spudich, 2000). The mammalian mitotic kinesin, CHO1/MKLP1, known to be involved in cleavage furrow formation is found in the nucleus of interphase cells, at the spindle poles during metaphase, and concentrates at the midbody and on the midzone microtubules during anaphase (Sellitto and Kuriyama, 1988; Nislow et al., 1992). Mammalian MKLP1 can crosslink microtubules in anti-parallel bundles via two microtubule-binding sites, one in the N-terminal motor domain and the other in the C-terminal tail (Nislow et al., 1992). Analysis of mutants in the fly and worm MKLP1 homologues, pavarotti and ZEN-4, respectively, has revealed that these show cytokinesis rather than spindle defects (Adams et al., 1998; Powers et al., 1998; Raich et al., 1998). In the Drosophila melanogaster mutant pavarotti various components required for cytokinesis, such as the septins and actin, fail to localize to the cleavage furrow (Adams et al., 1998), suggesting that this class of kinesins is needed at an early stage in cleavage furrow formation. Examination of the zen-4 mutant of Caenorhabditis elegans has shown that this is able to form a cleavage furrow that constricts, but then ceases contraction and undergoes a reversal of this process (Raich et al., 1998). Closer analysis of the zen-4 mutant revealed that the interdigitating central spindle microtubules are absent, raising the possibility that the MKLP1 family of kinesins is required to stabilize these microtubules. Furthermore, ZEN-4 does not participate in spindle elongation, since this is unaffected in RNA interference experiments (Raich et al., 1998). It has therefore been proposed that ZEN-4 is needed to specify the position at which the cleavage furrow will form, and to maintain it once it has formed (Powers et al., 1998).
Rab6-KIFL displays a similar subcellular localization to these mitotic kinesins, the endogenous protein participates in the same process of cleavage furrow formation and cytokinesis, and it is most closely related in terms of sequence homology with the MKLP1 kinesins. Like the MKLPs Rab6-KIFL also possess two microtubule-binding sites, one in the N-terminal kinesin motor domain and the other adjacent to the C-terminus (Echard et al., 1998). We interpret these observations as evidence of a role for Rab6-KIFL in cleavage furrow formation, similar to ZEN-4. Interestingly, it has recently been reported that MKLP1 is capable of binding to another GTPase involved in membrane traffic, Arf1 (Boman et al., 1999), providing further support for the view that kinesin motor protein-directed membrane traffic events might have specific functions during cell division. How our observations relate to the previous findings that Rab6-KIFL is involved in membrane traffic in interphase cells is not immediately obvious (Echard et al., 1998). These previous studies were performed using exogenously expressed Rab6-KIFL, whereas we have concentrated on an examination of the endogenous protein present in mitotic cells. The Golgi apparatus is known to require microtubules for its correct subcellular localization (reviewed in Burkhardt, 1998), and disruption of the normal microtubule cytoskeleton by Rab6-KIFL overexpression might in part explain the previous findings. However, we did observe that the Rab6-binding domain of Rab6-KIFL was able to localize partially to the Golgi apparatus, and in some cells overexpression of the full-length Rab6-KIFL triggered Golgi disassembly. Consistent with previous observations we were also able to detect a small pool of Rab6-KIFL on the Golgi apparatus in some interphase cells with the published antibody to this protein (Figure 2B; Echard et al., 1998). We have also been able to confirm using the yeast two-hybrid system that the GTP form of Rab6 binds to this domain in Rab6-KIFL (M.Clarke and F.A.Barr, unpublished observations; Echard et al., 1998). Therefore, we believe that Rab6-KIFL is a Rab6-binding kinesin and do not rule out that it plays a role in Rab6-mediated membrane traffic events to or from the Golgi apparatus. Indeed, one explanation for all these findings might come from the requirement for membrane traffic events during cell division.
In order for cells to divide there must be a membrane fusion event involving the plasma membrane, either directly between the inner leaflets of the plasma membrane, or indirectly by delivery of vesicles to the point of cytokinesis, which then fuse with the plasma membrane (Robinson and Spudich, 2000). All other cellular membrane fusion events are mediated by a common set of cytosolic and integral membrane components (Waters and Pfeffer, 1999). Syntaxins are small coiled-coil, integral membrane proteins involved directly in determining both the specificity and mechanism of membrane fusion (Weber et al., 1998). In organisms as diverse as flies, worms and plants there is evidence that plasma membrane syntaxins are required for membrane fusion events during cytokinesis, not just during the membrane traffic events in secretion and cell growth (Burgess et al., 1997; Lauber et al., 1997; Jantsch-Plunger and Glotzer, 1999). It is possible that one function of Rab6-KIFL is in the delivery of vesicles from the Golgi apparatus to the site of membrane fusion during cytokinesis. If this is the case then one or all of the known variants of Rab6 might be expected to localize to these vesicles or to membranes in the midzone region. Alternatively, it is possible that Rab6-KIFL binds to GTPases in addition to Rab6 in vivo. Candidates might be members of the Rho family of GTPases, which together with their regulatory proteins are known to be present in the cleavage furrow (Madaule et al., 1998; Tatsumoto et al., 1999). Although small GTPases of the Rho family are known to localize to the cleavage furrow, thus far none has been found in the spindle midzone like Rab6-KIFL (Prokopenko et al., 2000; Robinson and Spudich, 2000). We are currently investigating these and other possibilities.
Materials and methods
Molecular biology and protein expression
Human Rab6-KIFL was cloned from fetal human cDNA (Clontech) using specific primers HSR6K-F2 GCCGTCATGTCGCAAGGGATCCTTTCTCC and HSR6K-R2 CCATGACTGCTCTTCTCTTTCCCCACAGCC, and the pfu-turbo polymerase (Stratagene). The PCR products were purified from agarose gel slices using gel extraction spin columns (Qiagen), treated with Taq polymerase and dNTPs at 72°C for 20 min to add overhanging ‘A’ bases, then TA-cloned using the pCRII-TOPO vector system (Invitrogen). The Rab6-binding domain was amplified from the Rab6-KIFL clone using primers HSR6BD-F CGGATCCATAGTCTTCAGGTATCCCCCAGC and HSR6BD-R GCTCGAGGATGGGCCACTGACTGTTGTCTGGC. Mammalian cell expression constructs were made in the pEGFP-C vectors (Clontech), and those for expression in bacteria in the pQE30 vectors (Qiagen). DNA sequencing of all constructs was performed by PNACL UK Ltd. Bacterial strains XL1b and TOP10 were used for routine cloning, and JM109 for protein expression. JM109 cells expressing recombinant proteins were lysed by sonication in IMAC5 (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 5 mM imidazole), the extract was incubated with nickel–agarose (Qiagen), the nickel–agarose washed in IMC5 plus 20 mM imidazole, then the proteins eluted with IMAC5 plus 200 mM imidazole. Proteins were further purified by gel filtration in phosphate-buffered saline (PBS) over a Superose 6 HR10/30 column (Amersham–Pharmacia Biotech).
Antibodies
The recombinant Rab6-binding domain of Rab6-KIFL was used to produce a polyclonal antibody in sheep by Diagnostics Scotland. To eliminate any non-specific reactivity the antibody was affinity purified on Affigel-15 to which the specific antigen had been covalently coupled according to the manufacturer’s instructions (Bio-Rad). The purified immunoglobulin was dialysed against PBS, and 10 mg/ml bovine serum albumin (BSA) added to act as a stabilizing agent. The rabbit antibody to Rab6-KIFL, the MLO-7 antibody to GM130, and the antibody to human cyclin B2 were generous gifts of Dr Bruno Goud, Dr Martin Lowe and Dr Jonathon Pines, respectively. The Rab6 antibody was a rabbit polyclonal C-19 (Santa Cruz Biotech). Secondary antibodies for immunofluorescence were multiple labelling grade produced in donkey either conjugated to CY3 (Jackson Laboratories) or purchased as purified antibody from Jackson Laboratories for conjugation to Alexa-488 (Molecular Probes).
Cell culture
HeLa-S3 cells were cultured in RPMI plus 10% fetal calf serum (TCS Biologicals) in 175 cm2 plastic flasks (Nunc). For cell cycle arrests, cells from six flasks were trypsinized and resuspended in 1 l of medium, then grown for 24 h in a spinner flask of the appropriate size. The cells were recovered by centrifugation and resuspended in 1 l of medium containing either 2 mM hydroxyurea or 80 ng/ml nocodazole, then grown for a further 18 h (hydroxyurea) or 20 h (nocodazole) in glass spinner flasks. Extracts were prepared from nocodazole- and hydroxyurea-arrested cells or interphase cells as described previously (Misteli and Warren, 1994).
Transfection, microinjection and microscopy
HeLa cells grown on coverslips in six-well plates were transfected with 0.2 µg of DNA using Effectene (Qiagen) following the manufacturer’s instructions. Microinjection of HeLa cells grown on coverslips was performed on a Zeiss Axiovert S100 microscope equipped with an Eppendorf microinjection system and Zeiss LD-Acroplan 40×, 0.6 numerical aperture objective. Typically, 200 cells were microinjected using a 1/10 s injection time and 150–300 µPa pressure. Antibodies and pre-immune sera were diluted 1:10 in PBS to reduce sample viscosity and increase the reproducibility of microinjection. Cells were fixed in –20°C methanol for 4 min and processed for immunofluorescence. For Rab6 staining cells were fixed in 3% paraformaldehyde in PBS for 20 min at room temperature, washed in PBS, quenched in 50 mM ammonium chloride for 15 min, then permeabilized in 0.1% (v/v) Triton X-100 for 5 min at room temperature. All antibody incubations were carried out in PBS containing 2% (w/v) BSA (PBS–BSA) for 30 min at room temperature. Between antibody incubations the coverslips were washed three times for 5 min in PBS–BSA. Coverslips were mounted in Moviol 4-88 (Calbiochem) containing 0.25 µg/ml DAPI on glass slides. Standard cell imaging was performed on a Zeiss Axioplan microscope equipped with a Plan-Neofluor 100×, 1.3 numerical aperture oil immersion lens, a Hamamatsu Photonics C5985 camera, and using Apple Macintosh NIH Image 1.62 to control the camera and capture data. Confocal images were collected using a Bio-Rad MRC-600 confocal microscope using krypton–argon ion laser excitation at 488 and 568 nm, and collection data with 522/35 nm band pass and 585 nm long pass emission filters. Time-lapse imaging was performed on a Zeiss Axiovert S100 microscope equipped with a heated stage, a polychrome II light source (Till Photonics), a C4747-95 digital camera (Hamamatsu Photonics) and the Openlab 2.2.0 automated imaging system for Apple Macintosh (Improvision). All images were collected using a Zeiss LD-Acroplan 40×, 0.6 numerical aperture objective. Camera binning was 2×, and the white light illumination was set to the lowest possible intensity, imaging times were 40 ms for phase and 0.5 s for immunofluorescence images. All images were 640 × 480 pixels, with 256 levels of grey.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Liz Black for initial help with microinjection, Dave Gillespie for advice on cell cycle arrests and the elutriation of HeLa cells, Peter McHardy for assistance with confocal microscopy, and Elmar Schiebel, John Wyke, Vas Ponnambalam, Ben Short and Ulrike Grüneberg for reading this manuscript. We acknowledge the CRC–Beatson Institute for providing laboratory space and facilities, and Diagnostics Scotland for producing excellent antibodies. This work was supported by grants from the BBSRC and the Wellcome Trust.
References
- Adams R.R., Tavares,A.A., Salzberg,A., Bellen,H.J. and Glover,D.M. (1998) pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev., 12, 1483–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V.J. and Schroer,T.A. (1999) Membrane motors. Curr. Opin. Cell Biol., 11, 476–482. [DOI] [PubMed] [Google Scholar]
- Barr F.A. (1999) A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol., 9, 381–384. [DOI] [PubMed] [Google Scholar]
- Boman A.L., Kuai,J., Zhu,X., Chen,J., Kuriyama,R. and Kahn,R.A. (1999) Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil. Cytoskel., 44, 119–132. [DOI] [PubMed] [Google Scholar]
- Burgess R.W., Deitcher,D.L. and Schwarz,T.L. (1997) The synaptic protein syntaxin1 is required for cellularization of the Drosophila embryo. J. Cell Biol., 138, 861–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J.K. (1998) The role of microtubule-based motor proteins in maintaining the structure and function of the Golgi complex. Biochim. Biophys. Acta, 1404, 113–126. [DOI] [PubMed] [Google Scholar]
- Cuif M.H. et al. (1999) Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J., 18, 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Jollivet,F., Martinez,O., Lacapere,J.J., Rousselet,A., Janouiex-Lerosey,I. and Goud,B. (1998) Interaction of a Golgi associated kinesin-like protein with rab6. Science, 279, 580–585. [DOI] [PubMed] [Google Scholar]
- Heck M.M. (1999) Dr. Dolittle and the making of the mitotic spindle. BioEssays, 21, 985–990. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda,Y. and Okada,Y. (1998) Kinesin and dynein superfamily proteins in organelles transport and cell division. Curr. Opin. Cell Biol., 10, 60–73. [DOI] [PubMed] [Google Scholar]
- Jackman M., Firth,M. and Pines,J.(1995) Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J., 14, 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V. and Glotzer,M. (1999) Depletion of syntaxins in the early Caenorhabitis elegans embryo reveals a role for membrane fusion events during cytokinesis. Curr. Biol., 9, 738–745. [DOI] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger,I., Steinmann,T., Schwarz,H., Mayer,U., Hwang,I., Lukowitz,W. and Jurgens,G. (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol., 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P., Masatoshi,E., Watanabe,N., Fujisawa,K., Matsuoka,T., Bito,H., Ishizaki,T. and Narumiya,S. (1998) Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature, 394, 491–494. [DOI] [PubMed] [Google Scholar]
- Martinez O., Schmidt,A., Salaméro,J., Hoflack,B., Roa,M. and Goud,B. (1994) The small GTP-binding protein rab6 functions in intra-Golgi transport. J. Cell Biol., 127, 1575–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. and Warren,G. (1994) COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. J. Cell Biol., 125, 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. and Nichols,B.J. (1999) The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol., 9, 377–380. [DOI] [PubMed] [Google Scholar]
- Nislow C., Lombillo,V.A., Kuriyama,R. and McIntosh,J.R. (1992) A plus-end directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature, 359, 543–547. [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer,P., Khodjakov,A., Rieder,C.L. and Bornens,M. (2000) The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol., 149, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J., Bossinger,O., Rose,D., Strome,S. and Saxton,W. (1998) A nematode kinesin required for cleavage furrow advancement. Curr. Biol., 8, 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko S.N., Saint,R. and Bellen,H.J. (2000) Untying the gordian knot of cytokinesis: role of small G proteins and their regulators. J. Cell Biol., 148, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich W.B., Moran,A.N., Rothman,J.H. and Hardin,J. (1998) Cytokinesis and midzone microtubule organization in Caenorhabitis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell, 9, 2037–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt A. and Hubbard,T. (1998) Using neural networks for prediction of the subcellular location of proteins. Nucleic Acids Res., 26, 2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.N. and Spudich,J.A. (2000) Towards a molecular understanding of cytokinesis. Trends Cell Biol., 10, 228–237. [DOI] [PubMed] [Google Scholar]
- Sellitto C. and Kuriyama,R. (1988) Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J. Cell Biol., 106, 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima D.T. and Warren,G. (1998) Inheritance of the cytoplasm during cell division. In Endow,S.A. and Glover,D.M. (eds), Dynamics of Cell Division. Oxford University Press, Oxford, UK, pp. 248–269. [Google Scholar]
- Shima D.T., Cabrera-Poch,N., Pepperkok,R. and Warren,G. (1998) An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol., 141, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumoto T., Xie,X., Blumenthal,R., Okamoto,I. and Miki,T. (1999) Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases and involved in cytokinesis. J. Cell Biol., 147, 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos I., Raats,J., Hirano,T., Heasman,J., Karsenti,E. and Wylie,C. (1995) Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell, 81, 117–127. [DOI] [PubMed] [Google Scholar]
- Waters M.G. and Pfeffer,S.R. (1999) Membrane tethering in intracellular transport. Curr. Opin. Cell Biol., 11, 453–459. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman,B.V., McNew,J.A., Westermann,B., Gmachl,M., Parlati,F., Sollner,T.H. and Rothman,J.E. (1998) SNAREpins: minimal machinery for membrane fusion. Cell, 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz,R., Goldstein,L.S. and Cleveland,D.W. (1997) CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell, 91, 357–366. [DOI] [PubMed] [Google Scholar]