Abstract
Human ether-à-go-go-related gene (hERG) K+ channels have a critical role in cardiac repolarization. hERG channels close (deactivate) very slowly, and this is vital for regulating the time course and amplitude of repolarizing current during the cardiac action potential. Accelerated deactivation is one mechanism by which inherited mutations cause long QT syndrome and potentially lethal arrhythmias. hERG deactivation is highly dependent upon an intact EAG domain (the first 135 amino acids of the N terminus). Importantly, deletion of residues 2–26 accelerates deactivation to a similar extent as removing the entire EAG domain. These and other experiments suggest the first 26 residues (NT1–26) contain structural elements required to slow deactivation by stabilizing the open conformation of the pore. Residues 26–135 form a Per-Arnt-Sim domain, but a structure for NT1–26 has not been forthcoming, and little is known about its site of interaction on the channel. In this study, we present an NMR structure for the entire EAG domain, which reveals that NT1–26 is structurally independent from the Per-Arnt-Sim domain and contains a stable amphipathic helix with one face being positively charged. Mutagenesis and electrophysiological studies indicate that neutralizing basic residues and breaking the amphipathic helix dramatically accelerate deactivation. Furthermore, scanning mutagenesis and molecular modeling studies of the cyclic nucleotide binding domain suggest that negatively charged patches on its cytoplasmic surface form an interface with the NT1–26 domain. We propose a model in which NT1–26 obstructs gating motions of the cyclic nucleotide binding domain to allosterically stabilize the open conformation of the pore.
Keywords: Ion Channels, NMR, Potassium Channels, Protein Structure, Site-directed Mutagenesis, Kv11.1, EAG Domain, hERG, Solution Structure
Introduction
The human ether-à-go-go-related gene (hERG)5 encodes pore-forming subunits for channels that conduct the cardiac delayed rectifier potassium current (IKr) (1, 2). This current is essential for normal cardiac action potential repolarization (3). Altered hERG (Kv11.1) channel function, either due to pharmacological block by medications or inherited mutations, is a major cause of long QT syndrome (4–8), an abnormality of ventricular repolarization that causes prolongation of the QT interval and predisposes affected individuals to arrhythmia and sudden cardiac death (9, 10).
The critical role of hERG in cardiac repolarization derives from its unusual voltage-dependent gating properties, characterized by slow opening and closing of the activation gate but very fast onset and recovery from inactivation (1, 2, 11–16). hERG currents are relatively small during the plateau phase of the ventricular action potential, because most channels are in the nonconducting inactivated state. However, during repolarization, hERG current increases due to rapid recovery from inactivation followed by slow closure of the activation gate (deactivation). These distinctive gating properties make IKr highly responsive to membrane potential during the plateau of the action potential (17–19). Slow deactivation has a major influence on the physiological time course of IKr (20) and ensures hERG channels are available to oppose asynchronous and arrhythmogenic stimuli (19, 21). Long QT syndrome mutations that increase the rate of hERG deactivation reduce IKr amplitudes, decrease the contribution of this current to cardiac repolarization, and increase the potential for life-threatening arrhythmias (16, 22, 23).
The structural basis for the slow deactivation gating of hERG is only partially understood. Like other Kv channels, the pore and voltage sensing domains are formed by the co-assembly of four subunits each with six transmembrane-spanning helices. hERG channels also have large intracellular N and C termini (24). The C terminus includes a region homologous to the cyclic nucleotide binding domain (cNBD) of cyclic nucleotide-gated channels, although key residues for binding to nucleotides are not conserved and the functional role of this structural domain is unclear (25, 26). The N terminus is ∼400 residues long, with residues 1–135 referred to as the EAG domain because it is a highly conserved and defining feature of the EAG channel family (24). hERG channel deactivation is greatly accelerated by deleting either the whole N terminus or just the EAG domain (12–14, 16, 27). The crystal structure for residues 26–135 of the EAG domain has been solved and shown to fold into a Per-Arnt-Sim (PAS) domain (16). Unfortunately, the crystal structure did not resolve the N terminus 1–26 (NT1–26) region, which is now known to be the functionally critical domain for slowing deactivation gating (16, 27). Deletions within the NT1–26 domain result in accelerated deactivation rates similar to deleting the whole N terminus (13, 16, 27). Moreover, intracellular application of a peptide corresponding to residues 1–16 partially recovers deactivation kinetics to the N-terminal truncated (Δ2–354) channels and to a similar extent as dialyzing in the EAG domain (16, 27). These studies are consistent with the NT1–26 domain binding to a receptor site (that has yet to be identified) and stabilizing the open state of the pore. The precise role of the PAS domain remains uncertain, but it may control access of the NT1–26 domain to its binding site and dynamically regulate deactivation gating through interactions with the pore, cNBD, and voltage-sensing domain (28).
In this study, we have used NMR spectroscopy to resolve the structure of the whole EAG domain, including the NT1–26 domain. Our results indicate that NT1–26 is in two parts, with amino acids 1–10 being highly dynamic and flexible, whereas amino acids 11–24 form a stable amphipathic helix. One face of this newly resolved NT1–26 structural element is positively charged. Neutralizing basic residues profoundly accelerates deactivation and mimics deleting the N terminus. Charge reversal mutations of an acidic patch of the cNBD suggest this is where NT1–26 binds. We propose a model in which deactivation gating is mediated by the NT1–26 domain binding to negatively charged crevices in the cNBD, maintaining it an open conformation and thus allosterically stabilizing the open state of the pore.
EXPERIMENTAL PROCEDURES
Protein Sample Preparation and NMR Spectroscopy
The EAG domain (residues 1–135) was subcloned into an Escherichia coli expression vector with a C terminus hexahistidine tag as described previously (29). All NMR experiments were performed on Bruker DRX or AvanceII spectrometers operating at either 600 or 800 MHz that were fitted with cryogenically cooled probe heads. Experiments used 350-μl samples of the uniformly 15N- or 13C/15N-labeled EAG domain (0.5 mm) dissolved in 20 mm sodium acetate buffer, pH 5.0, containing 0.5 mm DTT, 0.5 mm EDTA, 0.2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 0.02% (w/v) sodium azide, at 25 °C. The samples were prepared in 95% H2O and 5% D2O. Detailed descriptions of the multidimensional experiments used to obtain the near complete sequence-specific 1H, 13C, and 15N assignments have been reported previously (29). Additionally, 15N-NOESY-HSQC (30) and 13C-NOESY-HSQC (31) spectra were acquired with an NOE mixing period of 100 ms. These spectra were recorded over ∼68 h, with acquisition times in the indirect dimensions of ∼16 ms for 1H and ∼7 and ∼11 ms for 13C and 15N, respectively, and for 80 ms in the real time domain. Water suppression was achieved using the gradient-based WATERGATE method (32). Two-dimensional 15N/1H-HSQC spectra were also acquired from a sample of the 15N-labeled EAG domain dissolved in D2O to identify any slowly exchanging amide protons in the protein (33). All NMR data were processed essentially as described previously (29) using Topspin (Bruker Biospin Ltd.), and the resulting spectra were analyzed using the program Sparky (T. D. Goddard and D. G. Kneller, Sparky 3, University of California, San Francisco).
Structure Calculations
The family of converged structures was determined using the program CYANA (34), as described previously (35, 36). Hydrogen bond constraints, involving 50 residues with slowly exchanging backbone amide signals and where the hydrogen bond acceptor was unambiguous in preliminary structures, were added to the final round of calculations. Backbone torsion angle constraints derived from the protein backbone dihedral angle prediction program TALOS+ (37) were included in both stages of the calculation. The 82 structures with no restraint violations (NOE violations <0.5 Å and <5° for dihedral angles) were refined with two cycles of restrained molecular dynamics simulated annealing with the AMBER 10 package (38) using the following protocol. Structures were initially energy-minimized (2000 steps), followed by 20 ps of simulated annealing in vacuum and then by 20 ps of simulated annealing using a generalized Born solvent model (39). The system was heated to 1200 K over the 1st ps, annealed at a constant temperature for 5 ps, and then cooled to 0 K during the remaining 14 ps. Force constants were 30 kcal mol−1 Å2 for distance constraints and 1000 kcal mol−1 rad−2 for dihedral angle constraints. The 20 structures with the lowest AMBER energy and with no distance constraint violation greater than 0.24 Å were selected. Analysis of the final family of structures obtained was carried out using PROCHECK-NMR (40) and MOLMOL (41).
Homology Modeling and Protein-Protein Docking
Homology models of the cNBD and C-linker of hERG were built using Modeler (42), with the HCN2 crystal structure as a template (Protein Data Bank code 1Q5O) (25). 2000 initial docking configurations for the N-terminal domain bound to the cNBD were generated using ZDOCK (43) for each of the 82 elucidated NMR conformers. The ZDOCK output was then re-scored and ranked using ZRANK (44). The top 10 complexes for each NMR conformer were saved for cluster analysis. Clustering of the 710 docked complexes was performed using the single linkage method in g_cluster, which is part of GROMACS molecular dynamics suite (45).
To optimize the interactions between NT1–10 and the cNBD C-helix acidic patch, Modeler was used to apply distance restraints between the Arg4/Arg5/His7 and Asp843/Glu847/Asp850/Glu857 clusters, for a set of docked solutions that bound to the side of the cNBD. An optimal model was then selected from this group based on good agreement with the known interaction data. To test the validity of this model as a docked conformation of the EAG domain, ZDOCK was used to re-dock the modeled structure onto the cNBD.
Site-directed Mutagenesis and Electrophysiology
Site-directed mutagenesis was performed using the QuikChange mutagenesis technique (Stratagene, La Jolla, CA) on the hERG pXoom construct (kindly provided by Dr. Thomas Jespersen) (46). Plasmid DNA was linearized with XbaI, and in vitro transcription was performed using T7 RNA polymerase (mMessage mMachine, Ambion, Austin, TX). Xenopus laevis oocytes were isolated, defolliculated, maintained in culture, and injected with wild type or mutant cRNA as described previously (47). Whole cell currents were recorded in Xenopus oocytes using a two-electrode voltage clamp (48). Microelectrodes were filled with 3 m KCl and the tips broken to give resistances of 1.1–1.5 megaohms. Recordings were made at room temperature 1–5 days after cRNA injection. Data were low pass filtered and sampled at 5 kHz and saved to computer for off-line analysis using a digidata 1320A data acquisition system (Molecular Devices, Sunnyvale, CA). Oocytes were perfused with a NaCl-based solution containing (in mm) NaCl 96, KCl 2, CaCl2 2, MgCl2 1, HEPES 5, pH 7.6. The properties of mutants were directly compared with WT hERG recorded in the same extracellular solution.
Voltage Protocols and Data Analysis
The time- and voltage-dependent kinetics of deactivation were measured using a fully activated I-V protocol. hERG currents were activated and inactivated with a 1-s prepulse to +40 mV, and tail currents were measured with 5-s pulses to potentials between −160 and +30 mV. The decay phase of tail currents were fit with a single exponential function as shown in Equation 1,
where τ is the time constant of deactivation; A is the amplitude of deactivating current, and C is the steady state current. Analyses of the kinetics of deactivation were performed using Clampfit software (Molecular Devices, Sunnyvale, CA). Although a double exponential function provides a more accurate fit, a monoexponential fit facilitates quantitative comparisons between WT hERG and hERG mutants. Data are presented as means ± S.E. (n = number of cells). Statistical comparisons were performed using unpaired Student's t test where appropriate. Differences were considered significant at p ≤ 0.05.
RESULTS AND DISCUSSION
Solution Structure of the EAG Domain
As 90.6% of the proton resonances were assigned by NMR spectroscopy, the combined automated nuclear Overhauser effect (NOE) assignment and structure determination protocol (CANDID) (49) were employed for structural calculations. The 20 structures with the lowest AMBER energy and with no distance constraint violations greater than 0.24 Å were selected for analysis. The NMR constraints and structural statistics are summarized in Table 1, and the structures have been deposited in the Protein Data Bank (accession code 2L1M).
TABLE 1.
NMR constraints and structural statistics
| Total NOE distance constraints | 2131 |
| Short range (i, i −1) | 1204 |
| Medium range (i, i− ≤4) | 414 |
| Long range (i, i − >5) | 513 |
| Total dihedral angle constraints | 241 |
| Ψ | 122 |
| ϕ | 119 |
| Violations | |
| Maximum distance violation | 0.24 Å |
| Maximum dihedral angle violation | 0° |
| Energies | |
| Mean AMBER energy | −5333.22 kcal mol−1 |
| Mean NOE energy | 1.259 kcal mol−1 |
| Deviations for idealized geometry | |
| Bond lengths | 0.0103 ± 5.98 × 10−5 Å |
| Bond angles | 2.103 ± 0.019° |
| r.m.s.d. from mean structurea | |
| Backbone atoms (N, Cα, C′) | 0.49 ± 0.08 Å |
| All heavy atoms | 1.00 ± 0.09 Å |
| Ramachandran plot | |
| Residues in most favorable regions | 84.2% |
| Residues in additionally allowed regions | 15.3% |
| Residues in generously allowed regions | 0.5% |
| Residues in disallowed regions | 0% |
a Residues are 26–135. r.m.s.d. means root mean square deviation.
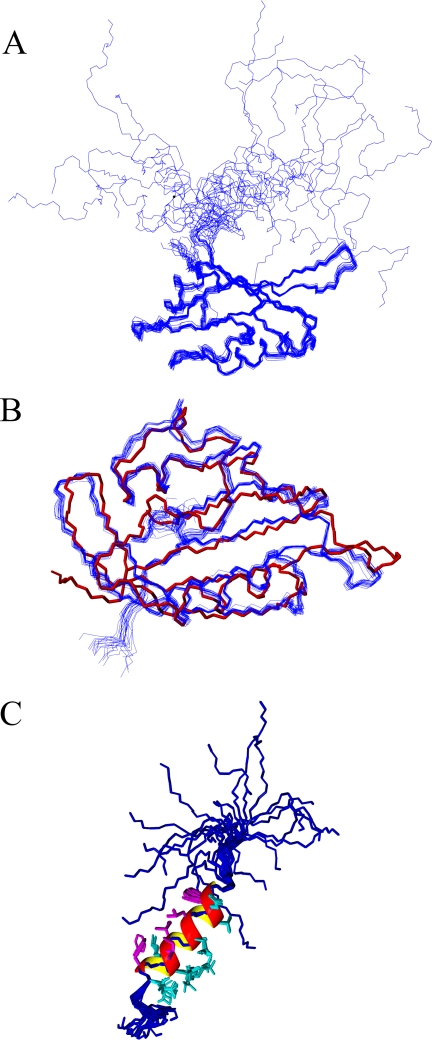
Superimposing the protein backbone for the family of solution structures demonstrates that the EAG domain is very well defined by the NMR data (Fig. 1A). The PAS domains in the NMR structures are in excellent agreement with the crystal structure by Morais Cabral et al. (16), with an average backbone root mean square deviation. of 0.92 ± 0.65 Å (Fig. 1B). The secondary structure locations in the PAS domain of the NMR structure (particularly the α-helices) are virtually identical, although the β-sheet regions are less regular. What is unique to the NMR structures are the coordinates for the NT1–26 domain that were not detectable in the electron density of the crystal structure (Fig. 1C). The NT1–26 region is structurally independent from the PAS domain and can be subdivided into two parts. Met1–Pro10 showed very sharp signals in the NMR spectrum, and these residues have only short range (i, i − 1) NOE information, indicating a highly dynamic, extended structure. On the other hand, Gln11–Gly24 exhibited a significant number (30%) of medium range (i, i− ≥2 ≤ 4) NOE assignments, and this coupled with the backbone shift data signify these amino acids form a well defined α-helix with equally well ordered side chains (Fig. 1C). The helix starts with the polar residues Gln11 and Asp12, but the remainder of the helix is amphipathic. One side is composed of entirely hydrophobic residues (Phe14, Leu15, Ile18, Ile19, and Phe22), and the other side is formed from either charged or polar residues (Thr13, Asp16, Thr17, Arg20, Lys21, and Glu23). NOE assignments for Met1–Gln25 do not show influences from the PAS domain. The dynamic nature of the NT1–26 domain, and lack of tertiary structure, may be important for its gating function and indicate it is highly unlikely to bind to the PAS domain. A search of the Protein Data Bank data base showed that of the 20 PAS domain structures with the most three-dimensional conformational similarity to hERG (Z scores >7.5), 8 started with a helix of four or more residues. Although many PAS domains form dimers and use an N-terminal amphipathic helix (50, 51) to stabilize this interaction, there was no evidence of dimerization in the data collected for the hERG PAS domain.
FIGURE 1.
Structure of the NT1–26 domain of hERG. A, superposition of the protein backbone (Ser26 to Val132) for the family of 20 lowest energy structures calculated using AMBER. B, comparison of the protein backbone of the PAS domain crystal structure (red, Protein Data Bank accession code 1BYW) with the family of NMR structures (blue). C, peptide backbones (blue) of Met1–Gln25 from the 20 NMR structures in A superimposed to illustrate the amphipathic helix extending from Glu11–Gly24. The helix is represented by a ribbon with the hydrophobic residues in magenta and the charged or polar residues in cyan. For clarity, only the side chains of residues in the helix are shown.
Deactivation Gating Requires a Positively Charged Surface on the NT1–26 Domain
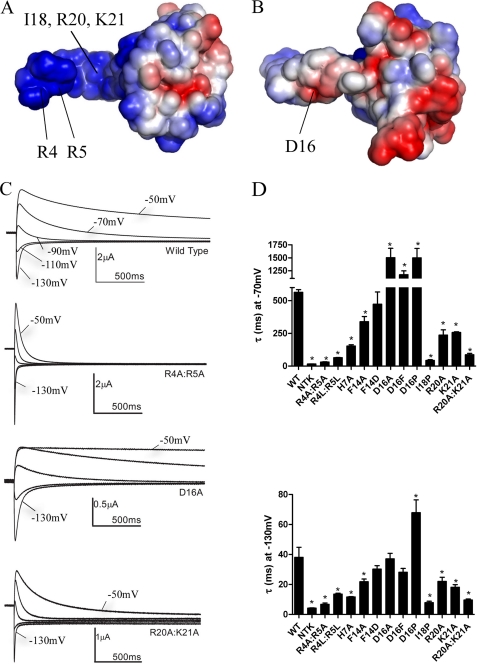
Calculation of the electrostatic surface of the NT1–26 domain revealed that it is highly positively charged on one side and neutral or slightly negatively charged on the opposite side (see Fig. 2, A and B). Basic residues are clustered within the unstructured region (Arg4, Arg5, and His7) and in the amphipathic helix (Arg20 and Lys21). Electrophysiological recordings were made from Xenopus oocytes injected with cRNA encoding either WT hERG or mutants in which basic residues were substituted with neutral Ala residues (see Fig. 2C). A two-step voltage protocol was applied in which the currents are fully activated (and inactivated) with a prepulse to +40 mV and then tail currents were elicited with test pulses to potentials between −50 and −130 mV. The decay of the tail currents is due to channel deactivation. The deactivation rate was substantially faster in R4A/R5A and R20A/K21A hERG than the WT channel (Fig. 2, C and D). The deactivation rates of WT and mutant hERG were compared by fitting the tail currents with an exponential function. Mutating basic residues to Ala either individually or in pairs significantly (p < 0.05, n = 5–9) accelerated deactivation rates (Fig. 2D). Mutating Arg4 and Arg5 to Leu also accelerated deactivation, demonstrating that it was the positive charge rather than residue size that was functionally critical. In contrast, mutation of the acidic residue Asp16 (on the opposite side of the amphipathic helix), to Ala or Phe, had no significant effect on deactivation at −130 mV and even slowed it at −70 mV. These mutations suggest that decreasing the negative charge (and thus extending the positive electrostatic surface) of the helix further stabilizes NT1–26 interactions with its binding site, but in a voltage-dependent manner, and implies that the interactions of the helix are subtly different at the two potentials. Mutation of Phe14 on the hydrophobic face of the helix had relatively minor effects on deactivation at −70 and −130 mV.
FIGURE 2.
A positively charged electrostatic surface on the NT1–26 domain is critical for normal deactivation. A, surface views of the EAG domain of hERG with the residues colored according to their electrostatic potential; areas of significant negative charge are shown in red and significant positive charge in blue and neutral in white. The NT1–26 domain extends out to the left of the molecule and is extensively positively charged on one side. B, 180° rotation of the structure in A about the horizontal axis. Labeled residues in A and B indicate those that when mutated significantly perturb deactivation gating. The position of the Ile18 residue is indicated with an arrow. Arg20 and Lys21 residues are located adjacent and to the right of Ile18 but are obscured in this orientation of the structure. C, representative current traces illustrating differences in rates of deactivation compared with WT hERG for channels in which charged NT1–26 residues have been mutated. Prepulses to +40 mV were applied before stepping down to a range of negative potentials. For clarity, only tail currents at potentials of −50 to −130 mV in 20-mV increments are shown. D, time constants for deactivation at −70 and −130 mV from single exponential fits of tail currents from NT1–26 mutants.* indicates time constants that are significantly different from WT hERG (p < 0.05, n ≥ 5).
We then asked if breaking the amphipathic helix by substituting Pro would disrupt deactivation gating. The deactivation rate of I18P (at a central position within the helix) was extremely rapid and similar to rates in NTK hERG, in which the N terminus has been deleted (Fig. 2D). Interestingly, the D16P mutation had the reverse effect and significantly slowed deactivation. Thus, breaking the helix or inserting a kink at position 16 appears to favor stronger interactions of NT1–26 with its binding site. Together, the NMR structure along with the functional studies indicate that NT1–26 forms an independent domain that is connected but not bound to the PAS domain. The positive face and the α-helical element of the NT1–26 domain form a surface that is necessary for stabilizing the open state of the channel.
Could the cNBD Form a Complex with NT1–26 to Regulate Deactivation?
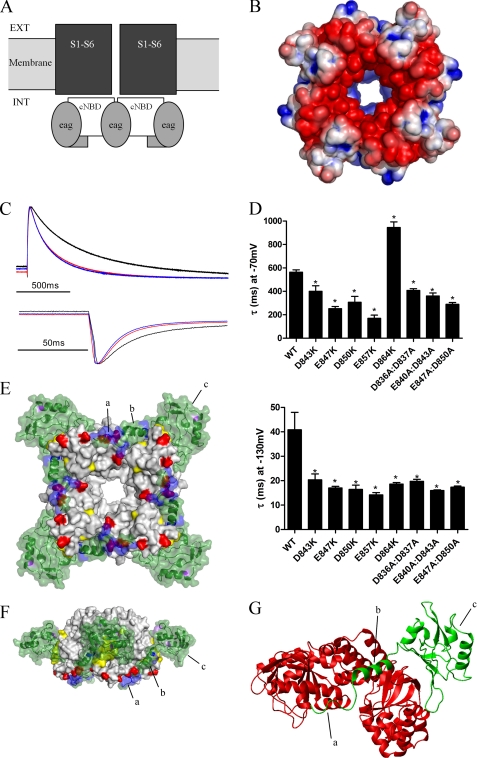
Previous studies have implicated the cNBD and S4-S5 linker as sites of interaction with the EAG domain of hERG (13, 23, 47). Given the functional importance of basic residues on the NT1–26 domain, we searched for acidic regions on the voltage sensor, pore, and cNBD and C-linker (a region connecting S6 of the pore to the cNBD) that could form charge-charge interactions with the N terminus. Fig. 3B shows the electrostatic surface of a homology model of the hERG channel cNBD and C-linker based on the crystal structure of HCN2 (25). The model shows four large acidic patches on or close to the cNBD C-helix (25), in a region symmetrically orientated around the central pore of the cNBD, and on the surface facing away from the membrane and presumed to be largely exposed to the cytoplasm. We hypothesized that these negatively charged regions could form interaction sites for the NT1–26 domains with a 1:1 stoichiometry. These acidic patches are due to a high density of Glu and Asp residues at positions 843, 847, 850, 857, and 864 in and around the cNBD C-helix. Within the central pore is an additional group of acidic residues at positions 840, 836, and 837. Acidic residues were systematically mutated to Lys, and the effects on deactivation rates were measured as described above. All the charge reversal mutants on surface positions of the cNBD significantly accelerated deactivation at −130 mV by a factor of 2.0–2.7-fold (p < 0.05, n = 5–8), with E857K and D847K having the most pronounced effects (Fig. 3, C and D). These results suggest that alterations to the electrostatic surface of the cNBD accelerate deactivation in a manner that is consistent with a charge-charge interaction with the NT1–26 domain.
FIGURE 3.
Deactivation is regulated by electrostatic interactions between the NT1–26 domain and the cNBD. A, schematic representation of structural domains of the hERG channel, illustrating the intracellular location of the cNBD (and C-linker) with respect to the transmembrane pore and voltage sensor domains (S1–S6). Ext, extracellular; Int, intracellular. B, surface view of the homology model of the cNBD of hERG with residues colored (as in Fig. 2A) according to their electrostatic potential. C, representative current traces (normalized to the peak tail current amplitude) to enable comparison of tail current time courses for WT hERG (black), E847K (red), and E857K (blue). D, time constants for deactivation for cNBD mutant currents, measured as described previously. Most mutations accelerated deactivation at both −70 and −130 mV. D864K was unusual in that deactivation at −70 mV was slowed. Asterisk indicates time constants that are significantly different from WT hERG (p < 0.05, n = 5). E, modeled complex of the EAG domain (green) and the cNBD and C-linker. Four molecules of the EAG domain interact with the tetrameric cNBD. Residues in purple and yellow are those mutated by Al-Owais et al. (23), and residues in red are acidic, and residues in blue are basic. Label a is the unstructured Met1–Pro10 region; b is the amphipathic helix (Gln11–Gly24), and c is the PAS domain. F, modeled complex rotated 90° about the horizontal axis and 45° about the central vertical axis relative to the image in E. G, expanded view of part of modeled complex showing interactions of one EAG domain (green) with two cNBDs (red). The C-linker is not shown. The NT1–26 amphipathic helix (b) sits in a cleft at the interface of two adjacent cNBDs. The unstructured Met1–Pro10 region interacts with the C-helix of one of the cNBDs.
A Modeled Complex for Domain Interaction
Using the NMR structure presented here and guided by the effects of mutations on the gating kinetics of the hERG channel, we built a model of the EAG-cNBD and C-linker interaction. The optimal docked conformation of the EAG domain to the cNBD is shown in Fig. 3, E–G. In this binding orientation, the PAS domain interacts with the outer surface of the β-roll component of the cNBD, in close contact with the residues identified by Al-Owais et al. (23) to perturb deactivation gating. The NT1–26 extends via a surface crevice toward the C-helix acidic patch, with Arg4, Arg5, and His7 able to interact with Asp843, Glu847, and Asp850.
In the apo-state crystal structures of the MlotiK1 channel, the cNBD C-helix is revealed to move in response to the absence of ligand (52, 53). This motion is in the direction of the crevice occupied by the NT1–26 helix in the docking (Fig. 3G). Thus, the binding of the deactivation domain to the cNBD may mimic the binding of cyclic nucleotides to the CNG or HCN channels by tethering NT1–10 to the C-helix and obstructing the gating motions of this helix. This would retain the channel in an active conformation, hence slowing channel deactivation. The exact mechanism for the transfer of the cNBD motions to the C-linker and channel pore remains uncertain.
Sequence alignment between hERG and the other EAG family members shows that although the acidic patch on the cNBD C-helix is well conserved in other ERG channels, it is only partially conserved in ELK channels and almost absent in EAG channels, except at the position corresponding to hERG Asp843. This may contribute to differences of deactivation gating between EAG family members despite the virtual sequence identity within the EAG domain.
The cNBD mutations did not perturb deactivation gating to the same extent as NT1–26 mutations. The reason is unclear, but it could occur because the cNBD negatively charged region is extensive, and multiple sites of interaction with NT1–26 may exist. Thus single or double mutations to the cNBD may not be sufficient to prevent binding with NT1–26, particularly as this domain is so flexible. We also cannot entirely rule out other sites of interaction. We analyzed our homology models of hERG to look for other potential interaction sites for the NT1–26 domain closer to the voltage sensor and pore, but no compelling sites were identified that were consistent with the NMR structure. Clearly, this may reflect the limitations of homology models, and further studies are required to elucidate the precise interactions.
A Mechanistic Model for hERG Channel Deactivation
We have provided evidence that the NT1–26 domain has a positively charged surface that is critical for deactivation gating. Mutation of basic residues profoundly accelerates channel closure. In addition, we have identified a negative surface on the cNBD that has a 4-fold symmetry, and when acidic residues are mutated, they result in qualitatively comparable effects on deactivation to mutation of the N terminus. Based on this, we propose a model in which the NT1–26 binds via electrostatic interactions to the cNBD to stabilize it in an open conformation. In HCN channels, the cNBD is proposed to be mechanically coupled to the activation gate on the channel pore by the C-linker domain (25). cAMP binding to the HCN cNBD results in conformational rearrangements that are transduced to the pore (25). There is good sequence and secondary structure homology between hERG and HCN channels in both the C-linker domain and cNBD (24.5% identical and 41.7% similar). Therefore, it is reasonable to expect comparable bidirectional coupling between the activation gate and cNBD in hERG. hERG channels are not gated directly by cyclic nucleotides (25); however, our results suggest the intriguing possibility that binding of the NT1–26 domain to the cNBD is transduced to the channel pore to maintain it in the open conformation. Thus, the stabilization of the open state of hERG by binding of the NT1–26 domain may be analogous to the similar effect of cyclic nucleotides on HCN and CNG channels (54).
Conclusions
Our study sheds significant new insight into the role of the EAG domain on deactivation gating. Mutations to hydrophobic patches on the surface of the EAG domain and the cNBD increase deactivation rates (16, 23). Homology modeling indicates that the EAG domain can interact with the cNBD at this hydrophobic interface without constraining the ability of the NT1–26 domain to form electrostatic interactions with the acidic patch of the cNBD (Fig. 3, E–G). The NMR data indicates that the NT1–26 domain functions as an independent functional domain and is unlikely to bind to the PAS domain. Mutations in the voltage sensor (55–57), S4-S5 linker (13, 47, 58), and C-terminal end of S6 (59–61) can also profoundly influence deactivation. It remains to be determined if the EAG domain regulates deactivation by directly, or allosterically, interacting with these other regions of the channel. Mutational analysis alone provides limited insights into protein-protein interactions, particularly if the interfaces are relatively large or involve multiple interacting partners. However, the model presented here is self-consistent and incorporates the work of others (16, 23). High resolution structures of EAG-domain complexes with other channel domains should shed further light on the mechanistic basis of deactivation gating in hERG.
Acknowledgments
We thank Prof. M. D. Carr for support, which made this project possible. We are grateful to Dr. M. Gosling for support. We also thank Prof. Mark Sansom for helpful discussions.
The atomic coordinates and structure factors (code 2L1M) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- hERG
- human ether-à-go-go-related gene
- cNBC
- cyclic nucleotide binding domain
- PAS
- Per-Arnt-Sim.
REFERENCES
- 1. Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T. (1995) Cell 81, 299–307 [DOI] [PubMed] [Google Scholar]
- 2. Trudeau M. C., Warmke J. W., Ganetzky B., Robertson G. A. (1995) Science 269, 92–95 [DOI] [PubMed] [Google Scholar]
- 3. Sanguinetti M. C., Jurkiewicz N. K. (1990) J. Gen. Physiol. 96, 195–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanguinetti M. C., Curran M. E., Spector P. S., Keating M. T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2208–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curran M. E., Splawski I., Timothy K. W., Vincent G. M., Green E. D., Keating M. T. (1995) Cell 80, 795–803 [DOI] [PubMed] [Google Scholar]
- 6. Roden D. M. (1996) Am. J. Cardiol. 78, 12–16 [DOI] [PubMed] [Google Scholar]
- 7. Spector P. S., Curran M. E., Keating M. T., Sanguinetti M. C. (1996) Circ. Res. 78, 499–503 [DOI] [PubMed] [Google Scholar]
- 8. Yang T., Snyders D. J., Roden D. M. (1995) Circulation 91, 1799–1806 [DOI] [PubMed] [Google Scholar]
- 9. Keating M. T., Sanguinetti M. C. (2001) Cell 104, 569–580 [DOI] [PubMed] [Google Scholar]
- 10. Sanguinetti M. C., Tristani-Firouzi M. (2006) Nature 440, 463–469 [DOI] [PubMed] [Google Scholar]
- 11. Wang S., Liu S., Morales M. J., Strauss H. C., Rasmusson R. L. (1997) J. Physiol. 502, 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schönherr R., Heinemann S. H. (1996) J. Physiol. 493, 635–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Trudeau M. C., Zappia A. M., Robertson G. A. (1998) J. Gen. Physiol. 112, 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spector P. S., Curran M. E., Zou A., Keating M. T., Sanguinetti M. C. (1996) J. Gen. Physiol. 107, 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith P. L., Baukrowitz T., Yellen G. (1996) Nature 379, 833–836 [DOI] [PubMed] [Google Scholar]
- 16. Morais Cabral J. H., Lee A., Cohen S. L., Chait B. T., Li M., Mackinnon R. (1998) Cell 95, 649–655 [DOI] [PubMed] [Google Scholar]
- 17. Zhou Z., Gong Q., Ye B., Fan Z., Makielski J. C., Robertson G. A., January C. T. (1998) Biophys. J. 74, 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hancox J. C., Levi A. J., Witchel H. J. (1998) Pflugers Arch. 436, 843–853 [DOI] [PubMed] [Google Scholar]
- 19. Lu Y., Mahaut-Smith M. P., Varghese A., Huang C. L., Kemp P. R., Vandenberg J. I. (2001) J. Physiol. 537, 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sale H., Wang J., O'Hara T. J., Tester D. J., Phartiyal P., He J. Q., Rudy Y., Ackerman M. J., Robertson G. A. (2008) Circ. Res. 103, e81–e95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hancox J. C., Witchel H. J., Varghese A. (1998) Biochem. Biophys. Res. Commun. 253, 719–724 [DOI] [PubMed] [Google Scholar]
- 22. Chen J., Zou A., Splawski I., Keating M. T., Sanguinetti M. C. (1999) J. Biol. Chem. 274, 10113–10118 [DOI] [PubMed] [Google Scholar]
- 23. Al-Owais M., Bracey K., Wray D. (2009) Eur. Biophys. J. 38, 569–576 [DOI] [PubMed] [Google Scholar]
- 24. Warmke J. W., Ganetzky B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., Gouaux E. (2003) Nature 425, 200–205 [DOI] [PubMed] [Google Scholar]
- 26. Brelidze T. I., Carlson A. E., Zagotta W. N. (2009) J. Biol. Chem. 284, 27989–27997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J., Myers C. D., Robertson G. A. (2000) J Gen. Physiol. 115, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gustina A. S., Trudeau M. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13082–13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muskett F. W., Mitcheson J. S. (2011) Biomol. NMR Assignments, in press [DOI] [PubMed] [Google Scholar]
- 30. Marion D., Kay L. E., Sparks S. W., Torchia D. A., Bax A. (1989) J. Am. Chem. Soc. 111, 1515–1517 [Google Scholar]
- 31. Muhandiram D. R., Farrow N. A., Xu G. Y., Smallcombe S. H., Kay L. E. (1993) J. Magn. Reson. Ser. B 102, 317–321 [Google Scholar]
- 32. Sklenar V., Piotto M., Leppik R., Saudek V. (1993) J. Magn. Reson. Ser. A 102, 241–245 [Google Scholar]
- 33. Muskett F. W., Frenkiel T. A., Feeney J., Freedman R. B., Carr M. D., Williamson R. A. (1998) J. Biol. Chem. 273, 21736–21743 [DOI] [PubMed] [Google Scholar]
- 34. Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 35. Waters L., Yue B., Veverka V., Renshaw P., Bramham J., Matsuda S., Frenkiel T., Kelly G., Muskett F., Carr M., Heery D. M. (2006) J. Biol. Chem. 281, 14787–14795 [DOI] [PubMed] [Google Scholar]
- 36. Muskett F. W., May F. E., Westley B. R., Feeney J. (2003) Biochemistry 42, 15139–15147 [DOI] [PubMed] [Google Scholar]
- 37. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsui V., Case D. A. (2000) J. Am. Chem. Soc. 122, 2489–2498 [Google Scholar]
- 40. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 41. Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 42. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 43. Chen R., Li L., Weng Z. (2003) Proteins 52, 80–87 [DOI] [PubMed] [Google Scholar]
- 44. Pierce B., Weng Z. (2007) Proteins 67, 1078–1086 [DOI] [PubMed] [Google Scholar]
- 45. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 46. Jespersen T., Grunnet M., Angelo K., Klaerke D. A., Olesen S. P. (2002) BioTechniques 32, 536–538, 540 [DOI] [PubMed] [Google Scholar]
- 47. Sanguinetti M. C., Xu Q. P. (1999) J. Physiol. 514, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perry M., de Groot M. J., Helliwell R., Leishman D., Tristani-Firouzi M., Sanguinetti M. C., Mitcheson J. (2004) Mol. Pharmacol. 66, 240–249 [DOI] [PubMed] [Google Scholar]
- 49. Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 50. Key J., Hefti M., Purcell E. B., Moffat K. (2007) Biochemistry 46, 3614–3623 [DOI] [PubMed] [Google Scholar]
- 51. McIntosh B. E., Hogenesch J. B., Bradfield C. A. (2010) Annu. Rev. Physiol. 72, 625–645 [DOI] [PubMed] [Google Scholar]
- 52. Clayton G. M., Silverman W. R., Heginbotham L., Morais-Cabral J. H. (2004) Cell 119, 615–627 [DOI] [PubMed] [Google Scholar]
- 53. Clayton G. M., Altieri S., Heginbotham L., Unger V. M., Morais-Cabral J. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Craven K. B., Zagotta W. N. (2006) Annu. Rev. Physiol. 68, 375–401 [DOI] [PubMed] [Google Scholar]
- 55. Liu J., Zhang M., Jiang M., Tseng G. N. (2003) J. Gen. Physiol. 121, 599–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang M., Liu J., Tseng G. N. (2004) J. Gen. Physiol. 124, 703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Subbiah R. N., Clarke C. E., Smith D. J., Zhao J., Campbell T. J., Vandenberg J. I. (2004) J. Physiol. 558, 417–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mitcheson J. S., Chen J., Sanguinetti M. C. (2000) J. Gen. Physiol. 115, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ficker E., Jarolimek W., Kiehn J., Baumann A., Brown A. M. (1998) Circ. Res. 82, 386–395 [DOI] [PubMed] [Google Scholar]
- 60. Wynia-Smith S. L., Gillian-Daniel A. L., Satyshur K. A., Robertson G. A. (2008) J. Gen. Physiol. 132, 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mitcheson J. S., Chen J., Lin M., Culberson C., Sanguinetti M. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12329–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]