Abstract
Spore formation in Bacillus subtilis begins with an asymmetric cell division, following which differential gene expression is established by alternative compartment-specific RNA polymerase σ factors. The spoIISAB operon of B. subtilis was identified as a locus whose mutation leads to increased activity of the first sporulation-specific sigma factor, σF. Inappropriate spoIISA expression causes lysis of vegetatively growing B. subtilis cells and Escherichia coli cells when expressed heterologously, effects that are countered by co-expression of spoIISB, identifying SpoIISA-SpoIISB as a toxin-antitoxin system. SpoIISA has three putative membrane-spanning segments and a cytoplasmic domain. Here, the crystal structure of a cytoplasmic fragment of SpoIISA (CSpoIISA) in complex with SpoIISB has been determined by selenomethionine-multiwavelength anomalous dispersion phasing to 2.5 Å spacing, revealing a CSpoIISA2·SpoIISB2 heterotetramer. CSpoIISA has a single domain α/β structure resembling a GAF domain with an extended α-helix at its N terminus. The two CSpoIISA protomers form extensive interactions through an intermolecular four-helix bundle. Each SpoIISB chain is highly extended and lacking tertiary structure. The SpoIISB chains wrap around the CSpoIISA dimer, forming extensive interactions with both CSpoIISA protomers. CD spectroscopy experiments indicate that SpoIISB is a natively disordered protein that adopts structure only in the presence of CSpoIISA, whereas surface plasmon resonance experiments revealed that the CSpoIISA·SpoIISB complex is stable with a dissociation constant in the nanomolar range. The results are interpreted in relation to sequence conservation and mutational data, and possible mechanisms of cell killing by SpoIISA are discussed.
Keywords: Bacterial Toxins, Crystal Structure, Membrane Proteins, Protein Structure, Protein-Protein Interactions, Native Disorder, Sporulation, Toxin-Antitoxin
Introduction
Toxin-antitoxin (TA)3 modules encoding a stable killer protein and a labile protein antidote were originally discovered through their association with the phenomenon of plasmid maintenance in bacteria (1, 2). In these systems, the toxin component has a longer cellular half-life than the antitoxin so that the host becomes addicted to the continuous expression of the antitoxin, and plasmid retention is driven by post-segregational cell killing. TA systems were subsequently found to be encoded on bacterial genomes where their biological function and physiological role have been the subject of widespread discussion and speculation (3–5). According to one line of thought, subpopulations of cells undergo altruistic toxin-mediated cell death for the benefit of the population as a whole. This might be a mechanism to contain the spread of phage infection or, under starvation conditions, to provide food for the survivors. A more widely accepted idea is that TA toxins induce a state of bacteriostasis rather than cell killing, enabling cells to survive temporary bouts of adversity such as starvation or to persist through exposure to antibiotics. In support of this idea is the observation that the actions of some toxins can be reversed by later expression of the antitoxin (6) and the finding that TA systems are most prevalent in organisms that are associated with slow growth and dormancy (4).
SpoIISA-SpoIISB is a chromosomally encoded TA system in Bacillus subtilis discovered as part of a study of cell differentiation in this organism. In sporulation, the cell division septum forms not at mid-cell as during vegetative growth but instead at a polar location dividing the cell into two compartments of unequal size: a larger mother cell and a smaller forespore. The two cells follow different patterns of gene expression orchestrated by cell type- and stage-specific RNA polymerase sigma factors. The mother cell goes on to engulf the forespore and nurture the latter as it matures into a resistant spore that is released upon mother cell autolysis. The spoIISAB operon was discovered following a screen for B. subtilis mutants in which sporulation is blocked after the formation of the polar septum (7).
Inactivation of spoIISB decreases the sporulation efficiency of wild type cells by 4 orders of magnitude, whereas inactivation of spoIISA in the same background has no effect on sporulation. In contrast, spoIISA inactivation in a spoIISB null mutant background fully restores sporulation, indicating that SpoIISB is required only to counteract the negative effect of SpoIISA on sporulation (7). Cells synthesizing SpoIISA in the absence of SpoIISB exhibit lethal damage of their cell envelopes shortly after asymmetric septation. It appears that the SpoIIS proteins are normally expressed in the mother cell because the defects associated with spoIISB deletion are corrected when SpoIISB is synthesized in the mother cell but not if it is synthesized in the forespore. Forced expression of SpoIISA in exponentially growing cells or in the forespore compartment during sporulation leads to similar morphological defects and cell death. Again protection is conferred by simultaneous synthesis of SpoIISB. These observations led to the conclusions that SpoIISA is a cell killer protein and that SpoIISB is its specific antidote (7).
TA systems, which have widespread distribution, have been grouped into families, with prominent examples being the CcdA-CcdB, HigB-HigA, MazE-MazF, ParD-ParE, PhD-Doc, RelB-RelE, VapB-VapC, and YoeB-YefM families (4) with new members continuing to emerge. SpoIISA-SpoIISB appears to define a new family of TA systems that is conserved among Bacilli. spoIISA encodes a 248-residue protein containing three putative transmembrane segments at its N terminus with the C-terminal two-thirds of the protein predicted to be located in the cytoplasm (7). The full-length protein is required for cell killing because neither the transmembrane domain nor the cytoplasmic domain, when expressed alone, are toxic. spoIISB encodes a basic hydrophilic protein of 56 amino acid residues, and neither SpoIISA nor SpoIISB has significant sequence similarity to proteins of known structure or function. As a step toward elucidating the biological role of the SpoIIS proteins and the mechanisms by which (i) SpoIISA mediates toxicity and (ii) this toxicity is overcome by SpoIISB, we describe here the characterization and structure of the complex between the cytoplasmic domain of SpoIISA and SpoIISB.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Culture Media, and Genetic Techniques
All bacterial strains, plasmids and PCR primers used in this study are listed in Tables 1 and 2. Unless otherwise indicated, Escherichia coli and B. subtilis cultures were grown in LB medium (8). Transformation of B. subtilis and assays of sporulation efficiency were performed as described previously (9). Sporulation was induced by nutrient exhaustion in liquid Difco sporulation medium. After 24 h of incubation at 37 °C, aliquots of the culture were serially diluted and plated on to LB plates before and after a 15-min incubation at 85 °C. The sporulation efficiency was defined in terms of colony-forming units.
TABLE 1.
PCR primers
| Primer | Sequencea | Source or reference |
|---|---|---|
| IISABamE | 5′-CTGTTTTGAAACGCAGGATCCATTATCCTTCA-3′ | This study |
| IISANcoS | 5′-GTGAGGGATTCGACCATGGTTTTATTCTTTCA-3′ | Ref. 30 |
| IISAPstE | 5′-TGTTTTGAAACGCTGCAGCCATTATCCTTCAC-3′ | This study |
| IISBamS | 5′-CAGTCAGTTTTGCTTTGATGGATCCTTTTATC-3′ | This study |
| IISBamE | 5′-CACCCTTTAAGAAAAGGATCCAGGACAAGAG-3′ | This study |
| IISBHindE | 5′-AACGAGTACCCGGAAAGCTTAAACCTTC-3′ | Ref. 30 |
| IISBNcoS | 5′-TGAAGGACCATGGAACGTGCGTTTC-3′ | Ref. 30 |
| IISBNdeS | 5′-AGGAAGGTGAAGGCATATGGAACGTGCGTT-3′ | This study |
| IISB22PstS | 5′-CGCCTGCAGCAAGAATCTTCAAAGAAAAC-3′ | This study |
| IISB31PstS | 5′-ACCACTGCAGCAAGCTATCAAGTCAGTC-3′ | This study |
| IISB40HindE | 5′-GTTCGTAAGCTTTCTAGATTCTTGCTGTATGC-3′ | This study |
| IISB44HindE | 5′-CAATCAAAGCTTAGTTTTCTTTGAAGATTCTTGC-3′ | This study |
| IISB46PstS | 5′-CCACTGCAGCGAAGCCGTTTAAAATC-3′ | This study |
| IISB48HindE | 5′-CGTTTAAGCTTCTAAATCAGCCGTTCGTTTTC-3′ | This study |
| IISB52HindE | 5′-TGCTAAGCTTCATTTATACTCGTCAATCAGCCGT-3′ | This study |
| IISB56PstS | 5′-GAAGGATAATGGCTGCAGCGTTTCAAAACAG-3′ | This study |
| PIISAA | 5′-GATGATGATGAATTCTCTCATTGTCGCAGTCA-3′ | This study |
| PIISAS | 5′-GATGATGATGAATTCTCTCATTGTCGCAGTCA-3′ | This study |
| PfrIISBA | 5′-GATGATGTAAAGCTTCTGCAAGAGTGG AACAA-3′ | Ref. 44 |
| PfrIISBS | 5′-GATGATGTACTGCAGACAAGGGAAAA CAGCTC-3′ | Ref. 44 |
| PKnA | 5′-GATGATGATGGATCCGAATGGCGAATGCGCATAC-3′ | Ref. 44 |
| PKnS | 5′-GATGATGATCTGCAGCCGCATCAGGCGATAAAC-3′ | Ref. 44 |
a Restriction endonuclease cleavage sites are underlined.
TABLE 2.
Plasmids and bacterial strains
| Plasmid | Description | Source or reference |
|---|---|---|
| pBAD24 | AmpRaraC; PBAD promoter | Ref. 51 |
| pBAD24-IISA(Pst) | AmpRaraC; PBAD promoter, spoIISA | This study |
| pET15-H-C-IISA | AmpRlacI; T7 promoter, sequence coding for His6 tag-fused 169-residue C-terminal part of SpoIIA | This study |
| pET15-IISB | AmpRlacI; T7 promoter, spoIISB | This study |
| pET26-C-IISA | KanRlacI; T7 promoter, sequence coding for non-His6-tag-fused 169-residue C-terminal part of SpoIIA | This study |
| pET-Duet-IISABB | AmpRlacI; 2xT7 promoter, sequence coding for His6 tag-fused 169-residue C-terminal part of SpoIIA, two copies of spoIISB | This study |
| pUS19 | AmpR SpcR | Ref. 52 |
| pUS19-FRIISB | AmpR SpcR; flanking regions upstream and downstream of spoIISB | This study |
| pUS19-DIISB | AmpR SpcR; flanking regions upstream and downstream of spoIISB; KanR | This study |
| pUS19-IISA(Pst) | AmpR SpcR; spoIISA | This study |
| pUS19-IISAB22 | AmpR SpcR; spoIISA spoIISB(C22) | This study |
| pUS19-IISAB31 | AmpR SpcR; spoIISA spoIISB(C31) | This study |
| pUS19-IISAB40 | AmpR SpcR; spoIISA spoIISB(N40) | This study |
| pUS19-IISAB44 | AmpR SpcR; spoIISA spoIISB(N44) | This study |
| pUS19-IISAB46 | AmpR SpcR; spoIISA spoIISB(C46) | This study |
| pUS19-IISAB48 | AmpR SpcR; spoIISA spoIISB(N48) | This study |
| pUS19-IISAB52 | AmpR SpcR; spoIISA spoIISB(N52) | This study |
| pUS19-IISAB56 | AmpR SpcR; spoIISA spoIISB(C56) | This study |
| E. coli | ||
| BL21(DE3) | F− ompT hsdS(rB−, mB−) gal dcm (DE3) | Novagen |
| B834(DE3) | F− ompT hsdS(rB−, mB−) gal dcm met (DE3) | Novagen |
| B. subtilis | ||
| PY79 | Prototrophic derivative of B. subtilis 168 | Ref. 53 |
| IB1252 | PY79 spoIISB::kan | This study |
Construction of Plasmids and Bacterial Strains
All of the PCR primers used in this study are listed in Table 1. Unless otherwise indicated, chromosomal DNA isolated from B. subtilis strain PY79 was used as a PCR template.
The plasmid pET15-H-C-IISA was constructed by inserting the 518-bp PCR fragment generated using the primers IISBamS and IISABamE. The PCR product was digested with BamHI and captured at the BamHI site of pET-15b. This plasmid encodes a His6-tagged fragment of SpoIISA (His6-CSpoIISA) encompassing the cytosolic domain. The plasmid pET26-C-IISA was prepared by cloning an 818-bp NdeI/HindIII fragment from partially cleaved pET15-H-C-IISA DNA into the corresponding site of plasmid pET-26b(+). For overproduction of SpoIISB (without a His6 tag), the 321-bp spoIISB PCR fragment generated with the primers IISBNcoS and IISBHindE was ligated at the NcoI/HindIII site of pET-15b to produce the plasmid pET15-IISB.
For co-expression, pET15-H-C-IISAB was constructed by inserting the 767-bp fragment amplified by PCR using the primers IISBamS and IISBamE, into the BamHI site of pET-15b. The cloned fragment encompasses the coding sequences for the soluble domain of spoIISA and spoIISB. Because spoIISB appeared to be underexpressed from this construct, we switched to the pETDuet system. First, the plasmid pET-Duet-IISB was prepared by inserting the 346-bp NdeI/EcoRV fragment from pET15-H-IISB into NdeI/EcoRV-digested pETDuet-1. The plasmid pET15-H-IISB was constructed by inserting the 210-bp PCR fragment (generated with primers IISBNdeS and IISBamE) into NdeI/BamHI-cut pET-15b Second, the 1127-bp NcoI/HindIII fragment from pET15-H-C-IISAB was ligated to correspondingly digested pET-Duet-IISB to generate pET-Duet-IISASB.
For constructing a B. subtilis spoIISB null mutant, the plasmid pUS19-ΔIISB was prepared as follows. A 577-bp sequence upstream and a 624-bp sequence downstream of spoIISB were PCR-amplified using the primer pairs PIISAS/PIISAA and PfrIISBS/PfrIISBA, respectively, and cloned into the EcoRI/BamHI and PstI/HindIII sites of plasmid pUS19, respectively, creating the plasmid pUS19-FRIISB. A 1541-bp fragment encoding resistance to kanamycin was PCR-amplified from the plasmid pUK19 (10) using the primers PKnS/PKnA and subsequently cloned into PstI/BamHI-cut pUS19-FRIISB to obtain pUS19-ΔIISB plasmid. Plasmid pUS19-ΔIISB was then integrated by double homologous recombination into the chromosome of B. subtilis PY79 giving rise to strain IB1252.
Integrative plasmids for assessing the ability of truncated SpoIISB proteins to cure SpoIISA toxicity were prepared in two steps. First, a 771-bp EcoRI/PstI DNA fragment encoding spoIISA was excised from plasmid pBAD24-IISA(Pst) and ligated into the corresponding sites of plasmid pUS19, resulting in plasmid pUS19-IISA(Pst). Plasmid pBAD24-IISA(Pst) was constructed by cloning a 756-bp PCR fragment encompassing spoIISA (using the primers IISANcoS and IISAPstE) into NcoI/PstI-cut pBAD24. In the next step, eight PCR fragments generated from the spoIISB gene using either the sense primers IISB22PstS, IISB31PstS, IISB46PstS, and IISB56PstS and the antisense primer IISBHindE or the sense primer IISB56PstS and the antisense primers IISB40HindE, IISB44HindE, IISB48HindE, and IISB52HindE were introduced into the PstI/HindIII sites of plasmid pUS19-IISA(Pst), giving rise to the plasmids pUS19-IISAB22, pUS19-IISAB31, pUS19-IISB46, pUS19-IISAB56, pUS19-IISB40, pUS19-IISB44, pUS19-IISB48, and pUS19-SB52. These pUS19 derivatives were integrated into the chromosome of the spoIISB null mutant by single homologous recombination.
Expression and Protein Purification
For production of the His6-CSpoIISA, CSpoIISA, and the His6-CSpoIISA2·SpoIISB2 complex, respectively, overnight cultures of E. coli BL21(DE3) harboring appropriate plasmids were diluted into fresh LB medium and incubated in 500-ml flasks with shaking at 37 °C until the culture reached an A600 of 0.6, when isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1 mm. The cells were shaken for a further 3 h prior to harvesting by centrifugation. For production of the selenomethionine-substituted protein complex, E. coli B834(DE3) cells harboring pETDuet-H-C-IISASB were grown in M9 minimal medium containing selenomethionine as described previously (11).
Prior to the isolation of proteins, the cells pellets were resuspended in appropriate solubilization buffers (the same buffers, which were used for equilibration of chromatography columns in the first purification step), and cells were lysed by sonication and clarified by centrifugation. After purification, protein fractions were analyzed by SDS-PAGE followed by Coomassie Blue staining, and their purity was judged by visual inspection.
For His6-CSpoIISA purification, the clarified supernatant was loaded onto a HisTrap HP Column (GE Healthcare) equilibrated with buffer containing 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, and 50 mm imidazole, and the proteins were eluted with a 50 to 500 mm imidazole gradient in the same buffer. Further fractionation on a HiPrep 16/60 Sephacryl S-100 HR (GE Healthcare) gel filtration column did not improve the sample purity. The sample was dialyzed against 20 mm Tris-Cl, pH 8.0, 50 mm NaCl and concentrated to ∼1.5 mg/ml. The protein was 95% pure with the overall yield ∼40 mg from 0.25 liters of culture.
For purification of CSpoIISA (His6 tag-free), the supernatant was loaded on HiPrep 16/10 Q XL column (GE Healthcare) equilibrated with buffer containing 20 mm Bis-Tris-HCl, pH 6.0, 50 mm NaCl, and after washing, the proteins were eluted with a 50–1000 mm NaCl gradient in 20 mm Bis-Tris-HCl, pH 6.0. The pooled CSpoIISA containing fractions were dialyzed against 20 mm Tris-HCl, pH 8.5, 150 mm NaCl and concentrated to ∼20 mg/ml and loaded onto a HiPrep 16/60 Sephacryl S-100 HR column (GE Healthcare) pre-equilibrated in the same buffer. The resulting protein sample was found to be ∼95% pure and was dialyzed against 20 mm phosphate buffer, pH 8.0, 50 mm NaCl. The final protein concentration was ∼1.5 mg/ml, and the overall yield was ∼20 mg from 0.25 liters of culture.
The clarified cell lysate-containing SpoIISB protein was loaded on HiPrep 16/10 SP XL column (GE Healthcare) equilibrated with 20 mm Hepes, pH 8.0, 50 mm NaCl, and after washing the proteins were eluted with a 50 to 1000 mm NaCl gradient in 20 mm Hepes, pH 8.0. The pooled SpoIISB-containing fractions were dialyzed against 20 mm Tris-HCl, pH 8.5, 150 mm NaCl and concentrated to ∼5 mg/ml and loaded onto a HiPrep 16/60 Sephacryl S-100 HR column (GE Healthcare) equilibrated in the same buffer. The resulting SpoIISB sample was judged to be at least 95% pure and was either supplemented with Tween 20 to final concentration 0.005% or dialyzed against 20 mm potassium buffer, pH 7.8. The overall yield was ∼20 mg from 0.5 liters of culture.
Supernatant containing the His6·CSpoIISA·SpoIISB complex was loaded on to a HisTrap HP Column (GE Healthcare) equilibrated with buffer containing 20 mm Tris-Cl, pH 9.0, 50 mm NaCl, pH 9.0, and 40 mm imidazole. After an extensive wash, the proteins were eluted with a 40 to 500 mm imidazole (pH adjusted to 9.0 to avoid complex precipitation) gradient in the same buffer. CSpoIISA·SpoIISB-containing fractions were pooled and passed through an HiPrep 16/60 Sephacryl S-100 HR (GE Healthcare) gel filtration column equilibrated in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, although this step did not noticeably improve the purity. The sample was dialyzed against 20 mm Tris-HCl, pH 8.0, 50 mm NaCl and concentrated to ∼20 mg/ml. The yield of complex was ∼40 mg from 0.25 liter of culture. The selenomethionine-substituted His6-C-SpoIISA·SpoIISB complex was purified using the same procedures as for the native complex yielding ∼40 mg/ml/0.5 liters of cells culture.
Circular Dichroism Spectroscopy
Circular dichroism spectra were recorded at 20 °C on a Jasco J-810 CD spectrophotometer using a 0.1-cm-path length quartz cell. The experiments were carried out in 20 mm potassium phosphate buffer, pH 7.8. The protein concentration in the samples varied from 0.1 mg/ml (for His6-C-SpoIISA) to 0.23 mg/ml (for His6-C-SpoIISA·SpoIISB at a molar ratio of 1:4). Random error and noise were reduced for each spectrum by averaging of three scans in the wavelength range 260–190 nm. The signal acquired for the buffer used for dilution of the proteins was subtracted from the spectra acquired for the proteins.
Surface Plasmon Resonance
A BIACORE 3000 instrument (GE Healthcare) was used for the surface plasmon resonance experiments which were carried out with a CM5 sensor chip. Amine-coupling reagents (N-ethyl-N′-(dimethylamino-propyl) carbodiimide, N-hydroxysuccinimide, ethanolamine pH 8.5), P20 detergent, and 10 mm sodium acetate, pH 4.0, were obtained from Biacore AB, other chemicals were from Sigma-Aldrich. As we observed oligomerization of a His6-tagged form of C-SpoIISA, for the SPR experiments, we used protein without the His6 tag. As judged by its mobility on size exclusion chromatography, SpoIISA exists as a dimer in solution (data not shown), and this dimeric preparation of SpoIISA was used for immobilization on the SPR chip. A C-SpoIISA stock solution in 20 mm phosphate buffer, pH 8.0, 50 mm NaCl was diluted to 15 μg/ml with sodium acetate buffer, pH 4.0. Subsequently, 850 response units of C-SpoIISA were immobilized via primary amines by 5-min injection of diluted protein over the activated analytical flow cell surface, and any remaining free active sites were blocked by ethanolamine. A reference flow cell was activated and blocked without protein coupling. Kinetic experiments were performed at 25 °C in phosphate-buffered saline, pH 7.4, with 0.005% P20 (PBS-P). Duplicates of SpoIISB (diluted from a stock solution containing 0.5 mg/ml protein in 20 mm Tris-HCl, pH 8.5, 150 mm NaCl, 0.005% Tween 20) at concentrations of 4.7, 18.8, and 75 nm together with a PBS-P control were injected at a flow rate of 100 μl/min over the sensor chip, with 120 and 150 s of association and dissociation time, respectively. Regeneration of the surface was accomplished by injection of 20 mm NaOH. Kinetic binding curves were double referenced (12) and fitted using the BIA evaluation software 4.1 (Biacore AB) to the 1:1 reaction model with correction for mass transport effects. Rate constants were approximated globally, maximal responses and mass transport were fitted locally, and the bulk response was set to zero.
Crystallization and Structure Determination
Crystals of the native and selenomethionine-substituted CSpoIISA·SpoIISB complex were obtained in hanging drops at 20 °C by mixing equal volumes of protein solution (20 mg/ml) with precipitant solution containing 0.4 m MES, 0.2 m malic acid, 0.43 m Tris-HCl, pH 9.0, and 21% polyethylene glycol 1500. The crystals belonged to space group P212121 with unit cell parameters a = 54.0 Å, b = 59.5 Å, c = 112.4 Å, and α = β = γ = 90° (Table 3). The crystals contain two heterodimers in the asymmetric unit, and the calculated Matthews co-efficient is 2.55 Å3 Da−1, which corresponds to a solvent content of 52%. The crystals were soaked in a solution of mother liquor containing 25% (v/v) glycerol for 10 s and then flash-cooled at 120 K in a stream of nitrogen gas.
TABLE 3.
Data collection and refinement statistics
| Native | Selenomethionine |
|||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Data collection | ||||
| X-ray source | Coppery rotating anode | European Synchrotron Radiation Facility Beamline ID29 | European Synchrotron Radiation FacilityBeamline ID29 | European Synchrotron Radiation Facility Beamline ID29 |
| Wavelength (Å) | 1.54189 | 0.9789 | 0.9790 | 0.9763 |
| Collection temperature (K) | 120 | 120 | 120 | 120 |
| Resolution range (Å) | 50.00–2.50 | 50.00–2.50 | 50.00–2.50 | 50.00–2.50 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell parameters (Å) | ||||
| a, b, c | 54.12, 61.15, 112.84 | 54.03, 59.47, 112.35 | 54.03, 59.47, 112.35 | 54.03, 59.47, 112.35 |
| a = b = g | 90° | 90° | 90° | 90° |
| Number of unique reflections, overall/outer shella | 11,829/470 | 12,777/524 | 12,353/358 | 12,529/403 |
| Completeness (%), overall/outer shella | 84.6/34.1 | 96.9/82.8 | 92.0/55.7 | 94.0/64.2 |
| Redundancy, overall/outer shella | 6.2/2.3 | 11.9/8.5 | 5.7/3.4 | 5.6/4.4 |
| I/s(I), overall/outer shella | 16.8/1.3 | 21.6/2.3 | 22.6/1.2 | 22.4/1.6 |
| Rmergeb (%), overall/outer shella | 10.3/50.8 | 11.8/53.4 | 11.2 /65.8 | 11.1 /58.3 |
| Refinement and model statistics | ||||
| Resolution range (Å) | 48.69–2.5 | |||
| R factorc (Rfreed) | 0.196 (0.258) | |||
| Reflections (working/free) | 12,265/650 | |||
| Outer shell R factorc (Rfreed) | 0.258 (0.305) | |||
| Outer shell reflections (working/free)e | 825/55 | |||
| Molecules/asymmetric unit | 4 | |||
| Number of protein non-hydrogen atoms | 3,118 | |||
| Number of water molecules | 18 | |||
| Root mean square deviation from targetf | ||||
| Bond lengths (Å) | 0.015 | |||
| Bond angles (°) | 1.537 | |||
| Average B factor (Å2) | 62.1 | |||
| Ramachandran plotg | 92.3/7.1/0.0/0.6 (Glu207A,B) | |||
a The outer shell corresponds to 2.59–2.50 Å for native and 2.54–2.50 Å for peak, inflection, and remote.
b Rmerge = ΣhklΣi|Ii − <I>|/ΣhklΣi <I>, where Ii is the intensity of the ith measurement of a reflection with indexes hkl, and <I> is the statistically weighted average reflection intensity.
c R factor = Σ||Fo| − |Fo||/Σ|Fo| where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
d Rfree is the R factor calculated with 5% of the reflections chosen at random and omitted from refinement.
e Outer shell for refinement corresponds to 2.565–2.500 Å.
f Root mean square deviation of bond lengths and bond angles from ideal geometry.
g Percentage of residues in most favored/additionally allowed/generously allowed/disallowed regions of the Ramachandran plot, according to PROCHECK. The residues in parentheses are Ramachandran outliers.
X-ray diffraction data from a native crystal were collected in-house on an image plate detector equipped with a MAR Research 345 scanner to a resolution of 2.6 Å using a Rigaku RUH3R x-ray generator (λCuKα = 1.5418 Å). Selenomethionine-derivative data sets were collected on Beamline ID29 at the European Synchrotron Radiation Facility (Grenoble, France). Three wavelength (0.9789, 0.9790, and 0.9763 Å) data were used for the multiwavelength anomalous dispersion experiment. Data processing and reduction were carried out using HKL2000 and SCALEPACK (13). The data collection statistics are summarized in Table 3. The determination of the positions of the eight anomalous scatterers was performed by solving the Patterson function at 2.6 Å resolution with the program SHELXD (14). Heavy atom parameters were refined, and multiwavelength anomalous dispersion phases with a mean figure of merit of 0.63 were calculated up to 2.25-Å resolution using the program MLPHARE (15). Calculated electron density maps allowed the building of an initial polyalanine model containing six helices and six β-strands and comprising 39% of the final model. Cycles of refinement with REFMAC (16) using input phases after solvent flattening, histogram matching and non-crystallographic symmetry averaging, eventually including model building using the program COOT (17), gave a final R factor of 19.9% (Rfree = 25.8%) at 2.5 Å resolution. TLS parameters were refined during the last steps of refinement (18). The refinement statistics are summarized in Table 3. The coordinates and structure factors have been deposited in the RCSB Protein Data Bank with the RCSB code rcsb060734 and Protein Data Bank code 3O6Q.
RESULTS
The Crystal Structure of CSpoIISA·SpoIISB
The coding sequence of the cytoplasmic domain of SpoIISA encompassing residues 80–248 fused to a sequence encoding a 21-residue N-terminal hexahistidine-containing peptide was co-overexpressed with the coding sequence of spoIISB from a pET-Duet-1 vector in E. coli BL21(DE3). The native CSpoIISA·SpoIISB complex and a complex of the selenomethionine-substituted proteins were purified by nickel chelation and gel filtration chromatography in high yields, ∼80 mg/liter of shaking culture. The crystals belonging to the orthorhombic space group P212121 were grown in hanging drops as described under “Experimental Procedures.”
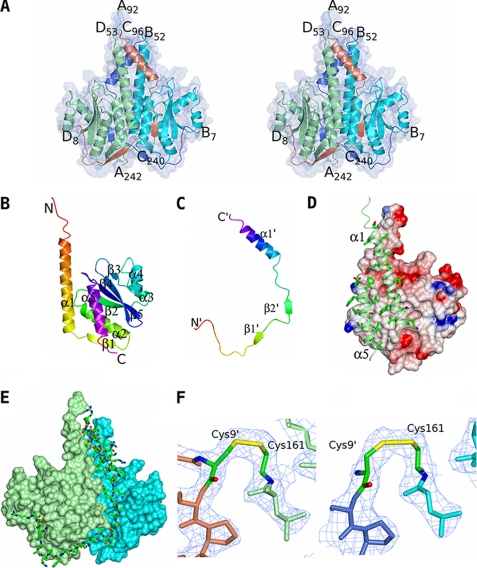
The crystal structure of the CSpoIISA·SpoIISB complex was solved by multiwavelength anomalous dispersion phasing of data collected from a selenomethionine-derivative crystal. The model has been refined against data extending to 2.5 Å spacing to give a crystallographic R factor of 19.6% (Rfree = 25.8%) with satisfactory geometry (Table 3). There are four molecules in the asymmetric unit: two CSpoIISA chains and two SpoIISB chains arranged in an A2B2 heterotetramer (Fig. 1A). Chains A and C comprise residues Gly92–Ile242 and Glu96–Leu240, respectively, of SpoIISA, and chains B and D comprise residues Asn7–Lys52 and Arg8–Arg53, respectively, of SpoIISB. Analysis of dissolved crystals by electrophoretic and mass spectrometric methods suggested that 37 residues at the N terminus of SpoIISA (the histidine tag, a linker, and residues 80–91 of the protein) had been cleaved from the chain during crystallization. We attribute the loss of these residues to the action of a contaminating protease over the extended period of time between purification of the protein and the appearance of crystals in the crystallization drops. It is assumed that the remaining missing residues at the N and C termini of the protein chains are disordered in the crystal.
FIGURE 1.
The crystal structure of CSpoIISA·SpoIISB. A, the structure of the toxin-antitoxin heterotetramer is shown in ribbon representation and colored by chain with a transparent molecular surface overlaid. The CSpoIISA chains are in light green and cyan, and the SpoIISB chains are colored light blue and coral. The N and C termini of the chains are labeled. B and C, ribbon tracings of the CSpoIISA and SpoIISB chains respectively colored ramped from their N termini (red) to their C termini (magenta). The secondary structure elements and the N and C termini are labeled with those of SpoIISB distinguished by apostrophes. D, the CSpoIISA dimer interface. CSpoIISA chain C is shown as an electrostatic surface with regions of positive and negative charge colored blue and red, respectively, and with apolar surfaces in white/gray. For clarity the CSpoIISA chain A has been truncated so that only the dimer-forming helices α1 and α5 are shown. These are shown as light green ribbons with side chains proximal to the dimer interface shown. The abundance of apolar interactions across the dimer interface is apparent. E, the interactions between SpoIISB and the CSpoIISA dimer. The CSpoIISA chains A and C are shown as surface representations and colored in light green and cyan, respectively. SpoIISB chain B is shown in cylinder representation and colored by atom type (carbon, green; nitrogen, blue; oxygen, red; sulfur, yellow). Its extensive contacts with both CSpoIISA chains are apparent. F, electron density in the vicinity of the disulfide bonds between Cys161 of CSpoIISA and Cys9′ of SpoIISB in the AD (left) and BC heterodimers (right). The chains are colored as in A, with the cysteines colored by atom type. The 2Fobs − Fcalc map is contoured at the 1σ level. These images and those in Fig. 3 were produced using the molecular graphics program CCP4MG (48).
The cytosolic domain of the killer protein consists of five α-helices and five segments of β-strand. A short segment of coil (residues 92–98) at the N terminus of the fragment precedes a seven-turn α-helix (α1) that extends the length of the molecule (Fig. 1B). There follows a β-strand (β1) and a second α-helix (α2) leading into the first strand of a four-stranded anti-parallel β-sheet (β2-β5-β4-β3), which is the central feature of the structure. β2 and β3 are separated by a pair of short α-helices, α3 and α4, that pack onto one face of the β-sheet with helices α2 and α5 packing onto the opposite face. The three anti-parallel β-strands β3, β4, and β5 are connected by short turns leading into the C-terminal α-helix α5, which runs anti-parallel to the first α-helix. The 145 equivalent Cα atoms of chains A and C can be superposed to give a root mean square deviation of 0.8 Å with the largest structural differences occurring at residues 166–174 and encompassing helix α3.
In contrast to the compact globular form of CSpoIISA, the antidote protein SpoIISB has a highly extended conformation (Fig. 1C). Residues 8–19, which are random coil, are followed by two segments that adopt β-strand conformation β1′ (residues 19–22), β2′ (residues 26–29), and an α-helix at the C terminus α1′ (residues 37–52). SpoIISB has no tertiary structure. The 45 equivalent Cα atoms of the SpoIISB chains B and D can be superposed to give a root mean square deviation of 0.8 Å with the largest deviations occurring at residues 14–18.
Quaternary Interactions in the CSpoIISA·SpoIISB Complex
The two SpoIISA subunits in the asymmetric unit associate in a dimer with the interaction interface constituted by a four-helix bundle formed by the coming together of the anti-parallel helical pair of α1 and α5 from each chain to form an ∼2-fold symmetric interface (Fig. 1D). In their interface, each subunit buries 1100 Å2 of what would otherwise be accessible surface area constituting some 12% of the molecular surface of each subunit and typical of dimer interfaces in proteins (19). Because the interaction surface is a helical bundle, the intermolecular interactions are formed by side chains from each subunit featuring just three polar interactions: reciprocal hydrogen bonds between Gln121 and Thr230 and a further hydrogen bond between the pair of Tyr120 residues. In protein-protein interfaces, there is typically one polar interaction/200 Å2 of buried surface area (19), so the interface in the SpoIISA dimer is unusually hydrophobic. There are substantial contributions to the hydrophobic interface made by residues Leu103, Asn106, Leu110, Leu114, Leu117, Gln121, and Leu124 from α1 and Phe224, Leu227, Ser231, Ser234, Ile235, Leu238, and Val239 from α5.
The interactions of SpoIISB and SpoIISA are extensive and span the full length of the SpoIISB chain (Fig. 1, A and E). Moreover, each SpoIISB chain makes an abundance of interactions with both subunits of the SpoIISA dimer. Thus, SpoIISB chain B buries 1250 Å2 or 22% of its accessible surface area through its interaction with SpoIISA chain A and a further 1450 Å2 or 26% of its accessible surface area in its interaction with SpoIISA chain C. The two segments of the SpoIISB chains that have β-strand conformations combine with similar segments in the SpoIISA chains (Fig. 1A). The first strand, β1′, forms a two-stranded intermolecular anti-parallel β-sheet with strand β1 of the first SpoIISA subunit. The second strand, β2′, pairs with the N-terminal portion of strand β4 of SpoIISA to lend the existing β-sheet in the second SpoIISA subunit an intermolecular character. So extensive are the interactions of SpoIISB with SpoIISA that only a handful of the 46 residues of SpoIISB observed in the structure do not form intermolecular interactions. Notably, the interactions include intermolecular disulfide bridges between Cys9′ of SpoIISB and Cys161 of SpoIISA (Fig. 1F). These disulfide bonds are clearly defined in the electron density maps, and their formation under the oxidizing environment of the crystallization experiments would be favored. The presence of these disulfide bonds suggests the possibility of redox control of toxicity. However, neither Cys residue is conserved in orthologous proteins; moreover, the significance of the disulfide bonds in the reducing intracellular environment is uncertain. The two SpoIISB subunits bind on opposite faces of the SpoIISA dimer, and there are negligible interactions between the two chains.
SpoIISB Is Unfolded in the Absence of Cytosolic Domain of SpoIISA
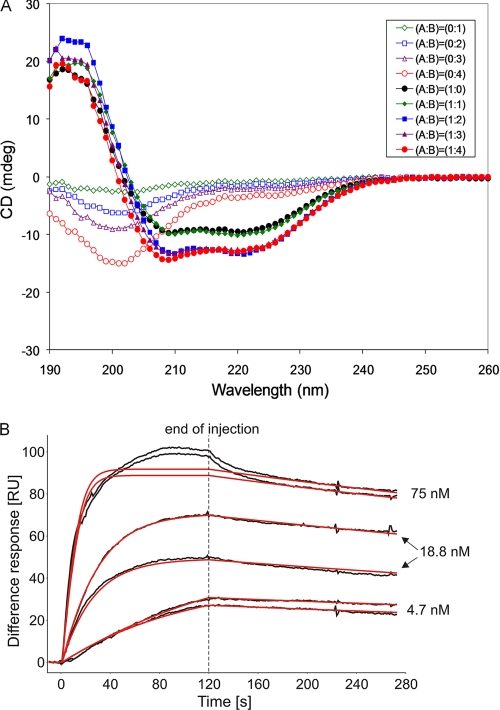
Circular dichroism spectroscopy was used to probe the structures of the SpoIIS proteins prior to complex formation. As shown in Fig. 2A, hexahistidine-tagged CSpoIISA (His6-CSpoIISA) exhibited strong features characteristic of a folded protein, a maximum in its CD spectrum at 190–200 nm and minima over the wavelength range 205–225 nm indicating the presence of secondary structure elements. By contrast the far-UV CD spectrum of SpoIISB has less pronounced features with a shallow minimum at 200 nm. This spectrum is barely distinct from the CD spectrum of a control sample containing buffer alone. We deduce from this spectrum that SpoIISB is natively disordered.
FIGURE 2.
Structural and functional assays. A, CD spectra of His6-C-SpoIISA and SpoIISB and their combination in various molar ratios. SpoIISA and SpoIISB are present at ratios of: 1:0 (filled black circles); 1:1 (filled green diamonds); 1:2 (filled blue squares); 1:3 (filled purple triangles); and 1:4 (filled red circles). The corresponding unfilled symbols represent the same concentrations of SpoIISB in the absence of His6-CSpoIISA. B, SPR analysis of binding of SpoIISB to the cytosolic domain of SpoIISA. Duplicate samples of SpoIISB at the concentrations indicated were injected over SpoIISA dimers immobilized on the sensor chip. The black lines are experimental binding sensorgrams, and the red lines are calculated binding curves fitted to a 1:1 model of interaction, assuming two independent binding sites on each SpoIISA dimer. The low overall chi square of fitting (1.8) indicated the appropriateness of the chosen interaction model. To further validate the approach, we simulated the SPR response for two nonequivalent binding sites on SpoIISA. The simulated curves largely diverged from the experimental data (not shown). Because of the incomplete regeneration of the CSpoIISA surface after the first injection of 18.8 nm SpoIISB, the second 18.8 nm SpoIISB replicate gives a lower response. Consequently, the maximal binding response parameter Rmax of SpoIISB was fitted locally during subsequent calculations of binding constants to take into account incomplete regeneration. Nonspecific binding occurred later in the injections of the highest SpoIISB concentration. The data from this region were not taken into account in the fitting.
To monitor the structural changes accompanying binding of the two proteins, we recorded a series of CD spectra in an experiment in which a sample of His6-CSpoIISA was titrated with aliquots of SpoIISB. As shown in Fig. 2A, the resultant spectra are not simply the sum of the spectra of the two proteins. The addition of SpoIISB leads to an increase in the molar ellipticity of the sample at 190–200 nm and a decrease in the molar ellipticity over the wavelength range 205–225 nm. This indicates that the addition of SpoIISB to His6-CSpoIISA leads to an increase in the amount of protein that has secondary structure. Because the CD spectrum of SpoIISB alone indicates that this protein is unstructured, we conclude that CSpoIISA induces structure in SpoIISB as the two proteins form a complex. These observations are consistent with the crystal structure, which shows that SpoIISB exhibits secondary structure in the complex that is dependent on extensive packing against SpoIISA. In the absence of the latter, SpoIISB could not adopt this structure, and we would expect the protein to be disordered.
SpoIISB Binds to CSpoIISA with High Affinity
Toxin binding by the anti-toxin was measured in surface plasmon resonance (SPR) experiments in which CSpoIISA was immobilized by covalent attachment to a CM5 sensor chip. The SPR signal accompanying the flow of SpoIISB over the chip is shown in Fig. 2B, from which it is evident that there is a relatively rapid rise in the response units as SpoIISB is introduced and a slow decrease in the response units when the flow of SpoIISB is stopped. Fitting of sensorgrams to a 1:1 Langmuir binding model (Fig. 2B) gives an association rate constant kon of 2.0 × 106 m−1 s−1, a dissociation rate constant koff of 9.1 × 10−4 s−1, and an equilibrium dissociation constant KD of 4.6 × 10−10 m. The subnanomolar affinity and binding in an extended conformation is reminiscent of the inhibition of the protease calpain by disordered calpastatin (20, 21) or of the inhibition of type 1 protein phosphatase by intrinsically disordered inhibitor-2 (22). High affinity binding of CSpoIISA to SpoIISB is largely conferred by the low rate of complex dissociation, reflected in the slow release of bound SpoIISB from the sensor chip after the end of the injection (Fig. 2B). This could have biological implications in effective blocking of SpoIISA by SpoIISB in vivo under conditions of limited renewal of the available SpoIISB pool in the cell.
CSpoIISA Has a GAF Domain Fold
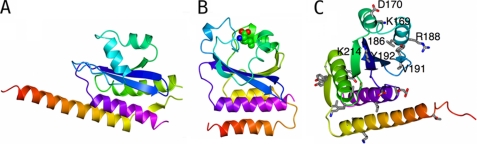
A search of the Protein Data Bank for entries with three-dimensional structural similarity to CSpoIISA using the program DALI (23) produced as the highest scoring hits a set of GAF domain-containing proteins. GAF (cGMP phospho-diesterase, adenylate cyclase, FhlA) domains are widespread in proteins of diverse function including cell signaling proteins, transcription factors, and proteins involved in light detection in bacteria, fungi, and plants (24). The GAF domain has three tiers: a basal layer of two or more α-helices, a middle tier of four or more strands that form a mixed β-pleated sheet, and a distal layer of more variable structure made up of segments connecting the strands of the β-sheet. Many GAF domains are dimers formed by the packing of α-helices from the basal layer to form a four helix bundle, a feature that is also reproduced in SpoIISA.
Among the highest scoring DALI matches is YebR (Protein Data Bank code 1vhm) (25), which gives a Z-score of 6.8 with 116 residues matching with a root mean square deviation of 3.7 Å and a sequence identity of 12%. YebR was later shown to have methionine-sulfoxide reductase activity (26). Another closely similar structure is that of the co-factor-binding domain of CodY (Protein Data Bank code 2gx5; Z-score = 5.2) (27), a GTP and branched chain amino acid responsive transcriptional regulator of stationary phase and virulence genes in low G/C Gram-positive bacteria (28). The similarity between SpoIISA and YebR and CodY extends over the three-helix bundle and four of the five strands of the β-sheet. For CodY, 106 of the 155 Cα atoms of the GAF domain superimpose with a root mean square deviation of 4.6 Å, and the similarities in the structures of SpoIISA and CodY are apparent from Fig. 3. The structurally equivalent residues have only 8% sequence identity.
FIGURE 3.
The GAF domain structure of CSpoIISA. A, CSpoIISA is shown as a ribbon color-ramped from residue 92 (red) to residue 242 (magenta). B, ribbon tracing of the co-factor binding domain of CodY (colored form residue 1 (red) to residue 155 (magenta)), which is similarly oriented and with its isoleucine ligand shown in space-filling representation and colored by atom. The similarities in the core elements of these structures are apparent. C, ribbon tracing of CSpoIISA colored as in A and with the side chains of invariant residues in the alignment shown in Fig. 4B displayed in cylinder format and colored by atom type (carbon, gray; nitrogen, blue; oxygen, red; sulfur, yellow). Invariant residues around the putative effector binding pocket are labeled.
In the GAF domains of cGMP phosphodiesterases and in that of CodY, the cyclic nucleotides and branched chain amino acid ligands respectively are bound in a pocket whose base is formed by the β-sheet and whose sides are formed by the loops connecting the strands of this sheet (Fig. 3B). In the case of CodY, where both unliganded and liganded structures have been determined (27, 29), branched chain amino acid binding is accompanied by dramatic refolding of these loops around the ligand. In SpoIISA, the segments connecting the strands of the sheet are short with the exception of β2-β3, which contains two α-helices that fold over the β-sheet. It is conceivable that these elements could bind a small molecule effector, and in this regard it may be significant that the highest atomic temperature factors are observed in helices 3 and 4, implying the mobility of these elements.
Deletion Mutagenesis of SpoIISB
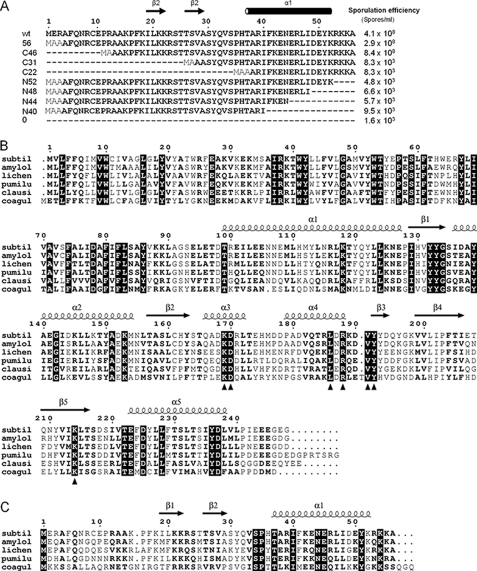
The early genetic analysis of spoIIS indicated that the C terminus of SpoIISB is important for SpoIISA neutralization. Deletion of the C-terminal four residues of SpoIISB and their replacement with an unrelated stretch of 23 residues led to a large drop in the sporulation frequency (7). To extend this study, we examined the capacity of a set of 5′- and 3′-deletants of spoIISB, encoding N- and C-terminally truncated proteins, respectively, to rescue SpoIISA toxicity as judged by sporulation efficiency measurements in B. subtilis (Fig. 4A). Cells expressing spoIISA and 5′ or 3′ deletants of spoIISB from their native promoters were assessed for their capacity to sporulate.
FIGURE 4.
Deletion mutagenesis of spoIISB and sequence alignment of SpoIISA and SpoIISB from B. subtilis and its relatives. A, deletion mutagenesis of SpoIISB. The sequences of SpoIISB and the deletion mutants with the numbers of residues in each protein are given on the left, with N and C denoting N- and C-terminal fragments, respectively. Residues additional to those in native SpoIISB are in the lighter font. The secondary structure elements of SpoIISB in the crystal structure of the TA complex are indicated above the sequences. Sporulation efficiencies of B. subtilis strains expressing spoIISA and the 5′ (upper) and 3′ (lower) deletants of spoIISB from their native promoters are shown at the right. The data presented are the means of three independent experiments. B and C, the sequences of six close SpoIISA homologues (corresponding NCBI accession numbers are NP_389166, YP_001420862, YP_078550, YP_001486404, YP_175438, and ZP_04433244) (B) and five close SpoIISB homologues (NCI accession numbers NP_389165, YP_001420861, YP_078549, YP_001486403, and ZP_04433245) (C), respectively, were aligned in the program CLUSTAL-W (49), and the secondary structural elements from chains A and D of the CSpoIISA2-SpoIISB2 structure were overlaid using ESPript (50). In both alignments, identical residues are depicted white on a black background, and conserved residues are in bold. subtil, sequences from B. subtilis subsp. subtilis str. 168; amylol, proteins from Bacillus amyloliquefaciens FZB42; lichen, proteins from Bacillus licheniformis ATCC 14580; pumilu, proteins from Bacillus pumilus SAFR-032; clausii, SpoIISA homologue from Bacillus clausii KSM-K16; coagul, protein sequences from Bacillus coagulans 36D1. The filled triangles below the SpoIISA alignment denote the invariant residues around the putative effector binding site highlighted in Fig. 3C.
As shown previously (7), sporulation of cells expressing spoIISA alone decreased to ∼1.6 × 103 spores/ml in comparison with wild type cells, which produced ∼4.1 × 108 spores/ml (Fig. 4A). Similar decreases in the sporulation efficiency were observed when spoIISA was co-expressed with spoIISB deletants encoding proteins with 27 or 36 N-terminal residues replaced by Met-Ala-Ala. The sporulation efficiencies of the corresponding strains were 8.3 × 103 and 8.3 × 103 spores/ml, respectively (Fig. 4A). However, when spoIISA was co-expressed with spoIISB deletants encoding proteins in which 3 or 12 N-terminal residues were replaced by Met-Ala and Met-Ala-Ala, respectively, the sporulation efficiencies of the strains were similar to the wild type strain (2.9 × 108 and 8.4 × 108 spores/ml; Fig. 4A). These results suggest that the N-terminal 12 residues are dispensable for the antitoxin activity of SpoIISB in vivo. Because the missing residues include Cys9′, the disulfide bridge is probably not critical for the antidote function. The sporulation efficiencies of strains encoding SpoIISB proteins with C-terminal deletions of 16, 12, 8, and 4 residues were 9.5 × 103, 5.7 × 103, 6.6 × 103, and 4.8 × 103 spores/ml, respectively. These are similar to the sporulation efficiency of cells in which the entire spoIISB gene has been deleted. Thus, C-terminal deletion of four or more residues leads to a loss of the capacity of SpoIISB to overcome SpoIISA toxicity. It seems probable therefore that the sporulation defect associated with the mut14 allele is caused by the four missing C-terminal residues rather than the additional 23 residues.
DISCUSSION
Sequence Conservation and Mutational Considerations
The sequences of SpoIISA and SpoIISB orthologues from Bacilli were identified and aligned so that conserved residues could be mapped onto the structure. The sequences of the SpoIISA orthologues from the Bacillus cereus group were excluded because they are surprisingly divergent from those of the B. subtilis group such that there are only four invariant residues (Ile37, Arg38, Tyr42, and Asp78) between the groups, all of which map to the membrane-spanning region (30). Orthologous SpoIISB sequences are even more divergent, and proximity to spoIISA on the chromosome was used to guide the identification of the appropriate coding sequences for the B. cereus family proteins. In view of the divergence, it was necessary to demonstrate functionally that the identified SpoIISA and SpoIISB orthologues from B. cereus and Bacillus anthracis do indeed act as toxin and antidote, respectively, following heterologous expression in E. coli (30).
As is apparent from Fig. 4B, there is lower sequence conservation in the cytosolic GAF domain of SpoIISA than in the putative membrane-spanning region. Among the conserved residues of the latter is Arg38, whose mutation to Gln is associated with loss of toxin function in vivo (7). Of the 24 invariant residues in the CSpoIISA sequences aligned in Fig. 4B, seven (Lys169, Asp170, Leu186, Arg188, Val191, Tyr192, and Lys214) are associated with the interface between the α3-α4 segment and the β-sheet that corresponds to the ligand-binding site in other GAF domains (Fig. 3C). In CodY, ligand binding is associated with a large structural reorganization of the segments of structure above the β-sheet when viewed as in Fig. 3B. These conserved residues may therefore be involved in the binding of an as yet unidentified effector molecule to SpoIISA. The residues that constitute the SpoIISA-SpoIISA dimer interface (helices α1 and α5) are generally well conserved, although few of these residues are invariant in Fig. 4B. Because this interface is very hydrophobic, this is not so surprising. The residues in B. subtilis SpoIISA that hydrogen bond across this interface appear to co-vary, with Gln121 and Thr230 being replaced by small and large hydrophobic residues, respectively, whereas Tyr120 can be replaced by Phe/Met. Thus, polar interactions would be replaced by apolar ones. This analysis suggests that the dimeric SpoIISA structure is conserved.
For the SpoIISB orthologues, the highest sequence conservation is observed in the C-terminal α-helix and in the segment connecting the two β-strands. The cysteine at position 9 in SpoIISB from B. subtilis is not conserved in any of the orthologues, so the disulfide bond in the CSpoIISA·SpoIISB complex is clearly dispensable for their interaction. Its disulfide bond-forming partner Cys161 in SpoIISA is conserved in some but not all of the sequences, indicating that the cysteine is stable and functional without forming a disulfide bond.
Conserved interactions between SpoIISA and SpoIISB are harder to pinpoint partly because the interface is so extensive and partly because the sequence conservation in SpoIISB is so low. In addition, there are a series of intermolecular main chain hydrogen bonds as SpoIISA and SpoIISB form intermolecular β-sheet interactions. Conservation of this feature would not be apparent in sequence alignments. The Ser34′-Glu223 charge-dipole interaction, however, is conserved (Fig. 4, B and C).
The mut9 and mut14 alleles that led to the identification of the spoIIS operon are associated, respectively, with a point mutation that leads to a Leu103 to Phe substitution in SpoIISA and a frameshift mutation in spoIISB that leads to replacement of the C-terminal four residues of SpoIISB with a random sequence of 23 residues. Leu103 in SpoIISA is situated at the heart of the molecular interface. As well as packing in the SpoIISA dimer interface, it packs closely with the Leu47′, Ile48′, and Tyr51′ side chains from SpoIISB. Its replacement with Phe would add additional bulk at a tightly packed interface, which would be expected to weaken the interactions with the antitoxin. It would seem that this weakening of the interaction is significant in vivo because mut9 has a 104-fold lower sporulation efficiency than wild type B. subtilis. In relation to the mut14 allele, the four missing C-terminal residues of SpoIISB are largely disordered in the structure. The data in Fig. 4A suggest, however, that the mut14 phenotype is due to the loss of a critical residue among these four C-terminal residues rather than the addition of the 23 random residues. Thus, simple deletion of residues 53–56 leads to almost complete loss the SpoIISA neutralizing function (Fig. 4A).
Comparison with Other TA Systems
The membrane-attached GAF domain structure of SpoIISA sets this toxin apart from the toxins of other TA systems characterized to date, many of which have structural and functional links to ribonucleases (31–36). The dominant target of these chromosomal TA system toxins is the translation apparatus.
SpoIISB is a typical antitoxin, exhibiting no structure as monitored by CD spectroscopy. The complete absence of structure in the isolated antidote is reminiscent of YefM and RelB from the YoeB-YefM and RelBE systems, respectively. This intrinsically unstructured character is thought to contribute to the functionally important susceptibility of the antidotes to proteolysis, although it has yet to be demonstrated that SpoIISB is vulnerable to proteolysis.
The structures of several toxins have been determined in complex with the cognate antitoxin (31, 32, 36–42). These complexes exhibit some variety in their quaternary structures ranging from TA dimers (VapC-VapB), TA2 trimers (YoeB-YefM2), T2A2 tetramers (RelE-RelB), to T4A2 hexamers (MazF-MazE). A feature of the majority of these complexes is the extended conformation and absence of tertiary structure in the toxin-binding domain of the antitoxin moiety that is draped around the toxin, forming extensive quaternary interactions. It is apparent from the CSpoIISA·SpoIISB complex structure that these characteristics are exaggerated in this system. Almost all of the SpoIISB chain is involved in toxin binding, with half of its accessible surface area buried by this interaction. The arrangement of the antitoxins is, however, distinct. Whereas in most TA complexes, each antitoxin chain interacts either exclusively or predominantly with one subunit of the toxin, each SpoIISB chain distributes its interactions almost equally between the two SpoIISA chains, such that SpoIISB resembles a bracket holding the two SpoIISA chains together.
Role of SpoIISA
In the absence of SpoIISB and in an otherwise wild type background, SpoIISA toxicity is not manifest during vegetative growth or stationary phase but is delayed until after the completion of the asymmetric septum and confined to the mother cell. One explanation for this would be that expression of the spoIISAB operon is regulated in a temporal and compartment-specific manner, under the control of a mother cell-specific transcription factor. The PAB and PB promoters directing expression of the spoIIS region were shown to be transcribed by RNA polymerase containing the housekeeping sigma factor, σA (7). More recently, a transcriptome study has suggested that spoIISA is up-regulated by the later-acting mother cell-specific sigma factor, σK (43). Because σK is not produced until 4 h after the initiation of sporulation when engulfment of the forepore by the mother cell is complete (stage III), the significance of this observation in relation to the stage II phenotype of spoIIS mutations is not clear.
SpoIISA is present in vegetatively growing cells, and protein levels are increased by growth on sporulation medium, although not in a Spo0A-dependent manner. Because the protein would be expected to partition into both compartments upon asymmetric cell division, its selective toxicity in the mother cell may be due to an as yet unidentified compartment-specific activator or inhibitor of SpoIISA activity. Immunolocalization studies as well as studies of GFP fusions to SpoIISA have confirmed that SpoIISA is targeted to the cell membrane (44). The sporulation septum has a distinct structure appearing thinner than the vegetative septum. If, as is probable, SpoIISA interferes with membrane integrity, the sporulation septum would be especially vulnerable. It should be noted that the cytoplasmic membrane does become susceptible to SpoIISA, if the protein is overproduced in vegetatively growing B. subtilis or even in E. coli (7, 30).
In B. subtilis cells, SpoIISA production in the absence of SpoIISB leads to large plasmolysis zones and the observation of holes in the peptidoglycan layer in cells with complete or incomplete septa (7). Disruption of the cytoplasmic membrane was also seen in E. coli cells (30), suggesting the possibility that the SpoIISA toxin could act as a holin (7). Like SpoIISA, the bacteriophage λ holin (S105) has three transmembrane helices with an Nout Cin topology. S105 accumulates in the host cytoplasmic membrane without causing defects because it interacts with a holin-antagonist (45). Once its activity is triggered, it forms large assemblies that create holes in the membrane large enough to allow the release of folded endolysins that then disrupt the cell wall (46). Intact SpoIISA is required for cell killing because neither the cytosolic region nor the transmembrane segments alone are toxic (30). The cytosolic domain of SpoIISA may regulate the further assembly of a toxic multimer in which the membrane spanning helices form a hole in the cell membrane. The possibility that SpoIISA toxicity might be driven mostly through its transmembrane domains is supported by the loss-of-function mutation R38Q, which is located between the first and the second membrane spanning helices (7). Alternatively, the toxic action may be intrinsic to the cytosolic domain with the membrane spanning segments localizing it to its site of action. In these models, the antidote SpoIISB would act by masking surfaces required for oligomerization and activity, respectively.
It is intriguing that SpoIISA was discovered through studies of sporulation, a process that involves programmed cell death and that is also associated with cannibalism (47). Cell death in these instances is mediated by defined alternative mechanisms, and there is no evidence for the involvement of SpoIISA. Thus, the circumstances under which SpoIISB inhibition of SpoIISA is overcome and toxicity is manifested are unknown. This may prove hard to unravel if killing occurs in only in a subpopulation of cells during growth or development.
Acknowledgments
We are grateful to Emília Chovancová for technical assistance and Dr. Katarína Muchová and other members of IB laboratory for valuable advice.
This work was supported by Grant 2/0016/10 from the Slovak Academy of Sciences, by grants from the Slovak Research and Development Agency under Contracts APVT-51-0278 and LPP-0218-06, by a European Science Foundation Grant ESF-EC-0106, and by Grant 082829/Z/07/Z from the Wellcome Trust.
The atomic coordinates and structure factors (code 3O6Q) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TA
- toxin-antitoxin
- MES
- 4-morpholineethanesulfonic acid
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Ogura T., Hiraga S. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gerdes K., Rasmussen P. B., Molin S. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 3116–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Masuda Y., Miyakawa K., Nishimura Y., Ohtsubo E. (1993) J. Bacteriol. 175, 6850–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005) Trends Biochem. Sci. 30, 672–679 [DOI] [PubMed] [Google Scholar]
- 5. Aizenman E., Engelberg-Kulka H., Glaser G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pedersen K., Christensen S. K., Gerdes K. (2002) Mol. Microbiol. 45, 501–510 [DOI] [PubMed] [Google Scholar]
- 7. Adler E., Barák I., Stragier P. (2001) J. Bacteriol. 183, 3574–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ausubel F. M., Brent R., Kingston R., Moore D., Seidman J. G., Smith J. A., Struhl K. (eds) (2003) Current Protocols in Molecular Biology, John Wiley & Sons, New York [Google Scholar]
- 9. Harwood C. R., Cutting S. M. (1990) Molecular Biological Methods for Bacillus, John Wiley & Sons; Chichester, UK [Google Scholar]
- 10. Ju J., Luo T., Haldenwang W. G. (1998) J. Bacteriol. 180, 1673–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramakrishnan V., Biou V. (1997) Methods Enzymol. 276, 538–557 [PubMed] [Google Scholar]
- 12. Myszka D. G. (1999) J. Mol. Recognit. 12, 279–284 [DOI] [PubMed] [Google Scholar]
- 13. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. Sect. D 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 15. Otwinowski Z. (1991) Proceedings of the CCP4 Study Weekend Isomorphous Replacement and Anomolous Scattering (Wolf W., Evans P. R., Leslie A. G. W. eds) pp. 80–86, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 16. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta. Crystallogr. Sect. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. (2004) Acta. Crystallogr. Sect. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Winn M., Isupov M., Murshudov G. N. (2001) Acta. Crystallogr. Sect. D 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 19. Janin J., Bahadur R. P., Chakrabarti P. (2008) Q. Rev. Biophys. 41, 133–180 [DOI] [PubMed] [Google Scholar]
- 20. Hanna R. A., Garcia-Diaz B. E., Davies P. L. (2007) FEBS Lett. 581, 2894–2898 [DOI] [PubMed] [Google Scholar]
- 21. Hanna R. A., Campbell R. L., Davies P. L. (2008) Nature 456, 409–412 [DOI] [PubMed] [Google Scholar]
- 22. Hurley T. D., Yang J., Zhang L., Goodwin K. D., Zou Q., Cortese M., Dunker A. K., DePaoli-Roach A. A. (2007) J. Biol. Chem. 282, 28874–28883 [DOI] [PubMed] [Google Scholar]
- 23. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aravind L., Ponting C. P. (1997) Trends Biochem. Sci. 22, 458–459 [DOI] [PubMed] [Google Scholar]
- 25. Badger J., Sauder J. M., Adams J. M., Antonysamy S., Bain K., Bergseid M. G., Buchanan S. G., Buchanan M. D., Batiyenko Y., Christopher J. A., Emtage S., Eroshkina A., Feil I., Furlong E. B., Gajiwala K. S., Gao X., He D., Hendle J., Huber A., Hoda K., Kearins P., Kissinger C., Laubert B., Lewis H. A., Lin J., Loomis K., Lorimer D., Louie G., Maletic M., Marsh C. D., Miller I., Molinari J., Muller-Dieckmann H. J., Newman J. M., Noland B. W., Pagarigan B., Park F., Peat T. S., Post K. W., Radojicic S., Ramos A., Romero R., Rutter M. E., Sanderson W. E., Schwinn K. D., Tresser J., Winhoven J., Wright T. A., Wu L., Xu J., Harris T. J. (2005) Proteins 60, 787–796 [DOI] [PubMed] [Google Scholar]
- 26. Lin Z., Johnson L. C., Weissbach H., Brot N., Lively M. O., Lowther W. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9597–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levdikov V. M., Blagova E., Colledge V. L., Lebedev A. A., Williamson D. C., Sonenshein A. L., Wilkinson A. J. (2009) J. Mol. Biol. 390, 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sonenshein A. L. (2005) Curr. Opin. Microbiol. 8, 203–207 [DOI] [PubMed] [Google Scholar]
- 29. Levdikov V. M., Blagova E., Joseph P., Sonenshein A. L., Wilkinson A. J. (2006) J. Biol. Chem. 281, 11366–11373 [DOI] [PubMed] [Google Scholar]
- 30. Florek P., Muchová K., Pavelcíková P., Barák I. (2008) FEMS Microbiol. Lett. 278, 177–184 [DOI] [PubMed] [Google Scholar]
- 31. Takagi H., Kakuta Y., Okada T., Yao M., Tanaka I., Kimura M. (2005) Nat. Struct. Mol. Biol. 12, 327–331 [DOI] [PubMed] [Google Scholar]
- 32. Kamada K., Hanaoka F. (2005) Mol. Cell 19, 497–509 [DOI] [PubMed] [Google Scholar]
- 33. Mauguen Y., Hartley R. W., Dodson E. J., Dodson G. G., Bricogne G., Chothia C., Jack A. (1982) Nature 297, 162–164 [DOI] [PubMed] [Google Scholar]
- 34. Sevcík J., Dodson E. J., Dodson G. G. (1991) Acta. Crystallogr. Sect. B 47, 240–253 [PubMed] [Google Scholar]
- 35. Bernard P., Couturier M. (1992) J. Mol. Biol. 226, 735–745 [DOI] [PubMed] [Google Scholar]
- 36. Miallau L., Faller M., Chiang J., Arbing M., Guo F., Cascio D., Eisenberg D. (2009) J. Biol. Chem. 284, 276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schumacher M. A., Piro K. M., Xu W., Hansen S., Lewis K., Brennan R. G. (2009) Science 323, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garcia-Pino A., Christensen-Dalsgaard M., Wyns L., Yarmolinsky M., Magnuson R. D., Gerdes K., Loris R. (2008) J. Biol. Chem. 283, 30821–30827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown B. L., Grigoriu S., Kim Y., Arruda J. M., Davenport A., Wood T. K., Peti W., Page R. (2009) PLoS Pathog. 5, e1000706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kamada K., Hanaoka F., Burley S. K. (2003) Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 41. Mattison K., Wilbur J. S., So M., Brennan R. G. (2006) J. Biol. Chem. 281, 37942–37951 [DOI] [PubMed] [Google Scholar]
- 42. Meinhart A., Alonso J. C., Sträter N., Saenger W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Steil L., Serrano M., Henriques A. O., Völker U. (2005) Microbiology 151, 399–420 [DOI] [PubMed] [Google Scholar]
- 44. Rešetárová S., Florek P., Muchová K., Wilkinson A. J., Barák I. (2010) Res. Microbiol. 161, 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Savva C. G., Dewey J. S., Deaton J., White R. L., Struck D. K., Holzenburg A., Young R. (2008) Mol. Microbiol. 69, 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dewey J. S., Savva C. G., White R. L., Vitha S., Holzenburg A., Young R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. González-Pastor J. E., Hobbs E. C., Losick R. (2003) Science 301, 510–513 [DOI] [PubMed] [Google Scholar]
- 48. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan P., Emsley K., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta. Crystallogr. Sect. D 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 49. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic. Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 51. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Benson A. K., Haldenwang W. G. (1993) J. Bacteriol. 175, 2347–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Youngman P., Perkins J. B., Losick R. (1984) Plasmid 12, 1–9 [DOI] [PubMed] [Google Scholar]