Abstract
Pathogenic Streptococcus agalactiae produces polysaccharide lyases and unsaturated glucuronyl hydrolase (UGL), which are prerequisite for complete degradation of mammalian extracellular matrices, including glycosaminoglycans such as chondroitin and hyaluronan. Unlike the Bacillus enzyme, streptococcal UGLs prefer sulfated glycosaminoglycans. Here, we show the loop flexibility for substrate binding and structural determinants for recognition of glycosaminoglycan sulfate groups in S. agalactiae UGL (SagUGL). UGL also degraded unsaturated heparin disaccharides; this indicates that the enzyme released unsaturated iduronic and glucuronic acids from substrates. We determined the crystal structures of SagUGL wild-type enzyme and both substrate-free and substrate-bound D175N mutants by x-ray crystallography and noted that the loop over the active cleft exhibits flexible motion for substrate binding. Several residues in the active cleft bind to the substrate, unsaturated chondroitin disaccharide with a sulfate group at the C-6 position of GalNAc residue. The sulfate group is hydrogen-bonded to Ser-365 and Ser-368 and close to Lys-370. As compared with wild-type enzyme, S365H, S368G, and K370I mutants exhibited higher Michaelis constants toward the substrate. The conversion of SagUGL to Bacillus sp. GL1 UGL-like enzyme via site-directed mutagenesis demonstrated that Ser-365 and Lys-370 are essential for direct binding and for electrostatic interaction, respectively, for recognition of the sulfate group by SagUGL. Molecular conversion was also achieved in SagUGL Arg-236 with an affinity for the sulfate group at the C-4 position of the GalNAc residue. These residues binding to sulfate groups are frequently conserved in pathogenic bacterial UGLs, suggesting that the motif “R-//-SXX(S)XK” (where the hyphen and slash marks in the motif indicate the presence of over 100 residues in the enzyme and parentheses indicate that Ser-368 makes little contribution to enzyme activity) is crucial for degradation of sulfated glycosaminoglycans.
Keywords: Bacterial Metabolism, Enzyme Kinetics, Glycosaminoglycan, Hydrolases, Protein Structure, X-ray Crystallography
Introduction
Glycosaminoglycans (e.g. hyaluronan, chondroitin, and heparin) are mammalian extracellular matrix polysaccharides with a repeating disaccharide unit that consists of a uronic acid residue, such as d-glucuronic acid (GlcUA)2 or l-iduronic acid (IdoUA), and an amino sugar residue, such as d-glucosamine (GlcN), N-acetyl-d-glucosamine (GlcNAc), or N-acetyl-d-galactosamine (GalNAc) (1, 2). These extracellular matrices, which are present in various mammalian tissues, play an important role in cell signaling, growth and differentiation, and cell-to-cell association to maintain the architecture of connective tissues (3). A large number of glycosaminoglycans, with the exception of hyaluronan, are frequently sulfated (4), and the sulfate groups together with uronic acids enhance the negative charge of the polysaccharides. For instance, chondroitin has sulfate group(s) at the C-4 and/or C-6 positions of the GalNAc residue and/or at the C-2 position of the GlcUA residue (5).
Degradation of glycosaminoglycans has been studied previously from the viewpoint of bacterial infections and neural regenerations. Bacterial pathogens, such as streptococci, degrade glycosaminoglycans to invade host cells by producing polysaccharide lyases (6). Distinct from polysaccharide hydrolases, glycosaminoglycan lyases recognize the uronic acid residue, cleave the linkage between sugars through the β-elimination reaction, and produce unsaturated oligosaccharides, resulting in the unsaturated uronic acid residue having a C=C double bond at the nonreducing terminus (7) (see Fig. 1A). Many pathogenic streptococci produce hyaluronate lyases as a spreading factor; these enzymes are capable of degrading both sulfated and unsulfated chondroitin as well as hyaluronan (8–12). Researchers have extensively studied the structure and function of streptococcal hyaluronate lyases to ultimately facilitate the development of therapeutic agents for inhibition of the lyases (13, 14). On the other hand, bacterial polysaccharide lyases (e.g. chondroitin lyase ABC) have been demonstrated to promote neural regeneration (15). Neurons, especially their axons, experience difficulty in regeneration due to the presence of some inhibitory molecules, such as chondroitin sulfate proteoglycans (16). Enzymatic degradation of chondroitin sulfate at the site of an injury enables neurons to regenerate axons and to restore postsynaptic activity. Therefore, elucidation of the bacterial mechanism underlying glycosaminoglycan degradation is important for establishment of therapy against bacterial infections and neural injury.
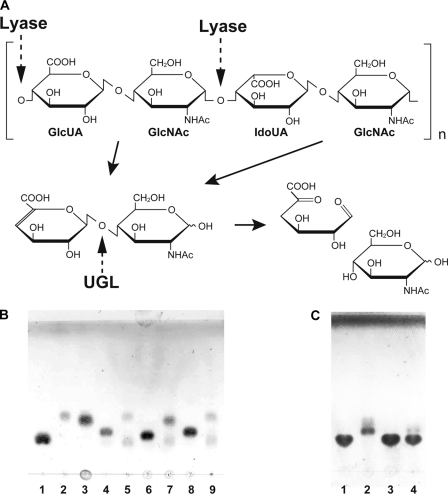
FIGURE 1.
UGL reaction. A, scheme of heparin degradation by UGL. The heparin molecule contains GlcUA and IdoUA. Dotted and solid arrows indicate the cleavage sites for polysaccharide lyases and UGL and the degradation pathway for polysaccharides, respectively. B, unsaturated glycosaminoglycan disaccharides were incubated at 30 °C with SagUGL, and the resultant products were detected on the TLC plate. Lane 1, GlcUA (70 nmol); lane 2, GlcNAc (70 nmol); lane 3, GalNAc (70 nmol); lane 4, unsaturated heparin disaccharide (52.5 nmol); lane 5, unsaturated heparin disaccharide (52.5 nmol) with SagUGL; lane 6, Δ0S (52.5 nmol); lane 7, Δ0S (52.5 nmol) with SagUGL; lane 8, unsaturated hyaluronan disaccharide (52.5 nmol); and lane 9, unsaturated hyaluronan disaccharide (52.5 nmol) with SagUGL. C, Δ4S (150 nmol) was incubated at 30 °C with SagUGL or BacillusUGL, and the resultant products were detected on the TLC plate. Lane 1, Δ4S (150 nmol); lane 2, Δ4S (150 nmol) with SagUGL; lane 3, Δ4S (150 nmol) with BacillusUGL WT; and lane 4, Δ4S (150 nmol) with BacillusUGL H210R.
Unsaturated glucuronyl hydrolase (UGL) acts on unsaturated glycosaminoglycan oligosaccharides that are produced by polysaccharide lyases and catalyzes the hydrolytic release of unsaturated GlcUA (ΔGlcUA) from saccharides (17) (see Fig. 1A). UGL is peculiar among general glycoside hydrolases in that it triggers hydration of vinyl ether groups specifically present in unsaturated saccharides but not of glycoside bonds (18). On the basis of its primary structure, UGL was categorized as a member of the glycoside hydrolase family 88 in the CAZy database (19, 20) after we first identified the gene for Bacillus sp. GL1 UGL (BacillusUGL) (21). The putative genes for UGL are present in the genome of various pathogenic bacteria, such as streptococci, enterococci, and vibrios. We recently clarified enzymatic characteristics of UGLs from pathogenic streptococci, including Streptococcus agalactiae, Streptococcus pneumoniae, and Streptococcus pyogenes, and determined the crystal structure of S. agalactiae UGL (SagUGL) (22). The enzyme gene is inducibly transcribed in S. agalactiae cells grown in the presence of glycosaminoglycan, indicating that the bacterium produces UGL as well as polysaccharide lyase for complete degradation of glycosaminoglycans. All of the streptococcal UGLs that have been characterized to date actively degrade unsaturated chondroitin and hyaluronan disaccharides and exhibit a preference for sulfated chondroitin disaccharide, especially for Δ6S, an unsaturated chondroitin disaccharide with a sulfate group at the C-6 position of the GalNAc residue.
This study deals with the identification of loop movement for substrate binding and structural determinants for substrate specificity in streptococcal UGL through x-ray crystallography of SagUGL in complex with Δ6S, kinetics of site-directed mutants, and molecular conversion of bacterial UGLs with altered substrate specificity.
EXPERIMENTAL PROCEDURES
Materials
Unsaturated glycosaminoglycan disaccharides were purchased from Seikagaku Biobusiness or Sigma-Aldrich. Silicagel 60/Kieselguhr F254 TLC plates were obtained from Merck. DEAE-Toyopearl 650M was from Tosoh. HiLoad 16/60 Superdex 75pg and Mono Q 10/100 GL were from GE Healthcare. Restriction endonucleases and DNA-modifying enzymes were from Toyobo. Other analytical grade chemicals were obtained from commercial sources.
Microorganisms and Culture Conditions
For expression of SagUGL and BacillusUGL, Escherichia coli strain HMS174(DE3) cells transformed with pET21b-SagUGL (22) and E. coli strain BL21(DE3) cells transformed with pET3a-BacillusUGL (19) were aerobically cultured at 30 °C in LB medium (23) supplemented with sodium ampicillin (0.1 mg/ml). When the turbidity at 600 nm reached 0.3–0.7, isopropyl β-d-thiogalactopyranoside was added to the culture at a final concentration of 0.1 mm, and the cells were further cultured at 16 °C for 44 h.
Purification
E. coli cells harboring pET21b-SagUGL or pET3a-BacillusUGL were grown in LB medium, collected by centrifugation at 6,700 × g and 4 °C for 5 min, and resuspended in 20 mm potassium phosphate (pH 7.0). The cells were ultrasonically disrupted (Insonator model 201M, Kubota) at 9 kHz and 0 °C for 5 min, and the clear solution obtained by centrifugation at 28,000 × g and 4 °C for 20 min was used as a cell extract. SagUGL and BacillusUGL were purified from the cell extract to homogeneity by several steps of column chromatography (19, 22). Briefly, SagUGL and BacillusUGL were purified by anion exchange chromatography (DEAE-Toyopearl 650M) followed by gel filtration chromatography (HiLoad 16/60 Superdex 75 pg) and finally by anion exchange chromatography (Mono Q 10/100 GL). The degree of purification was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (24).
TLC
The products derived from unsaturated glycosaminoglycan disaccharides through the reaction of bacterial UGLs were separated by TLC using a solvent system of 1-butanol/acetic acid/water (3:2:2, v/v). The products were visualized by heating the TLC plates at 130 °C for 5 min after spraying with 10% (v/v) sulfuric acid in ethanol.
Crystallization and X-ray Diffraction
Purified SagUGL enzymes of wild type (WT) and mutant D175N with Asp-175 replaced by Asn were concentrated by ultrafiltration using Centriprep (10,000 molecular weight cutoff) (Millipore) to 10 and 5 mg/ml, respectively. Both WT and D175N were crystallized by sitting drop vapor diffusion. The 3 μl of proteins were mixed with an equal volume of a reservoir solution. The reservoir solution for WT crystallization contained 30% (w/v) polyethylene glycol 200, 1% (w/v) polyethylene glycol 3000, and 0.1 m Hepes (pH 7.0). WT crystals grew up at 20 °C for a week. The reservoir solution for D175N crystallization included 40% (w/v) ethylene glycol, 5% (w/v) polyethylene glycol 3000, and 0.1 m Hepes (pH 7.5). D175N was crystallized at 20 °C for a month. To prepare a complex form of SagUGL and Δ6S, the D175N crystal was soaked at 20 °C for 10 min in a reservoir solution containing 0.2 m Δ6S before x-ray diffraction experiments. Crystals were placed in a cold nitrogen gas stream at −173 °C. X-ray diffraction images of crystals were collected using a Jupiter 210 charged-coupled device detector (Rigaku) for the WT crystal or a Quantum 210 charged-coupled device detector (Area Detector Systems Corp.) for D175N crystals with synchrotron radiation at a wavelength of 1.00 Å at the BL-38B1 station of SPring-8 (Hyogo, Japan). The data were processed and scaled with the HKL2000 program (25).
Structure Determination and Refinement
The structure was determined through molecular replacement with the Molrep program (26) supplied in the CCP4 interface program package (27) by using previously determined coordinates of SagUGL WT (Protein Data Bank code 2ZZR) as an initial model. Structure refinement was conducted with the Refmac5 program (28). Randomly selected 5% reflections were excluded from refinement and used to calculate Rfree. After each refinement cycle, the model was adjusted manually using the winCoot program (29). Water molecules were incorporated where the difference in density exceeded 3.0 σ above the mean and the 2Fo − Fc map showed a density of more than 1.0 σ. The structure of the enzyme-sugar complex was refined using the Refmac5 and the winCoot program with the chondroitin disaccharide parameter file constructed using PRODRG (50). Protein models were superimposed, and their root mean square deviation was determined with the LSQKAB program (30), a part of the CCP4 program package. Final model quality was checked with the PROCHECK program (31). Figures for protein structures were prepared using the PyMOL program (32). Electric charge on the molecular surface of bacterial UGLs was calculated using the APBS program (33). Coordinates used in this work were taken from the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics (34).
Site-directed Mutagenesis
Thr-235, Ser-365, Ser-368, and Lys-370 of SagUGL were substituted with Ala, His, Gly, and Ile, respectively, and His-210, His-339, Gly-342, and Ile-344 of BacillusUGL were substituted with Arg, Ser, Ser, and Lys, respectively. UGL mutants were constructed using a QuikChange site-directed mutagenesis kit (Stratagene). The plasmid pET21b-SagUGL (22) or pET3a-BacillusUGL (19) was used as a PCR template, and the synthetic oligonucleotides used as sense and antisense primers are shown in supplemental Table S1. PCR was carried out using KOD-FX polymerase (Toyobo) in place of Pfu polymerase. Mutations were confirmed by the dideoxy chain termination method (35) using automated DNA sequencer model 3730xl (Applied Biosystems). The cells of the E. coli host strain (HMS174(DE3)) were transformed with the mutant plasmids. Expression and purification of the mutants were conducted by using the same procedures as for SagUGL or BacillusUGL WT as described above. DNA manipulations such as plasmid isolation, subcloning, transformation, and gel electrophoresis were performed as described (23).
Kinetic Analysis
Kinetics parameters of SagUGLs (WT, T235A, S365H, S368G, and K370I) and BacillusUGLs (WT, H339S, G342S, and I344K) toward Δ6S or sulfate-free unsaturated chondroitin disaccharide (Δ0S) were determined as follows. The activity of UGLs was assayed at 30 °C by monitoring the decrease in absorbance at 235 nm arising from the double bond (molar extinction coefficient ϵ235 = 4,800 m−1 cm−1) in the substrates. The reaction mixtures consisted of substrate, 20 mm Tris-HCl (pH 7.5), and enzyme. The range of substrate concentration was fixed at 0.05–1.0 mm because the absorbance at 235 nm of the substrate at over 1.0 mm exceeded measurement limitations on the spectrometer. Km and kcat were calculated using the Michaelis-Menten equation with KaleidaGraph software (Synergy Software).
RESULTS AND DISCUSSION
Streptococcal UGL Involved in Heparin Degradation
In our previous studies (21, 22), Bacillus and streptococcal UGLs have been identified to degrade unsaturated chondroitin and hyaluronan disaccharides. Both chondroitin and hyaluronan include β-GlcUA as a component, whereas α-IdoUA, a C-5 epimer of GlcUA, is predominant (ratio in uronates, >70%) in heparin molecules (3). There is also a difference in glycoside bond pattern between chondroitin/hyaluronan and heparin/heparan sulfate. The 1→3 glycoside bond is present between uronate and amino sugar residues in chondroitin and hyaluronan, although both heparin and heparan sulfate contain the 1→4 glycoside bond between uronate and amino sugar residues. Thus, the enzyme activity of SagUGL was investigated using unsaturated heparin disaccharides with and without sulfate group(s) as a substrate. The sulfate-free unsaturated heparin disaccharide was completely degraded to unsaturated uronic acid and GlcNAc by SagUGL (Fig. 1B, lane 5). In combination with previously reported data, this result indicates that the enzyme acts on unsaturated α-IdoUA (ΔIdoUA) as well as unsaturated β-GlcUA residues in substrates and cleaves both glycoside bonds 1→3 and 1→4. The formation of a double bond between C-4 and C-5 atoms leads to loss of epimerization in GlcUA and IdoUA. In fact, C-3, C-4, C-5, and C-6 atoms of ΔGlcUA were determined to be located in a single plane through structural analysis of UGL and unsaturated glycosaminoglycan disaccharide (18). Thus, the degradation of both unsaturated chondroitin and heparin disaccharides by UGL was chemically appropriate because each nonreducing terminus showed the same conformation.
The activity of UGL on ΔGlcUA- and ΔIdoUA-containing glycosaminoglycan disaccharides suggests that each glycosaminoglycan in mammalian extracellular matrices is first depolymerized to unsaturated disaccharides by a specific polysaccharide lyase, and the resultant unsaturated disaccharides are degraded to the constituent monosaccharides by a single enzyme, UGL. Sulfate-bound disaccharides, such as unsaturated heparin disaccharides with sulfate group(s) at either or both the C-6 position of the GlcNAc residue or the nitrogen position of the GlcN residue, were also degraded by SagUGL (supplemental Fig. S1). Similar to chondroitin and heparin, heparan sulfate and dermatan sulfate include IdoUA and/or GlcUA and sulfate group(s) in their molecules (3). Therefore, streptococcal UGLs are considered as one of the key enzymes for degradation of all uronate-including glycosaminoglycans, chondroitin, hyaluronan, heparin, heparan sulfate, and dermatan sulfate after reactions of polysaccharide lyases.
Enzyme Affinity for Sulfated Substrate
Streptococcal UGLs, including SagUGL, showed a preference for sulfate-bound unsaturated disaccharides from glycosaminoglycans (e.g. chondroitin and heparin) (22). Because this preference of streptococcal UGLs was thought to be dependent on substrate binding, catalytic action, or both, SagUGL and BacillusUGL were kinetically analyzed using sulfate-free (Δ0S) and -bound (Δ6S) unsaturated chondroitin disaccharides as substrates (Table 1). We interpreted the kinetic parameters listed in Table 1 as follows. We treated kinetic parameters with higher Michaelis constants (Km over 1 mm) as estimations because kinetic studies of bacterial UGLs with the higher concentrations (over 1 mm) of substrate were difficult to perform because of their high absorbance at 235 nm. We determined kinetics parameters of SagUGL toward Δ0S and Δ6S. The Km value (1.27 mm) toward Δ0S was over 10-fold higher than that (0.100 mm) toward Δ6S, whereas turnover number (kcat) toward Δ0S was about 3.8-fold smaller than that toward Δ6S. In comparison with the great difference in the affinity for substrate (Km), the difference in kcat was considered to be insignificant. This result indicates that the enzyme exhibits a higher affinity for Δ6S than Δ0S, and its substrate specificity mostly depends on substrate binding. In contrast, BacillusUGL showed a preference for the unsulfated substrate rather than the sulfated substrate because of its high affinity (around 50-fold), although the differences in kcat of the enzyme toward Δ0S and Δ6S were less than 4-fold.
TABLE 1.
Kinetic parameters of bacterial UGLs
| Δ6S |
Δ0S |
|||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| mm | s−1 | s−1mm−1 | mm | s−1 | s−1mm−1 | |
| SagUGL | ||||||
| WT | 0.100 ± 0.0283 | 10.2 ± 0.662 | 102 | 1.27 ± 0.0941 | 2.69 ± 0.128 | 2.12 |
| T235A | 1.75 ± 0.344 | 3.96 ± 0.554 | 2.26 | NDa | ND | ND |
| S365H | 2.36 ± 0.160 | 3.85 ± 0.199 | 1.63 | 0.763 ± 0.215 | 1.69 ± 0.255 | 2.21 |
| S368G | 0.191 ± 0.0269 | 24.8 ± 1.12 | 130 | 1.40 ± 0.332 | 10.8 ± 1.71 | 7.71 |
| K370I | 1.47 ± 0.191 | 41.6 ± 3.84 | 28.2 | 0.371 ± 0.0692 | 3.97 ± 0.297 | 10.7 |
| BacillusUGL | ||||||
| WT | 18.6 ± 6.61 | 54.9 ± 18.3 | 2.95 | 0.381 ± 0.0394 | 14.1 ± 0.625 | 36.8 |
| H339S | 7.19 ± 1.54 | 24.5 ± 4.70 | 3.41 | 0.861 ± 0.147 | 16.9 ± 1.40 | 19.6 |
| G342S | 7.00 ± 2.65 | 20.9 ± 7.09 | 2.99 | 0.504 ± 0.0405 | 18.2 ± 0.665 | 36.1 |
| I344K | 2.20 ± 0.105 | 39.8 ± 1.41 | 18.1 | 0.566 ± 0.0928 | 23.8 ± 1.86 | 42.0 |
a ND, kinetic parameters could not be determined due to the very low activity.
Based on the kinetics of SagUGL and BacillusUGL, we conclude that the activity on the sulfated substrate was mainly dependent on the affinity for the substrate rather than turnover number. This lesser effect of kcat on substrate specificity was likely due to the multiple actions of UGL strictly on ΔGlcUA but not on amino sugar (18), i.e. (i) proton donation to the C-4 atom of ΔGlcUA, (ii) deprotonation of the water molecule, (iii) addition of the water molecule to the C-5 atom of ΔGlcUA, (iv) cleavage of the glycoside bond, and (v) conversion of ΔGlcUA to α-keto acid. Hereafter, substrate specificity was analyzed based on the affinity (Km) for the substrate.
The substrate specificity of SagUGL is suggestive of its structural features for specific binding to sulfate groups in the substrate. To clarify structural determinants for sulfate binding, we performed x-ray crystallography of the enzyme-substrate complex.
Loop Movement over Active Cleft
In a previous study, SagUGL was crystallized in a drop solution using ammonium sulfate as the major precipitant, and the crystal structure of the ligand-free enzyme was determined at 1.75-Å resolution (22). In this study, we conducted many experiments to prepare the enzyme-substrate complex through a soaking treatment of crystals in the substrate solution but failed to obtain the complex. Thus, crystallization conditions were rescreened to accommodate the substrate at the active site of the enzyme. Another crystal of SagUGL WT formed in a drop solution using polyethylene glycol as the major precipitant, and it showed crystallographic properties different from the previously reported crystal (22). This crystal belongs to the C2 space group with unit cell dimensions of a = 104.8 Å, b = 53.2 Å, c = 70.1 Å, and β = 96.6°. Furthermore, we subjected the crystal to structure determination at 1.95-Å resolution via molecular replacement using the previous structure (Protein Data Bank code 2ZZR) as an initial model. The details of data collection and model refinement statistics are summarized in supplemental Table S2.
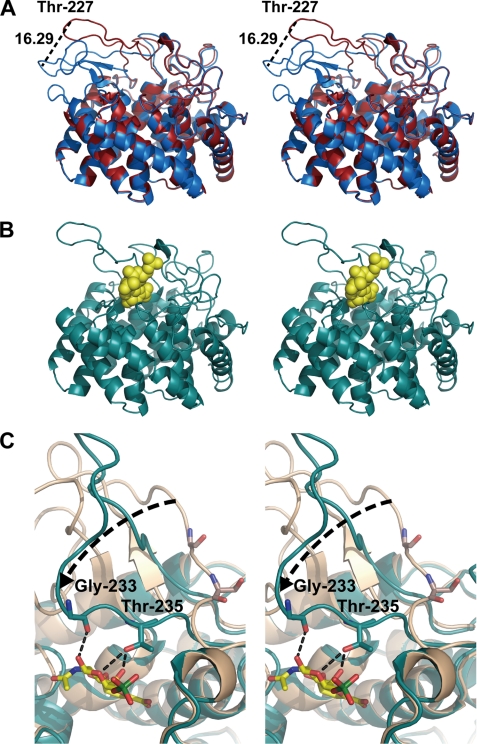
The overall structure, (α/α)6-barrel, of SagUGL in the C2 crystal was very similar to the previously determined structure (22) (Fig. 2A), although residues 151–171 were not assigned because of disorder. Furthermore, we observed a conformational change, i.e. loop movement over the active cleft, in this model that suggests that the loop (residues 219–236) has flexible motion. Because this loop covers the active cleft to interact with the substrate in the resulting structure of the enzyme-substrate complex (Fig. 2, B and C), the enzyme adopts a “closed form.” We estimated the oscillation length of the loop at the edge (Thr-227) to be 16.3 Å (Fig. 2A). The loop flexibility was also evidenced based on the average B-factor. The B-factor of the loop showed a high score (40.3 Å2), although the entire molecule of the enzyme had a B-factor of 21.9 Å2. The flexibility of the loop (residues 219–236) was found to be important for enzyme activity through the following structure determination of the enzyme-substrate complex and site-directed mutagenesis experiments. Briefly, we found that Gly-233 and Thr-235, both of which are located in the loop region, directly interact with the substrate (Fig. 2C), and the T235A mutant exhibited little enzyme activity (Table 1).
FIGURE 2.
Conformational change of SagUGL. A, superimposition of closed (red) and open (blue) forms of SagUGL WT (stereodiagram). The flexible loop moves with an oscillation width of 16.29 Å as indicated by the broken line. B, Δ6S (yellow) is accommodated at the active site of SagUGL D175N (cyan) in the closed form (stereodiagram). C, Gly-233 and Thr-235 binding to the substrate (stereodiagram).
Structure of Enzyme-Substrate Complex
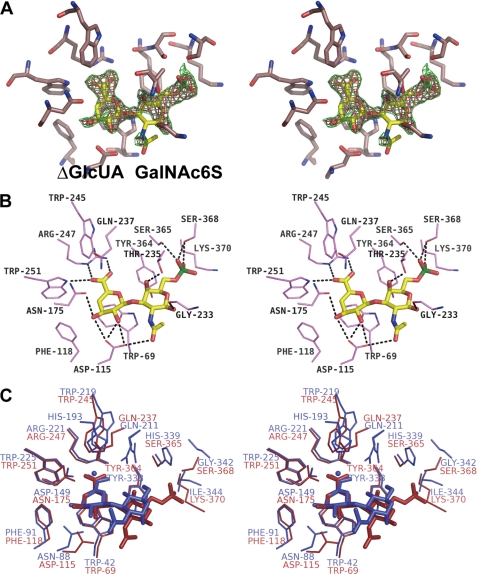
To analyze the interaction between SagUGL and substrate, we used the previously constructed mutant D175N with Asp-175 substituted with Asn for crystallization because the mutant exhibits no enzyme activity. The role of Asp-175 is as follows. Asp-175 acts as a general acid catalyst and donates a proton to the double bond (C-4 atom). Subsequently, Asp-175 also acts as a general base catalyst and deprotonates the water molecule. We treated the freshly prepared D175N crystals belonging to the C2 space group with and without Δ6S and subjected them to structure determination. Crystal structures of Δ6S-free and -bound D175N were determined by the molecular replacement method using the previous structure (Protein Data Bank code 2ZZR) as an initial model (Fig. 2B). The details of data collection and the model refinement statistics are summarized in supplemental Table S2. Both D175N and D175N-Δ6S models are structurally identical to the closed form of WT (D175N, root mean square deviation = 0.708 Å for 364 Cα; D175N-Δ6S, root mean square deviation = 0.644 Å for 370 Cα) and lacked about 20 residues (D175N, residues 153–171; D175N-Δ6S, residues 152–171) because of disorder. The substrate Δ6S is well fitted in the electron density map with an average B-factor of 42.6 Å2 (Fig. 3A) and bound to the active cleft (Fig. 2B).
FIGURE 3.
Active site structure of SagUGL D175N in complex with Δ6S. A, electron density of Δ6S in the omit (Fo − Fc) map calculated without the substrate and contoured at the 2.5 (green) and 3.0 σ (red) levels. B, interaction of SagUGL D175N with Δ6S (stereodiagram). Several residues bind to Δ6S through the formation of hydrogen bonds (broken lines). Atoms carbon, oxygen, nitrogen, and sulfur of Δ6S are colored yellow, pink, blue, and green, respectively. C, superimposition of SagUGL D175N-Δ6S complex (red) and BacillusUGL D88N-Δ0S complex (blue).
The crystal structure of D175N-Δ6S revealed the binding mode of the substrate Δ6S molecule to the active site cleft (Fig. 3B). There are several interactions between the enzyme and substrate through hydrogen bonds and van der Waals contacts (Table 2). Subsites are defined such that −n represents the nonreducing terminus and +n represents the reducing terminus, and cleavage occurs between the −1 and +1 sites (36). Because the glycoside bond in unsaturated glycosaminoglycan disaccharides is cleaved through the UGL reaction, uronate and amino sugar residues are positioned at −1 and +1 subsites, respectively. ΔGlcUA residue is accommodated at the −1 subsite via six hydrogen bonds by four residues (Asp-115, Asn-175, Arg-247, and Trp-251) and stacking with six residues (Trp-69, Asp-115, Phe-118, Asn-175, Trp-245, and Arg-247). In particular, a Trp-69 residue is parallel to the pyranose ring of ΔGlcUA through a stacking interaction. These interactions of SagUGL with ΔGlcUA are comparable with those of BacillusUGL with ΔGlcUA (D88N-Δ0S) (18).
TABLE 2.
Interaction between SagUGL D175N and Δ6S
| Hydrogen bonds (<3.3 Å) |
van der Waals contact (C-C distance <4.5 Å) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Sugar | Atom | Protein | Atom | Distance | Sugar | Atom | Protein | Atom |
| Å | ||||||||
| ΔGlcUA | O-2 | Asp-115 | Oδ2 | 2.6 | ΔGlcUA | C-1 | Trp-69 | Cδ2, Cζ2, Cγ, Cδ1, Cϵ2 |
| O-3 | Asn-175 | Oδ1 | 3.1 | C-2 | Asp-115 | Cγ | ||
| Asp-115 | Oδ2 | 3.2 | C-3 | Asn-175 | Cγ | |||
| O-6A | Arg-247 | Nη2 | 2.7 | Asp-115 | Cγ | |||
| O-6B | Trp-251 | Nϵ1 | 3.1 | Trp-69 | Cδ2, Cϵ3, Cζ3, Cη2, Cζ2, Cϵ2 | |||
| Arg-247 | Nϵ | 2.9 | Phe-118 | Cδ1, Cϵ1 | ||||
| GalNAc6S | O-1 | Gly-233 | O | 2.8 | C-4 | Asn-175 | Cγ | |
| O-4 | Thr-235 | Oγ1 | 2.5 | Trp-69 | Cδ2, Cϵ3, Cζ3, Cη2, Cζ2, Cϵ2 | |||
| O-5 | Thr-235 | Oγ1 | 3.2 | Phe-118 | Cϵ1 | |||
| O-7 | Asp-115 | Oδ1 | 3.0 | C-5 | Trp-69 | Cζ3, Cη2, Cζ2, Cϵ2 | ||
| O-1S | Ser-365 | Oγ | 2.9 | C-6 | Trp-245 | Cη2, Cζ2 | ||
| Ser-368 | Oγ | 2.6 | Trp-69 | Cη2, Cζ2 | ||||
| O-2S | Ser-368 | Oγ | 3.0 | Arg-247 | Cζ | |||
| GalNAc6S | C-3 | Trp-69 | Cδ1 | |||||
| C-4 | Tyr-364 | Cϵ1, Cζ | ||||||
| C-5 | Tyr-364 | Cϵ1, Cζ | ||||||
| C-6 | Thr-235 | Cγ2 | ||||||
| Tyr-364 | Cϵ1, Cϵ2, Cζ | |||||||
| Ser-365 | Cβ | |||||||
| C-7 | Trp-69 | Cδ1 | ||||||
| C-8 | Trp-69 | Cδ1 | ||||||
At subsite +1, seven hydrogen bonds by five residues (Asp-115, Gly-233, Thr-235, Ser-365, and Ser-368) as well as stacking and electrostatic interactions with five residues (Trp-69, Thr-235, Tyr-364, Ser-365, and Lys-370) are formed between the enzyme and the GalNAc residue with a sulfate group at the C-6 position (GalNAc6S). Distinct from those interactions between the enzyme and ΔGlcUA, there is a difference in the binding mode between SagUGL (D175N-Δ6S) and BacillusUGL (D88N-Δ0S) to the amino sugar residue (18), although the difference is partly generated from the presence of a sulfate group in the amino sugar (GalNAc6S). Of note, Gly-233 and Thr-235 bound to the pyranose frame of GalNAc6S are located in the flexible loop.
The carbonyl oxygen atom of Gly-233 in the flexible loop is directly hydrogen-bonded to O-1 of GalNAc6S (Fig. 2C and Table 2). In addition, the side chain of Thr-235 in the loop also binds to GalNAc6S through formation of both hydrogen bonds and van der Waals contacts. To clarify the role of Thr-235 in the enzyme reaction, we constructed a site-directed mutant, T235A, and subjected it to an enzyme assay (Table 1). In comparison with WT (Km = 0.100 mm), T235A exhibited less affinity (Km = 1.75 mm) for the substrate Δ6S. Furthermore, in the case of Δ0S as the substrate, no enzyme activity of T235A was detected. These results suggest that interaction of the flexible loop with GalNAc6S in Δ6S or GalNAc in Δ0S is essential to the enzyme reaction.
Sulfate Group-binding Residues
Because SagUGL exhibits a maximal activity toward Δ6S among various glycosaminoglycan disaccharides mostly due to its high affinity for the substrate (Table 1), hereafter we focused on examination of SagUGL recognition of the sulfate group at the C-6 position of the GalNAc residue in the substrate. There are three direct hydrogen bonds between the sulfate group and the enzyme (Table 2): O-1S···Ser-365 Oγ (2.9 Å), O-1S···Ser-368 Oγ (2.6 Å), and O-2S···Ser-368 Oγ (3.0 Å). In addition, a positively charged residue, Lys-370, is situated around the negatively charged sulfate group in the substrate. No water molecules are included around the sulfate group in the D175N-Δ6S complex. The residues Ser-365, Ser-368, and Lys-370 are completely conserved in UGLs of other pathogenic streptococcal species (S. pneumoniae and S. pyogenes) that are highly active on the sulfated substrate Δ6S but not in BacillusUGL, which indicates a preference for the unsulfated substrate Δ0S. In the case of BacillusUGL, Ser-365, Ser-368, and Lys-370 instead correspond to His-339, Gly-342, and Ile-344, respectively. Thus, we constructed three SagUGL mutants, S365H (Ser-365 to His), S368G (Ser-368 to Gly), and K370I (Lys-370 to Ile), which were overexpressed in E. coli cells, and purified each of them to homogeneity. Subsequently, we kinetically analyzed the enzyme characteristics of these mutants (Table 1). Compared with SagUGL WT, all three of these mutants, especially S365H and K370I, showed greatly increased Km values for Δ6S, indicating that the affinity of the mutants for the substrate was drastically reduced. In contrast, the binding affinity of S365H and K370I for Δ0S increased, and their Km values toward Δ0S were determined to be lower than those for the mutants toward Δ6S. These results indicate that S365H and K370I are converted to a BacillusUGL-like enzyme with a preference for unsulfated substrates.
To verify that the Ser-365, Ser-368, and Lys-370 residues are responsible for binding to the sulfate group at the C-6 position of the GalNAc residue in Δ6S, we also carried out the sequence conversion of BacillusUGL to a SagUGL-like enzyme. Three mutants of BacillusUGL, H339S (His-339 to Ser), G342S (Gly-342 to Ser), and I344K (Ile-344 to Lys), were constructed and subjected to an enzyme assay. We determined that the Km values of the three mutants toward Δ6S were lower than that of BacillusUGL WT, demonstrating that the mutants showed higher affinity for Δ6S than WT. To the contrary, the affinity of the mutants for Δ0S was slightly lower than that of WT, although the mutants still exhibited a preference for the unsulfated substrate rather than the sulfated substrate.
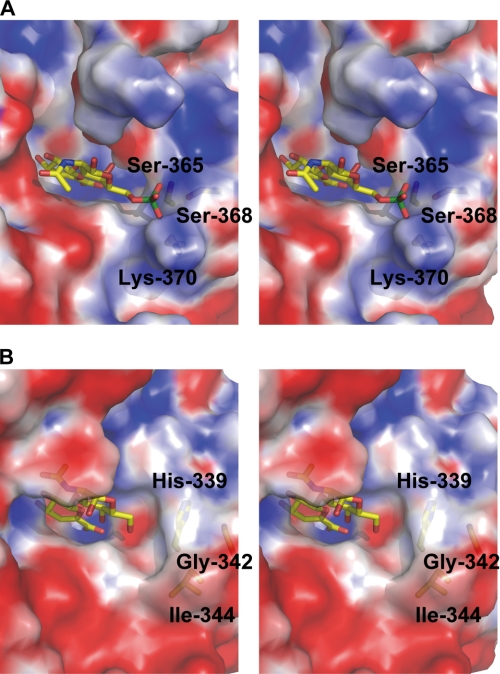
Because sulfate groups are negatively charged, we examined the electrostatic level on the molecular surface of both SagUGL and BacillusUGL. Although the active pocket binding to acidic ΔGlcUA was positively charged on both enzymes, the SagUGL region that interacted with the sulfate group showed a highly positive charge mainly due to the presence of Lys-370 (Fig. 4). This positive charge, which is crucial for sulfate binding, was also supported by mutational analysis of K370I. In contrast, the corresponding region of BacillusUGL was determined to be negatively charged.
FIGURE 4.
Electric charge on molecular surface of UGL active site. A, SagUGL D175N-Δ6S complex. B, BacillusUGL D88N-Δ0S complex. Positive and negative charges at pH 7.0 are colored blue and red, respectively.
In our recent study (22), Arg-236 was suggested to be one of the key residues involved in degradation of unsaturated chondroitin disaccharide with a sulfate group at the C-4 position of the GalNAc residue (Δ4S) through site-directed mutagenesis and in silico modeling. Because Arg-236 corresponds to His-210 in BacillusUGL, we constructed and characterized a mutant of BacillusUGL, H210R. The substrate Δ4S was degraded by BacillusUGL H210R similarly to SagUGL but not by BacillusUGL WT (Fig. 1C). This molecular conversion indicates that Arg-236 of SagUGL is responsible for recognition of the sulfate group at the C-4 position of the GalNAc residue in Δ4S.
In this study, SagUGL was found to act on unsaturated chondroitin, hyaluronan, and heparin disaccharides, demonstrating that this enzyme is involved in complete degradation of various uronate-containing glycosaminoglycans after treatment with polysaccharide lyases. Other than Bacillus and streptococcal enzymes, previous studies have also characterized two UGLs of a soil isolate, Flavobacterium heparinum (37, 38). One of the flavobacterial UGLs was found to be specific for unsaturated heparin disaccharides with a 1→4 glycoside bond, and the other was determined to prefer unsaturated chondroitin and hyaluronan disaccharides with a 1→3 glycoside bond. A single copy of UGL is included for each Streptococcus in the CAZy database, whereas Flavobacterium johnsoniae UW101, a probable close relative of F. heparinum, has two mutually homologous genes that code for UGL in the genome. Both the broad activity of streptococcal UGL toward various glycosaminoglycans and its single gene copy in the genome provide evidence that the enzyme is unique in streptococci responsible for degrading uronate (ΔGlcUA and ΔIdoUA)-containing glycosaminoglycan disaccharides.
In addition to Arg-236, three other residues, Ser-365, Ser-368, and Lys-370, were shown to be involved in binding to sulfate groups in unsaturated chondroitin disaccharides, although the mutant (S368G) exhibited sufficient enzyme activity for binding. Therefore, the arrangement of four residues is identified to form the motif “R-//-SXX(S)XK” that is crucial for degradation of sulfated glycosaminoglycans. The hyphen and slash marks in the motif indicate the presence of over 100 residues in the enzyme. The parentheses indicate that Ser-368 makes little contribution to enzyme activity. This motif is completely conserved in various UGLs from pathogenic species, including Clostridium perfringens, Enterococcus faecalis, Erysipelothrix rhusiopathiae, Mycoplasma fermentans, and streptococci such as S. agalactiae, Streptococcus equi, S. pneumoniae, S. pyogenes, and Streptococcus suis. In contrast, nonpathogenic soil isolates Bacillus sp. GL1 and F. heparinum include no such motif (supplemental Fig. S2). Some bacteria such as F. heparinum (39, 40) and Bacteroides thetaiotaomicron (41) can assimilate glycosaminoglycans as a sole carbon source. Glycosaminoglycan lyases and UGL are also prerequisites for cell growth in these bacteria. Although the motif for degradation of sulfated glycosaminoglycans is not induced in F. heparinum UGL, this bacterium produces some sulfatases involved in desulfation of glycosaminoglycans (42, 43).
The adhesion of pathogenic bacteria to mammalian cells is regarded as a primary mechanism of bacterial infection and plays an important role in the various secondary effects of the infectious process. Glycosaminoglycans present as an important component of the cell surface matrix are typical targets for microbial pathogens that invade host cells (44). Many specific interactions between pathogens and glycosaminoglycans have been described previously (45). Among these bacteria, pathogenic enterococci and streptococci have been shown to interact with various human glycosaminoglycans as follows. E. faecalis binds to heparin and heparan sulfate (46). A cell surface protein, alpha C protein, of S. agalactiae recognizes glycosaminoglycans, likely heparin and heparan sulfate (47). S. pneumoniae binds to chondroitin sulfate, heparin, and heparan sulfate (48). S. pyogenes binds to dermatan sulfate, heparin, and heparan sulfate (49). These targeted glycosaminoglycans (i.e. chondroitin sulfate, heparin, and heparan sulfate) are frequently sulfated; the sulfation level per repeating disaccharide unit was found to be 2.4 for heparin, 1.0 for chondroitin sulfate, and 0.8–1.4 for heparan sulfate (45). Therefore, sulfate groups in glycosaminoglycans are important for interactions between pathogenic bacteria and human cells. These interactions play a crucial role in promoting bacterial adhesion and invasion into human host cells. Although a large number of bacteria encode genes for UGL as well as for glycosaminoglycan lyases classified to the polysaccharide lyase family 8 in the CAZy database, it seems that pathogenic bacteria with the motif R-//-SXX(S)XK in the UGL sequence interact with sulfated glycosaminoglycans. Little knowledge has been accumulated on the molecular mechanism for the interaction between streptococci and glycosaminoglycans with the exception of the alpha C protein; therefore, further studies are necessary for elucidation of the involvement of glycosaminoglycan degradation in streptococcal adhesion to mammal cells.
In conclusion, this is, to our knowledge, the first report on the substrate recognition mechanism of streptococcal UGL through determination of crystal structure of the enzyme-substrate complex. This bacterial UGL can act on unsaturated heparin disaccharides in addition to unsaturated chondroitin and hyaluronan disaccharides because the enzyme triggers hydration of the vinyl ether group in unsaturated uronate residues (ΔGlcUA and ΔIdoUA). Examination of the active site structure of the sulfated substrate-bound UGL mutant and subsequent site-directed mutagenesis suggest that the motif R-//-SXX(S)XK in UGL is prerequisite for degradation of sulfated substrate.
Supplementary Material
Acknowledgments
We thank Drs. S. Baba and N. Mizuno of the Japan Synchrotron Radiation Research Institute (JASRI) for kind help in data collection. Diffraction data for crystals were collected at the BL-38B1 station of SPring-8 (Hyogo, Japan) with the approval of JASRI.
This work was supported in part by grants-in-aid from the Japan Society for the Promotion of Science (to K. M. and W. H.) and by the Targeted Proteins Research Program (to W. H.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. Part of this work was supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to Y. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3ANJ, 3ANI, and 3ANK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GlcUA
- d-glucuronic acid
- IdoUA
- l-iduronic acid
- GlcN
- d-glucosamine
- GlcNAc
- N-acetyl-d-glucosamine
- UGL
- unsaturated glucuronyl hydrolase
- ΔGlcUA
- unsaturated GlcUA
- BacillusUGL
- Bacillus sp. GL1 UGL
- Δ6S
- unsaturated chondroitin disaccharide sulfated at C-6 position of GalNAc residue
- SagUGL
- S. agalactiae UGL
- WT
- wild type
- D175N
- SagUGL mutant with Asp-175 substituted with Asn
- ΔIdoUA
- unsaturated iduronic acid
- T235A
- SagUGL mutant with Thr-235 substituted with Ala
- R236H
- SagUGL mutant with Arg-236 substituted with His
- S365H
- SagUGL mutant with Ser-365 substituted with His
- S368G
- SagUGL mutant with Ser-368 substituted with Gly
- K370I
- SagUGL mutant with Lys-370 substituted with Ile
- H210R
- BacillusUGL mutant with His-210 substituted with Arg
- H339S
- BacillusUGL mutant with His-339 substituted with Ser
- G342S
- BacillusUGL mutant with Gly-342 substituted with Ser
- I344K
- BacillusUGL mutant with Ile-344 substituted with Lys
- Δ0S
- unsaturated sulfate-free chondroitin disaccharide
- Δ4S
- unsaturated chondroitin disaccharide sulfated at C-4 position of GalNAc residue.
REFERENCES
- 1. Ernst S., Langer R., Cooney C. L., Sasisekharan R. (1995) Crit. Rev. Biochem. Mol. Biol. 30, 387–444 [DOI] [PubMed] [Google Scholar]
- 2. Hascall V., Esko J. D. (2009) Essentials of Glycobiology, 2nd Ed., Vol. 15, pp. 219–228, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 3. Gandhi N. S., Mancera R. L. (2008) Chem. Biol. Drug Des. 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 4. Lindahl U., Höök M. (1978) Annu. Rev. Biochem. 47, 385–417 [DOI] [PubMed] [Google Scholar]
- 5. Lauder R. M. (2009) Complement. Ther. Med. 17, 56–62 [DOI] [PubMed] [Google Scholar]
- 6. Jedrzejas M. J. (2007) Cell. Mol. Life Sci. 64, 2799–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Linhardt R. J., Avci F. Y., Toida T., Kim Y. S., Cygler M. (2006) Adv. Pharmacol. 53, 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paton J. C., Andrew P. W., Boulnois G. J., Mitchell T. J. (1993) Annu. Rev. Microbiol. 47, 89–115 [DOI] [PubMed] [Google Scholar]
- 9. Gase K., Ozegowski J., Malke H. (1998) Biochim. Biophys. Acta 1398, 86–98 [DOI] [PubMed] [Google Scholar]
- 10. Berry A. M., Lock R. A., Thomas S. M., Rajan D. P., Hansman D., Paton J. C. (1994) Infect. Immun. 62, 1101–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hynes W. L., Dixon A. R., Walton S. L., Aridgides L. J. (2000) FEMS Microbiol. Lett. 184, 109–112 [DOI] [PubMed] [Google Scholar]
- 12. Pritchard D. G., Lin B., Willingham T. R., Baker J. R. (1994) Arch. Biochem. Biophys. 315, 431–437 [DOI] [PubMed] [Google Scholar]
- 13. Li S., Kelly S. J., Lamani E., Ferraroni M., Jedrzejas M. J. (2000) EMBO J. 19, 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mello L. V., De Groot B. L., Li S., Jedrzejas M. J. (2002) J. Biol. Chem. 277, 36678–36688 [DOI] [PubMed] [Google Scholar]
- 15. Bradbury E. J., Moon L. D., Popat R. J., King V. R., Bennett G. S., Patel P. N., Fawcett J. W., McMahon S. B. (2002) Nature 416, 636–640 [DOI] [PubMed] [Google Scholar]
- 16. Kwok J. C., Afshari F., García-Alías G., Fawcett J. W. (2008) Restor. Neurol. Neurosci. 26, 131–145 [PubMed] [Google Scholar]
- 17. Itoh T., Akao S., Hashimoto W., Mikami B., Murata K. (2004) J. Biol. Chem. 279, 31804–31812 [DOI] [PubMed] [Google Scholar]
- 18. Itoh T., Hashimoto W., Mikami B., Murata K. (2006) J. Biol. Chem. 281, 29807–29816 [DOI] [PubMed] [Google Scholar]
- 19. Mori S., Akao S., Nankai H., Hashimoto W., Mikami B., Murata K. (2003) Protein Expr. Purif. 29, 77–84 [DOI] [PubMed] [Google Scholar]
- 20. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic. Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashimoto W., Kobayashi E., Nankai H., Sato N., Miya T., Kawai S., Murata K. (1999) Arch. Biochem. Biophys. 368, 367–374 [DOI] [PubMed] [Google Scholar]
- 22. Maruyama Y., Nakamichi Y., Itoh T., Mikami B., Hashimoto W., Murata K. (2009) J. Biol. Chem. 284, 18059–18069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27. Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Kabsch W. (1976) Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 31. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 32. DeLano W. L. (2004) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 33. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic. Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanger F., Nicklen S., Coulson A. R. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davies G. J., Wilson K. S., Henrissat B. (1997) Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Myette J. R., Shriver Z., Kiziltepe T., McLean M. W., Venkataraman G., Sasisekharan R. (2002) Biochemistry 41, 7424–7434 [DOI] [PubMed] [Google Scholar]
- 38. Gu K., Linhardt R. J., Laliberté M., Gu K., Zimmermann J. (1995) Biochem. J. 312, 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nader H. B., Porcionatto M. A., Tersariol I. L., Pinhal M. A., Oliveira F. W., Moraes C. T., Dietrich C. P. (1990) J. Biol. Chem. 265, 16807–16813 [PubMed] [Google Scholar]
- 40. Tkalec A. L., Fink D., Blain F., Zhang-Sun G., Laliberte M., Bennett D. C., Gu K., Zimmermann J. J., Su H. (2000) Appl. Environ. Microbiol. 66, 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng Q., Yu M. C., Reeves A. R., Salyers A. A. (1995) J. Bacteriol. 177, 3721–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Myette J. R., Soundararajan V., Shriver Z., Raman R., Sasisekharan R. (2009) J. Biol. Chem. 284, 35177–35188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Myette J. R., Soundararajan V., Behr J., Shriver Z., Raman R., Sasisekharan R. (2009) J. Biol. Chem. 284, 35189–35200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sawitzky D. (1996) Med. Microbiol. Immunol. 184, 155–161 [DOI] [PubMed] [Google Scholar]
- 45. Rostand K. S., Esko J. D. (1997) Infect. Immun. 65, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sava I. G., Zhang F., Toma I., Theilacker C., Li B., Baumert T. F., Holst O., Linhardt R. J., Huebner J. (2009) J. Biol. Chem. 284, 18194–18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aupérin T. C., Bolduc G. R., Baron M. J., Heroux A., Filman D. J., Madoff L. C., Hogle J. M. (2005) J. Biol. Chem. 280, 18245–18252 [DOI] [PubMed] [Google Scholar]
- 48. Tonnaer E. L., Hafmans T. G., Van Kuppevelt T. H., Sanders E. A., Verweij P. E., Curfs J. H. (2006) Microbes Infect. 8, 316–322 [DOI] [PubMed] [Google Scholar]
- 49. Frick I. M., Schmidtchen A., Sjöbring U. (2003) Eur. J. Biochem. 270, 2303–2311 [DOI] [PubMed] [Google Scholar]
- 50. Schuettelkopf A. W., Van Aalten D. M. F. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.