Abstract
IFNγ exhibits potent antitumor effects and plays important roles in the innate immunity against cancer. However, the mechanisms accounting for the antiproliferative effects of IFNγ still remain to be elucidated. We examined the role of Mnk1 (MAPK-interacting protein kinase 1) in IFNγ signaling. Our data demonstrate that IFNγ treatment of sensitive cells results in engagement of Mnk1, activation of its kinase domain, and downstream phosphorylation of the cap-binding protein eIF4E on Ser-209. Such engagement of Mnk1 plays an important role in IFNγ-induced IRF-1 (IFN regulatory factor 1) gene mRNA translation/protein expression and is essential for generation of antiproliferative responses. In studies aimed to determine the role of Mnk1 in the induction of the suppressive effects of IFNs on primitive hematopoietic progenitors, we found that siRNA-mediated Mnk1/2 knockdown results in partial reversal of the suppressive effects of IFNγ on human CD34+-derived myeloid (CFU-GM) and erythroid (BFU-E) progenitors. These findings establish a key role for the Mnk/eIF4E pathway in the regulatory effects of IFNγ on normal hematopoiesis and identify Mnk kinases as important elements in the control of IFNγ-inducible ISG mRNA translation.
Keywords: Antiviral Agents, Interferon, MAP Kinases (MAPKs), Serine-Threonine Protein Kinase, Signal Transduction
Introduction
The only known member of the Type II IFN family, IFNγ, plays an important role in the innate and adaptive immunity against microbial and viral infections and exhibits potent antitumor effects (1–4). IFNγ is a cytokine mainly secreted by T lymphocytes, activated natural killer cells, and antigen-presenting cells such as macrophages and dendritic cells (5, 6) and is known to elicit pleiotropic biological effects on cells and tissues. This cytokine enhances the activity of natural killer cells, facilitates class switching, and regulates immunoglobulin production by B cells (5–7). In addition, it regulates survival and proliferation of T cells, modulates the activity of antigen presenting cells and, under certain circumstances, can promote differentiation of several distinct cell types (5–7). Importantly, IFNγ facilitates immune responses to tumor cells, although it also inhibits angiogenesis and exerts direct anti-proliferative effects on a number of tumor cells (8). Thus, considering the broad effects of IFNγ, understanding the cellular mechanisms that regulate its biological effects is highly relevant in advancing our overall understanding of the mechanisms of innate immunity against cancer and viral infections.
Previous studies have established that IFNγ transduces signals by binding to its cell surface receptor, which is composed of two distinct subunits; the IFNγ receptor 1 and 2 chains, which are constitutively associated with the JAK family members JAK1 and JAK2 (reviewed in Refs. 2 and 3). Binding of IFNγ to its receptor results in interactions between the receptor chains leading to the phosphorylation of the STAT1 transcriptional activator, followed by its dimerization, translocation to the nucleus, and activation of gene transcription by IFNγ-activated sequences (GAS)2 (2, 3). Beyond the classic JAK-STAT pathway, the transcriptional response to IFNγ also involves IFNγ-activated transcription elements (9) that are controlled by the transcription factor CCAAT enhancer-binding protein-β. Notably, the activity of CCAAT enhancer-binding protein-β is positively regulated by the MAP kinases Erk1 and Erk2 (10). There has been also some previous evidence implicating protein kinase pathways in the generation of cellular responses to IFNγ. The phosphatidylinositol 3-kinase regulates transcriptional regulation by IFNγ (11, 12), whereas the Akt/mTOR pathway plays an important role downstream of phosphatidylinositol 3-kinase, promoting mRNA translation of ISGs (13, 14). PKC family members PKCδ, PKCθ, and PKCϵ have been also shown to play important roles in IFNγ signaling (11, 15, 16). Additionally there is evidence for important functional roles for MAPK pathways in the induction of IFNγ responses (17–20).
We determined whether Mnks (MAPK-interacting protein kinases) 1 and 2 are activated during engagement of the Type II IFN receptor and participate in the generation of IFNγ responses. Mnk1 is a downstream effector for both the p38 MAPK and Erk1/2 pathways and along with the related Mnk2 regulates phosphorylation of eIF4E (21, 22). Our data show that IFNγ treatment results in activation of Mnk1 and its downstream target eIF4E in an Mek/Erk-dependent manner. In studies using dual Mnk1/Mnk2 knock-out cells, we found that Mnk activity is essential for IFNγ-dependent mRNA translation of IRF-1 (interferon regulatory factor 1) and plays a critical role in the generation of growth inhibitory responses by the Type II IFN receptor. Altogether, our findings identify Mnk1 as a novel element required for mRNA translation of ISGs and generation of IFNγ antiproliferative responses.
MATERIALS AND METHODS
Antibodies, Cell Lines, and Reagents
The antibodies against p-Mnk1 (Thr-197/202), Mnk1, p-eIF4E (Ser-209), eIF4E, p-Erk1/2 (Thr-202/Tyr-204), Erk1//2, pSTAT1 (Tyr-701), and p-STAT1 (Ser-727) were obtained from Cell Signaling Technology (Danvers, MA). The antibodies against STAT1 and IRF-1 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The antibody against GAPDH was obtained from Millipore (Billerica, MA). Human and mouse IFNγ were obtained from PBL Interferon Source (Piscataway, NJ). U937 cells were grown in RPMI-1060 supplemented with 10% (v/v) fetal bovine serum and antibiotics. CD34+ cells were obtained from either Lonza (Basel, Switzerland) or Stemcell Technologies (Vancouver, Canada). Immortalized Mnk1−/− MEFs, Mnk2−/− MEFs, and Mnk1/Mnk2−/− MEFs were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum and antibiotics. The Mnk inhibitor CGP57380 and the Mek1/2 inhibitor U0126 were obtained from Calbiochem (Darmstadt, Germany). The siRNAs targeting human Mnk1 and Mnk2 as well as nontargeting siRNAs were obtained from Dharmacon.
Cell Lysis and Immunoblotting
The cells were treated with IFNγ (103 or 104 IU/ml) for the indicated times and were then lysed in phosphorylation lysis buffer as described in our previous studies (23, 24). In experiments using pharmacological inhibitors, the cells were pretreated with CGP57380 (5–10 μm) or U0126 (10 μm) for 1 h followed by IFNγ treatment for the indicated time in the continuous presence of the inhibitors; the cells were then lysed in phosphorylation lysis buffer. Immunoblotting was performed using an ECL method, as in our previous studies (25–27).
Luciferase Reporter Assay
Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were transfected with an 8× GAS luciferase construct (containing a luciferase reporter gene with eight GAS elements linked to a minimal prolactin promoter) and a constitutive β-galactosidase expression vector using the SuperFect transfection reagent according to the protocol of the manufacturer (Qiagen). The 8× GAS construct was kindly provided by Dr. Christofer Glass (University of California, San Diego, CA) (28). 48 h post-transfection, the triplicate cultures were either left untreated or treated with mouse IFNγ (1,000 units/ml) for 6 h. The cells were then lysed, and the luciferase activity was measured as per the manufacturer's instructions (Promega, Madison, WI) described in previous studies (23). The luciferase activity was then normalized utilizing the β-galactosidase activity for each sample.
Quantitative RT-PCR
The Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were either left untreated or were with IFNγ (1,000 units/ml) for 6 h and RNA was isolated using the RNeasy kit (Qiagen). Cellular mRNA was reverse transcribed into cDNA using the Omniscript RT kit and oligo(dT) primer (Qiagen) as described previously (16). Quantitative PCR was carried out as described previously (14). Commercially available FAM-labeled probes and primers (Applied Biosystems) to determine Irf-1 mRNA expression were used. GAPDH was used for normalization. The mRNA amplification was calculated as described previously (14), and the data were plotted as the fold increase as compared with untreated samples.
Isolation of Polysomal Fractions
The Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were treated with mouse IFNγ (1,000 units/ml) for 48 h, and polysomal fractionation was performed as in our previous studies (13, 24).
Hematopoietic Progenitor Cell Assays
CD34+ cells were transfected with either control nontargeting siRNA or siRNA specific to human Mnk1 and/or Mnk2 (Dharmacon, Lafayette, CO). In some experiments the cells were also treated with the Mnk inhibitor CGP57380 (5 μm) or diluent control DMSO. The cells were then cultured in a methylcellulose assay system in the absence or presence of human IFNγ (1,000 units/ml) for 14 days, and erythroid (BFU-E) or myeloid (CFU-GM) colonies were scored as described previously (27, 29). In the experiments to assess the effects of Mnk inhibition on leukemic CFU-L progenitors, U937 cells were transfected with either control nontargeting siRNA or siRNAs targeting Mnk1, Mnk2, or both or treated with either DMSO or CGP57380 (2.5 μm). The cells were then cultured in a methylcellulose assay system in the absence or presence of human IFNγ (1,000 units/ml) for 7 days, and colony-forming units were scored as described previously (30).
RESULTS
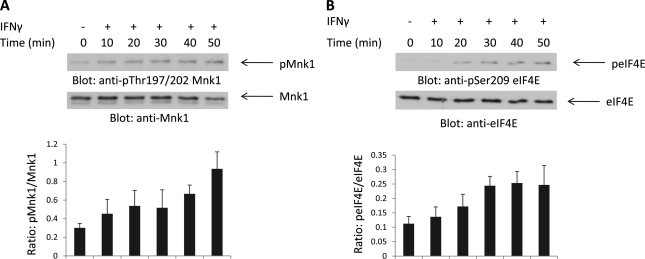
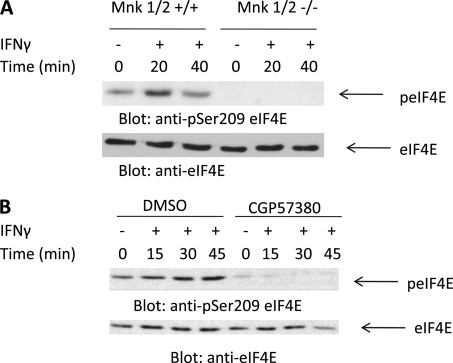
In initial studies we examined whether IFNγ induces phosphorylation/activation of Mnk1. For these, sensitive U937 cells were treated with human IFNγ for different times, and cell lysates were analyzed by SDS-PAGE and immunoblotted with an antibody that recognizes the phosphorylated/activated form of Mnk1. IFNγ treatment resulted in rapid phosphorylation/activation of Mnk1, which was noticeable at 10 min and was still detectable 50 min post-IFNγ treatment (Fig. 1A), suggesting the involvement of this kinase in IFNγ signaling. We also examined whether IFNγ treatment regulates phosphorylation of the downstream effector of Mnk1, eIF4E, at Ser-209, which is the Mnk phosphorylation site in other systems (31–34). As shown in Fig. 1B, IFNγ treatment of U937 cells resulted in phosphorylation of eIF4E (Fig. 1B). To determine whether Mnk activity is essential for Type II IFN-dependent phosphorylation of eIF4E, we examined whether such induction is blocked in immortalized MEFs from mice with targeted disruption of both the Mnk1 and Mnk2 genes (34). Mnk1/2+/+ and Mnk1/2−/− MEFs were serum-starved and then treated with mouse IFNγ for different times. As seen in Fig. 2A, IFNγ treatment resulted in strong phosphorylation of eIF4E in Mnk1/2+/+ MEFs, whereas this phosphorylation was not inducible in the Mnk1/2−/− MEFs (Fig. 2A). Consistent with this, in experiments in which the effects of pharmacological inhibition of Mnk were evaluated in U937 leukemic cells, we found that the IFNγ-dependent phosphorylation of eIF4E is Mnk1/2-dependent (Fig. 2B).
FIGURE 1.
IFNγ-mediated engagement of Mnk1 and eIF4E. A, U937 cells were treated with human IFNγ for the indicated times. Total lysates were separated by SDS-PAGE and immunoblotted with an antibody against phosphorylated Mnk1 (Thr-197/202). The same blot was stripped and reprobed with an antibody against total Mnk1. The signals for pMnk1 and total Mnk1 from three independent experiments (including the one shown in A) were quantitated by densitometry, and the intensity of pMnk1 relative to total Mnk1 expression was calculated. The data are expressed as the means of ratios of pMnk1 to Mnk1 levels ± S.E. for each experimental condition. B, U937 cells were treated with human IFNγ for the indicated times. Total lysates were separated by SDS-PAGE and immunoblotted with an antibody against phosphorylated eIF4E (Ser-209). The same blot was stripped and reprobed with an antibody against total eIF4E. The signals for peIF4E and total eIF4E from four independent experiments (including the one shown in B) were quantitated by densitometry, and the intensity of peIF4E relative to total eIF4E expression was calculated. The data are expressed as the means of ratios of peIF4E to eIF4E levels ± S.E. for each experimental condition.
FIGURE 2.
Mnk1/2 is required for IFNγ-mediated engagement of eIF4E. A, Mnk1/2+/+ and Mnk1/2−/− MEFs were serum-starved overnight and treated with mouse IFNγ for the indicated times. Equal amounts of lysates were separated by SDS-PAGE followed by immunoblotting with an antibody against phosphorylated eIF4E (S209). The same blot was stripped and reprobed with an antibody against total eIF4E. B, U937 cells were incubated with either DMSO or CGP57380 for 60 min and were then treated with human IFNγ for the indicated times. Equal amounts of lysates were separated by SDS-PAGE and then immunoblotted with an antibody against phosphorylated eIF4E (Ser-209). The blot was then stripped and reprobed with an antibody against eIF4E.
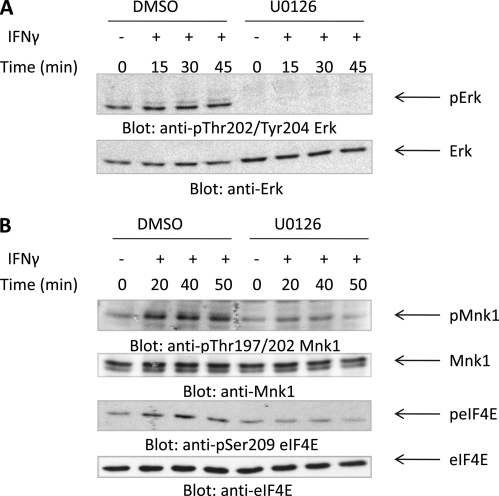
We subsequently sought to define upstream signaling events required for activation of Mnk1 and eIF4E during engagement of the Type II IFN receptor. Mnk1 has been shown to be phosphorylated by the Erk1 and Erk2 kinases in response to various stimuli (31–33). We examined the phosphorylation of Mnk1 and Mnk2 in U937 cells in the presence of the Mek/Erk inhibitor U0126. U937 cells were pretreated with either DMSO (control) or U0126 and then treated with IFNγ for the indicated times. IFNγ has been shown to result in the engagement of Erk1 and Erk2 in various systems (35, 36). As expected, U0126 inhibited IFNγ-dependent phosphorylation/activation of Erk1/2 (Fig. 3A). In DMSO pretreated cells, Mnk1 and eIF4E were phosphorylated by IFNγ, whereas in U0126-treated cells, the activation of Mnk1 and eIF4E was suppressed (Fig. 3B), indicating that the Mek/Erk pathway is required for IFNγ-mediated activation of Mnk1/eIF4E.
FIGURE 3.
IFNγ-mediated engagement of Mnk1 and eIF4E is Mek/Erk-dependent. A, U937 cells were incubated with either DMSO or U0126 for 60 min and were then treated with human IFNγ for the indicated times. Equal amounts of protein were separated by SDS-PAGE and then immunoblotted with antibodies against phosphorylated Erk1/2 (Thr-202/Tyr-204). The blot was then stripped and reprobed with an antibody against total Erk1/2. B, U937 cells were incubated with either DMSO or U0126 for 60 min and were then treated with human IFNγ for the indicated times. Equal amounts of lysates were separated by SDS-PAGE and then immunoblotted with antibodies against phosphorylated Mnk1 (Thr-197/202) or against phosphorylated eIF4E (Ser-209). The respective blots were then stripped and reprobed with antibodies against total Mnk1 or total eIF4E.
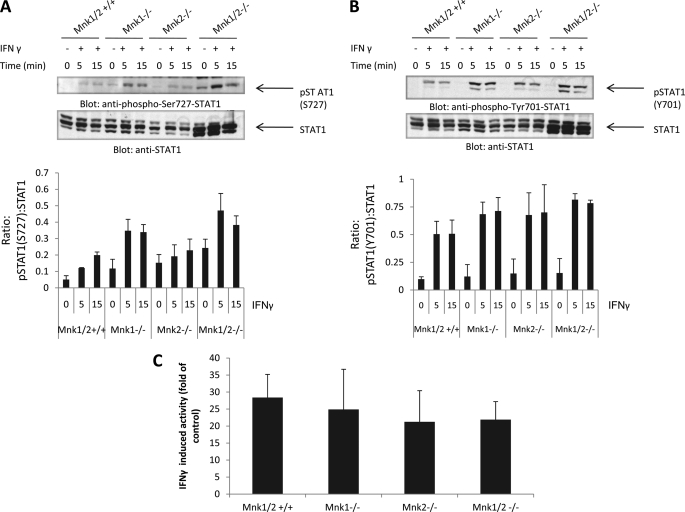
We next examined the role of Mnk1 as a putative mediator of IFNγ signaling events. We initially determined whether Mnk1 plays a role in the regulation of phosphorylation/activation of STAT1 and IFNγ-regulated gene transcription. STAT1 phosphorylation by IFNγ on both Tyr-701 and Ser-727 was intact in the absence of either Mnk1 or Mnk2 and in the absence of both Mnk1 and Mnk2 (Fig. 4, A and B). Consistent with this, transcriptional activation via GAS elements was intact in luciferase promoter assays (Fig. 4C), establishing that Mnk kinases do not play roles in the control of IFNγ-induced STAT1 activation or gene transcription. Interestingly, there was some increase seen in the IFNγ-induced serine phosphorylation of STAT1 (Fig. 4A), suggesting a compensatory effect, but the precise mechanism and relevance of this finding remains to be defined in future studies.
FIGURE 4.
Mnk1 and Mnk2 are not required for IFNγ-mediated engagement of STAT1 or activation of transcription via GAS elements. A, Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were treated with IFNγ for the indicated times. Equal amounts of total cell lysates were separated by SDS-PAGE and immunoblotted with antibodies against phosphorylated STAT1 (Ser-727). The blot was stripped and reprobed with antibody against total STAT1. The signals for pSTAT1 (Ser-727) and total STAT1 from three independent experiments (including the one shown in A) were quantitated by densitometry, and the intensity of pSTAT1 (S727) relative to total STAT1 expression was calculated. The data are expressed as the means of ratios of pSTAT1(S727) to STAT1 levels ± S.E. for each experimental condition. B, Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were treated with IFNγ for the indicated times. Equal amounts of total cell lysates were separated by SDS-PAGE and immunoblotted with antibodies against phosphorylated STAT1 (Tyr-701). The blot was stripped and reprobed with antibody against total STAT1. The signals for pSTAT1 (Tyr-701) and total STAT1 from three independent experiments (including the one shown in B) were quantitated by densitometry, and the intensity of pSTAT1 (Tyr-701) relative to total STAT1 expression was calculated. The data are expressed as the means of ratios of pSTAT1 (Tyr-701) to STAT1 levels ± S.E. for each experimental condition. C, Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were transfected with an 8× GAS luciferase construct. 48 h post-transfection, the cells were incubated with or without IFNγ for 6 h. The cells were then harvested and assayed for luciferase activity. The data are expressed as fold increases in luciferase activity in response to IFNγ treatment over control untreated cells and represent the means ± S.E. of four independent experiments.
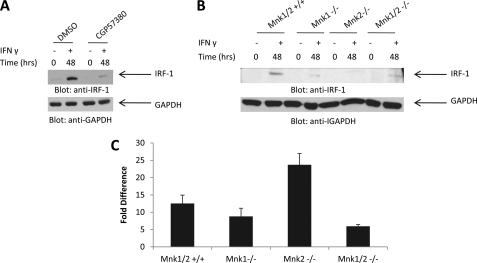
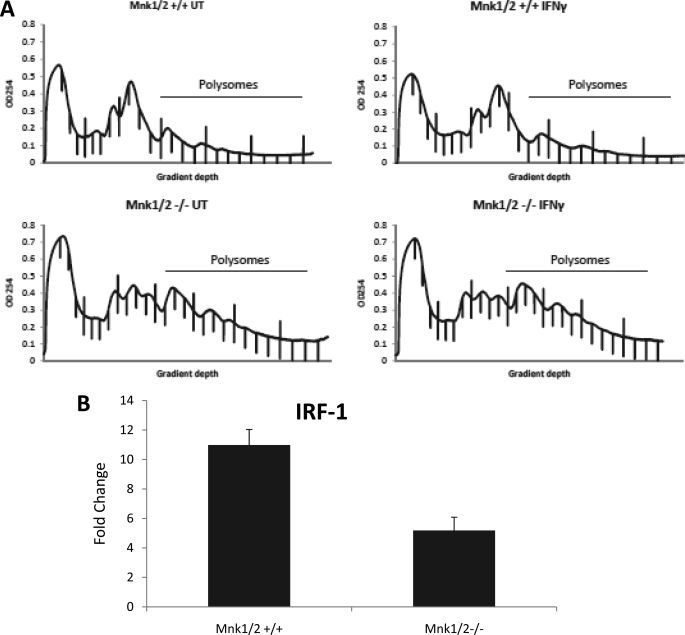
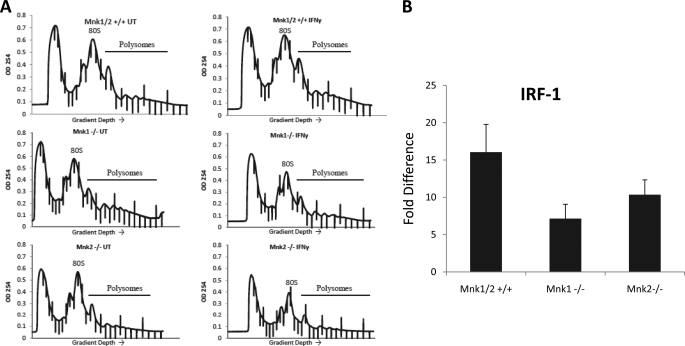
IFNγ up-regulates the expression of the IRF-1, which plays an important role in mediating the biological effects of IFNγ (7). To better understand the relevance of the Mnk pathway in the induction of IFNγ responses, we examined and compared the effects of IFNγ on IRF-1 gene transcription and protein expression in single or double Mnk1 and Mnk2 knock-outs and parental cells. IRF-1 protein was clearly IFNγ-inducible in parental MEFs, but such induction was attenuated in the Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs (Fig. 5B). Similarly, IFNγ-inducible IRF-1 protein expression was defective in cells treated with the Mnk inhibitor CGP57380 (Fig. 5A). These results indicate that Mnk1/2 expression/activity plays an important role in IFNγ-mediated protein expression of IRF-1. Interestingly, when IFNγ-dependent mRNA expression for IRF-1 was assessed in the different Mnk knock-out MEFs, we noticed a significant decrease in Mnk1/2−/− MEFs, whereas there were minimal effects in Mnk1 single knock-out MEFs (Fig. 5C). On the other hand, there was an increase in Mnk2−/− MEFs (Fig. 5C). Thus, although IRF-1 protein expression in response to IFNγ is defective in single or double Mnk1/Mnk2 knock-outs, there are variable profiles of IRF-1 transcriptional activation/mRNA expression seen in the different Mnk knock-out cells, suggesting that a different mechanism, possibly regulation of mRNA translation, primarily accounts for defective protein expression. To directly determine the role of the Mnk pathway in IRF-1 mRNA translation, Mnk1/2+/+ and Mnk1/2−/− MEFs were either left untreated or treated with IFNγ for 48 h. The cells were then subjected to hypotonic lysis and resolved on a sucrose gradient followed by RNA extraction from the polysomal fractions, and IRF-1 polysomal mRNA induction was analyzed by quantitative RT-PCR. As seen in Fig. 6, IRF-1 mRNA translation was attenuated in the Mnk1/2−/− MEFs, indicating that Mnk1 and Mnk2 play important roles in translation of IRF-1 mRNA. Similarly, defective IRF-1 mRNA expression was noticeable in studies using single Mnk1 or single Mnk2 knock-out MEFs, indicating involvement of both kinases in this regulation (Fig. 7).
FIGURE 5.
Requirement of Mnk1 and Mnk2 for IFNγ-induced IRF1 protein expression. A, U937 cells were incubated with either DMSO or CGP57380 for 60 min followed by treatment with human IFNγ for the indicated times. Equal amounts of lysates were separated by SDS-PAGE and immunoblotted with an antibody against IRF-1, and the same blot was also probed with an antibody against GAPDH. B, Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were treated with mouse IFNγ for the indicated times. Equal amounts of lysates were separated by SDS-PAGE and immunoblotted with an antibody against IRF-1, and the same blot was also probed with an antibody against GAPDH. C, Mnk1/2+/+, Mnk1−/−, Mnk2−/−, and Mnk1/2−/− MEFs were treated with mouse IFNγ. The expression of IRF-1 mRNA was assessed by quantitative RT-PCR, using GAPDH as a control. The data are expressed as the fold induction over corresponding untreated samples and represent the means ± S.E. of five independent experiments.
FIGURE 6.
Mnk kinases are required for IFNγ-induced IRF1 mRNA translation. A, Mnk1/2+/+ and Mnk1/2−/− MEFs were either left untreated or treated with mouse IFNγ. The cells were subjected to hypotonic lysis followed by separation on a 10–50% sucrose gradient, and the optical density at 254 nm (OD 254) was recorded. The optical density at 254 nm is shown as a function of gradient depth for each treatment. B, IRF-1 mRNA expression in the polysomal fractions was determined by quantitative RT-PCR, using GAPDH for normalization. The data are expressed as fold increases in the IFNγ-treated samples over untreated samples and represent the means ± S.E. of three independent experiments.
FIGURE 7.
Mnk1 and Mnk2 in IFNγ-induced mRNA translation. A, Mnk1/2+/+, Mnk1−/−, and Mnk2−/− MEFs were either left untreated or treated with mouse IFNγ. The cells were subjected to hypotonic lysis followed by separation on a 10–50% sucrose gradient, and the optical density at 254 nm (OD 254) was recorded. The optical density at 254 nm is shown as a function of gradient depth for each treatment. B, IRF-1 mRNA expression in the polysomal fractions was determined by quantitative RT-PCR, using GAPDH for normalization. The data are expressed as fold increases in the IFNγ-treated samples over untreated samples and represent the means ± S.E. of four independent experiments.
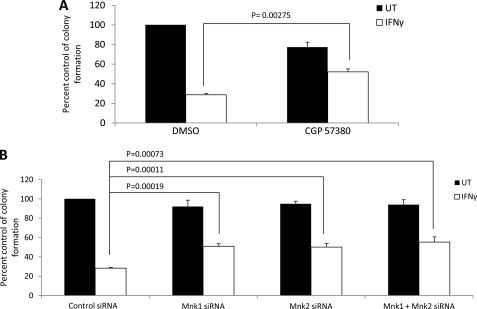
In subsequent studies, we directly examined the effects of Mnk1 and Mnk2 in the generation of IFNγ-dependent growth inhibitory responses. Leukemic U937 cells were treated with IFNγ, in the presence or absence of the Mnk inhibitor CGP57380, and leukemic progenitor (CFU-L) colony formation was assessed. As shown in Fig. 8A, simultaneous treatment with the Mnk inhibitor partially reversed the antiproliferative effects of IFNγ, suggesting a role for Mnk kinases in the generation of IFNγ-dependent antiproliferative responses. To confirm these results, we also used specific siRNAs targeting Mnk1 and/or Mnk2 and determined the effects of these knockdowns on IFNγ-mediated suppression of leukemic progenitor colony formation. There was partial reversal of the suppressive effects of IFNγ on leukemic progenitor colony formation (Fig. 8B), definitively establishing a requirement for Mnk1 in the process.
FIGURE 8.
Mnk kinases mediate the antiproliferative effects of IFNγ on U937 cells. A, U937 cells were incubated in clonogenic assays in methylcellulose with or without human IFNγ, in the presence of DMSO or CGP57380, as indicated. Leukemic CFU-L colonies were scored, and the data are expressed as percentages of control DMSO treated colonies and represent the means ± S.E. of three independent experiments. Paired t test analysis showed p = 0.00275 for the combination of DMSO and IFNγ versus the combination of CGP57380 and IFNγ. B, U937 cells were transfected with the indicated siRNAs and plated in a methylcellulose assay system in the absence or presence of human IFNγ. The data are expressed as percentages of control siRNA transfected cell-derived colony formation and represent the means ± S.E. of six independent experiments. Paired t test analysis showed p = 0.00019 for the combination of control siRNA and IFNγ versus the combination of Mnk1-specific siRNA and IFNγ; p = 0.00011 for the combination of control siRNA and IFNγ versus the combination of Mnk2-specific siRNA and IFNγ; and p = 0.00073 for the combination of control siRNA and IFNγ versus the combination of Mnk1- and Mnk2-specific siRNAs and IFNγ. UT, untreated.
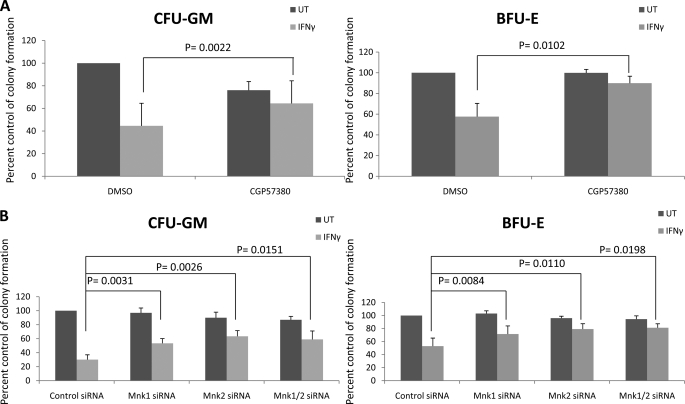
We also performed studies aimed at examining the roles of Mnk1 and Mnk2 as mediators of the suppressive effects of IFNγ on normal human hematopoiesis. Human CD34+ bone marrow cells were treated with IFNγ in the presence or absence of CGP57380, and normal myeloid (CFU-GM) or erythroid (BFU-E) colony formation was assessed. The suppressive effects of IFNγ on either CFU-GM and/or BFU-E (Fig. 9A) colonies were partially reversed by the CGP57380. Importantly, such reversal of myelossuppressive responses was also seen when CD34+ cells were transfected with siRNAs targeting Mnk1, Mnk2, or both (Fig. 9B), definitively establishing a role for the Mnk pathway as a mediator of the suppressive effects of IFNγ on normal hematopoiesis.
FIGURE 9.
Mnk kinases are essential for the generation of the myelossuppressive effects of IFNγ. A, CD34+ cells derived from normal bone marrow were incubated in clonogenic assays in methylcellulose with or without human IFNγ, in the presence of DMSO or CGP57380, as indicated. CFU-GM and BFU-E progenitor colonies were scored after 14 days in culture. The data are expressed as percentages of control colony formation from DMSO treated cells and represent the means ± S.E. of five independent experiments. Paired t test analysis showed p = 0.0022 for the combination of DMSO and IFNγ versus the combination of CGP57380 and IFNγ for CFU-GM colonies. Paired t test analysis showed p = 0.0102 for the combination of DMSO and IFNγ versus the combination of CGP57380 and IFNγ for BFU-E colonies. B, CD34+ cells derived from normal bone marrow were transfected with the indicated siRNAs and were then plated in a methylcellulose assay system, in the absence or presence of human IFNγ, as indicated. CFU-GM and BFU-E progenitor colonies were scored after 14 days in culture. The data are expressed as percentages of control colony formation from control siRNA transfected cells and represent the means ± S.E. of five independent experiments. Paired t test analysis showed p = 0.0031 for the combination of control siRNA and IFNγ versus the combination of Mnk1 siRNA and IFNγ for CFU-GM colonies; and p = 0.0084 for the combination of control siRNAs and IFNγ versus the combination of Mnk1 siRNA and IFNγ for BFU-E colonies. Paired t test analysis showed p = 0.0026 for the combination of control siRNA and IFNγ versus the combination of Mnk2 siRNA and IFNγ for CFU-GM colonies; and p = 0.011 for the combination of control siRNAs and IFNγ versus the combination of Mnk2 siRNA and IFNγ for BFU-E colonies. Paired t test analysis showed p = 0.0151 for the combination of control siRNA and IFNγ versus the combination of Mnk1 and Mnk2 siRNAs and IFNγ for CFU-GM colonies and p = 0.0198 for the combination of control siRNAs and IFNγ versus the combination of Mnk1 and Mnk2 siRNA and IFNγ for BFU-E colonies. UT, untreated.
DISCUSSION
Extensive work over the years has established the relevance of Mnk kinases in stress-activated signaling cascades and as mediators of growth factor and pro-inflammatory signals (37). A major target for Mnk kinases is the initiation factor eIF4E, which undergoes Mnk-mediated phosphorylation on serine 209 (34, 37). Phosphorylation of eIF4E at this site has been shown in different studies to be of importance in the initiation of mRNA translation for certain genes, as well as for oncogenic transformation and malignant cell proliferation (38–42). Mnk kinases have been also implicated in the production of TNF, IL-6, and monocyte chemoattractant protein-1 in response to LPS (43, 44), whereas more recent studies have provided evidence that, under certain circumstances, Mnk1 is involved in cap-independent translation (45, 46). The importance of the Mnk/eIF4E pathway in tumorigenesis was definitively established in recent work using knock-in mice expressing a mutant form of eIF4E, which cannot undergo phosphorylation on serine 209 (47). These studies demonstrated that phosphorylation on this site is required for tumorigenesis in a prostate cancer mouse model (47). Remarkably, eIF4E phosphorylation on serine 209 was also found to correlate with a high Gleason score, high levels of MMP3 expression, and disease progression in prostate cancer patients (47). Other recent studies demonstrated that Mnk1/2 activity is required for tumor development in the Lck-Pten mouse model (48), underscoring the relevance of the Mnk/eIF4E pathway in malignant tumor development.
The only Type II IFN, IFNγ, exhibits pleiotropic biological functions, including immunomodulatory, antitumor, and antiviral activities (49). This cytokine plays key roles in the generation of antineoplastic activities and in the immune surveillance against tumors (49). Interestingly, IFNγ has also been implicated in diverse pathophysiological states, ranging from bone marrow failure (50) to arteritis (51) or atherosclerosis (52). Such a functional diversity of responses suggests the existence and coordination of multiple cellular pathways activated by the Type II IFN receptor. It should be noted that the Type II IFN receptor is structurally and functionally distinct from the Type I IFN receptor, and IFNγ has only minimal identity with the family of Type I IFNs (53).
In the current study, we examined whether Mnk kinases are engaged in signaling by the Type II (IFNγ) receptor and their functional relevance in the induction of Type II IFN-mediated mRNA translation of regulated genes and generation of IFNγ responses. Our data demonstrate that Mnk1 is phosphorylated/activated in an IFNγ-inducible manner in sensitive cells and regulates downstream phosphorylation of eIF4E on serine 209. In studies using double knock-out MEFs for both Mnk1 and Mnk2, we identified a requirement for Mnks in the phosphorylation/activation of eIF4E. Moreover, mRNA translation of the IRF-1 gene and expression of the IRF-1 protein was defective in Mnk1 and/or Mnk2 MEFs, indicating a requirement for the Mnk pathway in IRF-1 protein expression. Notably, IRF-1 has been shown to promote induction of anti-tumor activities in a variety of tumors (54–56) and to play an important role in mediating the antiproliferative effects of IFNγ in malignant mesothelioma cell lines (57) and in oligodendrocyte progenitor cells (58).
Our findings establish that the function of Mnk kinases is essential for generation of the suppressive effects of IFNγ in normal human CD34+-derived erythroid (BFU-E) and myeloid (CFU-GM) progenitors, defining a critical and essential role for the pathway in the regulation of normal hematopoiesis by IFNγ. Based on our data, these regulatory effects of the Mnk pathway may reflect the requirement for Mnks in IRF-1 mRNA translation, because previous studies have also shown that IRF-1 plays an important role in mediating IFNγ-induced inhibitory responses in normal human hematopoietic CD34+ progenitor cells (59). Thus, as in the case of Type I IFNs (60), Mnk kinases appear to play key and essential roles in mRNA translation of certain ISGs and generation of signals required for antiproliferative responses and the suppression of hematopoiesis. The requirement of the Mnk/eIF4E pathway in the generation of IFN-dependent antiproliferative responses and suppressive effects on normal and leukemic hematopoiesis is in some ways surprising, because there is extensive evidence implicating this pathway in tumorigenesis and malignant cell proliferation (38–42, 47, 48). It is possible that, in contrast to growth factors and oncogenes, this pathway is used in a selective way by the Type II IFN receptor for mRNA translation of genes, such as IRF-1, that mediate growth inhibitory responses. Coordination of gene transcription by IFN-activated JAK-STAT pathways and IFN-dependent engagement of the Mnk/eIF4E pathway may lead to expression of proteins that mediate growth suppression. Importantly, it is possible that the Type II IFN receptor competes with growth factor receptors for use of this pathway, depriving them of a pathway essential for mitogenic responses and tumorigenesis.
In addition to eIF4E, Mnks are known to regulate the function of several other signaling proteins and effectors. Mnk1 is implicated in the phosphorylation of the heterogenous ribonuclear protein A1 (61), as well as PSF (the polypyrimidine tract-binding protein-associated splicing factor), both of which are AU-rich element-binding proteins that interact with the TNFα mRNA (62). In addition, Mnk1 phosphorylates Sprouty 2, a negative regulator of Erk signaling (63). The phosphorylation of Sprouty 2 by Mnk1 regulates its stability and prevents its degradation, providing an important control point for activation of the Erk pathway by FGF (63). Additionally, Mnk1 has been shown to phosphorylate cytosolic phospholipase A2 on serine 727, resulting in its activation and subsequent arachidonate release (64). The potential regulation of such Mnk-controlled pathways by the IFNγ receptor and implications that such pathways may have in IFNγ-induced, Mnk-dependent, hematopoietic suppression remains to be directly examined in future studies. Nevertheless, independent of the identity of putative downstream effectors that may be involved in the process, our data suggest a central role for this kinase in myelossuppression. Beyond its involvement in Type I (60) and II IFN signaling, Mnk activity is essential for mRNA translation of the gene for TNFα (62), a cytokine that exhibits potent myelossuppressive effects (65, 66). Thus, it is possible that, as is the case for p38 MAPK (17, 25, 66–68), Mnk is a central integrator of signals for the generation of myelossuppressive responses in the regulation of hematopoiesis. If this hypothesis proves to be correct, it would raise the possibility of studies to target this pathway for the treatment of bone marrow failure syndromes involving overproduction of myelossuppressive cytokines (50), and this should be addressed in future studies.
This work was supported, in whole or in part, by National Institutes of Health Grants CA77816, CA121192, AG029138, and HL08294. This work was also supported by a Merit Review grant from the Department of Veterans Affairs and a Malkin Scholars Award.
- GAS
- IFNγ-activated sequences
- MEF
- mouse embryonic fibroblast
- CFU-GM
- colony forming unit-granulocyte monocyte
- BFU-E
- burst forming unit-erythroid.
REFERENCES
- 1. Isaacs A., Lindenmann J. (1957) Proc. R. Soc. Lond. B Biol. Sci. 147, 258–267 [PubMed] [Google Scholar]
- 2. Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 3. Borden E. C., Sen G. C., Uze G., Silverman R. H., Ransohoff R. M., Foster G. R., Stark G. R. (2007) Nat. Rev. Drug. Discov. 6, 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schoenborn J. R., Wilson C. B. (2007) Adv. Immunol. 96, 41–101 [DOI] [PubMed] [Google Scholar]
- 5. Schroder K., Hertzog P. J., Ravasi T., Hume D. A. (2004) J. Leukocyte Biol. 75, 163–189 [DOI] [PubMed] [Google Scholar]
- 6. Boehm U., Klamp T., Groot M., Howard J. C. (1997) Annu. Rev. Immunol. 15, 749–795 [DOI] [PubMed] [Google Scholar]
- 7. Saha B., Jyothi Prasanna S., Chandrasekar B., Nandi D. (2010) Cytokine 50, 1–14 [DOI] [PubMed] [Google Scholar]
- 8. Ikeda H., Old L. J., Schreiber R. D. (2002) Cytokine Growth Factor Rev. 13, 95–109 [DOI] [PubMed] [Google Scholar]
- 9. Weihua X., Kolla V., Kalvakolanu D. V. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu J., Roy S. K., Shapiro P. S., Rodig S. R., Reddy S. P., Platanias L. C., Schreiber R. D., Kalvakolanu D. V. (2001) J. Biol. Chem. 276, 287–297 [DOI] [PubMed] [Google Scholar]
- 11. Choudhury G. G. (2004) J. Biol. Chem. 279, 27399–27409 [DOI] [PubMed] [Google Scholar]
- 12. Nguyen H., Ramana C. V., Bayes J., Stark G. R. (2001) J. Biol. Chem. 276, 33361–33368 [DOI] [PubMed] [Google Scholar]
- 13. Kaur S., Sassano A., Dolniak B., Joshi S., Majchrzak-Kita B., Baker D. P., Hay N., Fish E. N., Platanias L. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaur S., Lal L., Sassano A., Majchrzak-Kita B., Srikanth M., Baker D. P., Petroulakis E., Hay N., Sonenberg N., Fish E. N., Platanias L. C. (2007) J. Biol. Chem. 282, 1757–1768 [DOI] [PubMed] [Google Scholar]
- 15. Srivastava K. K., Batra S., Sassano A., Li Y., Majchrzak B., Kiyokawa H., Altman A., Fish E. N., Platanias L. C. (2004) J. Biol. Chem. 279, 29911–29920 [DOI] [PubMed] [Google Scholar]
- 16. Deb D. K., Sassano A., Lekmine F., Majchrzak B., Verma A., Kambhampati S., Uddin S., Rahman A., Fish E. N., Platanias L. C. (2003) J. Immunol. 171, 267–273 [DOI] [PubMed] [Google Scholar]
- 17. Verma A., Deb D. K., Sassano A., Kambhampati S., Wickrema A., Uddin S., Mohindru M., Van Besien K., Platanias L. C. (2002) J. Immunol. 168, 5984–5988 [DOI] [PubMed] [Google Scholar]
- 18. Horiuchi M., Itoh A., Pleasure D., Itoh T. (2006) J. Biol. Chem. 281, 20095–20106 [DOI] [PubMed] [Google Scholar]
- 19. Roy S. K., Hu J., Meng Q., Xia Y., Shapiro P. S., Reddy S. P., Platanias L. C., Lindner D. J., Johnson P. F., Pritchard C., Pagés G., Pouyssegur J., Kalvakolanu D. V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salmenperä P., Hämäläinen S., Hukkanen M., Kankuri E. (2003) Am. J. Physiol. Cell Physiol. 284, C1133–C1139 [DOI] [PubMed] [Google Scholar]
- 21. Fukunaga R., Hunter T. (1997) EMBO J. 16, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waskiewicz A. J., Flynn A., Proud C. G., Cooper J. A. (1997) EMBO J. 16, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uddin S., Lekmine F., Sharma N., Majchrzak B., Mayer I., Young P. R., Bokoch G. M., Fish E. N., Platanias L. C. (2000) J. Biol. Chem. 275, 27634–27640 [DOI] [PubMed] [Google Scholar]
- 24. Kaur S., Sassano A., Joseph A. M., Majchrzak-Kita B., Eklund E. A., Verma A., Brachmann S. M., Fish E. N., Platanias L. C. (2008) J. Immunol. 181, 7316–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uddin S., Yenush L., Sun X. J., Sweet M. E., White M. F., Platanias L. C. (1995) J. Biol. Chem. 270, 15938–15941 [DOI] [PubMed] [Google Scholar]
- 26. Ahmad S., Alsayed Y. M., Druker B. J., Platanias L. C. (1997) J. Biol. Chem. 272, 29991–29994 [DOI] [PubMed] [Google Scholar]
- 27. Verma A., Deb D. K., Sassano A., Uddin S., Varga J., Wickrema A., Platanias L. C. (2002) J. Biol. Chem. 277, 7726–7735 [DOI] [PubMed] [Google Scholar]
- 28. Horvai A. E., Xu L., Korzus E., Brard G., Kalafus D., Mullen T. M., Rose D. W., Rosenfeld M. G., Glass C. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1074–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayer I. A., Verma A., Grumbach I. M., Uddin S., Lekmine F., Ravandi F., Majchrzak B., Fujita S., Fish E. N., Platanias L. C. (2001) J. Biol. Chem. 276, 28570–28577 [DOI] [PubMed] [Google Scholar]
- 30. Dolniak B., Katsoulidis E., Carayol N., Altman J. K., Redig A. J., Tallman M. S., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Platanias L. C. (2008) J. Biol. Chem. 283, 12034–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X., Flynn A., Waskiewicz A. J., Webb B. L., Vries R. G., Baines I. A., Cooper J. A., Proud C. G. (1998) J. Biol. Chem. 273, 9373–9377 [DOI] [PubMed] [Google Scholar]
- 32. Waskiewicz A. J., Johnson J. C., Penn B., Mahalingam M., Kimball S. R., Cooper J. A. (1999) Mol. Cell. Biol. 19, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scheper G. C., Morrice N. A., Kleijn M., Proud C. G. (2001) Mol. Cell. Biol. 21, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meng Q., Raha A., Roy S., Hu J., Kalvakolanu D. V. (2005) J. Immunol. 174, 6203–6211 [DOI] [PubMed] [Google Scholar]
- 36. Valledor A. F., Sánchez-Tilló E., Arpa L., Park J. M., Caelles C., Lloberas J., Celada A. (2008) J. Immunol. 180, 4523–4529 [DOI] [PubMed] [Google Scholar]
- 37. Buxade M., Parra-Palau J. L., Proud C. G. (2008) Front. Biosci. 13, 5359–5373 [DOI] [PubMed] [Google Scholar]
- 38. Flynn A., Proud C. G. (1995) J. Biol. Chem. 270, 21684–21688 [DOI] [PubMed] [Google Scholar]
- 39. Pyronnet S., Dostie J., Sonenberg N. (2001) Genes. Dev. 15, 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishida M., Ishida T., Nakashima H., Miho N., Miyagawa K., Chayama K., Oshima T., Kambe M., Yoshizumi M. (2003) Circ. Res. 93, 1218–1224 [DOI] [PubMed] [Google Scholar]
- 41. Wheater M. J., Johnson P. W., Blaydes J. P. (2010) Cancer Biol. Ther. 10, 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bianchini A., Loiarro M., Bielli P., Busà R., Paronetto M. P., Loreni F., Geremia R., Sette C. (2008) Carcinogenesis 29, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 43. Andersson K., Sundler R. (2006) Cytokine 33, 52–57 [DOI] [PubMed] [Google Scholar]
- 44. Rowlett R. M., Chrestensen C. A., Nyce M., Harp M. G., Pelo J. W., Cominelli F., Ernst P. B., Pizarro T. T., Sturgill T. W., Worthington M. T. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G452–G459 [DOI] [PubMed] [Google Scholar]
- 45. Origanti S., Shantz L. M. (2007) Cancer Res. 67, 4834–4842 [DOI] [PubMed] [Google Scholar]
- 46. Goetz C., Everson R. G., Zhang L. C., Gromeier M. (2010) Mol. Ther. 18, 1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Furic L., Rong L., Larsson O., Koumakpayi I. H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L. A., Pandolfi P. P., Saad F., Sonenberg N. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ueda T., Sasaki M., Elia A. J., Chio II., Hamada K., Fukunaga R., Mak T. W. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13984–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller C. H., Maher S. G., Young H. A. (2009) Ann. N.Y. Acad. Sci. 1182, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Young N. S., Calado R. T., Scheinberg P. (2006) Blood 108, 2509–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weyand C. M., Younge B. R., Goronzy J. J. (2011) Curr. Opin. Rheumatol. 23, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McLaren J. E., Ramji D. P. (2009) Cytokine Growth Factor Rev. 20, 125–135 [DOI] [PubMed] [Google Scholar]
- 53. Pestka S., Krause C. D., Walter M. R. (2004) Immunol. Rev. 202, 8–32 [DOI] [PubMed] [Google Scholar]
- 54. Bouker K. B., Skaar T. C., Riggins R. B., Harburger D. S., Fernandez D. R., Zwart A., Wang A., Clarke R. (2005) Carcinogenesis 26, 1527–1535 [DOI] [PubMed] [Google Scholar]
- 55. Bowie M. L., Ibarra C., Seewalt V. L. (2008) Adv. Exp. Med. Biol. 617, 367–374 [DOI] [PubMed] [Google Scholar]
- 56. Kröger A., Dallügge A., Kirchhoff S., Hauser H. (2003) Oncogene 22, 1045–1056 [DOI] [PubMed] [Google Scholar]
- 57. Buard A., Vivo C., Monnet I., Boutin C., Pilatte Y., Jaurand M. C. (1998) Cancer Res. 58, 840–847 [PubMed] [Google Scholar]
- 58. Wang Y., Ren Z., Tao D., Tilwalli S., Goswami R., Balabanov R. (2010) Glia 58, 195–208 [DOI] [PubMed] [Google Scholar]
- 59. Sato T., Selleri C., Young N. S., Maciejewski J. P. (1995) Blood 86, 3373–3380 [PubMed] [Google Scholar]
- 60. Joshi S., Kaur S., Redig A. J., Goldsborough K., David K., Ueda T., Watanabe-Fukunaga R., Baker D. P., Fish E. N., Fukunaga R., Platanias L. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12097–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Buxadé M., Parra J. L., Rousseau S., Shpiro N., Marquez R., Morrice N., Bain J., Espel E., Proud C. G. (2005) Immunity 23, 177–189 [DOI] [PubMed] [Google Scholar]
- 62. Buxadé M., Morrice N., Krebs D. L., Proud C. G. (2008) J. Biol. Chem. 283, 57–65 [DOI] [PubMed] [Google Scholar]
- 63. DaSilva J., Xu L., Kim H. J., Miller W. T., Bar-Sagi D. (2006) Mol. Cell. Biol. 26, 1898–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hefner Y., Borsch-Haubold A. G., Murakami M., Wilde J. I., Pasquet S., Schieltz D., Ghomashchi F., Yates J. R., 3rd, Armstrong C. G., Paterson A., Cohen P., Fukunaga R., Hunter T., Kudo I., Watson S. P., Gelb M. H. (2000) J. Biol. Chem. 275, 37542–37551 [DOI] [PubMed] [Google Scholar]
- 65. Broxmeyer H. E., Williams D. E., Lu L., Cooper S., Anderson S. L., Beyer G. S., Hoffman R., Rubin B. Y. (1986) J. Immunol. 136, 4487–4495 [PubMed] [Google Scholar]
- 66. Katsoulidis E., Li Y., Yoon P., Sassano A., Altman J., Kannan-Thulasiraman P., Balasubramanian L., Parmar S., Varga J., Tallman M. S., Verma A., Platanias L. C. (2005) Cancer Res. 65, 9029–9037 [DOI] [PubMed] [Google Scholar]
- 67. Platanias L. C. (2003) Pharmacol. Ther. 98, 129–142 [DOI] [PubMed] [Google Scholar]
- 68. Platanias L. C. (2003) Blood 101, 4667–4679 [DOI] [PubMed] [Google Scholar]