Abstract
The inhibitory T-cell surface-expressed receptor, cytotoxic T lymphocyte-associated antigen-4 (CTLA-4), which belongs to the class of cell surface proteins phosphorylated by extrinsic tyrosine kinases that also includes antigen receptors, binds the related ligands, B7-1 and B7-2, expressed on antigen-presenting cells. Conformational changes are commonly invoked to explain ligand-induced “triggering” of this class of receptors. Crystal structures of ligand-bound CTLA-4 have been reported, but not the apo form, precluding analysis of the structural changes accompanying ligand binding. The 1.8-Å resolution structure of an apo human CTLA-4 homodimer emphasizes the shared evolutionary history of the CTLA-4/CD28 subgroup of the immunoglobulin superfamily and the antigen receptors. The ligand-bound and unbound forms of both CTLA-4 and B7-1 are remarkably similar, in marked contrast to B7-2, whose binding to CTLA-4 has elements of induced fit. Isothermal titration calorimetry reveals that ligand binding by CTLA-4 is enthalpically driven and accompanied by unfavorable entropic changes. The similarity of the thermodynamic parameters determined for the interactions of CTLA-4 with B7-1 and B7-2 suggests that the binding is not highly specific, but the conformational changes observed for B7-2 binding suggest some level of selectivity. The new structure establishes that rigid-body ligand interactions are capable of triggering CTLA-4 phosphorylation by extrinsic kinase(s).
Keywords: Cell Surface Receptor, Crystal Structure, Phosphotyrosine Receptor, Receptor Structure-Function, Signal Transduction, Conformational Change, Receptor Triggering
Introduction
Interactions of leukocyte cell surface proteins dominate the earliest stages of adaptive immune responses. The key elements of the T-cell antigen recognition apparatus are now characterized (1) and include the T-cell antigen receptor (TCR),6 the co-receptors CD4 and CD8, the adhesion protein CD2, the phosphatase CD45 and the CD28 family proteins. CD28-related proteins comprise a set of small, T-cell expressed receptors that bind a distinct group of small, related, cross-reactive ligands (reviewed in Refs. 2–4). The best studied of these, CD28 and CTLA-4, cross-react with shared ligands, B7-1 (CD80) and B7-2 (CD86) expressed on antigen presenting cells (3, 4), forming a complex system whose signaling outcomes are determined by the affinity and stoichiometry of the interactions, by the timing of expression of the proteins, and by competition between the receptors for ligands (5). CD28 is constitutively expressed on most resting human T-cells and B7-2 is rapidly induced on antigen presenting cells early in immune responses, whereas the expression of B7-1 and CTLA-4 generally occurs after a delay of 24–48 h (6).
In addition to sharing binding partners, the two pairs of genes encoding CTLA-4 and CD28, and B7-1 and B7-2, are each very closely linked on separate chromosomes in humans and mice (7, 8). This, and the similar domain organization and limited sequence homology of the proteins reveal an evolutionary history involving gene duplication. CTLA-4, like CD28, is expressed at the cell surface as a covalent homodimer of single V-set immunoglobulin superfamily (IgSF) domains. B7-1 and B7-2 are comprised of single pairs of V-set and C1-set IgSF domains (reviewed in Ref. 4). CTLA-4 binds bivalently to B7-1 and B7-2 (9, 10), whereas CD28 is monovalent (11). In solution, B7-2 binds CD28 and CTLA-4 more weakly than B7-1, whereas relative to its CTLA-4-binding affinity, B7-2 binds CD28 more effectively than B7-1 (11). Studies suggesting (12) and then confirming (13, 14) that B7-1 dimerizes at the cell surface indicate that inhibitory signaling by CTLA-4 is likely to be avidity enhanced by the formation of one-dimensional arrays of B7-1 and CTLA-4 homodimers. Simulations of complex assembly suggest that B7-2 is the dominant ligand of CD28 and B7-1 ligates most of the CTLA-4 (5), in broad agreement with observations of the behavior of the proteins at the synapse (15).
In the absence of CD28-dependent signaling, naive T-cells enter an anergic state when triggered via their TCR (reviewed in Refs. 3 and 16). CTLA-4, on the other hand, delivers powerful inhibition of T-cell responses as illustrated by the development of a lethal lymphoproliferative disorder in CTLA-4-deficient mice (17). Neither CTLA-4 nor CD28 have any intrinsic catalytic activity but, along with antigen receptors and numerous other immune signaling proteins, belong to the class of receptors phosphorylated on cytoplasmic tyrosine residues by extrinsic tyrosine kinases, such as the Src kinases Lck, Fyn, and Lyn, and resting lymphocyte kinase (18–20). Tyrosines that initiate inhibitory signaling when phosphorylated generally have a motif referred to as an immunoreceptor tyrosine-based inhibition motif (ITIM), which has the characteristic pattern (I/V/L)XYXX(L/V), whereas activating motifs (ITAMs) are defined by the sequence YXX(L/I)-(X)6–8-YXX(L/I) (reviewed in Refs. 21 and 22). A third motif, i.e. the immunoreceptor tyrosine-based switch motif (ITSM) has the sequence TXYXX(V/I) and is considered to have intermediate signaling character. CTLA-4 and CD28 are unusual in that their tyrosine phosphorylation sites do not conform to any of these consensus sequences. The downstream signaling consequences of ligand binding by CTLA-4 are unclear. It is proposed that phosphorylation by Lck or Fyn (23) might stabilize CTLA-4 expression at the cell surface by preventing association with clathrin-coated pits (18, 19, 24) or allow the binding of downstream mediators such as phosphatidylinositol 3-kinase (PI 3-kinase) (20) or the phosphatase SHP-2 (23, 25), although this is controversial (4). How phosphorylation of the receptor is “triggered” by ligand binding in the first instance is also unknown but broadly expected to involve conformational changes in the receptor or its oligomerization (4). Phosphorylation of CD28 results in the recruitment of PI 3-kinase, propagating activator signaling (26, 27).
Because of their importance in the immune response and, increasingly, in the context of immunotherapy (28, 29), the structures of costimulatory proteins have been studied in considerable detail. The human CTLA-4 monomer was first studied at low resolution using NMR methods (30), and this was followed by crystallographic analyses of B7-1 homodimers (12), B7-1 homodimers and B7-2 monomers complexed with CTLA-4 (9, 10), and superagonistic antibody-bound CD28 monomers (31). Taken at face value, the NMR structure of the monomer versus the complexes implied that CTLA-4 undergoes structural changes local to the ligand-binding region upon ligand binding, whereas the crystal structure of murine CTLA-4 monomers (32) raised the possibility that the CTLA-4 homodimer undergoes radical rearrangements. Missing from the analysis has been a high-resolution structure of native apo-CTLA-4 homodimers. Our analysis at 1.8-Å resolution now reveals that both CTLA-4 and B7-1 are remarkably unaffected by complex formation in contrast to B7-2, which undergoes induced fit. We also produce further confirmation of the shared evolutionary history of the CTLA-4/CD28 subgroup of the IgSF and antigen receptors, first noted in structural analyses of CD28 (31). Consideration of all the structural data for the CTLA-4/CD28 subgroup suggests that neither conformational rearrangements nor receptor oligomerization are part of the triggering mechanism for receptors dependent on extrinsic tyrosine kinases.
EXPERIMENTAL PROCEDURES
Protein Expression and Crystallization
Details of the crystallization of the human CTLA-4 dimer are to be published elsewhere.7 Briefly, chimeric cDNA encoding, in the following order, residues 1–161 of the extracellular region of human CTLA-4(CD152), including the signal peptide sequence, a thrombin cleavage site, the heavy chain constant domains 2 and 3 of murine IgG1 (residues 103–323 of the secreted protein; Fc), and a C-terminal Lys-(His)6 tag, was cloned into the pEE14 expression vector as described previously (33). The correct sequence of the construct was confirmed by dideoxy sequencing. The expression vector encoding the chimeric CTLA-4Fc construct was then transfected into Chinese hamster ovary (CHO)-K1 cells using the FuGENE 6 transfection reagent (Roche Applied Science). Clones resistant to 25 μm methionine sulfoximine were selected and screened for the secretion of CTLA-4Fc into the tissue culture supernatant using dot blots and/or Western blots. The best clone was selected for large scale production of protein using large scale (2–5 liter) tissue culture flasks (Cell Factories; Nunc, Roskilde, Denmark) in the presence of 10 μm kifunensine (Ref. 34; Toronto Research Chemicals, North York, Ontario, Canada). The CTLA-4Fc secreted into the tissue culture supernatant was harvested after ∼4 weeks and the protein was extracted by metal-chelate chromatography using nickel-nitrilotriacetic acid-agarose (Qiagen, West Sussex, UK) and further purified by size exclusion chromatography. Removal of the Fc from CTLA-4Fc was achieved by cleaving the protein with thrombin and re-extracting with nickel-nitrilotriacetic acid-agarose to deplete free Fc and uncleaved CTLA-4Fc. The homodimer was then deglycosylated with endo Hf (New England Biolabs, Hitchin, UK) at room temperature for 3 h and purified by lectin affinity chromatography and gel filtration as described (35) prior to crystallization.
Initial conditions for the crystallization of the CTLA-4 homodimer in the nanoliter scale involved sitting-drop vapor diffusion screening using a sparse matrix crystallization screening kit at 295 K (Hampton Research, Laguna Niguel, CA). Crystals appeared after 20 h under a variety of conditions, most containing low molecular weight polyethylene glycols.
Crystals were transferred to a freezing solution made by adding glycerol to the precipitant solution to a final concentration of 30% before being cooled to 100 K for data collection. Data were collected at beamline I04 at the Diamond Light Source, UK. X-ray data were processed and scaled with the HKL suite (36). Phases were determined by molecular replacement using Phaser (37) and the coordinates of CTLA-4 (mol C of Stamper et al. (9)). Initially a single domain was identified, following which a second round of rotation and translation searches found the second domain of the homodimer. The structure was subsequently refined in Refmac5 (38) and Phenix (39). Refinement was carried out with 5% of the data set aside for Rfree cross-validation. Water molecules were located in Coot (40). Structural superpositions were performed also using Coot (40). Figures were prepared using PyMol (41).
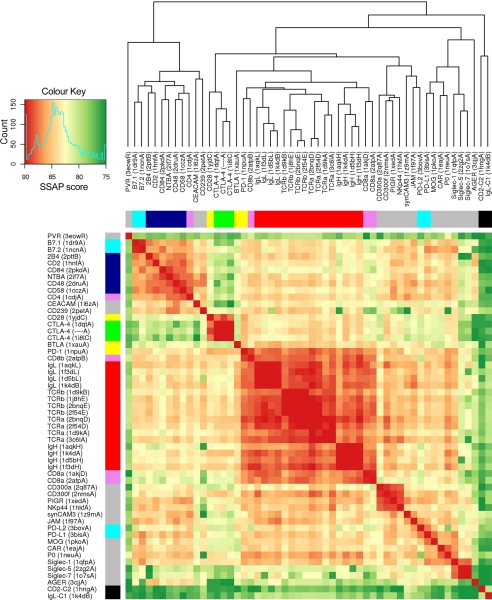
Structural Comparisons of V-set IgSF Domains
52 V-set IgSF domains of known structures from the Protein Data Bank (PDB) were carefully selected to represent all major functional subgroups in this set. Of these, 40 had pre-defined boundaries available from the CATH data base, a public resource that provides definitions and classifications of individual domains from PDB structures (42). For the remaining 12 entries in the list, immunoglobulin superfamily domain boundaries were identified manually. An outgroup, consisting of the C2-set IgSF domain of CD2 and the C1-set IgSF domain of an immunoglobulin light chain, was also included. All possible pairwise structural alignments involving the 54 domains were calculated using the structure comparison program SSAP (43). SSAP produces a score that measures the similarity between two aligned structures. SSAP scores were used to construct a dissimilarity matrix of the 54 IgSF domains, which was in turn used to generate the structural similarity heatmap shown in Fig. 3. Domains in columns and rows of the heatmap were ordered according to a structural similarity dendrogram, which was computed by complete hierarchical clustering of the dissimilarity matrix. Both heatmap and dendrogram were created using the R statistical package (r-project.org). The same pairwise structural alignments were also used to produce a similar heatmap (supplemental Fig. S2) based on another structural similarity score, called SIMAX, computed as follows: SIMAX is a normalized root mean square (r.m.s.) deviation score, where the r.m.s. deviation of the superposition, max(L1,L2) is the length in amino acids of the largest domain in the superposition, and Nmat is the number of aligned residue pairs (44). All data are provided as standard output of SSAP.
FIGURE 3.
Structural similarity dendrogram and heatmap for a selection of V-set IgSF domains, based on SSAP scores. Domains are identified by the common short names of the proteins in which they occur, with the identifier and chain letter for the PDB structure used for comparison in brackets. Domains were ordered according to a dendrogram constructed using complete hierarchical clustering based on scores obtained from pairwise structural comparisons using SSAP (43). Colors in the heatmap range from red to dark green for high and low structural similarity, respectively (see color key). A histogram in the color key panel shows the frequency distribution of SSAP scores in the set. Color bars on the left and upper sides of the heatmap indicate known functional or family groupings: green, CTLA-4; yellow, other members of the costimulatory/inhibitory family; red, antigen receptors; pink, coreceptors; dark blue, CD2 family; light blue, B7 family; black, outgroup (C1 and C2 set IgSF domains); gray, other V-set proteins.
Isothermal Titration Calorimetry
The extracellular regions of B7-1 (sB7-1) and CTLA-4 (CTLA-4Fc) were expressed as a histidine-tagged and IgG1-Fc fusion protein in CHO-K1 cells, respectively (45), and the V-set ligand-binding domain of B7-2 (B7-2v) was refolded from inclusion bodies expressed in bacteria (46). Isothermal titration calorimetry experiments were performed using the MCS or VP-ITC systems (MicroCal Inc., MA) as described (47, 48). In a typical experiment, sB7-1 or B7-2v at 0.2 mm were added in 20 × 15-μl injections to a 0.02 mm solution of CTLA-4 in the 1.463-ml calorimeter cell at the temperatures indicated. The resulting data were fitted as described (47) after subtracting the heats of dilution resulting from addition of sB7-1 or B7-2v to buffer and buffer to CTLA-4Fc, determined in separate control experiments. Titration data were fitted using a non-linear least squares curve-fitting algorithm with three floating variables: stoichiometry, association constant (Ka), and change of enthalpy on binding (ΔHobs). All binding data were analyzed by fitting the binding isotherm to a single independent binding site model using Origin software provided with the ITC (MicroCal Inc.). ITC gives a complete thermodynamic characterization of an interaction based on equation: ΔG = −RT × ln(Ka) = ΔHobs − TΔS, where R is the gas constant, T is the absolute temperature, and ΔG, ΔHobs, and ΔS are the standard free energy, observed enthalpy, and entropy changes going from unbound to bound states, respectively. All experiments were done in triplicate.
RESULTS
Structure Determination
Apo-CTLA-4 monomers (residues 1–110, mature polypeptide numbering) were initially expressed in Escherichia coli following the method of Chang et al. (49). We obtained crystals with the same cell dimensions and space group as those obtained by Chang et al. (49), but prior to or during crystallization the monomer underwent a strand swap creating a non-physiological, non-covalent dimer (50). Residues 1–125 of CTLA-4 were therefore expressed as a fusion protein with thrombin-cleavable murine IgG Fc in Chinese hamster ovary cells in the presence of the glycosylation inhibitor kifunensine (34). Following deglycosylation with endoglycosidase H, the homodimer yielded orthorhombic crystals with unit cell dimensions of a = 43.86 Å, b = 51.46 Å, and c = 102.85 Å, and space group P212121, which diffracted to Bragg spacings of 1.8 Å.7 Phases were determined by molecular replacement using CTLA-4 monomer (PDB code 1i8l, mol C; Ref. 9) and Phaser (37), and the structure was refined in Refmac5 (38) and Phenix (39). Water molecules were located in Coot (40). Sugar residues at the glycosylation sites were readily modeled as β-d-N-acetylglucose linkages. Model refinement and processing statistics are summarized under supplemental Table S1.
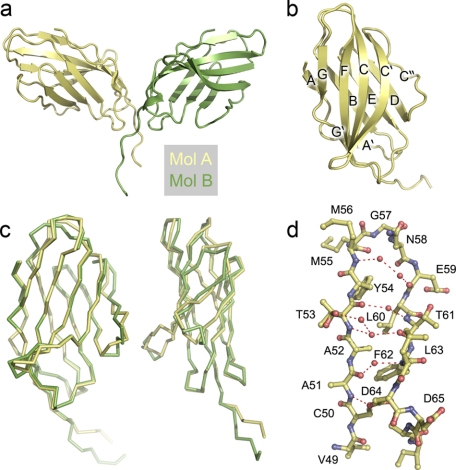
The asymmetric unit of the unit cell is comprised of the disulfide-bonded CTLA-4 homodimer (Fig. 1a). Electron density was continuous from Met3 to Pro123 for both monomers and in mol B it extended to Ser125, i.e. to the penultimate residue of the extracellular region. The higher average temperature factors in mol B (45.423 versus 22.257 Å2) reflected, at least to some degree, anisotropy and mosaicity in the diffraction data and differences in crystal packing. Electron density from the final model, showing the networked water spanning the C′ and C″ strands, is presented in supplemental Fig. S1. In the following we compare the apo-CTLA-4 monomers with four copies of the monomer in the CTLA-4/B7-1 (10) and CTLA-4/B7-2 (9) crystals, and with the single copy in the lipocalin/CTLA-4 crystals (51).
FIGURE 1.
The structure of apo-CTLA-4. a, the asymmetric unit of the CTLA-4 crystals containing the apo-CTLA-4 homodimer. Mol A, which had the lowest B factors is colored yellow and mol B, green. b, secondary structure of the IgSF V-set domain of CTLA-4 (mol A). c, two orthogonal views of mol A and mol B of CTLA-4 shown as α-carbon representations following superposition with Coot (40). The position of the domains is similar to b. d, detail of the structural water in the region of the C′ and C″ strands and the C′C″ loop of mol A. The residue numbers are shown.
V-set IgSF Domain of Apo-CTLA-4
The two apo human CTLA-4 monomers in the asymmetric unit exhibit the two-layer AGFCC′ and ABED IgSF domain topology initially assigned by NMR analysis (Fig. 1b) (30). The IgSF domains are attached to the membrane via a short eight-residue “stalk” that starts at Pro119 (the first residue beyond the G strand) and is disulfide-bonded at Cys122. Overall, the two halves of the apo-CTLA-4 homodimer are less similar to one another than they are similar to their complexed counterparts (Fig. 1c; discussed also below). Mol B of the apo structure exhibits greater overall similarity with molecules D and C of the B7-1 complex (r.m.s. differences = 0.91 Å and 0.91 Å, 116 and 117 residues, respectively) and molecules C and D of the B7-2 complex (r.m.s. deviation = 0.95 Å and 1.02 Å for 116 and 108 residues, respectively) than with mol A of apo-CTLA-4 (r.m.s. deviation = 1.13 Å for 117 residues). Mol A is also more similar to the complexed forms of CTLA-4 than to apo mol B (supplemental Table S2). The principal sites of mol A and B differences are in the BC and C′C″ loops and position of the C″ strand, and in the organization of the stalk region beyond Pro119 (Fig. 1c). Minor differences in position are seen in the CC′ and FG loops. Tyr100 in chain A occupies a different position to that in chain B (discussed below). Tyr100 is adjacent to Pro101, which is involved in the cis-trans isomerization seen in the strand-swapped structure of bacterially expressed CTLA-4 (50).
As expected, apo human CTLA-4 exhibits much less similarity with the murine CTLA-4 monomer (r.m.s. deviation = 1.46 Å for 116 amino acids of mol A of the murine structure). In particular, in murine CTLA-4, rather than H bonding exclusively with the C′ strand, the C″ strand forms two classical and two weaker inter-strand hydrogen (H) bonds with strand D, whereas in human CTLA-4 the shortest distance of any C″ amide to neighboring strand D carbonyl oxygens (5.0 Å in mol A) is beyond H-bonding distance. Strand C″ of apo human CTLA-4 is connected to main chain atoms of strand C′ over distances of ∼5 Å or larger by a network of five water molecules (Fig. 1d and supplemental Fig. S1), and to strand D by two water molecules and H bonds between the side chain of Glu59 and main chain nitrogen of Ser72, and between the side chain of Ser72 and main chain oxygen and nitrogen of Asn58 and Leu60, respectively. This is the most variable region of monomer structure (Fig. 1c).
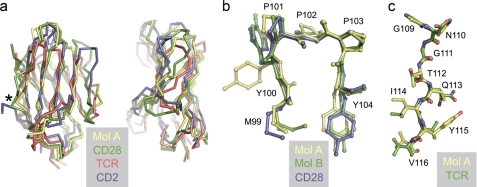
Automated structure comparisons with mol A of apo-CTLA-4 using DALI (52) identified CD28 (PDB code 1yjd; r.m.s. deviation = 1.62 Å for 107 residues) and programmed death-1 (PD-1; PDB code 1npu; r.m.s. deviation = 1.82 Å) as the structures most similar to CTLA-4; similar results were obtained for mol B (supplemental Table S2). The important differences between CTLA-4 and CD28 are: (i) the insertion in CD28 of Glu108, and the replacement of Gly107 in CTLA-4 with Asn, creating the G-strand kink of CD28; (ii) the longer AB, BC, and CC′ loops of CTLA-4 and shorter C″D loop; and (iii) the positioning of the C′C″ loop in CD28 allowing H-bonding of the C′ and C″ strands (Fig. 2a). The most important similarity is the largely identical conformation of the Met99-Tyr-Pro-Pro-Pro-Tyr (MYPPPY) sequence of the FG loop forming the core of the ligand-binding surface of both proteins (Fig. 2b). We have located 228 waters in the entire structure and find that the ligand-binding surface of CTLA-4 is unusually dry. Only a single water molecule, forming an H-bond with the main chain carbonyl of Met99, is common to the binding sites of mol A and B (not shown). The conformation of the MYPPPY sequence is such that relatively few atoms are available for H-bonding: the carbonyls of Pro101 and Pro103 are buried, and the main chain amide and carbonyl of Tyr100 form intramolecular H-bonds. There are no other mol B-associated waters in the region of the binding face. In mol A, an additional water molecule forms an H-bond with Tyr104 (not shown). It should be noted, however, that the MYPPPY sequences at both ends of the homodimer are involved in hydrophobic lattice interactions with neighboring molecules that may exclude water.
FIGURE 2.
The CTLA-4 IgSF V-set domain is more similar to that of antigen receptors than adhesion molecules. a, α-carbon representations of the IgSF V-set domains of apo-CTLA-4 (mol A; yellow), CD28 (PDB code 1yjd; green), TCR V domain (PDB code 3c6l; red), and CD2 (PDB code 1hnf; blue) in two orthogonal views, after superposition with Coot. The conserved G-strand β-bulge is indicated by the asterisk. b, superpositions, using Coot, of the apo-CTLA-4 mol A (yellow) and mol B (green) FG loop 99MYPPPY104 sequences with that of CD28 (blue), all shown in ball-and-stick representation. The view of the FG loop is similar to that in a. c, superimposed ball-and-stick representations of the G strands of apo-CTLA-4 (mol A; yellow) and of the TCR (red) in the region of the β-bulge following superposition of the entire domains with Coot. CTLA-4 residues are numbered.
Following CD28 and PD-1 in the DALI-based structure comparisons, the next 215 most similar structures all contain antibody or TCR variable domains, partly reflecting the large numbers of antigen receptor structures in the PDB. The r.m.s. differences for the superposition of CTLA-4 with the most similar TCR variable (Vα) domain (PDB code 3c6l) are 2.09 Å for 102 residues, contrasting with an r.m.s. deviation of 2.89 Å for 86 residues for comparison of apo-CTLA-4 with the V-set IgSF domain of the adhesion protein, CD2 (PDB code 1hnf). Differences between CTLA-4 and the Vα domain are restricted to loop-length variation only: the BC, C′C″, and FG loops of CTLA-4 are 3, 4, and 2 residues longer, respectively, and the CC′ loop is one residue shorter (Fig. 2a). Important shared features are the long CC′ loop and conserved β-bulges in the C′ and G strands in TCR and CTLA-4, which together impose a pronounced twist on the AGFCC′ sheet and are important, in antigen receptors, for variable domain packing (53). Most noteworthy is the very high degree of sequence conservation and Cα conformation in the region of the G strand β-bulge (GNGTQIYV for CTLA-4 using the single letter code and GDGTQLVV for Vα, PDB code 3c6l; Fig. 2c). The Gly-X-Gly motif is conserved in all antibody VL and 98% of VH domain sequences (53) and is significant because the second Gly adopts (ϕ,ψ) values that are energetically unfavorable for non-glycine residues, creating the bulge. The loops of CD2 and CTLA-4 have comparable lengths and the key domain differences are instead: (i) that the short A strand of CD2 does not H-bond with strand B; (ii) that CD2 lacks the longer CC′ strand of CTLA-4 and antigen receptors that imposes the twist on the A′GFCC′ sheet; (iii) the position of the C″ strand in CD2 allowing canonical H-bonding with C′, extending the AGFCC′C″ sheet; (iv) the shorter DE loop that allows repositioning of the BC loop and the top of the G strand of CD2, inducing a twist in the FG loop that is shared with, e.g. B7-1; and (v) repositioning of the C′ and G β-bulges in CD2 versus CTLA-4 and antigen receptors (Fig. 2a).
Comparisons of CTLA-4 with Other IgSF V-set Domains
Because the DALI analysis suggested that the CTLA-4/CD28/PD-1 subset of V-set IgSF domains are more related to antigen receptor V-set domains than those of adhesion molecules, as initially implied by similar comparisons of CD28 (31), the structural and evolutionary relationships among V-set IgSF domains were investigated further. Fifty-two V-set IgSF domains from the PDB, plus single C1-set and C2-set domains used as an outgroup were subjected to pairwise structural comparisons using the sequential structure alignment program (SSAP) (43). The SSAP scores were used to create a dendrogram of structural similarities (Fig. 3), which indicates that the CD28/CTLA-4 family form a subgroup of V-set domains distinct from the two main subgroups, i.e. the antigen receptors and CD8, and the “adhesion molecules” including the CD2 and CEA subfamilies, CD4, B7-1, and B7-2. The CD28/CTLA-4 subgroup is, however, much more closely related to antigen receptors than to adhesion molecules and more divergent from the adhesion molecule subgroup than the antigen receptors themselves. PD-1 represents an intermediate, having most structural homology to antigen receptors, but significantly higher similarity to the CTLA-4/CD28 subgroup than does any antigen receptor. PD-1 is most closely related to CD8β, but CD8β is not more similar to CTLA-4 or CD28 than to antigen receptors. The B and T lymphocyte attenuator, another molecule proposed to be related to CTLA-4 and PD-1 (54), is most similar to the antigen receptors and PD-1 and quite divergent from CTLA-4 and CD28; in fact, it scores more highly with the “adhesion” subset. B and T lymphocyte attenuator is proposed to have an I-set rather than a V-set domain (55, 56), so it is unsurprising that it is clearly structurally distinct from the other costimulatory receptors.
Many of the V-set IgSF domains are not easily assigned to one of the major subgroups, reflecting the considerable diversity across the IgSF. Some domains appear to be almost as different from the rest of the V-set group as C1- or C2-set IgSF domains, e.g. CD155 (PVR), and to a lesser extent RAGE (AGER). Others appear to form structural subgroups whose relationship to the larger subgroups is not obvious, e.g. the Siglecs, which are distant from all the other main subgroups and three other groups equidistant from the antigen receptor and adhesion subgroups, i.e. (i) CD300a/f, PIGR, and NKp44; (ii) synCAM3, JAM and PD-L2; and (iii) PD-L1, MOG, CAR, and P0. Similar results were obtained using SIMAX scores, which corresponds to the r.m.s. deviation normalized by the length of the largest domain in a pairwise alignment (57), the main differences being the positioning of the domains outside the major groups identified with SSAP (supplemental Fig. S2). The retention of the major groupings, however, and the conserved position of CTLA-4, CD28, and PD-1 within them, strongly supports the conclusion that the CD28/CTLA-4 family but not B and T lymphocyte attenuator, and the antigen receptors shared a common ancestor possibly similar to PD-1, and that the two families diverged early.
Apo-CTLA-4 Homodimer
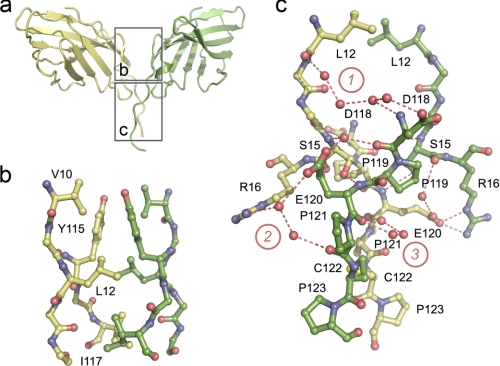
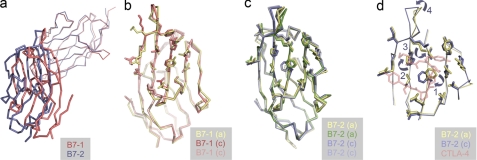
Apo-CTLA-4 comprises a non-crystallographically related homodimer held together by a disulfide bond at Cys122 and by a series of symmetric interactions involving the A′ strand and AB loop of one monomer and the G′ strand of the other, burying in total 1,113 Å2 of protein surface. Electron density was clearly discernible for the disulfide bond. The observed arrangement is very different to that proposed for the murine CTLA-4 homodimer (32). As discussed previously (10), the proposed murine homodimer is very probably incorrect given the limited conservation of interface residues, that dimerization would be precluded by glycosylation, that co-ligation of sB7-1 molecules around an axis orthogonal to the membrane (as observed in crystals of CTLA-4/B7-1 complexes) would not be possible, and that disulfide bonding of the monomers would also be precluded. Because the human CTLA-4 apo homodimer is within the asymmetric unit of the new crystals, we conclude that it very likely represents the native homodimer structure.
The apo-CTLA-4 interface divides naturally into two regions, one hydrophobic and the other hydrophilic and water-mediated (Fig. 4a). The core of the first region consists of reciprocal, symmetric hydrophobic interactions of Tyr115 of strand G both with itself and with Val10 and Leu12 in the A′ strand of the other chain. This region is extended by interactions of Leu12 with Ile117 of strand G, and Ile117 with its symmetry mate (Fig. 4b). In the second region located closer to the disulfide, reciprocal interchain H-bonds join the side chain of Ser15 and the main chain amide of Glu120, and, via a water, the main chain nitrogen of Ser15 and main chain carbonyl of Asp118 (Fig. 4c). Non-reciprocal H-bonds also form between the side chains of Glu120 of mol A and Arg16 of mol B (Fig. 4c). Three networks of water molecules also mediate H-bonding in this region. In network 1, Leu12 of mol A non-reciprocally contacts Asp118 of mol B via four water molecules. Network 2 consists of two water molecules mediating non-reciprocal contact between the side chain of Arg16 of mol A and the main chain oxygen of Pro121 and the side chain of Glu120 of mol B. Finally, in network 3, the carbonyls of Glu120 form reciprocal contacts mediated by three waters. A single water mediates H-bonds between the side chain of Glu120 of mol A and the main chain carbonyl of Ser15 of mol B. Although there are differences in the conformations of mol A and B between Asp118 and Cys122, these are the result of rigid-body movements originating at Pro119, suggesting that the stalk-like segment preceding the disulfide bond is relatively rigid, stabilized by the Pro119-Glu-Pro-Cys-Pro sequence.
FIGURE 4.
The interface region of the apo-CTLA-4 homodimer. a, the CTLA-4 homodimer is shown in ribbon format: yellow, mol A; green, mol B. The interface is divided into two regions (outlined), i.e. top and bottom, which are expanded in b and c. In b, the hydrophobic region at the “top” of the interface is shown in ball-and-stick format. Residues forming the hydrophobic core are labeled. In c the hydrophilic, water-mediated interaction is shown also in ball-and-stick format. The three principle water networks are identified with circled numbers. Residues participating in the network of interactions are labeled. Dashed red lines identify H-bonds involved in the intermolecular association.
Formation of the homodimer interface is likely disulfide bond-dependent given that forms of CTLA-4 lacking Cys122 are monomeric (30). The stalk structure is broadly conserved in the complexes except in the case of the B7-2d1·CTLA-4 complex for which strand G is interrupted by a “kink” at Ile117 (9). Thereafter, the assignment of the CTLA-4 strand G in this complex falls out of register with the other structures. This difference notwithstanding, the monomers in the apo structure adopt a preformed arrangement favoring bivalent interactions with B7-related ligands positioned orthogonally on the opposite cell surface. However, a limited degree of rotational flexibility at the interface is also possible (discussed below). Although it buries similar surface area (991 versus 1113 Å2), the CD28 interface is substantially different in that it involves 10 rather than four residues, all of which constitute a small, three-stranded A′G′F β sheet present in IgSF V-set domains (31). CD28-like dimerization of CTLA-4 is prevented by substitution of His116 by Tyr, by the disruption of the hydrophobic core of the interface by substitution of Ile114 by Gln, and by the intrusion of the longer CC′ loop into the plane formed by the small A′G′F β sheet of CTLA-4.
Ligand Binding-dependent Structural Changes in CTLA-4
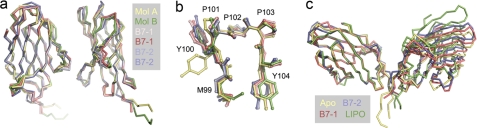
Comparison of the apo-CTLA-4 monomers in the asymmetric unit with the monomer pairs in the B7-1 and B7-2 complexes and the monomer in the lipocalin complex reveals remarkably few, if any, changes attributable to ligand binding (Fig. 5a and supplemental Table S2). The ϕ angle for Met99 of CTLA-4 in the CTLA-4·B7-2 complex (−150°) differs from that in apo-CTLA-4 (−40°), perhaps reflecting the limited resolution of the B7-2·CTLA-4 complex (3.2 Å) given that the apo conformation is identical to that of the B7-1 and lipocalin complexes. The only other regions that differ at all are the positions of the BC and CC′ loops, and the C″ strand and its adjacent loops, and in the conformation of the COOH-terminal end of the G strand (in the case of the B7-2 complex only), all of which are well separated from the ligand binding region. In no instance do both apo structures differ from all five complexed CTLA-4 monomers, i.e. there are no systematic, binding-dependent changes. The main chain atom positions of the ligand-binding MYPPPY sequence of the complexes are essentially identical to those for the apo structure (Fig. 5b; r.m.s. deviation = 0.25–0.78 Å and 0.37–0.75 Å for comparisons with the MYPPPY sequences of mol B and mol A of apo-CTLA-4, respectively). The side chain positions are also very similar except for Tyr100 of mol A, which is rotamer shifted by ∼120° due to lattice contacts. Variation in the relative positions of the two monomers within the homodimer is also minor and non-systematic. Rotations about the non-crystallographic 2-fold axis and about an axis parallel to the cell surface passing through the hydrophobic region of the interface (centered on Tyr115) are each less than ∼5° from the positions observed in the apo-CTLA-4 homodimer, except in the case of the lipocalin complex for which the rotation around the second axis is ∼15° (Fig. 5c). All these effects, which slightly distort the position of the disulfide bond in the stalk, are explicable by crystal packing only.
FIGURE 5.
The rigid-body interaction of CTLA-4 with its ligands. In a the two halves of the apo homodimer, i.e. mol A (yellow) and mol B (green), are superimposed using Coot with two copies of the CTLA-4 monomer seen in the crystals of the CTLA-4/B7-1 complex (red, light red; Ref. 10) and in the crystals of the CTLA-4·B7-2 complex (blue, light blue; Ref. 9). The only regions of significant variation are in the positions of the BC and CC′ loops, the C″ strand and its adjacent loops, with the largest differences being those between mol A and mol B of the apo structure. Structures are shown as α-carbon representations. In b the structures of the 99MYPPPY104 FG loops of the apo and complex structures, shown in ball-and-stick format, are compared following superposition of the loops with Coot. The color scheme is the same as that used in a; the largest differences are observed for the two apo structures. In c, the structure of the apo-CTLA-4 homodimer (yellow) is compared with that of the homodimer from the B7-1 (red), B7-2 (blue), and lipocalin (green; Ref. 51) complexes, all shown as α-carbon representations following superposition on mol A of apo-CTLA-4.
CTLA-4 Recognition by B7-1 and B7-2
The poor sequence conservation but similar chemistry of the ligand-binding surfaces of B7-1 and B7-2 has been noted previously (58). The structures of the complexes formed by CTLA-4 with B7-1 and B7-2 have not been compared in detail, however, and are substantially different. Arg29 of B7-1, which forms an electrostatic contact with Glu33 of CTLA-4, is replaced with valine in B7-2, creating a larger binding pocket in B7-2 that is only partially filled by CTLA-4. The B7-1/CTLA-4 interface therefore exhibits much greater geometric complementarity than the B7-2·CTLA-4 complex (shape complementarity values of 0.754–0.784 versus 0.584–0.663, respectively; Ref. 59). Partially compensating for this, three extra residues inserted in the C″ strand of B7-2 are accommodated by a twist that allows Lys49 to H-bond with Tyr100 of CTLA-4, extending the surface area buried upon binding (367–390 Å2 versus 324–353 Å2 for the B7-2·CTLA-4 and B7-1·CTLA-4 complexes, respectively) and repositioning B7-2 relative to B7-1 in the respective complexes (Fig. 6a).
FIGURE 6.
Recognition of CTLA-4 by B7-1 and B7-2. In a, the shift in the positioning of the ligand-binding domain of B7-1 (red) versus that of B7-2 (blue) is illustrated after superimposing the CTLA-4 monomers from each structure (thin red and blue lines). The structures are shown as α-carbon representations. In b and c, the side chain positions of CTLA-4-contacting residues of B7-1 and B7-2, respectively, are shown for the apo (a) structures (B7-1, yellow; B7-2, yellow and green) and the complexed (c) structures (B7-1, red, light red; B7-2, blue and light blue). Only one copy of apo-B7-1 is shown because there was just the one molecule in the asymmetric unit of the apo-B7-1 crystals (12). In d, the systematic movements in Phe31 (labeled 1), His88 (labeled 2), and Met95 (labeled 3) that lead to repositioning of the FG loop of B7-2 (labeled 4) are illustrated for one copy each of the ligand-binding domains of B7-2 before (yellow) and after (blue) complex formation. The MYPPPY sequence of CTLA-4 is shown transparently in red.
B7-1 binds CTLA-4 in an even more rigid-body-like manner than CTLA-4 engages B7-1. The Cα differences of complexed versus apo-B7-1 (r.m.s. deviation = 0.80 Å for 197 residues) are small and comparable with those of the two copies of B7-1 in the complex crystals (r.m.s. deviation = 0.39 Å for 199 residues; Fig. 6b). The only substantial change involves Arg94 of the FG loop, which moves 1–1.5 Å in forming an H-bond with Tyr104 of CTLA-4 in both copies of B7-1 in the complex. For B7-2 the situation is more complicated (Fig. 6, c and d). In apo versus liganded B7-2 (r.m.s. deviation = ∼0.7 Å for all main chain residues in both copies in each lattice), the FG loop rotates away from the ligand 2.3–5.5 Å in a concerted “induced fit” rearrangement initiated by a ∼120° rotation of Phe31 of B7-2 that allows a stacking interaction with Pro102 of the MYPPPY sequence of CTLA-4. Rotation of Phe31 induces a 90° rotation of the side chain of His88 that in turn drives away the side chain of Met95 by up to 2.7 Å, inducing a rigid-body shift in the FG loop (i.e. from Lys91 to Met95). Extension and rotation of the side chains of Lys49 and Arg97 also allows H-bonds to form with Tyr100 and Tyr104 of CTLA-4, respectively. At the resolution of the complexes it is not possible to visualize the waters in the binding face.
Thermodynamics of Ligand Recognition by CTLA-4
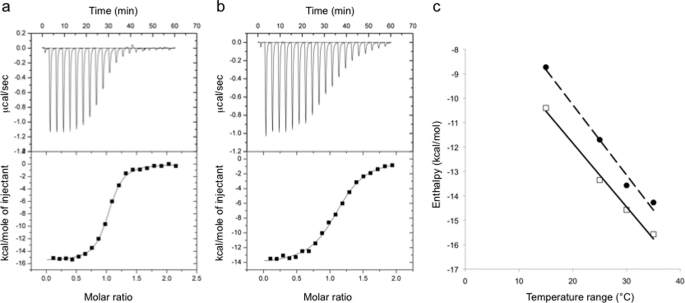
For thermodynamic analysis the extracellular regions of B7-1 (sB7-1) and CTLA-4 (CTLA-4Fc) were expressed as a histidine-tagged and an IgG1-Fc fusion protein in CHO-K1 cells, respectively (45), and the V-set ligand-binding domain of B7-2 (B7-2v) was refolded from inclusion bodies expressed in bacteria (46). Representative data for the isothermal calorimetric titration of the CTLA-4Fc homodimer with sB7-1 and B7-2v at 35 °C are shown (Fig. 7, a and b); these results are representative of titrations performed at other temperatures (Table 1). The affinities of the interactions being in the micromolar range meant that they were well suited to measurement by isothermal calorimetry. At the temperatures investigated (15–35 °C) the affinity of sB7-1 for CTLA-4 was ∼4–8-fold higher than that of B7-2v for CTLA-4. In surface plasmon resonance-based assays of the binding of CHO cell-expressed sB7-2 to CTLA-4Fc (11), the B7-2/CTLA-4 affinity was slightly lower and the 13-fold B7-1/B7-2 differential somewhat higher. These differences cannot be attributed to glycosylation effects because the affinity of glycosylated CHO cell-derived sB7-2 for CTLA-4Fc measured by calorimetry at 25 °C was identical to that of B7-2v (data not shown). The binding stoichiometry for both ligands is 1:1 calculated using the concentration of CTLA-4 monomers, consistent with the crystal structures. Thermodynamic parameterization of the binding of sB7-1 and B7-2v to CTLA-4Fc (Table 1) reveals that the interactions are energetically very similar. The interactions are ΔH driven and accompanied by unfavorable ΔS contributions at all temperatures from 15 to 35 °C. Across these temperatures there is almost complete enthalpy-entropy compensation for the interactions of sB7-1 and B7-2v. The higher affinity of sB7-1 for CTLA-4Fc versus B7-2v is largely attributable to a more favorable ΔHobs. The ΔHobs for the association of CTLA-4Fc with sB7-1 and B7-2v declines linearly from −10.4 kcal mol−1 at 15 °C to −15.57 kcal mol−1 at 35 °C, and from −8.73 to −14.27 kcal mol−1, respectively (Fig. 7c), yielding ΔCp terms of −261 and −288 cal mol−1 K−1. The similarities in the values of ΔCp for sB7-1 and B7-2v binding suggest that there is little difference in the overall changes in degrees of freedom of the atoms in formation of each complex. Because this term is usually dominated by solvent effects it suggests that the surface areas buried in the formation of the two complexes are very similar. The slightly more negative value of ΔCp obtained for B7-2v binding might result from the Glu/Val substitution given that these residues will be differentially hydrated (i.e. charged versus hydrophobic exposed surface). This might in turn be compensated for by the burial of an extra ∼40 Å2 of protein surface in the B7-2 interaction.
FIGURE 7.
ITC measurements of CTLA-4 binding to B7-1 and B7-2. Example of raw data titrations of sB7-1 (a) and B7-2v (b) at 0.2 mm into an isothermal calorimetry cell containing a 0.02 mm solution of CTLA-4Fc at pH 7.4 and 35 °C in a buffer containing 150 mm NaCl. Similar titrations were undertaken at various temperatures, the results of which are summarized in Table 1. c, plots of observed enthalpy versus temperature for the interaction of sB7-1 (open squares) and B7-2v (closed circles) with CTLA-4Fc. The slopes of these plots give the change in heat capacity (ΔCp) on binding of B7-1 and B7-2 to CTLA-4, the values of which are −261 and −288 cal mol−1 K−1, respectively.
TABLE 1.
Summary of thermodynamic data for the binding of sB7-1 and B7-2v to CTLA-4Fc
| Temperature | Kda | ΔGcalcb | ΔHobsc | TΔScalcd | ne |
|---|---|---|---|---|---|
| °C | μm | kcal/mol | |||
| sB7-1/CTLA-4Fc | |||||
| 15 | 0.08 | −9.33 | −10.40 ± 0.08 | −1.07 | 0.93 |
| 25 | 0.12 | −9.41 | −13.35 ± 0.06 | −3.94 | 1.08 |
| 30 | 0.14 | −9.48 | −14.57 ± 0.06 | −5.09 | 1.1 |
| 35 | 0.23 | −9.37 | −15.57 ± 0.09 | −6.20 | 0.99 |
| B7-2v/CTLA-4Fc | |||||
| 15 | 0.45 | −8.36 | −8.73 ± 0.08 | −0.37 | 1.13 |
| 25 | 0.56 | −8.52 | −11.70 ± 0.09 | −3.18 | 1.08 |
| 30 | 1.10 | −8.26 | −13.57 ± 0.11 | −5.31 | 1.13 |
| 35 | 0.83 | −8.57 | −14.27 ± 0.14 | −5.70 | 1.11 |
a Kd = 1/Ka.
b ΔGcalc = −RT × ln(Ka).
c Errors for fits of ΔHobs are shown, for titrations done 2–3 times each. The fitting errors on the Kd values are of the order of <10% of the values shown. All experiments have “C values” between 10 and 250.
d TΔScalc = ΔHobs − ΔGcalc.
e Stoichiometry calculated using [CTLA-4 monomer].
DISCUSSION
The importance of CTLA-4 in human disease is illustrated by genome-wide association studies showing that polymorphism in the CTLA4 gene confers susceptibility to autoimmunity, and by increasing efforts to target this receptor in cancer immunotherapy (discussed in Ref. 60). Nevertheless, relatively little is known about the cell-intrinsic functions of this molecule. With regard to its structure and how it signals, the unusual situation arose that crystal structures of ligand-bound but not apo-CTLA-4 had been published, despite the success of two groups in obtaining crystals of apo-CTLA-4 (9, 49). We have found (50) that apo-CTLA-4 monomers expressed in bacteria according to the method of Chang et al. (49) form strand-exchanged dimers during crystallization. Mammalian cell-expressed material yielded very high quality crystals of an apo-CTLA-4 homodimer we assume to be natively folded because of its similarities to ligand-bound CTLA-4. Schönfeld et al. (51) may have succeeded in crystallizing well folded, bacterially expressed CTLA-4 because they prepared a slightly longer form of the protein and because, during crystallization, the FG loop was “capped” with a high affinity ligand, lipocalin. B7-2 may also have stabilized CTLA-4 in the complex of Schwartz et al. (9). The structure of the apo homodimer reveals the nature of ligand recognition by CTLA-4 and offers new insights into the likely evolution of the CD28 family of proteins and antigen receptors. It also places new constraints on how signaling is initiated by receptors of this type.
The structural analysis reveals that the ligand-bound and apo-CTLA-4 monomers are remarkably similar, exhibiting fewer differences than the two halves of the apo homodimer, confirming that CTLA-4 binds its ligands in a largely rigid-body-like manner. Unexpectedly, in contrast to the rigid-body binding of B7-1, B7-2 binding to CTLA-4 exhibits elements of induced fit. Despite these differences and the differences in the geometric fits of the respective interfaces, the thermodynamic signatures of the interactions are surprisingly similar. Both interactions are enthalpically driven, with the 4–8-fold differences in affinity being attributable mostly to enthalpic effects perhaps resulting from the additional polar interactions made by Glu33 present in B7-1 but replaced by Val in B7-2. Measurements of ΔCp also failed to identify differences attributable to conformational effects during binding, but this is perhaps not surprising as the B7-2 rearrangements do not bury substantially more protein surface beyond that already present in the apo binding site. Thus, despite being based on different mechanisms both interactions are accompanied by similar unfavorable entropic contributions to ΔG. Typically this unfavorable entropy results from a conformational change resulting in the restriction of degrees of freedom of the atoms or a trapping of otherwise bulk-solvent waters. Because the mechanisms of binding of B7-1 and B7-2 are so different, it seems likely that both mechanisms of producing unfavorable entropic contribution are occurring. For example, the induced fit involves ordering of structure and a net increase in the non-covalent bonding between atoms of the system (as reflected in the favorable ΔH). In the case of the rigid-body B7-1 interaction the most likely cause of the entropic effect is water sequestration at the interface (see Ref. 61). The structures of the complexes are presently not of sufficiently high resolution to confirm this observation.
Physiologically, the similarity of the overriding energetics of the interactions resulting from enthalpy and entropy compensation could be important. The interactions are not highly specific but do show some selectivity because it is likely that only recognition of CTLA-4 by B7-2 induces the required conformational changes required for complex formation. Selectivity (as opposed to specificity, which is based on the absolute affinity) has been seen in a number of protein interactions wherein a particular ligand is required to undergo a structural change to provide the appropriate cognate binding site (62, 63). Selectivity therefore does not necessarily require a tighter interaction to ensure the appropriate protein binds, but requires that the interacting binding partner is able to induce the required fit. An additional point is that many functional studies suggest that the primary functional ligand of B7-2 is CD28 (discussed in Ref. 11), so it is conceivable that the binding site of B7-2 is “optimized” for CD28 rather than CTLA-4 binding, in which case the conformational change in B7-2 may represent a compromise that allows B7-2 to contribute to inhibitory signaling by CTLA-4. The on-rates for B7-1 and B7-2 association are indistinguishable and very high (∼2 × 106 m−1; Ref. 11), suggesting that the conformational rearrangement in B7-2 occurs after formation of an initial encounter complex, as in the case of TCR recognition of conformationally distinct MHC/peptide variants (64).
DALI analysis showed that the CTLA-4 V-set IgSF domain is more similar to that of antigen receptor V-set domains than adhesion molecules, as also noted for CD28 (31). These findings were extended using SSAP (43), which indicates that the CD28/CTLA-4 family forms a subgroup of V-set domains distinct from both antigen receptors (and CD8), and the adhesion molecule grouping that includes CD2, CD4, and B7-2. Because the CD28/CTLA-4 subgroup is more closely related to antigen receptors than adhesion molecules but more divergent from the adhesion molecules than the antigen receptors themselves, it seems likely that a CD28/CTLA-4 family precursor separated from the lineage giving rise to antigen receptors relatively early, with the distinctive mode of binding focused on the proline-rich FG loop perhaps allowing rapid divergence of the rest of the domain. Very early on in the analysis of the IgSF, Williams and Barclay (65) suggested that the entire family might have derived by gene duplication and divergence from a primordial molecule with a single V-set domain, the function of which could have been homophilic recognition. Sequence comparisons (66) and, later, the structure of the CD8αα homodimer (67) implied initially that a CD8-like molecule was a precursor of the antigen receptors. Our comparisons suggest that PD-1 is structurally intermediate between the CTLA-4/CD28 subgroup and the antigen receptor/CD8 grouping, consistent with the precursor being a signaling protein with tyrosine phosphorylation motifs. It is noteworthy that the receptor/ligand interaction of PD-1 and PD-L1 V-set domains is reminiscent of the interactions of both CD8 and antigen receptor V-set domain dimers (68), suggesting how in trans receptor/ligand interactions might have yielded in cis interacting dimers. A transition of this type involving dimerization of a PD-1-like molecule would have the effect of at least doubling its signaling capacity via duplication of its cytoplasmic signaling motifs. The transient association of, e.g. Src kinases with these signaling motifs may have been superseded in the case of CD8 by a more stable one comprised of cysteine-coordinated zinc (69).
Many V-set IgSF domains are not easily assigned to the adhesion, CTLA-4/CD28, or antigen receptor/CD8 subgroups, emphasizing the considerable structural diversity across the IgSF and the difficulties associated with reconstructing evolutionary relationships. The sequence, structural, and genetic evidence, nevertheless, overwhelmingly suggests that CTLA-4 and CD28 evolved via duplication of a shared precursor. Because CTLA-4 and CD28 are both reliant on tyrosine phosphorylation of their cytoplasmic domains by extrinsic kinases, a feature they share with antigen receptors, it is highly likely that the common ancestor of these molecules was a signaling protein.
Assuming that the signaling mechanism has been preserved, what can be learnt about it from the CTLA-4 and CD28 structural data? Signaling by these proteins was expected to involve conformational changes or ligand-induced dimerization (4). CTLA-4 now joins the set of just two other signaling receptors dependent on extrinsic tyrosine kinases for which pre- and post-binding structures are known. Structures of ligand-bound and unbound TCRs show that ligand engagement is not generally accompanied by conformational changes within the αβ subunits that could transmit a signal intracellularly (reviewed in Ref. 70), although this has been disputed (71). Similarly, structures of single TCRs complexed with MHC-peptide ligands that generate very different signals in T-cells are largely identical (72). It needs to be noted, however, that all of these structures lacked the signaling CD3 subunits of the TCR complex, leaving open the possibility that inter-molecular rearrangements contribute to signaling. Binding of the inhibitory receptor SIRPα to CD47 is of the rigid-body type on the CD47 side, but substantial loop rearrangements of up to 5 Å accompany CD47 binding by SIRPα (73). The remarkable similarities between the liganded and apo forms of intact CTLA-4 homodimers thus provide the clearest evidence yet that conformational rearrangements in their extracellular domains are not an essential requirement for signaling by receptors dependent on extrinsic tyrosine kinases. The observation that, like CTLA-4, CD28 is a homodimer that would otherwise be well suited to oligomeric interactions but is instead monovalent and binds a monomeric ligand (11), strongly implies that receptor oligomerization is also unlikely to be an essential part of the shared signaling mechanism. Rigid-body interactions and the likely dimensions of CD28- and CTLA-4-ligand complexes (∼150 Å; Ref. 10) are, however, compatible with a topological model of signaling in which the physical segregation of small receptors from large phosphatases enhances their phosphorylation, as proposed for the TCR (74, 75).
Supplementary Material
Acknowledgments
We are grateful to the staff of beamline IO4 at the Diamond Light Source (Didcot, United Kingdom) for assistance with crystallographic data collection.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 3OSK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
C. Yu et al., manuscript in preparation.
- TCR
- T cell receptor
- SSAP
- sequential structure alignment program
- endo
- endoglycosidase H
- r.m.s.
- root mean square
- PDB
- Protein Data Bank.
REFERENCES
- 1. Evans E. J., Hene L., Sparks L. M., Dong T., Retiere C., Fennelly J. A., Manso-Sancho R., Powell J., Braud V. M., Rowland-Jones S. L., McMichael A. J., Davis S. J. (2003) Immunity 19, 213–223 [DOI] [PubMed] [Google Scholar]
- 2. van der Merwe P. A., Davis S. J. (2003) Annu. Rev. Immunol. 21, 659–684 [DOI] [PubMed] [Google Scholar]
- 3. Greenwald R. J., Freeman G. J., Sharpe A. H. (2005) Annu. Rev. Immunol. 23, 515–548 [DOI] [PubMed] [Google Scholar]
- 4. Teft W. A., Kirchhof M. G., Madrenas J. (2006) Annu. Rev. Immunol. 24, 65–97 [DOI] [PubMed] [Google Scholar]
- 5. Jansson A., Barnes E., Klenerman P., Harlén M., Sørensen P., Davis S. J., Nilsson P. (2005) J. Immunol. 175, 1575–1585 [DOI] [PubMed] [Google Scholar]
- 6. Lenschow D. J., Walunas T. L., Bluestone J. A. (1996) Annu. Rev. Immunol. 14, 233–258 [DOI] [PubMed] [Google Scholar]
- 7. Lafage-Pochitaloff M., Costello R., Couez D., Simonetti J., Mannoni P., Mawas C., Olive D. (1990) Immunogenetics 31, 198–201 [DOI] [PubMed] [Google Scholar]
- 8. Howard T. A., Rochelle J. M., Seldin M. F. (1991) Immunogenetics 33, 74–76 [DOI] [PubMed] [Google Scholar]
- 9. Schwartz J. C., Zhang X., Fedorov A. A., Nathenson S. G., Almo S. C. (2001) Nature 410, 604–608 [DOI] [PubMed] [Google Scholar]
- 10. Stamper C. C., Zhang Y., Tobin J. F., Erbe D. V., Ikemizu S., Davis S. J., Stahl M. L., Seehra J., Somers W. S., Mosyak L. (2001) Nature 410, 608–611 [DOI] [PubMed] [Google Scholar]
- 11. Collins A. V., Brodie D. W., Gilbert R. J., Iaboni A., Manso-Sancho R., Walse B., Stuart D. I., van der Merwe P. A., Davis S. J. (2002) Immunity 17, 201–210 [DOI] [PubMed] [Google Scholar]
- 12. Ikemizu S., Gilbert R. J., Fennelly J. A., Collins A. V., Harlos K., Jones E. Y., Stuart D. I., Davis S. J. (2000) Immunity 12, 51–60 [DOI] [PubMed] [Google Scholar]
- 13. Bhatia S., Edidin M., Almo S. C., Nathenson S. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15569–15574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. James J. R., Oliveira M. I., Carmo A. M., Iaboni A., Davis S. J. (2006) Nat. Methods 3, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 15. Pentcheva-Hoang T., Egen J. G., Wojnoonski K., Allison J. P. (2004) Immunity 21, 401–413 [DOI] [PubMed] [Google Scholar]
- 16. Sansom D. M., Walker L. S. (2006) Immunol. Rev. 212, 131–148 [DOI] [PubMed] [Google Scholar]
- 17. Waterhouse P., Penninger J. M., Timms E., Wakeham A., Shahinian A., Lee K. P., Thompson C. B., Griesser H., Mak T. W. (1995) Science 270, 985–988 [DOI] [PubMed] [Google Scholar]
- 18. Bradshaw J. D., Lu P., Leytze G., Rodgers J., Schieven G. L., Bennett K. L., Linsley P. S., Kurtz S. E. (1997) Biochemistry 36, 15975–15982 [DOI] [PubMed] [Google Scholar]
- 19. Shiratori T., Miyatake S., Ohno H., Nakaseko C., Isono K., Bonifacino J. S., Saito T. (1997) Immunity 6, 583–589 [DOI] [PubMed] [Google Scholar]
- 20. Schneider H., Schwartzberg P. L., Rudd C. E. (1998) Biochem. Biophys. Res. Commun. 252, 14–19 [DOI] [PubMed] [Google Scholar]
- 21. Long E. O. (2008) Immunol. Rev. 224, 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Daëron M., Jaeger S., Du Pasquier L., Vivier E. (2008) Immunol. Rev. 224, 11–43 [DOI] [PubMed] [Google Scholar]
- 23. Chuang E., Lee K. M., Robbins M. D., Duerr J. M., Alegre M. L., Hambor J. E., Neveu M. J., Bluestone J. A., Thompson C. B. (1999) J. Immunol. 162, 1270–1277 [PubMed] [Google Scholar]
- 24. Zhang Y., Allison J. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9273–9278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marengère L. E., Waterhouse P., Duncan G. S., Mittrücker H. W., Feng G. S., Mak T. W. (1996) Science 272, 1170–1173 [DOI] [PubMed] [Google Scholar]
- 26. Prasad K. V., Cai Y. C., Raab M., Duckworth B., Cantley L., Shoelson S. E., Rudd C. E. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2834–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Truitt K. E., Hicks C. M., Imboden J. B. (1994) J. Exp. Med. 179, 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Linsley P. S., Nadler S. G. (2009) Immunol. Rev. 229, 307–321 [DOI] [PubMed] [Google Scholar]
- 29. Peggs K. S., Quezada S. A., Allison J. P. (2008) Immunol. Rev. 224, 141–165 [DOI] [PubMed] [Google Scholar]
- 30. Metzler W. J., Bajorath J., Fenderson W., Shaw S. Y., Constantine K. L., Naemura J., Leytze G., Peach R. J., Lavoie T. B., Mueller L., Linsley P. S. (1997) Nat. Struct. Biol. 4, 527–531 [DOI] [PubMed] [Google Scholar]
- 31. Evans E. J., Esnouf R. M., Manso-Sancho R., Gilbert R. J., James J. R., Yu C., Fennelly J. A., Vowles C., Hanke T., Walse B., Hünig T., Sørensen P., Stuart D. I., Davis S. J. (2005) Nat. Immunol. 6, 271–279 [DOI] [PubMed] [Google Scholar]
- 32. Ostrov D. A., Shi W., Schwartz J. C., Almo S. C., Nathenson S. G. (2000) Science 290, 816–819 [DOI] [PubMed] [Google Scholar]
- 33. Davis S. J., Ward H. A., Puklavec M. J., Willis A. C., Williams A. F., Barclay A. N. (1990) J. Biol. Chem. 265, 10410–10418 [PubMed] [Google Scholar]
- 34. Chang V. T., Crispin M., Aricescu A. R., Harvey D. J., Nettleship J. E., Fennelly J. A., Yu C., Boles K. S., Evans E. J., Stuart D. I., Dwek R. A., Jones E. Y., Owens R. J., Davis S. J. (2007) Structure 15, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davis S. J., Davies E. A., Barclay A. N., Daenke S., Bodian D. L., Jones E. Y., Stuart D. I., Butters T. D., Dwek R. A., van der Merwe P. A. (1995) J. Biol. Chem. 270, 369–375 [DOI] [PubMed] [Google Scholar]
- 36. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 39. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 42. Cuff A. L., Sillitoe I., Lewis T., Redfern O. C., Garratt R., Thornton J., Orengo C. A. (2009) Nucleic Acids Res. 37, D310–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Orengo C. A., Taylor W. R. (1996) Methods Enzymol. 266, 617–635 [DOI] [PubMed] [Google Scholar]
- 44. Kolodny R., Koehl P., Levitt M. (2005) J. Mol. Biol. 346, 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Merwe P. A., Bodian D. L., Daenke S., Linsley P., Davis S. J. (1997) J. Exp. Med. 185, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang X., Schwartz J. C., Almo S. C., Nathenson S. G. (2002) Protein Expr. Purif. 25, 105–113 [DOI] [PubMed] [Google Scholar]
- 47. Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 48. Ladbury J. E., Chowdhry B. Z. (1996) Chem. Biol. 3, 791–801 [DOI] [PubMed] [Google Scholar]
- 49. Chang C. Y., Fenderson W. H., Lavoie T. B., Peach R. J., Einspahr H. M., Sheriff S. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1468–1469 [DOI] [PubMed] [Google Scholar]
- 50. Sonnen A. F., Yu C., Evans E. J., Stuart D. I., Davis S. J., Gilbert R. J. (2010) J. Mol. Biol. 399, 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schönfeld D., Matschiner G., Chatwell L., Trentmann S., Gille H., Hülsmeyer M., Brown N., Kaye P. M., Schlehuber S., Hohlbaum A. M., Skerra A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8198–8203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Holm L., Park J. (2000) Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 53. Chothia C., Novotný J., Bruccoleri R., Karplus M. (1985) J. Mol. Biol. 186, 651–663 [DOI] [PubMed] [Google Scholar]
- 54. Watanabe N., Gavrieli M., Sedy J. R., Yang J., Fallarino F., Loftin S. K., Hurchla M. A., Zimmerman N., Sim J., Zang X., Murphy T. L., Russell J. H., Allison J. P., Murphy K. M. (2003) Nat. Immunol. 4, 670–679 [DOI] [PubMed] [Google Scholar]
- 55. Compaan D. M., Gonzalez L. C., Tom I., Loyet K. M., Eaton D., Hymowitz S. G. (2005) J. Biol. Chem. 280, 39553–39561 [DOI] [PubMed] [Google Scholar]
- 56. Nelson C. A., Fremont M. D., Sedy J. R., Norris P. S., Ware C. F., Murphy K. M., Fremont D. H. (2008) J. Immunol. 180, 940–947 [DOI] [PubMed] [Google Scholar]
- 57. Dessailly B. H., Nair R., Jaroszewski L., Fajardo J. E., Kouranov A., Lee D., Fiser A., Godzik A., Rost B., Orengo C. (2009) Structure 17, 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X., Schwartz J. C., Almo S. C., Nathenson S. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2586–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 60. Paterson A. M., Sharpe A. H. (2010) Nat. Immunol. 11, 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ladbury J. E. (1996) Chem. Biol. 3, 973–980 [DOI] [PubMed] [Google Scholar]
- 62. O'Brien R., Rugman P., Renzoni D., Layton M., Handa R., Hilyard K., Waterfield M. D., Driscoll P. C., Ladbury J. E. (2000) Protein Sci. 9, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arold S. T., Ulmer T. S., Mulhern T. D., Werner J. M., Ladbury J. E., Campbell I. D., Noble M. E. (2001) J. Biol. Chem. 276, 17199–17205 [DOI] [PubMed] [Google Scholar]
- 64. Lee J. K., Stewart-Jones G., Dong T., Harlos K., Di Gleria K., Dorrell L., Douek D. C., van der Merwe P. A., Jones E. Y., McMichael A. J. (2004) J. Exp. Med. 200, 1455–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Williams A. F., Barclay A. N. (1988) Annu. Rev. Immunol. 6, 381–405 [DOI] [PubMed] [Google Scholar]
- 66. Johnson P., Williams A. F. (1986) Nature 323, 74–76 [DOI] [PubMed] [Google Scholar]
- 67. Leahy D. J., Axel R., Hendrickson W. A. (1992) Cell 68, 1145–1162 [DOI] [PubMed] [Google Scholar]
- 68. Lin D. Y., Tanaka Y., Iwasaki M., Gittis A. G., Su H. P., Mikami B., Okazaki T., Honjo T., Minato N., Garboczi D. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Boggon T. J., Eck M. J. (2004) Oncogene 23, 7918–7927 [DOI] [PubMed] [Google Scholar]
- 70. Garcia K. C., Teyton L., Wilson I. A. (1999) Annu. Rev. Immunol. 17, 369–397 [DOI] [PubMed] [Google Scholar]
- 71. Beddoe T., Chen Z., Clements C. S., Ely L. K., Bushell S. R., Vivian J. P., Kjer-Nielsen L., Pang S. S., Dunstone M. A., Liu Y. C., Macdonald W. A., Perugini M. A., Wilce M. C., Burrows S. R., Purcell A. W., Tiganis T., Bottomley S. P., McCluskey J., Rossjohn J. (2009) Immunity 30, 777–788 [DOI] [PubMed] [Google Scholar]
- 72. Ding Y. H., Baker B. M., Garboczi D. N., Biddison W. E., Wiley D. C. (1999) Immunity 11, 45–56 [DOI] [PubMed] [Google Scholar]
- 73. Hatherley D., Graham S. C., Turner J., Harlos K., Stuart D. I., Barclay A. N. (2008) Mol. Cell 31, 266–277 [DOI] [PubMed] [Google Scholar]
- 74. Davis S. J., van der Merwe P. A. (1996) Immunol. Today 17, 177–187 [DOI] [PubMed] [Google Scholar]
- 75. Davis S. J., van der Merwe P. A. (2006) Nat. Immunol. 7, 803–809 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.