Abstract
During neurite outgrowth, Rho small G protein activity is spatiotemporally regulated to organize the neurite sprouting, extension, and branching. We have previously identified a potent Rho GTPase-activating protein (GAP), RA-RhoGAP, as a direct downstream target of Rap1 small G protein in the neurite outgrowth. In addition to the Ras-associating (RA) domain for Rap1 binding, RA-RhoGAP has the pleckstrin homology (PH) domain for lipid binding. Here, we showed that phosphatidic acid (PA) bound to the PH domain and enhanced GAP activity for Rho. RA-RhoGAP induced extension of neurite in a diacylglycerol kinase-mediated synthesis of the PA-dependent manner. Knockdown of RA-RhoGAP reduced the diacylglycerol kinase-induced neurite extension. In contrast to the effect of the RA domain, the PH domain was specifically involved in the neurite extension, not in the sprouting and branching. These results indicate that PA and Rap1 cooperatively regulate RA-RhoGAP activity for promoting neurite outgrowth.
Keywords: Cytoskeleton, G Proteins, Lipid-binding Protein, Rho, Signal Transduction
Introduction
Formation and extension of axons and dendrites, the so-called neurite outgrowth, are crucial events in neuronal differentiation and maturation during development of the nervous system (1, 2). As neurites extend further and acquire their final axonal and dendritic identities, neurons establish synaptic contacts and reach full maturation (3–5). These morphological changes require remodeling of the actin cytoskeleton (6). Rho small G protein is a key regulator of the actin cytoskeleton organization in neurons and has been shown to play important roles in several aspects of neurite outgrowth, such as sprouting, extension, and branching (7, 8). An increase in Rho activity results in reduction of neurite sprouting, extension, and branching, whereas a decrease in Rho activity enhances neurite sprouting, extension, and branching (9–12). The Rho activity is positively regulated by guanine nucleotide exchange factors (GEFs)2 and negatively regulated by GTPase-activating proteins (GAPs) (13). Several RhoGEFs and RhoGAPs have been shown to be important for the neurite outgrowth (14). Among them, we have previously identified RA-RhoGAP as a novel downstream target of the Rap1 small G protein, which has the PH, RA, GAP, and annexin-like repeat domains (15). RA-RhoGAP is activated by Rap1 and induces inactivation of Rho, resulting in the neurite outgrowth. Upon stimulation, Rap1 is transiently activated at the tips of the sprouting neurites (16). However, sustained inactivation of Rho is required for the neurite extension and branching after sprouting (11, 12, 17). Thus, other factors may alternatively suffice to activate RA-RhoGAP at the leading edges of neurites.
In the cellular signaling mechanism, the lipid-mediated second messenger system is one of the major machineries in various kinds of cells (18–20). Following activation of Gq protein-coupled receptors, such as metabotropic glutamate receptor 1 (mGluR1) and mGluR5, in response to external stimuli, phospholipase C yields a pair of second messengers, diacylglycerol (DAG) and inositol 1,4,5-triphosphate (21). In this system, diacylglycerol kinase (DGK) phosphorylates DAG to produce another second messenger, phosphatidic acid (PA) (22, 23). One of the best known functional roles of DGK is the negative regulation of protein kinase C, for which DAG acts as an allosteric activator and whose activity plays a central role in many different cell types (18). In addition, recent studies have revealed that PA also acts as second messenger to regulate a number of signaling molecules (19, 20). Therefore, DGK is thought to mediate signal transduction by modulating levels of DAG and PA, attenuation of DAG and production of PA (22, 23). PA is a negatively charged phospholipid that can function as a lipid anchor by binding directly to positively charged sites on effector proteins (20). Among the various cellular functions controlled by PA, cell proliferation and survival signaling are mediated by specific interactions with mSOS and mammalian target of rapamycin (24, 25), respectively. DOCK2, a GEF for Rac, binds PA through polybasic regions to mediate neutrophil chemotaxis (26). In addition, the cytosolic tyrosine kinase Fer binds PA through the FX domain to mediate cell migration (27). Although DGKβ has been implicated in neurite outgrowth (28), other protein targets that interact with PA to promote neurite outgrowth are largely unknown.
We have identified here RA-RhoGAP as a downstream target of PA. PA activates RA-RhoGAP and induces inactivation of Rho more potently than Rap1. RA-RhoGAP is involved in a DGKβ-PA signaling pathway that mediates neurite outgrowth. Rap1 activates RA-RhoGAP and enhances the number, length, and branching of neurites. In contrast, PA activates RA-RhoGAP and enhances the length of neurites preferentially. Thus, PA and Rap1 cooperatively regulate RA-RhoGAP activity and promote neurite outgrowth.
EXPERIMENTAL PROCEDURES
Construction of Expression Vectors
Mammalian expression vectors for RA-RhoGAP were constructed by subcloning the cDNAs into pCA vectors (29) with N-terminal HA tags. The constructs of RA-RhoGAP contained the following aa: pCA-HA-RA-RhoGAP, aa 1–1194; pCA-HA-PH-RA domain, aa 1–370. The cDNA for the GAP activity-defective mutant, R399A, was reported previously (15). The cDNA for a PH domain mutant of RA-RhoGAP (PH MT) was generated by replacing lysine at position 95, arginine at position 96, arginine at position 100, and histidine at position 104 with alanines using the QuikChange site-directed mutagenesis kit (Stratagene, Loyola, CA). The cDNA for an RA domain mutant of RA-RhoGAP (RA MT) was similarly generated by replacing lysine at position 199 and arginine at position 240 were replaced with glutamic acids. The cDNAs of the mutants were subcloned into the pCA vectors with N-terminal HA tags. The cDNA of DGKβ was provided by Dr. K. Goto (Yamagata University, Yamagata, Japan) and subcloned into the pCA vector with a C-terminal Myc tag. The cDNAs of Rap1B-G12V and Rap1B-Q63E were provided by Dr. M. Matsuda (Kyoto University, Kyoto, Japan) and subcloned into the pCA vectors with N-terminal Myc tags. Bacterial expression vectors for GST-fused PH domains (aa 1–273) of RA-RhoGAP and PH MT were constructed by subcloning the cDNAs into pGEX vectors.
Antibodies
A mouse anti-HA monoclonal Ab (mAb) was purchased from Babco (Richmond, CA). A rabbit anti-c-Myc polyclonal Ab (pAb) and a mouse anti-GST mAb were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). A rabbit anti-RA-RhoGAP pAb was purchased from LifeSpan Biosciences, Inc. (Seattle).
Protein-Lipid Overlay Assay
GST-fused PH domain (GST-PH domain WT or GST-PH domain MT) was generated in Escherichia coli and purified. Protein-lipid overlay assays were performed using PIP StripsTM (Echelon Biosciences, Salt Lake City, UT). PIP StripsTM were blocked in 1% skim milk in PBS-T (0.1% Tween 20 in PBS) at room temperature for 1 h. The strips were then incubated with 0.4 mg of GST-PH domain WT or GST-PH domain MT at 4 °C overnight. Then the strips were washed three times in PBS-T and incubated with the anti-GST mAb (1:2000) at room temperature for 1 h. The strips were then incubated with a horseradish peroxidase-conjugated anti-mouse IgG antibody (1:2000; GE Healthcare) at room temperature for 1 h. The signals were detected by enhanced chemiluminescence (ECL) (GE Healthcare).
Liposome Co-sedimentation Assay
Liposomes composed of phosphatidylcholine (PC), both PC and PA, both PC and PI(3)P, or both PC and PI(3,4)P2 (weight ratio 6:4), were incubated with 1 μg of the purified GST-PH domain WT or GST-PH domain MT in 100 μl of buffer (50 mm Tris/HCl, pH 7.4, and 100 mm NaCl). After incubation at 30 °C for 30 min, the reaction mixtures were centrifuged at 10,000 × g for 10 min. Pellets were then subjected to SDS-PAGE and immunoblotted with the anti-GST mAb.
RhoGAP Assay
RhoGAP assay was performed as described previously (15, 30). Lipid-modified Rap1B, RhoA, and His-RA-RhoGAP proteins were generated in Sf9 cells using the baculovirus expression system as described previously (15, 31, 32). The unprenylated form of Rap1B-G12V was generated in E. coli and purified. Lipid-modified RhoA (3 pmol) was incubated in 10 μl of reaction mixture containing 20 mm Tris/HCl, pH 8.0, 10 mm EDTA, 5 mm MgCl2, 0.4 mm dithiothreitol, 0.3% CHAPS, and 1.67 μm [γ-32P]GTP (1 × 104 cpm/pmol) at 30 °C for 10 min. The GTP loading was terminated by adding 5 μl of 80 mm MgCl2. For dose-dependent effects of PA on the GAP activity, His-RA-RhoGAP (0.12 pmol) and liposomes composed of PC (12 nmol); PC (11.78 nmol) and PA (0.22 nmol); PC (11.45 nmol) and PA (0.55 nmol); PC (10.9 nmol) and PA (1.1 nmol); PC (9.8 nmol) and PA (2.2 nmol); or PC (7.6 nmol) and PA (4.4 nmol) were incubated at 30 °C for 30 min. In some instances, water-soluble dioctanoylphosphatidic acid was incubated instead of the PA-containing liposomes. The samples were mixed with 12.5 μl of [γ-32P]GTP-loaded RhoA in a total volume of 50 μl and further incubated at 25 °C for 3 min. For dose-dependent effects of unprenylated Rap1 on the GAP activity, His-RA-RhoGAP (0.12 pmol) and various amounts of the GTPγS-bound form of unprenylated Rap1B-G12V (0.5, 1.0, 2.5, 5, 10, and 20 pmol) were incubated at 30 °C for 30 min, followed by incubation with [γ-32P]GTP-loaded RhoA at 25°C for 3 min. For comparison of the effects of PA and Rap1 on the GAP activity, His-RA-RhoGAP (0.12 pmol), His-RA-RhoGAP (0.12 pmol) and the GTPγS-bound form of lipid-modified Rap1B (Rap1) (5 pmol), His-RA-RhoGAP (0.12 pmol) and liposomes composed of PC (10.9 nmol) and PA (1.1 nmol), or His-RA-RhoGAP (0.12 pmol), Rap1 (5 pmol) and liposomes composed of PC (10.9 nmol) and PA (1.1 nmol) were incubated at 30 °C for 30 min. The samples were mixed with 12.5 μl of [γ-32P]GTP-loaded RhoA in a total volume of 50 μl and further incubated at 25 °C for the indicated periods of time. Then the mixture was applied to a nitrocellulose filter, and the radioactivity retained on the filter was measured by Cerenkov counting.
Knockdown of RA-RhoGAP by the RNA Interference (RNAi) Method
The mammalian expression vector, pSUPER (33), was used for expression of small interfering RNA (siRNA) in NG108 cells. RA-RhoGAP siRNA and the scrambled siRNA were designed as described previously (15). In some instances, stealth siRNAs (Invitrogen) were designed toward RA-RhoGAP (5′-AAUGCUGGCAGAAUGGCUGUCUCUG-3′ and 5′-UAAAUAAGCUAGGUGGCUGCUGGCC-3′) and used for knockdown.
Quantification of Neurites
NG108 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). To activate group I mGluRs, NG108 cells were cultured in DMEM supplemented with 1% FBS in the presence or absence of 10 μm DHPG for 1 or 2 days. To induce Rap1B-mediated neurite outgrowth, NG108 cells were cultured in DMEM supplemented with 1% FBS in the presence or absence of 0.6 mm 8CPT-cAMP for 2 days and processed for immunocytochemistry (15, 34). Images were captured using a confocal laser-scanning microscope (LSM 510-V3.2; Carl Zeiss, Inc., Jena, Germany) using a ×20 objective. Collected data were exported as 8-bit TIFF files and processed using Adobe Photoshop software. For the neurite outgrowth assay, 40–60 cells randomly chosen in each group were measured for morphological parameters in a single experiment, and the statistical values of the morphological parameters were obtained from three independent experiments. The number of primary neurites per cell was defined as the number of thin cell processes with a length longer than one cell diameter. The length of individual neurites for each cell was measured by use of the LSM Image Browser software (Zeiss). Neurite length was defined as the traced distance from the neurite tip to the point where the neurite emanated from the cell body. Neurite length encompasses all visible parts of a neurite without the length of its branches. Collaterals longer than one cell diameter were counted as branches. The statistical significance of differences between each group was analyzed by the two-tailed Student's t test.
RESULTS
Specific Binding of the PH Domain of RA-RhoGAP to Phosphatidic Acid
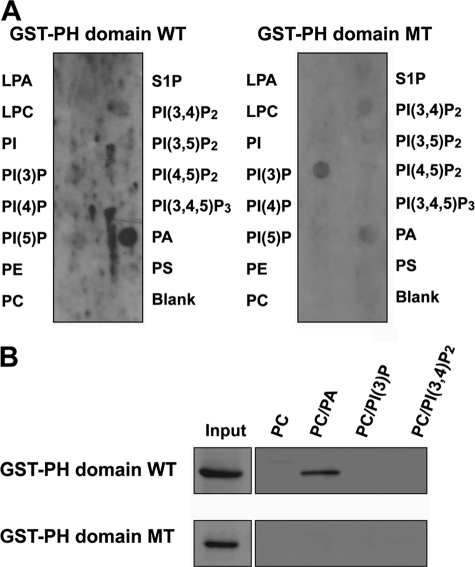
PH domains are well known as phospholipid-binding domains (35, 36). To examine which phospholipid preferentially bound to the PH domain of RA-RhoGAP, we performed a protein-lipid overlay assay using purified GST-fused PH domain of RA-RhoGAP (GST-PH domain WT). GST-PH domain WT bound strongly to PA (Fig. 1A). The faint bindings of GST-PH domain WT to PI(3)P and PI(3,4)P2 were also detected. Previous site-directed mutagenesis studies of the PH domains identified four conserved basic residues essential for the binding to phospholipids (37, 38). Therefore, we mutated the four residues of the PH domain of RA-RhoGAP to alanines (PH domain MT) and generated GST-fused PH domain MT (GST-PH domain MT). GST-PH domain MT very weakly bound to PA relative to GST-PH domain WT (Fig. 1A). On the other hand, the mutations did not affect the faint bindings to PI(3)P and PI(3,4)P2, suggesting that these bindings are dispensable. These results indicate that the PH domain of RA-RhoGAP binds to PA preferentially. To further confirm the binding to PA, we performed a liposome co-sedimentation assay. GST-PH domain WT was significantly co-sedimented with the liposome composed of PC and PA but not with the liposome composed of PC, PC and PI(3)P, or PC and PI(3,4)P2 (Fig. 1B). GST-PH domain MT was not co-sedimented with the liposome composed of PC, PC, and PA, PC and PI(3)P, or PC and PI(3,4)P2. In good agreement with the above protein-lipid overlay assay, these results indicate that the PH domain of RA-RhoGAP binds to PA.
FIGURE 1.
Specific binding of the PH domain of RA-RhoGAP to PA. A, phospholipid-binding ability of GST-PH domain WT or GST-PH domain MT. PIP StripsTM were incubated with GST-PH domain WT or GST-PH domain MT (0.1 mg/ml), and proteins bound to lipids were detected with the anti-GST mAb. LPA, lysophosphatidic acid; LPC, lysophosphocholine; PE, phosphatidylethanolamine; S1P, sphingosine 1-phosphate; PS, phosphatidylserine. B, liposome co-sedimentation assay. Liposomes composed of PC, both PC and PA, both PC and PI(3)P, or both PC and PI(3,4)P2 were incubated with 1 μg of GST-PH domain WT or GST-PH domain MT at 30 °C for 30 min. The reaction mixtures were then centrifuged, and the pellets, as well as input proteins, were analyzed by immunoblotting with the anti-GST mAb.
Enhancement of GAP Activity of RA-RhoGAP by PA
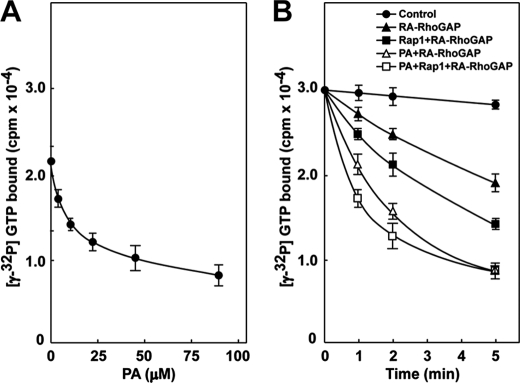
We previously demonstrated that RA-RhoGAP has a GAP activity toward RhoA, and the GAP activity is enhanced by prenylated Rap1B binding to the RA domain of RA-RhoGAP (15). To exclude the possibility that prenylated Rap1B might recruit RA-RhoGAP to liposomes accompanying RhoA, we examined the effect of unprenylated Rap1B on the GAP activity. Unprenylated Rap1B enhanced the GAP activity in a dose-dependent manner (supplemental Fig. 1A), indicating that Rap1 binding to the RA domain allosterically enhances the GAP activity. To examine the effect of PA binding on the GAP activity of RA-RhoGAP, we measured the GAP activity in the absence or presence of various combinations of PA and the GTPγS-bound form of prenylated Rap1B (Rap1). First, we titrated PA. [γ-32P]GTP-bound RhoA was incubated with His-RA-RhoGAP in the presence of PC-based liposomes containing various amounts of PA. The GAP activity of RA-RhoGAP was enhanced in a PA concentration-dependent manner (Fig. 2A). In addition, we also observed enhancement of the GAP activity by using dioctanoylphosphatidic acid, a soluble form of PA, instead of the PA liposomes (supplemental Fig. 1B). These results indicate that PA binding to the PH domain allosterically enhances the GAP activity of RA-RhoGAP. Next, we measured the GAP activity in the presence of PC-based liposomes containing PA, Rap1, or both PC-based liposomes containing PA and Rap1. Consistent with the above results and the previous report (15), the GAP activity was enhanced by PA and Rap1, respectively. The GAP activity was enhanced by both PA and Rap1 more than PA or Rap1 alone (Fig. 2B). These results indicate that PA binding to the PH domain enhances the GAP activity of RA-RhoGAP independently of Rap1.
FIGURE 2.
Enhancement of GAP activity of RA-RhoGAP by PA. A, enhancement of GAP activity of RA-RhoGAP by PA. [γ-32P]GTP bound to RhoA was incubated with 2.4 nm His-RA-RhoGAP and liposomes composed of PC (240 μm); PC (236 μm) and PA (4.5 μm); PC (229 μm) and PA (11 μm); PC (217.5 μm) and PA (22.5 μm); PC (195 μm) and PA (45 μm); or PC (150 μm) and PA (90 μm) for 3 min. After incubation, the hydrolysis of [γ-32P]GTP bound to RhoA was assayed by measuring the radioactivity of [γ-32P]GTP bound to RhoA using the nitrocellulose filtration method. B, enhancement of GAP activity of RA-RhoGAP by PA and Rap1. [γ-32P]GTP bound to RhoA was incubated with 2.4 nm His-RA-RhoGAP, His-RA-RhoGAP, and 100 nm GTPγS-bound form of Rap1B (Rap1), His-RA-RhoGAP, and liposomes composed of PC (195 μm) and PA (45 μm), His-RA-RhoGAP, 100 nm Rap1, and liposomes composed of PC (195 μm) and PA (45 μm) or buffer (Control) for the indicated periods of time. After incubation, the hydrolysis of [γ-32P]GTP bound to RhoA was assayed by measuring the radioactivity of [γ-32P]GTP bound to RhoA using the nitrocellulose filtration method. The results shown are representative of three experiments.
Involvement of PA Binding to the PH Domain in the Neurite Extension by Activation of GAP Activity of RA-RhoGAP
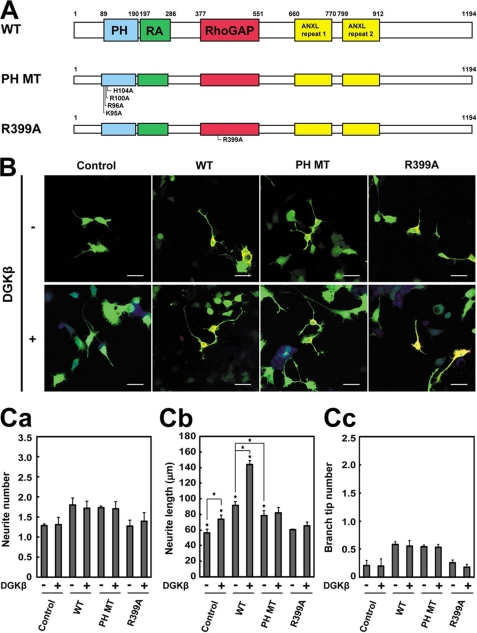
We have previously shown that RA-RhoGAP regulates the neurite outgrowth (15). It also has been reported that DGKβ, an enzyme to produce PA (22, 23), regulates the neurite outgrowth (28). Therefore, we transfected various constructs of RA-RhoGAP (Fig. 3A) and/or DGKβ into NG108 cells and examined the effect of PA binding to RA-RhoGAP on the neurite outgrowth. We first overexpressed DGKβ in NG108 cells. Overexpression of DGKβ increased the length of neurites (Fig. 3, B and C, panel b). However, overexpression of DGKβ did not affect the number and branching of neurites (Fig. 3C, panels a and c), indicating that overexpression of DGKβ preferentially induces neurite extension. We next overexpressed RA-RhoGAP, RA-RhoGAP-PH MT (the PA binding-defective mutant), or RA-RhoGAP-R399A (a GAP activity-defective mutant) or co-overexpressed each of them with DGKβ. Consistent with the previous report (15), overexpression of RA-RhoGAP increased the number, length, and branching of neurites relative to untransfected control cells (Fig. 3, B and C). Co-overexpression of RA-RhoGAP and DGKβ further increased the length of neurites but not the number and branching of neurites relative to overexpression of RA-RhoGAP (Fig. 3, B and C). These results indicate that DGKβ enhances RA-RhoGAP-mediated neurite extension. Overexpression of RA-RhoGAP-PH MT also increased the number, length, and branching of neurites relative to untransfected control cells (Fig. 3, B and C). However, overexpression of RA-RhoGAP-PH MT increased the length of neurites less than overexpression of RA-RhoGAP (Fig. 3C, panel b). Co-overexpression of RA-RhoGAP-PH MT and DGKβ did not increase the length of neurites as well as the number and branching, relative to overexpression of RA-RhoGAP-PH MT (Fig. 3, B and C). These results indicate that DGKβ requires PA binding to the PH domain of RA-RhoGAP to enhance RA-RhoGAP-mediated neurite extension. Consistent with the previous report that the GAP activity is essential for inducing neurite outgrowth (15), overexpression of RA-RhoGAP-R399A did not increase the length, number, and branching of neurites relative to untransfected control cells (Fig. 3, B and C). Indeed, co-overexpression of RA-RhoGAP-R399A and DGKβ did not increase the length, number, and branching of neurites relative to overexpression of RA-RhoGAP-R399A (Fig. 3, B and C). Overall, these results indicate that PA binding to RA-RhoGAP is preferentially involved in the neurite extension and suggest that DGKβ activates the GAP activity of RA-RhoGAP through provision of PA, leading to exclusive neurite extension.
FIGURE 3.
Involvement of PA binding to the PH domain in the RA-RhoGAP-mediated neurite extension. A, schematic structure of RA-RhoGAP and its point mutants. B, effects of RA-RhoGAP and its mutants on the neurite outgrowth in the DGKβ-untransfected or DGKβ-transfected NG108 cells. NG108 cells were transfected with GFP (Control) or co-transfected with GFP as a morphological marker along with HA-RA-RhoGAP (WT), HA-RA-RhoGAP-PH MT (PH MT), HA-RA-RhoGAP-R399A (R399A), myc-DGKβ (DGKβ), both myc-DGKβ and HA-RA-RhoGAP, both myc-DGKβ and HA-RA-RhoGAP-PH MT, or both myc-DGKβ and HA-RA-RhoGAP-R399A, cultured in DMEM supplemented with 10% FBS for 48 h, and allowed to extend neurites. The transfected cells were identified by the expression of GFP (green), and the expressions of HA-RA-RhoGAP and myc-DGKβ were examined by doubly staining with the anti-HA mAb (red) and the anti-Myc pAb (blue). Bars, 50 μm. C, quantitative analysis of the neurites outgrowth of the transfected NG108 cells. 40–60 neurons were measured for morphological parameters in a single experiment, and the statistical values of the morphological parameters were obtained from three independent experiments. Panel a, quantitative analysis of the number of neurites per transfected cell. Panel b, quantitative analysis of the length of neurites per transfected cell. Panel c, quantitative analysis of the number of branch tips per transfected cell. Error bars indicate S.D. Asterisks indicate statistical significance (Student's t test; *, p < 0.01).
Involvement of RA-RhoGAP in the Neurite Extension Downstream of DGKβ
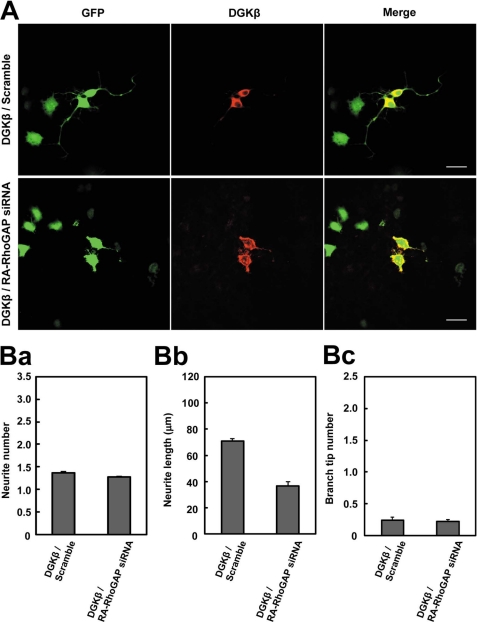
To further confirm that RA-RhoGAP preferentially regulated the neurite extension downstream of DGKβ, we performed knockdown of RA-RhoGAP in the DGKβ-overexpressing cells by using the RNAi method. Scramble RNA or RA-RhoGAP siRNA, the same one used in the previous study (15), was co-transfected with DGKβ in NG108 cells. Knockdown of RA-RhoGAP reduced the neurite extension of the DGKβ-overexpressing cells (Fig. 4, A and B, panel b). However, the knockdown of RA-RhoGAP did not affect the number and branching of neurites of DGKβ-overexpressing cells (Fig. 4B, panels a and c). We also designed two other stealth siRNAs targeting RA-RhoGAP (supplemental Fig. 2A) and obtained the similar results by using these siRNAs (supplemental Fig. 2, B and C). Human RA-RhoGAP gene is not a target of the above RA-RhoGAP siRNAs that are designed toward both mouse and rat genes. Indeed, expression of exogenous human RA-RhoGAP in the RA-RhoGAP knockdown cells overexpressing DGKβ rescued the RNAi-mediated perturbation of neurite outgrowth (supplemental Fig. 3), ruling out the possibility of the off-target effects. Therefore, these results indicate that RA-RhoGAP preferentially regulates the neurite extension downstream of DGKβ.
FIGURE 4.
Involvement of RA-RhoGAP in the neurite extension downstream of DGKβ. A, effect of RA-RhoGAP knockdown on the neurite outgrowth in the DGKβ-overexpressing NG108 cells. NG108 cells were co-transfected with GFP as a morphological marker along with both scramble RNA (Scramble) and myc-DGKβ or both RA-RhoGAP siRNA and myc-DGKβ, cultured in DMEM supplemented with 10% FBS for 48 h, and allowed to extend neurites. The cells expressing the RA-RhoGAP siRNA or the scramble RNA were identified by the expression of GFP (green). The expression of myc-DGKβ was examined by immunostaining with the anti-Myc pAb (red). It is noted that the RA-RhoGAP siRNA is the same one as used in the previous study (15). Bars, 50 μm. B, quantitative analysis of the neurite outgrowth of the transfected NG108 cells. Panel a, quantitative analysis of the number of neurites per transfected cell as in Fig. 3C, panel a. Panel b, quantitative analysis of the length of neurites per transfected cell as in Fig. 3C, panel b. Panel c, quantitative analysis of the number of branch tips per transfected cell as in Fig. 3C, panel c.
Involvement of RA-RhoGAP in the Neurite Extension Downstream of Metabotropic Glutamate Receptors (mGluRs)
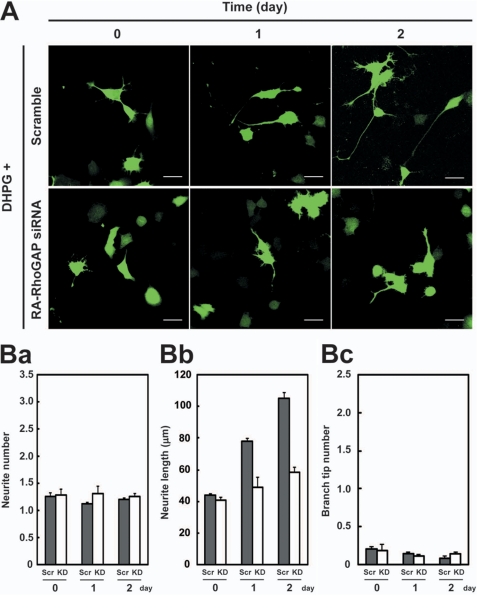
Previous studies show that activation of group I mGluRs (mGluR1 and mGluR5) induces production of DAG through phospholipase C (21, 39, 40). DAG is a substrate of DGKβ for PA production (22, 23). NG108 cells expressed mGluR1, mGluR5, and DGKβ as detected by RT-PCR analysis (supplemental Fig. 4). Therefore, we examined the effect of activation of mGluRs on the neurite outgrowth in NG108 cells. NG108 cells were cultured in the absence or presence of dihydroxyphenylglycine (DHPG), a group I mGluRs agonist (39). In the presence of DHPG, NG108 cells extended longer neurites than in the absence of DHPG (supplemental Fig. 5). The number and branching of the neurites did not increase in the presence of DHPG. These results indicate that activation of mGluRs preferentially induces neurite extension. Next, we examined whether RA-RhoGAP was involved in the mGluRs-mediated neurite extension. We knocked down RA-RhoGAP in NG108 cells by siRNA that was the same one used in the previous study (15) and cultured in the presence of DHPG. The knockdown of RA-RhoGAP reduced the mGluRs-mediated neurite extension (Fig. 5, A and B, panel b). However, the knockdown of RA-RhoGAP did not affect the number and branching of neurites (Fig. 5B, panels a and c). We also obtained similar results by using the two other stealth RA-RhoGAP siRNAs (supplemental Fig. 6). In addition, expression of exogenous human RA-RhoGAP in the knockdown cells rescued the RNAi-mediated perturbation of neurite outgrowth in the presence DHPG (supplemental Fig. 7), ruling out the possibility of the off-target effects. These results indicate that RA-RhoGAP preferentially regulates the neurite extension downstream of mGluRs.
FIGURE 5.
Involvement of RA-RhoGAP in the mGluR-mediated neurite extension. A, effect of RA-RhoGAP knockdown on the mGluR-mediated neurite outgrowth in NG108 cells. NG108 cells were co-transfected with GFP as a morphological marker along with the scramble RNA (Scramble) or RA-RhoGAP siRNA, cultured in DMEM supplemented with 1% FBS in the presence of 10 μm DHPG for the indicated periods of time, and allowed to extend neurites. The cells expressing the RA-RhoGAP siRNA or scramble RNA were identified by expression of GFP (green). It is noted that the RA-RhoGAP siRNA is the same one as used in the previous study (15). Bars, 50 μm. B, quantitative analysis of the neurites outgrowth of the transfected NG108 cells. Scr, scramble RNA; KD, RA-RhoGAP siRNA. Panel a, quantitative analysis of the number of neurites per transfected cell as in Fig. 3C, panel a. Panel b, quantitative analysis of the length of neurites per transfected cell as in Fig. 3C, panel b. Panel c, quantitative analysis of the number of branch tips per transfected cell as in Fig. 3C, panel c.
We next overexpressed various constructs of RA-RhoGAP in the absence or presence of DHPG and examined the effect of PA binding to RA-RhoGAP on the mGluRs-mediated neurite extension. Overexpression of RA-RhoGAP in the presence of DHPG increased the length of neurites but not the number and branching relative to overexpression of RA-RhoGAP in the absence of DHPG (supplemental Fig. 8, A and B), supporting that RA-RhoGAP preferentially regulates the neurite extension downstream of mGluRs. Overexpression of RA-RhoGAP-PH MT in the presence of DHPG did not increase the length of neurites as well as the number and branching, relative to overexpression of RA-RhoGAP-PH MT in the absence of DHPG (supplemental Fig. 8, A and B). These results indicate that PA binding to the PH domain is required for RA-RhoGAP-mediated neurite extension downstream of mGluRs. Overexpression of RA-RhoGAP-R399A in the presence of DHPG did not increase the length, number, and branching of neurites relative to overexpression of RA-RhoGAP-R399A in the absence of DHPG (supplemental Fig. 8, A and B), supporting that the GAP activity is essential for inducing neurite outgrowth. Activation of mGluRs had the same effect on the RA-RhoGAP-mediated neurite outgrowth as overexpression of DGKβ. Collectively, these results indicate that activation of mGluRs enhances the GAP activity of RA-RhoGAP through PA production by DGKβ, leading to exclusive neurite extension.
No Involvement of PA Binding to the PH Domain in the Rap1-mediated Neurite Outgrowth
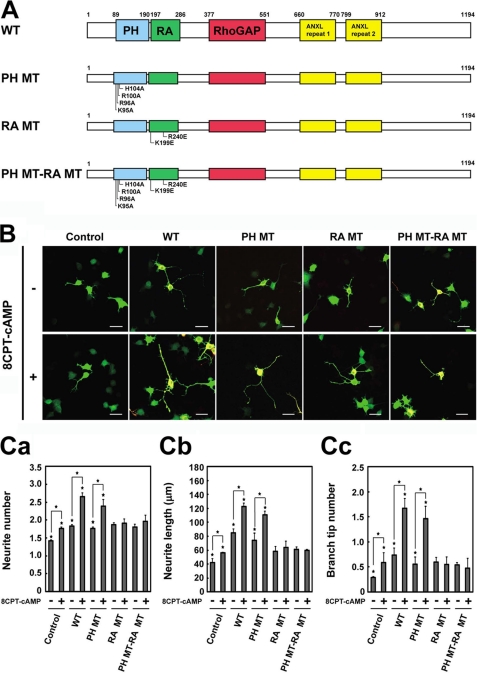
Upon GTP loading, Rap1 binds to the RA domain of RA-RhoGAP and activates the GAP activity, leading to neurite outgrowth (15). We have previously shown that 8-pCPT-2′-O-Me-cAMP (8CPT-cAMP), an activator of Epac, a Rap1GEF, induces neurite outgrowth of NG108 cells through activation of Rap1 followed by activation of RA-RhoGAP (15). Therefore, we overexpressed various constructs of RA-RhoGAP (Fig. 6A) in the absence or presence of 8CPT-cAMP in NG108 cells, and we examined whether PA binding to the PH domain was involved in the Rap1-RA-RhoGAP-mediated neurite outgrowth. Consistent with the previous report (15), NG108 cells in the presence of 8CPT-cAMP increased the length, number, and branching of neurites relative to those in the absence of 8CPT-cAMP (Fig. 6, B and C). Overexpression of RA-RhoGAP in the presence of 8CPT-cAMP further increased the length, number, and branching of neurites relative to overexpression of RA-RhoGAP in the absence of 8CPT-cAMP (Fig. 6, B and C), indicating that activation of Rap1 enhances RA-RhoGAP-mediated neurite outgrowth. Similarly, overexpression of RA-RhoGAP-PH MT in the presence of 8CPT-cAMP further increased the length, number, and branching of neurites relative to overexpression of RA-RhoGAP-PH MT in the absence of 8CPT-cAMP (Fig. 6, B and C). These results indicate the PA binding defect has no effect on the Rap1-RA-RhoGAP-mediated neurite outgrowth. Previous site-directed mutagenesis studies of RA domains identified two conserved basic residues essential for the binding to Rap1 (41, 42). Therefore, we generated RA-RhoGAP-RA MT by mutating the two residues of the RA domain of RA-RhoGAP to glutamic acids (Fig. 6A). These mutations abolished the binding ability of RA-RhoGAP to Rap1 (data not shown). Consistent with the previous report that the Rap1 binding is essential for the Rap1-RA-RhoGAP-mediated neurite outgrowth (15), overexpression of RA-RhoGAP-RA MT in the presence of 8CPT-cAMP did not increase the length, number, and branching of neurites relative to overexpression of RA-RhoGAP-RA MT in the absence of 8CPT-cAMP (Fig. 6, B and C). Furthermore, we generated RA-RhoGAP-PH MT-RA MT, where the PH and RA domains were double mutated. Overexpression of RA-RhoGAP-PH MT-RA MT in the presence of 8CPT-cAMP did not increase the length, number, and branching of neurites relative to overexpression of RA-RhoGAP-PH MT-RA MT in the absence of 8CPT-cAMP (Fig. 6, B and C). Therefore, these results suggest that PA binding to the PH domain is not involved in the Rap1-RA-RhoGAP-mediated neurite outgrowth.
FIGURE 6.
No involvement of PA binding to the PH domain in the 8CPT-cAMP-induced neurite outgrowth. A, schematic structure of RA-RhoGAP and its point mutants. B, effects of RA-RhoGAP and its mutants on the 8CPT-cAMP-induced neurite outgrowth in NG108 cells. NG108 cells were transfected with GFP (Control) or co-transfected with GFP as a morphological marker along with HA-RA-RhoGAP (WT), HA-RA-RhoGAP-PH MT (PH MT), HA-RA-RhoGAP-RA MT (RA MT), or HA-RA-RhoGAP-PH MT-RA MT (PH MT-RA MT), cultured in DMEM supplemented with 1% FBS in the presence or absence of 0.6 mm 8CPT-cAMP for 48 h, and allowed to extend neurites. The transfected cells were identified by the expression of GFP (green), and the expression of HA-RA-RhoGAP was examined by immunostaining with the anti-HA mAb (red). Bars, 50 μm. C, quantitative analysis of the neurite outgrowth of the transfected NG108 cells. Panel a, quantitative analysis of the number of neurites per transfected cell as in Fig. 3C, panel a. Panel b, quantitative analysis of the length of neurite per transfected cell as in Fig. 3C, panel b. Panel c, quantitative analysis of the number of branch tips per transfected cell as in Fig. 3C, panel c. Asterisks indicate statistical significance (Student's t test; *, p < 0.01).
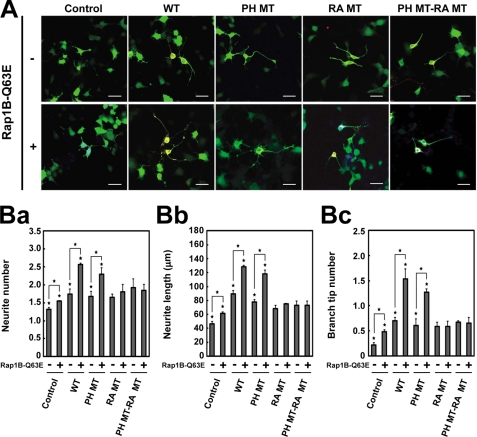
To confirm these results, we overexpressed Rap1B-Q63E, a constitutively active mutant of Rap1B and/or various constructs of RA-RhoGAP in NG108 cells. Consistent with the previous report (15), overexpression of Rap1B-Q63E increased the number, length, and branching of neurites relative to untransfected control cells (Fig. 7, A and B). Co-overexpression of Rap1B-Q63E and RA-RhoGAP further increased the number, length, and branching of neurites relative to overexpression of RA-RhoGAP (Fig. 7, A and B), indicating that constitutively active Rap1 enhances RA-RhoGAP-mediated neurite outgrowth. Similarly, co-overexpression of Rap1B-Q63E and RA-RhoGAP-PH MT further increased the number, length, and branching of neurites relative to overexpression of RA-RhoGAP-PH MT (Fig. 7, A and B). These results indicate that the PA-binding defect has no effect on the Rap1-RA-RhoGAP-mediated neurite outgrowth. By contrast, co-overexpression of Rap1B-Q63E and RA-RhoGAP-RA MT did not increase the number, length, and branching of neurites relative to overexpression of RA-RhoGAP-RA MT (Fig. 7, A and B). Co-overexpression of Rap1B-Q63E and RA-RhoGAP-PH MT-RA MT also did not increase the number, length, and branching of neurites relative to overexpression of RA-RhoGAP-PH MT-RA MT (Fig. 7, A and B). Furthermore, we obtained identical results by using Rap1B-G12V, an alternative constitutively active mutant (supplemental Fig. 9). Overall, these results indicate that PA binding to the PH domain is not involved in the Rap1-RA-RhoGAP-mediated neurite outgrowth.
FIGURE 7.
No involvement of PA binding to the PH domain in the Rap1B-Q63E-mediated neurite outgrowth. A, effects of RA-RhoGAP and its mutants on the Rap1B-Q63E-mediated neurite outgrowth in NG108 cells. NG108 cells were transfected with GFP (Control) or co-transfected with GFP as a morphological marker along with HA-RA-RhoGAP (WT), HA-RA-RhoGAP-PH MT (PH MT), HA-RA-RhoGAP-RA MT (RA MT), HA-RA-RhoGAP-PH MT-RA MT (PH MT-RA MT), myc-Rap1B-Q63E, both myc-Rap1B-Q63E and HA-RA-RhoGAP, both myc-Rap1B-Q63E and HA-RA-RhoGAP-PH MT, both myc-Rap1B-Q63E and HA-RA-RhoGAP-RA MT, or both myc-Rap1B-Q63E and HA-RA-RhoGAP-PH MT-RA MT, cultured in DMEM supplemented with 10% FBS for 48 h, and allowed to extend neurites. The transfected cells were identified by the expression of GFP (green), and the expressions of HA-RA-RhoGAP and myc-Rap1B-Q63E were examined by doubly staining with the anti-HA mAb (red) and the anti-Myc pAb (blue). Bars, 50 μm. B, quantitative analysis of the neurites outgrowth of the transfected NG108 cells. Panel a, quantitative analysis of the number of neurites per transfected cell as in Fig. 3C, panel a. Panel b, quantitative analysis of the length of neurites per transfected cell as in Fig. 3C, panel b. Panel c, quantitative analysis of the number of branch tips per transfected cell as in Fig. 3C, panel c. Asterisks indicate statistical significance (Student's t test; *, p < 0.01).
Involvement of PA Binding to the PH Domain in the Localization of RA-RhoGAP at Plasma Membrane
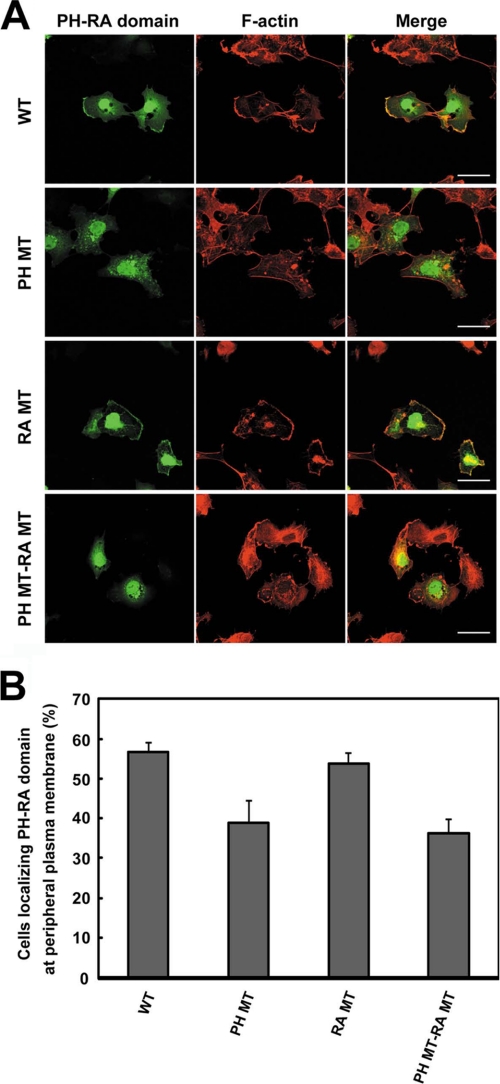
It has been reported that PH domain-containing proteins localize at the membrane by binding of the PH domains to phospholipids (35, 43). Therefore, we examined whether binding of the PH domain of RA-RhoGAP to PA was responsible for membrane localization. Although we first transfected full-length RA-RhoGAP into COS7 cells, exogenous RA-RhoGAP caused extensive morphological changes due to the excess GAP activity, complicating assessment of membrane localization (data not shown). To avoid the GAP activity-dependent morphological changes, we generated an N-terminal fragment of RA-RhoGAP encompassing both the PH and RA domains (PH-RA domain) but not the GAP domain, and we transfected it into COS7 cells. The exogenous overexpression of PH-RA domain did not cause any morphological changes and was dominantly localized at the peripheral plasma membrane as visualized by F-actin staining (Fig. 8). Next, we overexpressed the PH MT-RA domain (the PA binding-defective mutant). Notably, the localization of PH MT-RA domain at the peripheral plasma membrane significantly decreased as compared with that of the PH-RA domain. It has been reported that some of RA domain-containing proteins localize at the membrane by binding of the RA domains to Rap1 (44, 45). Therefore, we also overexpressed the PH-RA MT domain (the Rap1 binding-defective mutant) or the PH MT-RA MT domain (the mutant defective in both PA binding and Rap1 binding). PH-RA MT domain was dominantly localized at the peripheral plasma membrane similarly to the PH-RA domain, suggesting that binding of the RA domain of RA-RhoGAP to Rap1 is dispensable for the membrane localization. However, the localization of the PH MT-RA MT domain at the peripheral plasma membrane significantly decreased as compared with that of PH-RA domain or PH-RA MT domain. These results indicate that binding of the PH domain to PA is responsible for the localization of RA-RhoGAP at the plasma membrane.
FIGURE 8.
Requirement of PA binding to the PH domain for the localization of RA-RhoGAP at plasma membrane. A, localization of PH-RA domain and its mutants in COS7 cells. COS7 cells were transfected with HA-PH-RA domain (WT), HA-PH MT-RA domain (PH MT), HA-PH-RA MT domain (RA MT), or HA-PH MT-RA MT domain (PH MT-RA MT) and cultured for 36 h. The cells were immunostained with the anti-HA mAb (green) and phalloidin (red). The staining of F-actin was used for detection of peripheral plasma membrane. Bars, 50 μm. B, percentage of the cells localizing PH-RA domain at peripheral plasma membrane. The analysis of localization was performed on 40–60 transfected cells.
DISCUSSION
During neurite outgrowth, Rho GTPase is spatiotemporally regulated to organize the neurite sprouting, extension, and branching (1, 7, 8, 11, 12, 17). RA-RhoGAP is a key regulator of the Rho-mediated neurite outgrowth (15). GTP-bound Rap1 binds to RA-RhoGAP through the RA domain, leading to activation of the GAP activity (15). In this study, we have shown that PA binds to RA-RhoGAP through the PH domain, leading to activation of the GAP activity. Therefore, PA and Rap1 cooperatively regulate RA-RhoGAP activity for neurite outgrowth. PA and Rap1 have been shown to regulate morphological development of neurons (46–48). The dual activation mechanisms of RA-RhoGAP by PA and Rap1 will provide a possible link between PA signaling and Rap1 signaling in neuronal morphology.
Rho is inactivated in growth cones of neurites (17). In agreement, RA-RhoGAP exclusively localizes at the growth cones (15). Although we demonstrated that either PA or Rap1 binding to RA-RhoGAP was sufficient for inducing neurite outgrowth, the PA binding and the Rap1 binding showed distinct morphological effects. Activation of RA-RhoGAP by Rap1 binding induced neurite sprouting, extension, and branching, whereas activation of RA-RhoGAP by PA binding exclusively induced neurite extension without sprouting and branching. These results indicate that spatiotemporal regulation of Rho activity for neurite outgrowth is defined by the activation mode of RA-RhoGAP rather than subcellular localization of RA-RhoGAP. Rap1 is transiently activated at the tips of the sprouting neurites (16). However, sustained inactivation of Rho is required for the extension and branching of neurites after sprouting (11, 12, 17). Our present results raise an attractive possibility that sequential activation of RA-RhoGAP by Rap1 and PA will underlie the sustained inactivation of Rho, leading to morphological development of neurites.
Neurons initiate morphological development by sprouting several immature neurites (1–3, 8). The neurons moderately extend the immature neurites and select one of them as a prospective axon (2, 3, 8). Then, the neurons further extend the neurites and branch them, and finally acquire axonal and dendritic identities (2, 3, 8). Exclusive neurite extension as observed by PA-mediated activation of RA-RhoGAP is characteristic of axonal and dendritic growth in the late stage of the neuronal development (3, 8). On the other hand, neurite sprouting and extension as observed by Rap1-mediated activation of RA-RhoGAP appear to be important for the initial stage of the neuronal development. Indeed, Rap1 has been shown to localize at tips of immature growing neurites and direct axon specification (16). Therefore, developing neurons would undergo temporary inactivation of Rho by Rap1 binding to RA-RhoGAP for immature neurite sprouting, extension, and axon specification, followed by sustained inactivation of Rho by PA binding to RA-RhoGAP for axonal and dendritic growth. Growing axon and dendrite properly undergo branching and extension of the neurites, resulting in arborization (3, 8). Given that Rap1-mediated activation of RA-RhoGAP induces neurite branching, growing axon and dendrite would temporarily inactivate Rho by Rap1 binding to RA-RhoGAP for branching, followed by sustained inactivation of Rho by PA binding to RA-RhoGAP for extending the branched neurites. Further studies will be required to address the possible involvement of sequential activation of RA-RhoGAP by Rap1 and PA in spatiotemporal control of Rho activity for morphological development of neurons.
We demonstrated here that DGKβ produced PA responsible for activation of RA-RhoGAP, leading to neurite outgrowth. Spatiotemporal accumulation of specific lipids has been shown to be involved in neurite outgrowth (49, 50). Local accumulation of phosphatidylinositol 3,4,5-trisphosphate produced by PI3K induced neurite outgrowth by recruiting PH domain-containing GEFs followed by activation of Rac1/Cdc42 (51). Recently, it was reported that accumulation of PA produced by DGKβ induces neurite outgrowth (48). PA binding to the PH domain of RA-RhoGAP was involved in membrane localization (Fig. 8). Although PA binding to the PH domain significantly promoted membrane localization of RA-RhoGAP, the PH MT-RA MT domain still localized at the plasma membrane in ∼40% of the transfected cells (Fig. 8B). These results raise the possibility that the N-terminal region of RA-RhoGAP may have a weak activity to localize at the membrane independently of PA binding. In this case, PA binding to the PH domain would be involved in stabilizing membrane localization of RA-RhoGAP rather than targeting to membrane. Collectively, these results suggest that, in analogy with phosphatidylinositol 3,4,5-trisphosphate, local accumulation of PA produced by DGKβ particularly in growth cones will promote membrane localization of RA-RhoGAP through the PH domain, leading to inactivation of Rho for inducing neurite outgrowth. Genetic ablation of DGKβ causes a pronounced defect in dendritic spine formation (48). Therefore, the recruitment and activation of RA-RhoGAP by DGKβ-produced PA may be of particular importance in dendritic spine development. PA is also produced by phospholipase D (19), and evidence is accumulating that phospholipase D regulates neurite outgrowth (47). However, it remains to be elucidated whether phospholipase D regulates activity of RA-RhoGAP for neurite outgrowth. In addition to the biochemical roles of PA as a second messenger, PA has an intrinsic physical property to destabilize lipid packing and promote membrane curvature due to its cone-shaped geometry (20, 52). In biological systems, membrane curvature required for morphological change is supposed to be caused by coordinated action between cytoskeletal reorganization and lipid packing (53–55). Therefore, the activation mechanism of RA-RhoGAP by PA might underlie coordination between Rho-regulated actin reorganization and PA-induced membrane curvature for neurite outgrowth.
mGluRs are activated by glutamate released from presynaptic nerve terminals as a neurotransmitter and mediate synaptic transmission by yielding second messengers, including DAG, in the postsynapses (21). We demonstrate here that activation of group I mGluRs followed by provision of DAG were responsible for triggering the DGKβ-PA-RA-RhoGAP-Rho signaling pathway, leading to neurite outgrowth. These results strongly suggest that the DGKβ-PA-RA-RhoGAP-Rho pathway has a potential ability to convert synaptic activities to cytoskeletal reorganization, leading to activity-dependent morphological change of neurons. Neurons are known to frequently change their morphology in response to synaptic activities for synapse formation underlying synaptic plasticity and neuronal circuit reorganization (56, 57). The activity-dependent morphological change is particularly evident in dendritic spines (58–60), and mGluRs and DGKβ are reported to be the molecules that regulate spine formation (48, 61). Therefore, mGluR-triggered activation of the DGKβ-PA-RA-RhoGAP-Rho pathway may mediate activity-dependent spine remodeling. Evidence is accumulating that mGluRs and DGKβ are involved in synaptic plasticity (48, 62). mGluRs and DGKβ are expressed in the hippocampus (28, 63) and regulate long term potentiation in the CA1 area (48, 62). In our preliminary experiments, RA-RhoGAP was also expressed in the hippocampus and localized at the dendritic spines in cultured hippocampal neurons (data not shown). Therefore, the possible activity-dependent spine remodeling mediated by the mGluR-triggered DGKβ-PA-RA-RhoGAP-Rho pathway might be involved in regulation of synaptic plasticity such as hippocampal long term potentiation. Further studies, including generation of RA-RhoGAP-deficient mice, will be required to address these concerns.
In summary, we have described here that, in addition to the Rap1-RA-RhoGAP-Rho pathway, an alternative signaling pathway, the DGKβ-PA-RA-RhoGAP-Rho pathway, plays a pivotal role in the neurite outgrowth.
Supplementary Material
Acknowledgments
We thank Drs. K. Goto (Yamagata University, Yamagata, Japan) and M. Matsuda (Kyoto University, Kyoto, Japan) for kindly providing us with the cDNA of DGKβ and the cDNA of Rap1B, respectively. We also thank Dr. T. Nakamura (Tokyo University of Science, Chiba, Japan) for helpful discussion.
This work was supported by grants-in-aid for scientific research and for the Global COE Program F11 from the Ministry of Education, Culture, Sports, Science, and Technology, by a grant provided by The Japan Epilepsy Research Foundation, and by the grant provided by Hyogo Science and Technology Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–9.
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- mGluR
- metabotropic glutamate receptor
- DAG
- diacylglycerol
- DGK
- diacylglycerol kinase
- PA
- phosphatidic acid
- mAb
- monoclonal antibody
- pAb
- polyclonal antibody
- GST
- glutathione S-transferase
- siRNA
- small interfering RNA
- PI
- phosphatidylinositol
- PC
- phosphatidylcholine
- PH
- pleckstrin homology
- RA
- Ras-associating domain
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- DHPG
- dihydroxyphenylglycine
- 8CPT-cAMP
- 8-pCPT-2′-O-Me-cAMP
- aa
- amino acid
- PI(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PI(3)P
- phosphatidylinositol 3-phosphate.
REFERENCES
- 1. da Silva J. S., Dotti C. G. (2002) Nat. Rev. Neurosci. 3, 694–704 [DOI] [PubMed] [Google Scholar]
- 2. Horton A. C., Ehlers M. D. (2003) Neuron 40, 277–295 [DOI] [PubMed] [Google Scholar]
- 3. Dotti C. G., Sullivan C. A., Banker G. A. (1988) J. Neurosci. 8, 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goslin K., Banker G. (1989) J. Cell Biol. 108, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradke F., Dotti C. G. (2000) Curr. Biol. 10, 1467–1470 [DOI] [PubMed] [Google Scholar]
- 6. Mitchison T., Kirschner M. (1988) Neuron 1, 761–772 [DOI] [PubMed] [Google Scholar]
- 7. Luo L. (2000) Nat. Rev. Neurosci. 1, 173–180 [DOI] [PubMed] [Google Scholar]
- 8. Govek E. E., Newey S. E., Van Aelst L. (2005) Genes Dev. 19, 1–49 [DOI] [PubMed] [Google Scholar]
- 9. Jalink K., Eichholtz T., Postma F. R., van Corven E. J., Moolenaar W. H. (1993) Cell Growth Differ. 4, 247–255 [PubMed] [Google Scholar]
- 10. Hirose M., Ishizaki T., Watanabe N., Uehata M., Kranenburg O., Moolenaar W. H., Matsumura F., Maekawa M., Bito H., Narumiya S. (1998) J. Cell Biol. 141, 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sebök A., Nusser N., Debreceni B., Guo Z., Santos M. F., Szeberenyi J., Tigyi G. (1999) J. Neurochem. 73, 949–960 [DOI] [PubMed] [Google Scholar]
- 12. Da Silva J. S., Medina M., Zuliani C., Di Nardo A., Witke W., Dotti C. G. (2003) J. Cell Biol. 162, 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takai Y., Sasaki T., Matozaki T. (2001) Physiol. Rev. 81, 153–208 [DOI] [PubMed] [Google Scholar]
- 14. Dickson B. J. (2001) Curr. Opin. Neurobiol. 11, 103–110 [DOI] [PubMed] [Google Scholar]
- 15. Yamada T., Sakisaka T., Hisata S., Baba T., Takai Y. (2005) J. Biol. Chem. 280, 33026–33034 [DOI] [PubMed] [Google Scholar]
- 16. Schwamborn J. C., Püschel A. W. (2004) Nat. Neurosci. 7, 923–929 [DOI] [PubMed] [Google Scholar]
- 17. Picard M., Petrie R. J., Antoine-Bertrand J., Saint-Cyr-Proulx E., Villemure J. F., Lamarche-Vane N. (2009) Cell. Signal. 21, 1961–1973 [DOI] [PubMed] [Google Scholar]
- 18. Nishizuka Y. (1992) Science 258, 607–614 [DOI] [PubMed] [Google Scholar]
- 19. Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 20. Stace C. L., Ktistakis N. T. (2006) Biochim. Biophys. Acta 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 21. Hermans E., Challiss R. A. (2001) Biochem. J. 359, 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakane F., Imai S., Kai M., Yasuda S., Kanoh H. (2007) Biochim. Biophys. Acta 1771, 793–806 [DOI] [PubMed] [Google Scholar]
- 23. Cai J., Abramovici H., Gee S. H., Topham M. K. (2009) Biochim. Biophys. Acta 1791, 942–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. (2001) Science 294, 1942–1945 [DOI] [PubMed] [Google Scholar]
- 25. Zhao C., Du G., Skowronek K., Frohman M. A., Bar-Sagi D. (2007) Nat. Cell Biol. 9, 706–712 [DOI] [PubMed] [Google Scholar]
- 26. Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. (2009) Science 324, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Itoh T., Hasegawa J., Tsujita K., Kanaho Y., Takenawa T. (2009) Sci. Signal. 2, ra52 [DOI] [PubMed] [Google Scholar]
- 28. Hozumi Y., Watanabe M., Otani K., Goto K. (2009) BMC Neurosci. 10, 99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 30. Fukui K., Sasaki T., Imazumi K., Matsuura Y., Nakanishi H., Takai Y. (1997) J. Biol. Chem. 272, 4655–4658 [DOI] [PubMed] [Google Scholar]
- 31. Mizuno T., Kaibuchi K., Yamamoto T., Kawamura M., Sakoda T., Fujioka H., Matsuura Y., Takai Y. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Umikawa M., Obaishi H., Nakanishi H., Satoh-Horikawa K., Takahashi K., Hotta I., Matsuura Y., Takai Y. (1999) J. Biol. Chem. 274, 25197–25200 [DOI] [PubMed] [Google Scholar]
- 33. Hisata S., Sakisaka T., Baba T., Yamada T., Aoki K., Matsuda M., Takai Y. (2007) J. Cell Biol. 178, 843–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakisaka T., Baba T., Tanaka S., Izumi G., Yasumi M., Takai Y. (2004) J. Cell Biol. 166, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lemmon M. A., Ferguson K. M. (2000) Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 36. Lemmon M. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 37. Wakamatsu I., Ihara S., Fukui Y. (2006) Mol. Cell. Biochem. 293, 137–145 [DOI] [PubMed] [Google Scholar]
- 38. Jackson S. G., Zhang Y., Bao X., Zhang K., Summerfield R., Haslam R. J., Junop M. S. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 324–330 [DOI] [PubMed] [Google Scholar]
- 39. Conn P. J., Pin J. P. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 205–237 [DOI] [PubMed] [Google Scholar]
- 40. Bellone C., Lüscher C., Mameli M. (2008) Cell. Mol. Life Sci. 65, 2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vetter I. R., Linnemann T., Wohlgemuth S., Geyer M., Kalbitzer H. R., Herrmann C., Wittinghofer A. (1999) FEBS Lett. 451, 175–180 [DOI] [PubMed] [Google Scholar]
- 42. Liu C., Takahashi M., Li Y., Song S., Dillon T. J., Shinde U., Stork P. J. (2008) Mol. Cell. Biol. 28, 7109–7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haubert D., Gharib N., Rivero F., Wiegmann K., Hösel M., Krönke M., Kashkar H. (2007) EMBO J. 26, 3308–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao Y., Satoh T., Gao X., Jin T. G., Hu C. D., Kataoka T. (2001) J. Biol. Chem. 276, 28478–28483 [DOI] [PubMed] [Google Scholar]
- 45. Song C., Hu C. D., Masago M., Kariyai K., Yamawaki-Kataoka Y., Shibatohge M., Wu D., Satoh T., Kataoka T. (2001) J. Biol. Chem. 276, 2752–2757 [DOI] [PubMed] [Google Scholar]
- 46. Chen Y., Wang P. Y., Ghosh A. (2005) Mol. Cell. Neurosci. 28, 215–228 [DOI] [PubMed] [Google Scholar]
- 47. Kanaho Y., Funakoshi Y., Hasegawa H. (2009) Biochim. Biophys. Acta 1791, 898–904 [DOI] [PubMed] [Google Scholar]
- 48. Shirai Y., Kouzuki T., Kakefuda K., Moriguchi S., Oyagi A., Horie K., Morita S. Y., Shimazawa M., Fukunaga K., Takeda J., Saito N., Hara H. (2010) PLoS ONE 5, e11602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi S. H., Jan L. Y., Jan Y. N. (2003) Cell 112, 63–75 [DOI] [PubMed] [Google Scholar]
- 50. Ménager C., Arimura N., Fukata Y., Kaibuchi K. (2004) J. Neurochem. 89, 109–118 [DOI] [PubMed] [Google Scholar]
- 51. Aoki K., Nakamura T., Fujikawa K., Matsuda M. (2005) Mol. Biol. Cell 16, 2207–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cazzolli R., Shemon A. N., Fang M. Q., Hughes W. E. (2006) IUBMB Life 58, 457–461 [DOI] [PubMed] [Google Scholar]
- 53. Sheetz M. P. (2001) Nat. Rev. Mol. Cell Biol. 2, 392–396 [DOI] [PubMed] [Google Scholar]
- 54. Kooijman E. E., Chupin V., de Kruijff B., Burger K. N. (2003) Traffic 4, 162–174 [DOI] [PubMed] [Google Scholar]
- 55. McMahon H. T., Gallop J. L. (2005) Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 56. Zito K., Svoboda K. (2002) Neuron 35, 1015–1017 [DOI] [PubMed] [Google Scholar]
- 57. Yuste R., Bonhoeffer T. (2004) Nat. Rev. Neurosci. 5, 24–34 [DOI] [PubMed] [Google Scholar]
- 58. Engert F., Bonhoeffer T. (1999) Nature 399, 66–70 [DOI] [PubMed] [Google Scholar]
- 59. Maletic-Savatic M., Malinow R., Svoboda K. (1999) Science 283, 1923–1927 [DOI] [PubMed] [Google Scholar]
- 60. Yang Y., Wang X. B., Frerking M., Zhou Q. (2008) J. Neurosci. 28, 5740–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vanderklish P. W., Edelman G. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1639–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anwyl R. (2009) Neuropharmacology 56, 735–740 [DOI] [PubMed] [Google Scholar]
- 63. Lujan R., Nusser Z., Roberts J. D., Shigemoto R., Somogyi P. (1996) Eur. J. Neurosci. 8, 1488–1500 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.