Abstract
In alveolar epithelial cells (AECs), the membrane-anchored proteoglycan dystroglycan (DG) is a mechanoreceptor that transmits mechanical stretch forces to activate independently the ERK1/2 and the adenosine 5′-monophosphate-activated protein kinase (AMPK) signaling cascades in a process called pathway bifurcation. We tested the hypothesis that the cytoskeleton cross-linker plectin, known to bind both DG and AMPK in muscle cells, acts as a scaffold to regulate DG-mediated mechanical stimulation and pathway bifurcation. We demonstrate that plectin and DG form a complex in AECs and that this complex interacts with ERK1/2 and AMPK. Plectin knockdown reduces DG interaction with AMPK but not with ERK1/2. Despite this, mechanoactivation of both signaling pathways is significantly attenuated in AECs deficient in plectin. Thus, DG has the dual role of mechanical receptor and scaffold for ERK1/2, whereas plectin acts as a scaffold for AMPK signaling but is also required for DG-mediated ERK1/2 activation. We conclude that the DG-plectin complex plays a central role in transmitting mechanical stress from the extracellular matrix to the cytoplasm.
Keywords: Adaptor Proteins, AMP-activated Kinase (AMPK), Epithelial Cell, ERK, Signal Transduction, Dystroglycan, Equibiaxial Stretching, Mechanical Stretch, Pathway Bifurcation, Plectin
Introduction
In the lung, alveolar epithelial cells (AECs)3 not only mediate the exchange of gases between the circulation system of the host and its external environment but are also highly responsive to a number of mechanical forces (1). These forces include deformation and strain that occur during lung expansion and relaxation from breathing movements, and shear stress during the distension of the airway walls and blood vessels from bulk air and blood flow (2). Over the past few years, there has been increasing interest in identifying molecules that “sense” physical forces on the cell surface and in defining the signaling pathways activated by mechanical stimulation (1, 3–6). Stretch-activated ion channels, integrins, cell-cell adhesion molecules, cytoskeleton elements, and the extracellular matrix (ECM) have all been implicated in transducing mechanical signals in a manner that is detectable as chemical signals (e.g. protein phosphorylation) in the cytoplasm of the stimulated cell (1, 7).
We are interested in investigating the molecular underpinnings of cellular responses to physical force in rat AECs. In particular, we have previously tested the hypothesis that matrix molecules secreted by cultured AECs and transmembrane matrix receptors on the substratum surface of these cells are crucial molecular links in the process of “converting” a mechanical stimulus in the form of cyclic stretching into a cytoplasmic signal (8). Specifically, in prior studies we demonstrated that rat AECs assemble an ECM rich in fibers composed of the α3, β1, and γ1 subunits of laminin (laminin-311), complexed with perlecan and nidogen (8). This complex transmits mechanosignals in the form of stretch, via the matrix receptor dystroglycan (DG), to activate ERK1/2 (8). Moreover, we have also shown that DG is required for stretch-induced activation of the adenosine 5′-monophosphate-activated protein kinase (AMPK) signaling cascade in AECs in vitro and in vivo (9).
DG is a transmembrane cell surface protein expressed in muscle and the nervous system, as well as in epithelia and endothelia (10, 11). DG was first identified as a laminin-binding protein in the brain and also as a component of a multimeric transmembrane protein complex known as the dystrophin-glycoprotein complex in muscle (12–15). There is only one gene (dystrophin-associated glycoprotein 1) coding for DG in vertebrates, and its corresponding amino acid sequence is highly conserved (15, 16). Translation of the dystrophin-associated glycoprotein 1 mRNA gives rise to a polypeptide chain that is proteolytically cleaved into two noncovalently associated proteins, namely α-DG and β-DG (13, 17). On the cytoplasmic side of muscle cells, β-DG interacts with the actin cytoskeleton via utrophin and dystrophin, although in the extracellular milieu α-DG interacts with laminin (18–20). In addition, β-DG associates indirectly with desmin intermediate filaments via its interaction with plectin, a high molecular weight cytoskeletal cross-linker protein that has important roles in mechanical stabilization of cells and tissues (21, 22).
The associations listed above highlight the well documented important structural role that DG plays in mediating cytoskeleton-cell surface interactions in muscle cells. However, the functions of DG in non-muscle cells are just beginning to emerge (8, 9, 23–25). There is evidence that DG regulates matrix assembly in epithelial cells (26). In addition, the concept of outside-in signaling that is commonly associated with integrins (27–29) is also applicable to DG given the recent finding that laminin-DG engagement results in tyrosine phosphorylation of the β-DG cytoplasmic tail and thus disrupts DG binding to dystrophin/utrophin, thereby regulating DG-actin association (30). Moreover, our previous results directly implicate DG in two signal transduction pathways initiated by cyclic stretching of cultured rat AECs, namely the ERK1/2 and AMPK cascades (8, 9).
How DG regulates ERK1/2 and AMPK mechanoactivation is unknown. In this study, our goal was therefore to identify a molecular partner for DG in rat AECs that is involved in the transmission of mechanical signals to activate these two distinct signaling pathways. We surmised that a good candidate is a cytoskeletal component that is strategically positioned for both mechanical sensing and signaling. Thus, we hypothesized that the cytoskeletal cross-linker plectin may mediate mechanical stretch force transmission via DG. Our results indicate that plectin is expressed in AECs and not only forms a complex with DG but is also necessary for DG-mediated mechanical signaling.
EXPERIMENTAL PROCEDURES
Antibodies and Other Reagents
Polyclonal rabbit antibodies against lamin A/C, AMPK, ERK1/2, phosphorylated ERK1/2, phosphorylated acetyl-CoA carboxylase (ACC), and the monoclonal rabbit antibody against total ACC were purchased from Cell Signaling Technology Inc. (Beverly, MA). The rabbit monoclonal antibody against plectin was obtained from Epitomics (Burlingame, CA). The mouse monoclonal antibodies against β-DG (8D5 and MANDAG2/7D11) were obtained from Abcam Inc. (Cambridge, MA) and Dr. Glenn E. Morris (RJAH Orthopaedic Hospital, Oswestry, UK), respectively. Mouse monoclonal antibody against vimentin was purchased from Pharmingen. Cytochalasin D, the actin-disrupting drug, was purchased from Sigma. Pan-keratin rabbit polyclonal antibody was a gift from Dr. Robert D. Goldman (Northwestern University), and the microtubule-depolymerizing drug (colcemid) and mouse monoclonal antibody against tubulin (DM1α) were gifted by Dr. Vladimir Gelfand (Northwestern University). The function-blocking mouse monoclonal antibody (CM6) against the α3 laminin subunit was described previously (8). The specific AMPK inhibitor, compound C, was obtained from Calbiochem/EMD Biosciences Inc., and the MEK1/2 inhibitor, U0126, was purchased from Promega (Madison, WI).
Isolation of AECs
All studies using animals were approved by the Northwestern University Animal Care and Use Committee. Type II AECs were isolated from pathogen-free male Sprague-Dawley rats (200–225 g) as described previously (8, 31). Briefly, the lungs were perfused via the pulmonary artery, lavaged, and digested with elastase (3 units ml−1; Worthington). AECs were purified by differential adherence to immunoglobulin G, and cell viability was assessed by trypan blue exclusion (>95%). Cells were resuspended in Dulbecco's modified Eagle's medium (Sigma) containing 10% fetal bovine serum (Hyclone, Logan, UT) with 2 mm glutamine, 100 units ml−1 penicillin, 0.25 μg ml−1 amphotericin B, and 100 μg ml−1 streptomycin. For mechanical stretch experiments, ∼0.75–1.0 × 106 AECs were seeded in 6-well, 35-mm laminin, YIGSR peptide-coated and elastomer-bottomed Bioflex® plates (Flexcell International, Hillsborough, NC). For immunofluorescence assays on YIGSR-peptide-coated glass coverslips, about 0.15–0.2 × 106 cells were seeded. All cells were incubated in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. The day of isolation and plating was designated culture day 0. The purity of the AECs was determined to be 90 ± 5% by phase microscopic evaluation of epithelial cell morphology and by immunofluorescence microscopy using keratin antibodies as a marker of epithelial phenotype and vimentin antibody as a marker for mesenchymal cells (supplemental Fig. S1, A–F).
Cultured RLE-6TN Cells
RLE-6TN (rat lung epithelial-T-antigen negative) cells were a gift from Dr. Karen Ridge (Northwestern University) and were used as controls for immunofluorescence staining against rat vimentin protein. These cells are rat alveolar type II cells that have been immortalized and transformed with the SV40-T antigen gene as described previously (32). Cells were resuspended in Nutrient Mixture F-12 Ham's medium (Sigma) containing 10% fetal bovine serum with 2 mm glutamine, 100 units ml−1 penicillin, 0.25 μg ml−1 amphotericin B, and 100 μg ml−1 streptomycin. Cells were used after 8 passages in culture and seeded at a density of 0.05 × 106 for 3 days before being fixed and stained for immunofluorescence. All cells were incubated in a humidified atmosphere of 5% CO2 and 95% air at 37 °C.
Cyclic Stretching
AECs cultured for 3 days on Bioflex® culture plates were serum-starved for 24 h, and the cells were subjected to equibiaxial stretching (i.e. equal stretching in both x and y axis directions) the next day using the Cyclic Strain Unit FX-4000 (Flexcell International, Hillsborough, NC). This unit consists of a controlled vacuum unit and a base plate to hold the culture plates. A vacuum was cyclically applied to the Bioflex® plates using the Flexercell device with loading stations in place to impose equibiaxial stretching at 30 cycles per min and a stretching/relaxation ratio of 1:1, resulting in a 10% linear elongation of the membrane as measured microscopically. This is the same regimen that we have used in previous reports (8, 9, 33). Cells maintained on unstretched Bioflex® plates served as static controls.
Adenoviral Infection
We have previously described how recombinant adenoviruses encoding short hairpin RNAs (shRNAs) against rat DG were created and used (8), and rat plectin-specific shRNAs were created in a similar manner. Briefly, two sets of single-stranded oligonucleotides encoding the plectin shRNA and its complement were synthesized by Invitrogen as follows: 2736 forward, AAAAGCAACTGAATGAGTACAAAGGTTCGCCTTTGTACTCATTCAGTTGC, and 2736 reverse, CACCGCAACTGAATGAGTACAAAGGCGAACCTTGTAACTCATTCAGTTGC; 6615 forward, AAAAGCAGCAGACTCTTCAGCAAGATTCGTCTTGCTGAAGAGTCTGCTGC, and 6615 reverse, CACCGCAGCAGACTCTTCA-GCAAGACGAATCTTGCTGAAGAGTCTGCTGC.
The sequences were selected using an algorithm provided by Invitrogen. Each oligonucleotide pair was annealed, ligated into the pENTR/U6 vector, and used to transform competent Escherichia coli cells following an established protocol (Invitrogen). Plasmid DNA was isolated from the kanamycin-resistant colonies and sequenced. The pENTR/U6 construct was used in a recombination reaction with the adenoviral vector pAD/BLOCK-iT-DEST (Invitrogen), and the resulting shRNA adenoviral vector was linearized by PacI restriction digest and transfected into 293A cells using Lipofectamine following Invitrogen protocols. An adenoviral vector containing no DNA was used as a negative control. Approximately 12 days after transfection, the adenovirus-containing 293A cells were harvested and lysed to prepare a crude viral stock. The resultant viral stock was amplified, and viral concentration was determined. In experiments involving DG knockdown, the adenoviral shRNA against DG was added to cells at a multiplicity of infection of 1:10, similar to our previous studies (8, 9). However, for plectin knockdown experiments, we did not detect appreciable knockdown with each individual virus at a multiplicity of infection below 1:100. Therefore, AECs were infected with a mixture of the two viruses on day 2 at a total multiplicity of infection of 1:50 (1:25 of each). An adenoviral vector containing shRNA targeting human (but not rat) lamin A/C (supplied by Invitrogen) was processed the same way and used as a negative control for the adenoviral infection at a multiplicity of infection of 1:10 (for DG) or 1:50 (for plectin). Medium containing the virus was replaced the next day with fresh serum-free medium, and assays were performed 2 days after infection.
Immunofluorescence Microscopy
To immunostain for tubulin, keratin, and vimentin, the AECs or RLE-6TN cells on glass coverslips were extracted for 2 min in a 50:50 mixture of ice-cold acetone/methanol. To immunostain for actin, the AECs were fixed in a 3.7% formaldehyde/phosphate-buffered saline (PBS) solution for 15 min followed by 5 min of incubation in 25 mm ammonium chloride. To immunostain for DG/plectin, the AECs were fixed for 5 min in a solution of 40:10:5:45 acetone/formaldehyde (37% stock)/glacial acetic acid/water followed by an additional 5 min of wash in PBS. The cells were then incubated in blocking buffer (5% bovine serum albumin (BSA) in PBS) for an hour followed by incubation in appropriate primary antibodies in blocking buffer for another hour. After several washes with PBS, the cells were incubated in secondary antibodies in blocking buffer for another hour, washed again, and then mounted onto slides. All incubations involving glass coverslips were done at room temperature. In the case of immunofluorescence assays on Bioflex® plates, the cells were immediately washed in ice-cold PBS after mechanical treatment (static control or cyclic stretching). Cell fixation, immunostaining, and washing were all done on the Bioflex® plates at 4 °C, before the membranes were cut out and mounted onto slides. Prepared slides were viewed on a Zeiss LSM 510 laser-scanning confocal microscope (Carl Zeiss, Thornwood, NY) with either 40× Plan-NeoFluar 1.3 NA oil or 63× Plan-Apochromat 1.4 NA oil. The Zeiss Image Viewer software was used to export microscope images in the 8-bit TIFF format, which were subsequently imported into GIMP 2.6 (The GIMP Team) and Photoshop CS2 (Adobe Systems Inc., San Jose, CA) to generate figures.
Drug and Antibody Treatment
We first determined the minimum quantity of drug and time required for cytoskeleton disruption by processing AECs, 4 days post-isolation, for immunofluorescence using rhodamine-labeled phalloidin and tubulin antibody at various times after treatment. Treatment with 5 μm cytochalasin D for 1 h induced optimal disruption of the actin cytoskeleton, although 5 μm colcemid for 1 h induced disassembly of the microtubule network. For blocking AMPK activation (34) or ERK1/2 activation, AECs were treated with 200 nm of compound C or 10 μm of U0126, respectively, for 1 h.
Immunoprecipitation
AECs were seeded on 100-mm plastic dishes at a density of 5 × 106 per dish. On day 4 post-isolation, the cells were washed in cold PBS and lysed in RIPA buffer (0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 1 mm EGTA, 1 mm EDTA, 150 mm NaCl in 50 mm Tris-HCl, pH 7.5) containing a mixture of protease and phosphatase inhibitors. The cell lysate was sheared using a syringe fitted with a 25⅝-gauge needle to denature DNA, incubated in ice for 30 min, and centrifuged to pellet debris, and the supernatant was pre-cleared with agarose G beads. Supernatant was then divided into two portions with primary β-DG antibody (7D11) or plectin antibody being added to one portion of the supernatant and control isotype-matched antibody (mouse IgG for 7D11, rabbit IgG for plectin antibody) to the second portion. Both were incubated overnight at 4 °C. Agarose protein G beads were then used to pull down immunocomplexes from the supernatants, and the precipitated proteins were collected from the beads by incubation in sample buffer (80 mm Tris, pH 6.8, 3% SDS, 35% glycerol, 10% β-mercaptoethanol). The sample buffer was heated to 100 °C for 3 min and subsequently processed for SDS-PAGE and Western immunoblotting.
SDS-PAGE and Immunoblotting
Whole-cell extracts were prepared by solubilizing AECs in sample buffer consisting of 8 m urea, 1% SDS in 10 mm Tris-HCl, pH 6.8, and 15% β-mercaptoethanol (35). These extracts and immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose, and processed for immunoblotting as described previously (35–37). Immunoreactive antibodies against DG (8D5), AMPK, plectin, phospho-ERK1/2, phospho-JNK, or phospho-ACC (a downstream target of activated AMPK) were used and detected by enhanced chemiluminescence (Thermo Scientific, Rockford, IL). To control for protein loading, a set of gels was prepared in parallel, loaded with an equal volume of samples, and probed with antibodies against total ERK1/2, total ACC, total JNK, or lamin A/C. Immunoblots were scanned using Photoshop CS2 (Adobe Systems Inc., San Jose, CA) and quantified using MetaMorph (Molecular Devices, Downingtown, PA). Results from at least three independent experiments were combined and are presented as means ± S.E.
Data Analyses
All data sets were initially compared by analysis of variance. Significant differences between experimental conditions were explored with two-tailed, paired Student's t tests using STATA 10 (StataCorp LP, College Station, TX). A significant difference was prospectively identified as p < 0.05.
RESULTS
Cyclic Stretch Activates JNK and AMPK in a Laminin-311-dependent Manner in AECs
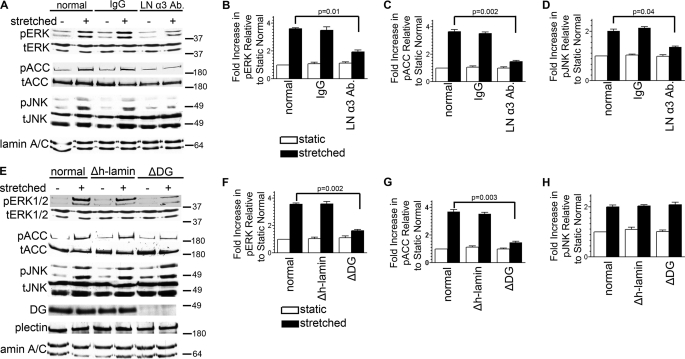
In a previous study we demonstrated that a function-blocking α3 laminin antibody (CM6) inhibits stretch activation of ERK1/2 in AECs (8). In addition to ERK1/2, several other pathways are activated in AECs by the stretch regimen we use, including the AMPK signaling cascade and the JNK pathway (Fig. 1A) (9, 38). To assess whether the α3 laminin subunit blocking antibody also inhibited these other pathways, AECs were subjected to stretch in the presence of the blocking antibody CM6 or control IgG, and then extracts of the cells were probed by immunoblotting with antibodies against phosphorylated ACC and JNK (Fig. 1A). The antibody against phosphorylated ACC is targeted against phosphoserine 79, a residue specifically phosphorylated by AMPK and not by protein kinase A (the alternative in vivo kinase for ACC) (39–42). Blockade of the laminin α3 subunit results in the attenuation of mechanical activation of both AMPK and JNK (Fig. 1, A–D).
FIGURE 1.
Laminin-311 and DG in activation of mechanosignaling in AEC. A–D, day 3 AECs were serum-starved for 24 h with untreated media (normal) or with media containing isotype-matched antibody (50 μg/ml) or the function-inhibiting antibody (CM6) against the α3 laminin subunit (LN α3 Ab) (50 μg/ml). E–H, day 2 AECs were incubated in untreated media (normal) or in media treated with adenovirus encoding either DG (ΔDG) or human lamin shRNA (Δh-lamin), as indicated. One day later, the cells were switched to serum-free media for 24 h. A–H, cells were exposed to equibiaxial stretch at 30 cycles per min for 10 min. Cell lysates were collected and immunoblotted against phosphorylated ACC (pACC) (as a marker for AMPK activation), total ACC (tACC), phosphorylated ERK (pERK), total ERK (tERK), phosphorylated JNK (pJNK) or total JNK (tJNK). In addition, extracts were probed with antibodies against DG and plectin to demonstrate knockdown of DG and that DG deficiency had no impact on plectin expression. Lamin A/C was used as a housekeeping loading control. The α3 laminin subunit antibody significantly reduced mechanical activation of the ERK1/2, AMPK, and JNK pathways. DG knockdown resulted in attenuated mechanoactivation of both AMPK and ERK1/2 but not JNK. All blots are representative of at least three independent experiments. p values in B, C, D, F, and G denote significance.
We have also previously shown that DG mediates stretch activation of the ERK1/2 and AMPK pathways in AECs (8, 9). Thus, we tested whether DG is required for JNK activation in mechanically stimulated AECs. To do so, we delivered shRNA against DG to AECs with adenovirus, which results in a knockdown in DG expression of ≥90% at 48 h post-infection compared with uninfected or control shRNA-treated cells (Fig. 1E) (8, 9). In extracts of AECs exhibiting DG knockdown, there is a dramatic attenuation of stretch-induced activation of ERK1/2 and AMPK, consistent with our previous studies (Fig. 1, E–G) (8, 9). However, DG knockdown does not impact JNK signaling induced by stretch (Fig. 1, E and H). These data indicate that DG is required for ERK1/2 and AMPK mechanoactivation in AECs, whereas a second mechanoreceptor, working through laminin-311/nidogen/perlecan, is required for activation of the JNK pathway in AECs. The identity of this receptor has yet to be determined.
Cyclic Stretch Activates Distinct, Independent Pathways in AECs
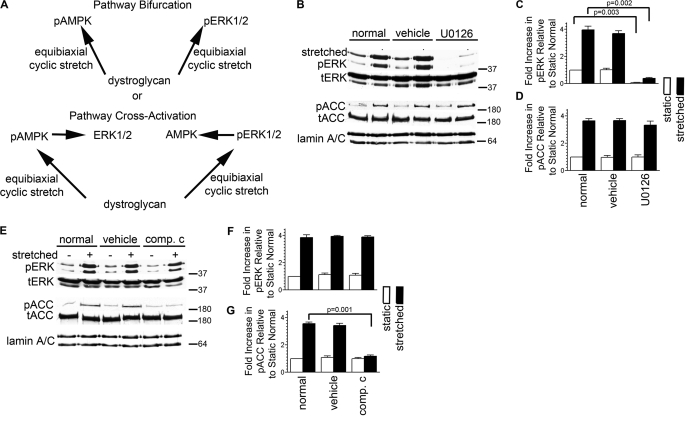
To assess whether DG in AECs activates the ERK1/2 and AMPK pathways independently or whether these pathways in some way cross-activate, we used a pharmacological approach involving U0126, used to block ERK1/2 activation by inhibiting MEK1/2, and compound C, which inhibits AMPK activation (Fig. 2A). We first determined the concentrations of each of these reagents needed to block the respective pathways (supplemental Fig. S2, A and B). U0126 shows no impact on stretch-induced activation of AMPK while blocking ERK1/2 mechanoactivation, consistent with our previous results (Fig. 2, B–D) (9). On the other hand, inhibiting AMPK activation using compound C does not block stretch-induced activation of ERK1/2 (Fig. 2, E–G). In other words, the mechanical stimulus transmitted through DG is bisected to activate independently two separate pathways. Given that the ERK1/2 and AMPK pathways share a common mechanical receptor yet they are distinct and have diverging downstream targets (43, 44), this is an example of signaling pathway bifurcation.
FIGURE 2.
DG mediates the bifurcation of mechanical signals to activate ERK1/2 and AMPK independently. A, model showing that a priori DG can signal in AECs to mediate either pathway bifurcation or cross-activation of ERK1/2 and AMPK pathways. B–G, day 3 AECs were serum-starved for 24 h and then they were incubated in untreated media (normal) or in media treated with vehicle (DMSO), U0126, or compound C (comp. c) for an hour prior to equibiaxial stretch, as indicated. Cell lysates were immunoblotted against phosphorylated ACC (pACC), total ACC (tACC), phosphorylated ERK (pERK), or total ERK (tERK). Lack of effect in one pathway whereas the other is inhibited demonstrates that DG mediates pathway bifurcation of mechanical signals in AECs. All blots are representative of at least three independent experiments. p values in C and G denote significance.
Is the Cytoskeleton Involved in Stretch Activation of Signal Pathways in AECs?
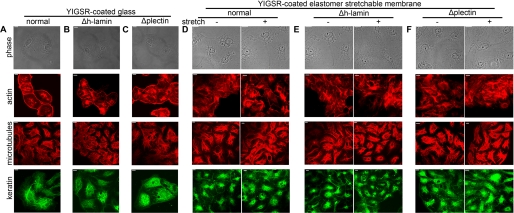
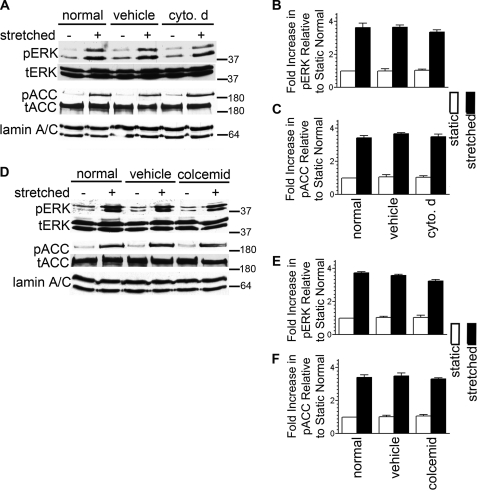
We next focused on identifying mediators of DG mechanosignaling in AECs. Our starting point was the ECM-receptor-cytoskeleton pathway, which Ingber (7) renamed “the tensegrity model” of signal transmission. In the model, mechanosignals are transduced through the structural organization and interconnectedness of the cytoskeleton. Microtubules act as struts, and the actin and intermediate filament cytoskeleton function as cables that provide a physical basis for translating mechanical forces into biochemical responses. Because this model predicts that transmembrane receptors such as DG physically couple the internal cytoskeleton networks to the ECM for mechanical signal transfer across the cell surface, we therefore investigated cytoskeleton involvement in transducing mechanical signals via DG in AECs. AECs possess networks of actin, tubulin, and keratin (Fig. 4A and supplemental Fig. S3, A, D, and G). They do not assemble a vimentin cytoskeleton network (supplemental Fig. S1B). We treated AECs with cytochalasin D and colcemid to disrupt the actin and microtubule cytoskeletons, respectively. Vehicle treatment does not affect the actin, microtubule, or keratin cytoskeletal networks (supplemental Fig. S3, A, D, and G). Collapse of the actin cytoskeleton is induced by cytochalasin D in AECs, but it has no effect on either the microtubule or keratin networks (supplemental Fig. S3, B, E, and H). Likewise, depolymerization of the microtubule cytoskeleton in AECs is induced by colcemid and has no effect on the actin network (supplemental Fig. S3, C and F). In addition, we were surprised to find that the keratin cytoskeleton is disrupted after colcemid treatment (supplemental Fig. S3I). Also surprising is the finding that both ERK1/2 and AMPK in AECs are robustly activated following stretch (Fig. 3, A–F) after cytochalasin D or colcemid treatment, suggesting that cytoskeleton association is not directly involved in DG-dependent mechanical signaling. This notion is further supported by the observation that the cytoskeletons of AEC do not undergo any dramatic reorganization following the stretch regimen we used in our studies (Fig. 4D).
FIGURE 4.
Plectin knockdown has little effect on cell shape or the cytoskeletal networks of AECs. A–F, day 2 AECs seeded on laminin YIGSR peptide-coated glass coverslips or elastomer membranes were either left untreated (normal) or were infected with adenovirus encoding human lamin shRNA (Δh-lamin) or plectin shRNA (Δplectin). The next day, cells were serum-starved, and 24 h later, cells on glass coverslips were fixed and immunostained for actin, tubulin, and keratin although those on elastomer membranes were either left static or were exposed to equibiaxial stretch at 30 cycles per min for 10 min before being fixed and immunostained for actin, tubulin, and keratin, as indicated. Phase contrast images of the cells (upper panels) reveal that cell shape or size was not perturbed either by plectin knockdown or stretch, and similarly there was no effect on cytoskeletal architecture (lower panels) under the same conditions. Scale bar, 10 μm.
FIGURE 3.
Cytoskeletal network disruption does not affect mechanical signaling in AECs. A–F, day 3 AECs were serum-starved. One day later they were incubated in untreated media (normal) or in media treated with vehicle (DMSO) or with the indicated drugs at 5 μm each for an hour. Cells were then exposed to equibiaxial stretch at 30 cycles per min for 10 min; cell lysates were collected and subsequently immunoblotted against phosphorylated ERK (pERK), total ERK (tERK), phosphorylated ACC (pACC). or total ACC (tACC). The results indicate that stretch-mediated pathway bifurcation and activation of ERK1/2 and AMPK are independent of an intact cytoskeleton. All blots are representative for at least three independent experiments. cyto. d, cytochalasin D.
Plectin Knockdown Has No Effect on the Cytoskeletal Networks of AECs
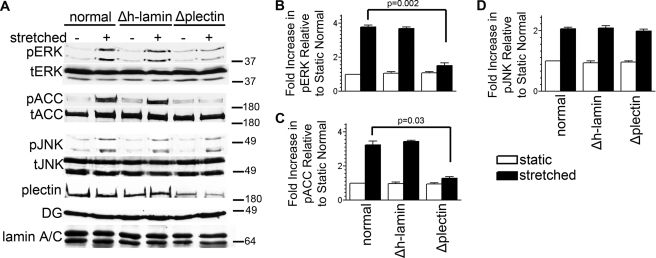
Because plectin interacts with DG in muscle cells and also has the ability to bind AMPK in these cells (22, 45), we hypothesized that plectin might mediate DG-dependent pathway bifurcation in AECs. To test this hypothesis, we first generated adenoviruses encoding plectin-specific shRNA. Two of our adenoviral stocks, encoding two distinct shRNAs targeting different regions of plectin, are able to induce knockdown of plectin expression in rat AECs. Extracts of AECs infected with a combination of these two viruses were processed for immunoblotting at 48 and 72 h after infection (supplemental Fig. S4A). Plectin expression is maximally reduced after 48 h by up to 72% compared with uninfected or human lamin shRNA-treated cells (supplemental Fig. S4B). We chose the 48-h time point for all subsequent experiments involving plectin knockdown because it matches our previous studies involving DG knockdown (8, 9) as well as studies outlined here (Fig. 1, E–H). Plectin knockdown has no obvious impact on the size and shape of AECs, when compared with untreated cells or AECs infected with human lamin shRNA (Fig. 4, A–F). Additionally, the actin, microtubule, and keratin networks in plectin knockdown cells are indistinguishable by immunofluorescence from those in control AECs (Fig. 4, A–C). More importantly, cyclic stretching has little discernible effects on the cytoskeletal architecture (Fig. 4, D–F).
Plectin Is Required for the Activation of ERK1/2 and AMPK in Response to Mechanical Stretch
We next assessed the impact of plectin deficiency on DG-mediated mechanical activation of the ERK1/2 and AMPK pathways in AECs. It has been reported that base-line activity of ERK1/2 is increased in mouse keratinocytes deficient in plectin (46). We do not see base-line activation of ERK1/2 or AMPK in plectin-deficient AECs (compare static controls with or without shRNA, Fig. 5, A–C). However, mechanoactivation of both ERK1/2 and AMPK in AECs expressing plectin shRNA is significantly reduced when compared with uninfected or control shRNA-treated cells (Fig. 5, A–C). In sharp contrast, plectin deficiency has no impact on JNK signaling in mechanically stimulated AECs (Fig. 5, A and D). These results suggest that plectin is not a global mechanosensor in AECs whose knockdown disrupts multiple signaling cascades. Rather, plectin seems to be involved in a specific cooperative and synergistic relationship with DG to mediate mechanical signaling of two particular pathways in AECs.
FIGURE 5.
Plectin is required for pathway bifurcation in AECs. A–D, day 2 AECs were left untreated (normal) or were treated with adenovirus encoding either human lamin shRNA (Δh-lamin) or plectin shRNA (Δplectin), as indicated. One day later, the cells were serum-starved and then 24 h later were exposed to equibiaxial stretch at 30 cycles per min for 10 min. Cell lysates were immunoblotted against phosphorylated ACC (pACC), total ACC (tACC), phosphorylated ERK (pERK), total ERK (tERK), phosphorylated JNK (pJNK), or total JNK (tJNK). Cell lysates were also immunoblotted using antibodies against DG and plectin to demonstrate that knockdown of plectin had no impact on DG expression. Plectin knockdown resulted in attenuated mechanoactivation of both AMPK and ERK1/2 but not JNK. All blots are representative of at least three independent experiments. p values in B and C denote significance.
Plectin and DG Assemble as a Scaffolding Complex of Signaling Molecules in AECs
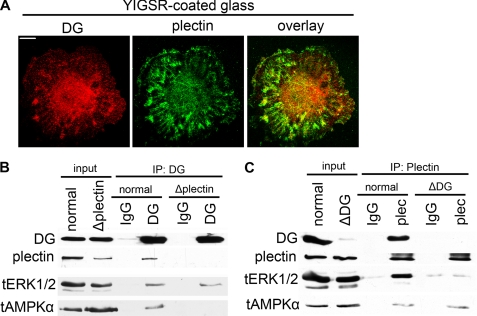
Although DG and plectin interact directly in muscle cells (22), their association in AECs has not been established. Immunofluorescence colocalization assays demonstrate that β-DG and plectin associate in AECs in perinuclear focal adhesion sites (Fig. 6A). To further confirm this association, we then performed immunoprecipitation assays on lysates of untreated, plectin-, or DG shRNA-treated AECs. Immunoblotting demonstrates that the DG antibody, but not control mouse IgG, is able to immunoprecipitate both DG and plectin from lysates of AECs (Fig. 6B). The DG antibody also coimmunoprecipitates both ERK1/2 and AMPK (Fig. 6B). In plectin-deficient AECs, the DG antibody immunoprecipitates only DG and ERK1/2 but not plectin and AMPK (Fig. 6B). In reciprocal immunoprecipitation assays, immunoblotting demonstrates that the plectin antibody, but not control rabbit IgG, immunoprecipitated both plectin and DG from lysates of AECs (Fig. 6C). Like the DG antibody, the plectin antibody also coimmunoprecipitates both ERK1/2 and AMPK (Fig. 6C). However, in DG-deficient AECs, only plectin and AMPK but not DG and ERK1/2 are immunoprecipitated (Fig. 6C). Thus, our results demonstrate that DG forms a scaffolding complex with plectin in AECs that recruits and tethers the kinases ERK1/2 and AMPK.
FIGURE 6.
Plectin forms a scaffolding complex with DG to recruit ERK1/2 and AMPK. A, day 3 AECs seeded on laminin YIGSR-peptide coated glass coverslips were serum-starved and then the next day fixed and immunostained for DG and plectin as indicated. The individual DG and plectin stains are overlaid in the 3rd panel. DG and plectin colocalize in perinuclear focal contacts. B and C, day 2 AECs were left untreated (normal) or were treated with adenovirus encoding either plectin shRNA (Δplectin) or DG shRNA (ΔDG), as indicated. One day later, the cells were serum-starved, and the next day cell lysates were then prepared for immunoprecipitation (IP) using anti-β-DG or anti-plectin antibodies as well as their respective species-matched IgG controls. Precipitated proteins were subjected to immunoblotting using antibodies against DG, plectin, ERK1/2, and AMPK. Results indicate that DG recruits ERK1/2 to the complex, whereas plectin recruits AMPK. Plectin knockdown blocks DG association with AMPK but not with ERK1/2, although DG knockdown blocks plectin association with ERK1/2 but not with AMPK. All blots are representative of at least three independent experiments.
DISCUSSION
Our data indicate that plectin is an important mediator of mechanosignaling in lung cells. In this regard, plectin is not traditionally associated with signaling. Rather, plectin is best known for its role binding to intermediate filaments, actin, and microtubules and as a mechanical stabilizer of cells and tissues by cross-linking cytoskeleton systems (21, 47). For example, humans with a mutation in the plectin gene that results in plectin deficiency develop skin blistering and muscular dystrophy (48, 49). The former is due to destabilization of keratinocyte adhesion to the ECM (50), although the latter is due to an absence of desmin interaction with DG in the dystrophin-associated glycoprotein complex of muscle cells (51). Thus, one explanation for our results is that plectin functions as a molecular Velcro that mechanically couples the cytoskeleton framework within AECs to the cell surface mechanical receptor(s) and their downstream signaling machinery. This possibility seems unlikely because we observe very little effect on the overall organization of the cytoskeleton in cells exhibiting plectin deficiency both before and after mechanical stretch. More importantly, disruption of the actin and microtubule cytoskeletons following treatment with drugs fails to impact stretch-activated signaling in AECs. We can also rule out a role for the keratin cytoskeleton because AMPK and ERK1/2 mechanoactivation are preserved despite the rather dramatic and unexpected effect of colcemid on keratin organization in these cells. We are somewhat surprised by these cytoskeleton studies because we had predicted that the cytoskeleton would be involved in mechanosignaling in AECs based on the tensegrity model.
In lung cells, we have demonstrated that plectin functions as a mechanosignaling scaffold, a finding consistent with other indications that plectin is an important signaling regulator. For example, plectin interacts with the nonreceptor tyrosine kinase Fer and attenuates its catalytic activity (52). Plectin also binds the scaffold for many kinases and receptors called the receptor for activated C kinase 1 (RACK1), thereby reducing the ability of RACK1 to scaffold and participate in signaling (53, 54). These examples involve signal inhibition by scaffold sequestration. In our system, plectin is a positive regulator of signaling. Indeed, this function is more akin to that of scaffolds that increase the efficiency of signal propagation through kinase cascades (55, 56), serving as adapters for kinase cascade activation via cross-linking kinases to their upstream or downstream partners (57, 58), and linking multiple components of a specific pathway by assembling unique signaling complexes (59). A classic example of such a scaffold is the yeast protein Ste5 involved in the ERK1/2 pathway. Ste5 is required for mating but not for the high osmolarity or starvation responses (60). Another scaffold protein Ste11 functions in at least three separate signaling pathways as follows: mating, the filamentous pathway, and the high osmolarity glycerol response (60). Because these three pathways share a common scaffold and yet are activated by different stimuli, Ste11 has evolved to activate only a single pathway in the presence of a specific stimulus, a phenomenon that is termed “pathway insulation” (60).
As we have reported here, mechanical signaling in AECs via plectin involves regulation of two signaling cascades that are activated by the same receptor (DG) from a single stimulus, a process termed pathway bifurcation. Plectin is clearly a molecular partner for DG to effect this bifurcation. Although the upstream ECM ligand for DG, namely laminin-311, appears to be involved in the activation of a third pathway, namely the JNK pathway, both DG and plectin appear to have no role in mechanoactivation of the JNK pathway but are specifically involved in pathway bifurcation and activation of ERK1/2 and AMPK. In addition, we have provided evidence that plectin serves as a molecular partner for DG using immunofluorescence colocalization assays and immunoprecipitation assays. Immunofluorescence colocalization images indicate that plectin and DG co-reside in perinuclear focal adhesion sites. Moreover, immunoprecipitation data indicate that plectin serves as a scaffold for AMPK, a finding consistent with evidence that plectin binds to AMPK in myocytes to regulate energy homeostasis (45). In addition, our data demonstrate that DG has a dual role by acting not only as the mechanical receptor to receive signals from the stretched ECM but also as a scaffold to recruit ERK1/2. This is in agreement with the finding that the β-DG cytoplasmic region has predicted ERK1/2 binding sequences in the juxtamembrane region (25). A combination of yeast two-hybrid and proteomic analyses also have demonstrated that components of the ERK pathway interact with the β-DG cytoplasmic tail (24).
Because plectin knockdown also results in the impairment of both AMPK and ERK1/2 activation, these findings imply the requirement of a molecular complex involving DG and plectin for proper mechanical signal transduction in AECs. In other words, DG, plectin, ERK1/2, and AMPK form a supramolecular complex that has the necessary steric and allosteric interactions to enable the mechanoactivation. Thus, at the cell surface of AECs, we propose that plectin and DG form a scaffold that regulates mechanical stimuli via tethering of signaling molecules, such as AMPK and ERK1/2, in the vicinity of their regulatory molecules and their upstream activators, and/or by determining access of specific intracellular substrates to their signaling binding partners. In the absence of one of the scaffolding partners, such as when DG or plectin expression is knocked down, we suggest that the remaining scaffold is conformationally deficient and unable to complete the requisite interactions for mechanical signal transduction.
In summary, our studies lead to a working model in which DG has a major role for mediating integrin-independent, matrix-driven mechanical signal transduction in AECs via plectin to activate the ERK1/2 and AMPK pathways. This model holds implications not only for the ways in which epithelial cells sense and respond to mechanical stimulation in general but also for the pathophysiology of certain lung diseases. Widespread use of positive pressure mechanical ventilation in the treatment of respiratory disorders is known to exacerbate existing lung damage (61–63). This injury might be exacerbated or mitigated by the activation of pro-inflammatory or protective signaling pathways in the lung. For example, our in vitro data suggest that the DG-mediated activation of AMPK signaling following mechanical stimulation of AECs may have some protective effects because it reduces generation of harmful reactive oxygen species (9). Likewise, activation of ERK1/2 has been reported to have beneficial effects at the cellular level (64, 65). It is clear that further dissection of the molecular underpinnings of the DG/plectin-mediated signaling may in the future help in the design of new approaches to ameliorate the negative effects of lung mechanical ventilation in vivo.
Supplementary Material
Acknowledgments
We are very grateful to Drs. Glenn E. Morris, Robert D. Goldman, Vladimir Gelfand, and Karen Ridge for their gifts of antibodies, reagents, and cells.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 HL092963 (to J. C. R. J. and G. R. S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- AEC
- alveolar epithelial cell
- DG
- dystroglycan
- AMPK
- adenosine 5′-monophosphate-activated protein kinase
- ECM
- extracellular matrix
- ACC
- acetyl-CoA carboxylase.
REFERENCES
- 1. Liu M., Tanswell A. K., Post M. (1999) Am. J. Physiol. 277, L667–L683 [DOI] [PubMed] [Google Scholar]
- 2. Wirtz H. R., Dobbs L. G. (2000) Respir. Physiol. 119, 1–17 [DOI] [PubMed] [Google Scholar]
- 3. Edwards Y. S. (2001) Comp. Biochem. Physiol. Part A, Mol. Inegr. Physiol. 129, 245–260 [DOI] [PubMed] [Google Scholar]
- 4. Martinac B. (2004) J. Cell Sci. 117, 2449–2460 [DOI] [PubMed] [Google Scholar]
- 5. Reichelt J. (2007) Eur. J. Cell Biol. 86, 807–816 [DOI] [PubMed] [Google Scholar]
- 6. Tsunozaki M., Bautista D. M. (2009) Curr. Opin. Neurobiol. 19, 362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ingber D. E. (1997) Annu. Rev. Physiol. 59, 575–599 [DOI] [PubMed] [Google Scholar]
- 8. Jones J. C., Lane K., Hopkinson S. B., Lecuona E., Geiger R. C., Dean D. A., Correa-Meyer E., Gonzales M., Campbell K., Sznajder J. I., Budinger S. (2005) J. Cell Sci. 118, 2557–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Budinger G. R., Urich D., DeBiase P. J., Chiarella S. E., Burgess Z. O., Baker C. M., Soberanes S., Mutlu G. M., Jones J. C. (2008) Am. J. Respir. Cell Mol. Biol. 39, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durbeej M., Campbell K. P. (1999) J. Biol. Chem. 274, 26609–26616 [DOI] [PubMed] [Google Scholar]
- 11. Shimizu H., Hosokawa H., Ninomiya H., Miner J. H., Masaki T. (1999) J. Biol. Chem. 274, 11995–12000 [DOI] [PubMed] [Google Scholar]
- 12. Smalheiser N. R., Schwartz N. B. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6457–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ibraghimov-Beskrovnaya O., Ervasti J. M., Leveille C. J., Slaughter C. A., Sernett S. W., Campbell K. P. (1992) Nature 355, 696–702 [DOI] [PubMed] [Google Scholar]
- 14. Gee S. H., Blacher R. W., Douville P. J., Provost P. R., Yurchenco P. D., Carbonetto S. (1993) J. Biol. Chem. 268, 14972–14980 [PubMed] [Google Scholar]
- 15. Smalheiser N. R., Kim E. (1995) J. Biol. Chem. 270, 15425–15433 [DOI] [PubMed] [Google Scholar]
- 16. Barresi R., Campbell K. P. (2006) J. Cell Sci. 119, 199–207 [DOI] [PubMed] [Google Scholar]
- 17. Bozzi M., Morlacchi S., Bigotti M. G., Sciandra F., Brancaccio A. (2009) Matrix Biol. 28, 179–187 [DOI] [PubMed] [Google Scholar]
- 18. Holt K. H., Crosbie R. H., Venzke D. P., Campbell K. P. (2000) FEBS Lett. 468, 79–83 [DOI] [PubMed] [Google Scholar]
- 19. Ervasti J. M., Ohlendieck K., Kahl S. D., Gaver M. G., Campbell K. P. (1990) Nature 345, 315–319 [DOI] [PubMed] [Google Scholar]
- 20. Ervasti J. M., Campbell K. P. (1991) Cell 66, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 21. Wiche G. (1998) J. Cell Sci. 111, 2477–2486 [DOI] [PubMed] [Google Scholar]
- 22. Rezniczek G. A., Konieczny P., Nikolic B., Reipert S., Schneller D., Abrahamsberg C., Davies K. E., Winder S. J., Wiche G. (2007) J. Cell Biol. 176, 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michele D. E., Campbell K. P. (2003) J. Biol. Chem. 278, 15457–15460 [DOI] [PubMed] [Google Scholar]
- 24. Spence H. J., Dhillon A. S., James M., Winder S. J. (2004) EMBO Rep. 5, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moore C. J., Winder S. J. (2010) Cell Commun. Signal. 8, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weir M. L., Oppizzi M. L., Henry M. D., Onishi A., Campbell K. P., Bissell M. J., Muschler J. L. (2006) J. Cell Sci. 119, 4047–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hynes R. O. (1992) Cell 69, 11–25 [DOI] [PubMed] [Google Scholar]
- 28. van der Flier A., Sonnenberg A. (2001) Cell Tissue Res. 305, 285–298 [DOI] [PubMed] [Google Scholar]
- 29. Legate K. R., Wickström S. A., Fässler R. (2009) Genes Dev. 23, 397–418 [DOI] [PubMed] [Google Scholar]
- 30. Ilsley J. L., Sudol M., Winder S. J. (2001) Cell. Signal. 13, 625–632 [DOI] [PubMed] [Google Scholar]
- 31. DeBiase P. J., Lane K., Budinger S., Ridge K., Wilson M., Jones J. C. (2006) J. Histochem. Cytochem. 54, 665–672 [DOI] [PubMed] [Google Scholar]
- 32. Driscoll K. E., Carter J. M., Iype P. T., Kumari H. L., Crosby L. L., Aardema M. J., Isfort R. J., Cody D., Chestnut M. H., Burns J. L., LeBoeuf R. A. (1995) In Vitro Cell Dev. Biol. Anim. 31, 516–527 [DOI] [PubMed] [Google Scholar]
- 33. Correa-Meyer E., Pesce L., Guerrero C., Sznajder J. I. (2002) Am. J. Physiol. Lung Cell. Mol. Physiol. 282, L883–L891 [DOI] [PubMed] [Google Scholar]
- 34. Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., Musi N., Hirshman M. F., Goodyear L. J., Moller D. E. (2001) J. Clin. Invest. 108, 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 36. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, pp. 471–510, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Klatte D. H., Kurpakus M. A., Grelling K. A., Jones J. C. (1989) J. Cell Biol. 109, 3377–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cohen T. S., Gray, Lawrence G., Khasgiwala A., Margulies S. S. (2010) PLoS One 5, e10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim K. H., López-Casillas F., Bai D. H., Luo X., Pape M. E. (1989) FASEB J. 3, 2250–2256 [DOI] [PubMed] [Google Scholar]
- 40. Davies S. P., Sim A. T., Hardie D. G. (1990) Eur. J. Biochem. 187, 183–190 [DOI] [PubMed] [Google Scholar]
- 41. Munday M. R., Campbell D. G., Carling D., Hardie D. G. (1988) Eur. J. Biochem. 175, 331–338 [DOI] [PubMed] [Google Scholar]
- 42. Winder W. W., Wilson H. A., Hardie D. G., Rasmussen B. B., Hutber C. A., Call G. B., Clayton R. D., Conley L. M., Yoon S., Zhou B. (1997) J. Appl. Physiol. 82, 219–225 [DOI] [PubMed] [Google Scholar]
- 43. Karagounis L. G., Hawley J. A. (2009) Int. J. Biochem. Cell Biol. 41, 2360–2363 [DOI] [PubMed] [Google Scholar]
- 44. Bodart J. F. (2010) J. Cell Biochem. 109, 850–857 [DOI] [PubMed] [Google Scholar]
- 45. Gregor M., Zeöld A., Oehler S., Marobela K. A., Fuchs P., Weigel G., Hardie D. G., Wiche G. (2006) J. Cell Sci. 119, 1864–1875 [DOI] [PubMed] [Google Scholar]
- 46. Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S., Wiche G., Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S., Wiche G. (2006) J. Cell Biol. 174, 557–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Na S., Chowdhury F., Tay B., Ouyang M., Gregor M., Wang Y., Wiche G., Wang N. (2009) Am. J. Physiol. Cell Physiol. 296, C868–C877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McLean W. H., Pulkkinen L., Smith F. J., Rugg E. L., Lane E. B., Bullrich F., Burgeson R. E., Amano S., Hudson D. L., Owaribe K., McGrath J. A., McMillan J. R., Eady R. A., Leigh I. M., Christiano A. M., Uitto J. (1996) Genes Dev. 10, 1724–1735 [DOI] [PubMed] [Google Scholar]
- 49. Smith F. J., Eady R. A., Leigh I. M., McMillan J. R., Rugg E. L., Kelsell D. P., Bryant S. P., Spurr N. K., Geddes J. F., Kirtschig G., Milana G., de Bono A. G., Owaribe K., Wiche G., Pulkkinen L., Uitto J., McLean W. H., Lane E. B. (1996) Nat. Genet. 13, 450–457 [DOI] [PubMed] [Google Scholar]
- 50. Koster J., van Wilpe S., Kuikman I., Litjens S. H., Sonnenberg A. (2004) Mol. Biol. Cell 15, 1211–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hijikata T., Murakami T., Ishikawa H., Yorifuji H. (2003) Histochem. Cell Biol. 119, 109–123 [DOI] [PubMed] [Google Scholar]
- 52. Lunter P. C., Wiche G. (2002) Biochem. Biophys. Res. Commun. 296, 904–910 [DOI] [PubMed] [Google Scholar]
- 53. McCahill A., Warwicker J., Bolger G. B., Houslay M. D., Yarwood S. J. (2002) Mol. Pharmacol. 62, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 54. Osmanagic-Myers S., Wiche G. (2004) J. Biol. Chem. 279, 18701–18710 [DOI] [PubMed] [Google Scholar]
- 55. Schaeffer H. J., Catling A. D., Eblen S. T., Collier L. S., Krauss A., Weber M. J. (1998) Science 281, 1668–1671 [DOI] [PubMed] [Google Scholar]
- 56. Whitmarsh A. J., Cavanagh J., Tournier C., Yasuda J., Davis R. J. (1998) Science 281, 1671–1674 [DOI] [PubMed] [Google Scholar]
- 57. Pryciak P. M., Huntress F. A. (1998) Genes Dev. 12, 2684–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feng Y., Song L. Y., Kincaid E., Mahanty S. K., Elion E. A. (1998) Curr. Biol. 8, 267–278 [DOI] [PubMed] [Google Scholar]
- 59. Whitmarsh A. J., Davis R. J. (1998) Trends Biochem. Sci. 23, 481–485 [DOI] [PubMed] [Google Scholar]
- 60. Harris K., Lamson R. E., Nelson B., Hughes T. R., Marton M. J., Roberts C. J., Boone C., Pryciak P. M. (2001) Curr. Biol. 11, 1815–1824 [PubMed] [Google Scholar]
- 61. Dreyfuss D., Soler P., Basset G., Saumon G. (1988) Am. Rev. Respir. Dis. 137, 1159–1164 [DOI] [PubMed] [Google Scholar]
- 62. Corbridge T. C., Wood L. D., Crawford G. P., Chudoba M. J., Yanos J., Sznajder J. I. (1990) Am. Rev. Respir. Dis. 142, 311–315 [DOI] [PubMed] [Google Scholar]
- 63. Vlahakis N. E., Hubmayr R. D. (2005) Am. J. Respir. Crit. Care Med. 171, 1328–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bonni A., Brunet A., West A. E., Datta S. R., Takasu M. A., Greenberg M. E. (1999) Science 286, 1358–1362 [DOI] [PubMed] [Google Scholar]
- 65. Arreola-Mendoza L., Del Razo L. M., Mendoza-Garrido M. E., Martin D., Namorado M. C., Calderon-Salinas J. V., Reyes J. L. (2009) Toxicol. Lett. 191, 279–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.