Abstract
Hepatocyte growth factor (HGF) is a heparin-binding cytokine that enhances growth, motility, and angiogenesis of many tumor types, including multiple myeloma where it is often highly expressed. However, little is known regarding what controls HGF level and activity in these tumors. Evaluation of bone marrow biopsies from myeloma patients revealed a strong positive correlation between the levels of HGF and heparanase, an endoglucuronidase known to promote aggressive tumor behavior. In vitro, addition of recombinant heparanase to myeloma cells or transfection of myeloma cell lines with the cDNA for heparanase significantly increased tumor cell expression and secretion of biologically active HGF. Shed syndecan-1, whose levels in myeloma are also enhanced by heparanase expression, binds to secreted HGF. This syndecan-1-HGF complex is active as shown by its ability to stimulate paracrine signaling via c-Met, the cell surface receptor for HGF. Surprisingly, heparanase enzyme activity was not required for up-regulation of HGF expression by the tumor cells. This is in contrast to the heparanase-mediated enhanced syndecan-1 shedding, which does require activity of the enzyme. This suggests that two different functional domains within the heparanase enzyme (the enzyme active site and a separate site) contribute to events leading to enhanced HGF signaling. These findings demonstrate a novel mechanism driving the HGF pathway whereby heparanase stimulates an increase in both HGF expression and syndecan-1 shedding to enhance HGF signaling. This work also provides further mechanistic insight into the dynamic role of heparanase in driving aggressive tumor progression.
Keywords: Heparan Sulfate, Heparin-binding Protein, Proteoglycan Structure, Tumor, Tumor Metastases, Heparanase, Hepatocyte Growth Factor, Myeloma, Syndecan
Introduction
Heparanase, an endo-β-d-glucuronidase capable of cleaving intact heparan sulfate chains from proteoglycans, is up-regulated in a wide variety of human cancers and has been associated with promoting an array of cellular events leading to enhanced tumor progression (1). In addition to its enzymatic glycosidase activity, various nonenzymatic roles of heparanase such as enhancement of AKT signaling have also been described (2). In multiple myeloma, data from in vitro and in vivo models coupled with analysis of clinical samples have identified heparanase as a promoter of tumor angiogenesis, growth, and metastasis (3–7). Together these findings indicate that heparanase is a key regulator of myeloma progression.
Many of the pro-tumorigenic effects of heparanase in myeloma have been traced to heparanase-stimulated up-regulation of syndecan-1 expression and shedding (2, 4). Syndecan-1, a heparan sulfate proteoglycan, is expressed on most myeloma tumor cells and is a critical determinant of myeloma cell survival and growth (8). Heparanase stimulates the synthesis and shedding of syndecan-1 via increased expression of two sheddases, MMP-9 and uPA3 (7). Syndecan-1 remains biologically active after it is shed from cells and can control the localization and availability of many heparin-binding growth factors (9). Using in vivo models of myeloma, our laboratory has demonstrated the synergistic action of heparanase and shed syndecan-1. For example, shed syndecan-1 binds to vascular endothelial growth factor, anchoring it close to the matrix and thereby promoting endothelial cell invasion (6).
Hepatocyte growth factor (HGF), a heparin-binding cytokine, is primarily expressed by mesenchymal cells and influences epithelial and endothelial cell behavior in a paracrine manner (10, 11). The pleiotropic effects of HGF are mediated via its binding to the proto-oncogenic c-met receptor (12). Uniquely in multiple myeloma, HGF is synthesized by tumor cells (13), and its gene expression is higher than other known growth factors, making it one of the most highly expressed soluble chemokines in myeloma patients (14). Elevated levels of HGF in the serum of myeloma patients are associated with poor prognosis (15) and have been shown to regulate tumor angiogenesis (16), cell migration, survival (17), and bone disease in myeloma (18). Surprisingly, very little is understood about the molecular mechanisms that control HGF expression in myeloma. Studies reveal that levels of soluble syndecan-1 correlate positively with levels of HGF expression and regulate its signaling in myeloma (19, 20). This observation, along with the established association between heparanase and increased shedding of syndecan-1, points to a novel role for heparanase in regulating HGF activity.
In this study, using both in vitro and in vivo models of myeloma, we find that heparanase significantly enhances HGF expression along with the elevation of syndecan-1 shedding. The secreted HGF binds to the shed syndecan-1 and enhances its bioactivity. Interestingly, although heparanase enzyme activity is required for enhanced syndecan-1 shedding, the active enzyme is not required for enhanced HGF synthesis. This indicates that heparanase activates HGF signaling via a novel dual mechanism that likely involves different functional domains of the enzyme. These findings provide unique insight into how HGF expression and activity are up-regulated in myeloma and further establish heparanase as a critical modulator of myeloma disease progression.
EXPERIMENTAL PROCEDURES
Cells and Transfections
CAG cells were established from the bone marrow aspirate of a patient with myeloma at the Arkansas Cancer Research Center as described previously (21). U266 cells were obtained from the American Type Culture Collection (Manassas, VA). MM.1S cells were a kind gift from Drs. Nancy Krett and Steven Rosen, Northwestern University. The human osteosarcoma cell line Saos-2 was provided by Dr. Majd Zayzafoon, University of Alabama at Birmingham. All the myeloma cell lines were cultured in RPMI 1640 growth medium supplemented with 10% fetal bovine serum (FBS). Saos-2 cells were cultured in DMEM supplemented with 10% FBS. CAG cells transfected with empty vector or vector containing the cDNA for human heparanase to generate heparanase low (HPSE-low) and heparanase high (HPSE-high) cells, respectively, have been previously described (4). Generation of CAG cells (M225 and M343) transfected with vectors carrying mutations in the enzyme active site of heparanase at Glu-225 or Glu-343 (4) and the stable knockdown of heparanase by shRNA (HPSE knockdown) in CAG cells along with control knockdown have been described previously (7).
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue of tumors formed from HPSE-high and HPSE-low cells were used for immunohistochemical staining. Briefly, sections were deparaffinized and hydrated through a series of xylene and graded alcohol washes, followed by antigen retrieval in 10 mm sodium citrate buffer, pH 6.0. Endogenous peroxidase activity was quenched by incubating the sections in 3% H2O2 and blocking nonspecific antigen-binding sites with 5% bovine serum albumin (BSA) in PBS. Sections were incubated overnight at 4 °C with primary antibody against human heparanase (antibody kindly provided by Dr. Israel Vlodavsky) or HGF (R&D Systems, Minneapolis, MN). After washing with PBS, sections were incubated in appropriate biotin-conjugated secondary antibodies (Vector Laboratories, Burlingame, CA). Antibody complexes were visualized using 3,3′-diaminobenzidine substrate (Vector Laboratories). All slides were counterstained with Gill's formulation number 2 hematoxylin. Photographic images were taken using a Nikon microscope equipped with a SPOT camera.
Paraffin-embedded bone marrow core biopsy specimens of patients with myeloma, obtained with approval from the University of Alabama at Birmingham Institutional Review Board, were immunostained for heparanase and HGF. Scoring for staining intensity was evaluated by two different readers, including a board-certified hematopathologist in a blinded fashion. Scoring was as follows: 0 for negative samples, 1+ for least intensely positive, and 4+ for most intensely positive. Each section was also compared with an adjacent section stained with an isotype-matched irrelevant primary antibody as a negative control. This level of staining was presumed to be negative and therefore 0. Samples previously identified as 4+ were used as positive controls.
RNA Extraction and Real Time PCR for Human HGF
5 × 105 cells of cultured HPSE-low, HPSE-intermediate, and HPSE-high CAG transfectants were seeded in 1 ml of serum-free medium and incubated overnight at 37 °C and 5% CO2. After incubation, cells were washed once with PBS, and RNA was extracted. In other experiments, 2.5 × 105 cells of different myeloma cell lines (CAG, U266, and MM.1S) were incubated with either 100 ng of active recombinant heparanase or 250 ng of the inactive recombinant heparanase in serum-free medium for 12 h (both the active and inactive forms of recombinant heparanase were kindly provided by Dr. Israel Vlodavsky). Cells were then washed once with PBS, and RNA was extracted using RNeasy columns (Qiagen, Valencia, CA). One microgram of total RNA was reverse-transcribed to cDNA using random hexamer primers and RevertAid First Strand cDNA synthesis kit (Fermentas, Hanover, MD) as per the manufacturer's protocol. 25 ng of diluted cDNA was mixed with 2× IQTM SYBR® green supermix (Bio-Rad), along with gene-specific primers (200 nm final concentration). The primers used for HGF were 5′-CAATAGCATGTCAAGTGGAG-3′ (forward) and 5′-CTGTGTTCGTGTGGTATCAT-3′ (reverse) and 5′-GTTGACCCACTAATAGGGAACG-3′ (forward) and 5′-GGATTCTGACTTAGAGGGGTTC-3′ (reverse) for 28 S rRNA. The cycle parameters included initial denaturation at 95 °C for 3 min followed by 45 cycles of 95 °C for 10 s and 60 °C for 60 s, and followed by 1 cycle of 95 °C for 60 s and 55 °C for 60 s. To ensure specific amplification, a melt curve was generated at the end of the PCR for each sample. The PCR cycle at which the fluorescence exceeded a set threshold (CT), for each sample was determined by the iCycler software. Data were analyzed according to the comparative CT method, as described previously (22), using internal control (28 S rRNA) transcript levels to normalize differences in sample loading and preparation.
Preparation and Treatment of Conditioned Media
The different CAG transfectants used in this study were seeded at a concentration of 5 × 105 cells/ml in complete growth medium and incubated for 48 h at 37 °C and 5% CO2 in a humidified chamber. Media conditioned by the cells were removed at the end of the incubation period and centrifuged at 1000 rpm twice to remove all the cells. The clarified media were then aliquoted and stored at −20 °C until further use. The levels of HGF in the conditioned medium were determined using human HGF DuoSet ELISA (R&D Systems) as per the manufacturer's instructions. In experiments performed for determining the activity of HGF, 1 ml of the conditioned medium was immunoprecipitated with 2.5 μg/ml of a polyclonal anti-human HGF antibody (R&D Systems) or isotype-specific control antibody bound to protein G-Sepharose beads (GE Healthcare). In other experiments, 1 ml of the conditioned medium was mixed with c-met inhibitor (1 μm) SU11274 (Sigma) or dimethyl sulfoxide (DMSO).
Quantification of IL-11
Cells from the osteosarcoma cell line Saos-2 were seeded in 24-well plates at 5 × 104 cells per well in complete growth medium. After overnight incubation, the medium was removed, and the monolayer was washed once with sterile PBS before treatments. In most of the experiments, conditioned media from transfectants after various treatments were mixed with an equal volume of RPMI 1640 media containing 2% FBS and incubated with a pre-washed Saos-2 monolayer for 24 h. At the end of the incubation, cell culture media were collected and centrifuged to remove cell debris, and the levels of IL-11 were determined using human IL-11 DuoSet ELISA (R&D Systems) as per the manufacturer's instructions.
To determine the role of shed syndecan-1 in HGF-driven IL-11 expression, soluble syndecan-1 in the conditioned media was immunodepleted using 2 μg/ml polyclonal anti-human syndecan-1 antibody (R&D Systems) or goat IgG as a control, bound to protein G-Sepharose beads. Heparan sulfate chains from the shed syndecan-1 were removed by addition of 5 milliunits/ml heparinase III (Seikagaku, Kogyo, Japan) at 37 °C for 2 h. Levels of shed syndecan-1 were determined by ELISA as described previously (4). Experiments also included depletion of heparin-binding growth factors such as HGF from the conditioned media by incubating overnight at 4 °C with 25 μl of heparin immobilized on agarose (MP Biomedicals, Solon, OH). Clarified conditioned media after heparin-agarose treatment were mixed with 1 ng of recombinant human HGF (R&D Systems) alone or along with 250 ng of partially purified soluble syndecan-1, added to Saos-2 cells in culture, and incubated for 24 h. Isolation and purification of soluble syndecan-1 were performed as described previously (23).
Determination of RANKL Levels
Medium conditioned by HPSE-high and HPSE-low cells prepared as described above was added to Saos-2 cells and incubated for 30 h. In some cultures, 2.5 μm SU11274 was added in addition to HPSE-high medium. At the end of the incubation period, media were removed, and Saos-2 cells were washed once with PBS, and the RNA was extracted and cDNA synthesized as described above. Real time PCR analysis was performed using primers specific for human RANKL and cycle parameters as described previously (24). Data were analyzed according to the comparative CT method described above, using internal control (28 S rRNA) transcript levels to normalize differences in sample loading and preparation.
Statistical Analyses
Statistical significance between each two experimental groups was analyzed by Student's t test, and the correlations between heparanase and HGF expression in multiple myeloma patient samples were assessed using Spearman correlation coefficient. p < 0.05 was considered statistically significant.
RESULTS
Heparanase Up-regulates Expression of HGF Protein by Myeloma Cells
Previous studies have shown that heparanase enhances the expression and shedding of syndecan-1 (4) and that high levels of shed syndecan-1 correlate positively with the high levels of HGF in the bone marrow of myeloma patients (20). To determine whether there was a correlation between levels of heparanase and HGF in myeloma patients, we examined their expression in serial sections of bone marrow core biopsy specimens from 19 myeloma patients (Table 1). Sections were scored for intensity of heparanase and HGF staining, revealing a strikingly significant positive correlation between heparanase and HGF levels (Table 1). This correlative data from myeloma patients led us to investigate whether heparanase was driving up-regulation of HGF expression in myeloma cells.
TABLE 1.
Data show staining intensity for heparanase and HGF on plasma cells from 19 myeloma patients
Intensity was scored by two independent readers, including a board-certified hematopathologist (scored from 1 to 4 with 4 being the strongest intensity of staining observed). The correlation between the scores of staining intensity of heparanase and HGF was calculated using a Spearman correlation coefficient program and was found to be significant (p < 0.0002).
| Patient | Heparanase | HGF |
|---|---|---|
| 1 | 4 | 4 |
| 2 | 4 | 3 |
| 3 | 4 | 3 |
| 4 | 4 | 3 |
| 5 | 4 | 4 |
| 6 | 4 | 4 |
| 7 | 4 | 3 |
| 8 | 4 | 3 |
| 9 | 3 | 2 |
| 10 | 3 | 1 |
| 11 | 3 | 2 |
| 12 | 3 | 3 |
| 13 | 2 | 2 |
| 14 | 2 | 2 |
| 15 | 2 | 1 |
| 16 | 2 | 1 |
| 17 | 2 | 2 |
| 18 | 2 | 3 |
| 19 | 2 | 2 |
To examine this in vitro, a panel of CAG human myeloma cells transfected with cDNA for heparanase were utilized. This panel includes cells having low, intermediate, or high levels of heparanase expression (4). HPSE-high cells prepared by transfection with vector containing the cDNA for human heparanase express a 4-fold higher level of heparanase than HPSE-low cells, which carry the empty vector (3). Furthermore, the elevated levels of enzyme activity present in HPSE-high cells are in the same range as that present in the bone marrow of many myeloma patients (3). Thus the cells utilized represent an appropriate model for examining the effects of high heparanase expression on myeloma tumors.
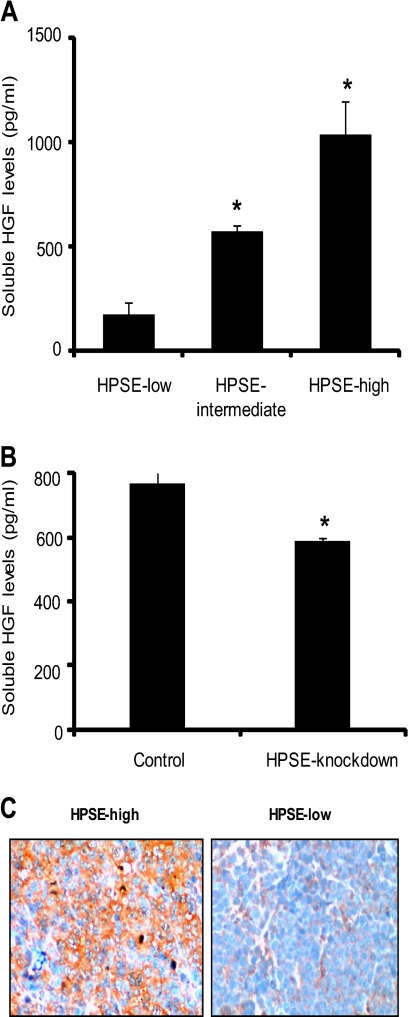
Analysis of media conditioned by these cells demonstrated that when heparanase expression was enhanced, the level of HGF increased (Fig. 1A). Similarly, HGF protein expression decreased when heparanase was knocked down in wild-type CAG cells (Fig. 1B). It is important to note that either increasing or knocking down heparanase expression in CAG myeloma cell lines does not cause any change in their mean doubling time.4 Therefore, the observed change in HGF expression is not a consequence of a difference in growth rates.
FIGURE 1.
HGF protein expression is up-regulated when heparanase expression is increased. A, stable transfectants of CAG cells expressing heparanase at either high, intermediate, or low levels were seeded at equal density and grown for 48 h. The level of HGF secreted into the conditioned media was determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.05. B, human CAG myeloma cells were transduced with lentiviral vectors delivering either control or heparanase shRNA sequences. Control cells or cells with stable knockdown of heparanase were plated at equal density and grown for 48 h, and soluble HGF levels were determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.05. C, immunohistochemical staining of HGF (brown staining) in sections from tumors formed by either HPSE-low or HPSE-high cells growing subcutaneously in the flanks of male SCID mice (original magnification, ×400).
We next examined whether heparanase-mediated up-regulation of HGF in vitro is mimicked in myeloma tumors growing in mice. Immunostaining of tumors formed by CAG cells expressing high levels of heparanase clearly revealed a strong staining for HGF compared with much weaker staining in tumors formed by cells expressing low levels of heparanase (Fig. 1C).
Heparanase Controls HGF Expression by Regulating Its Level of Transcription in Myeloma Cell Lines
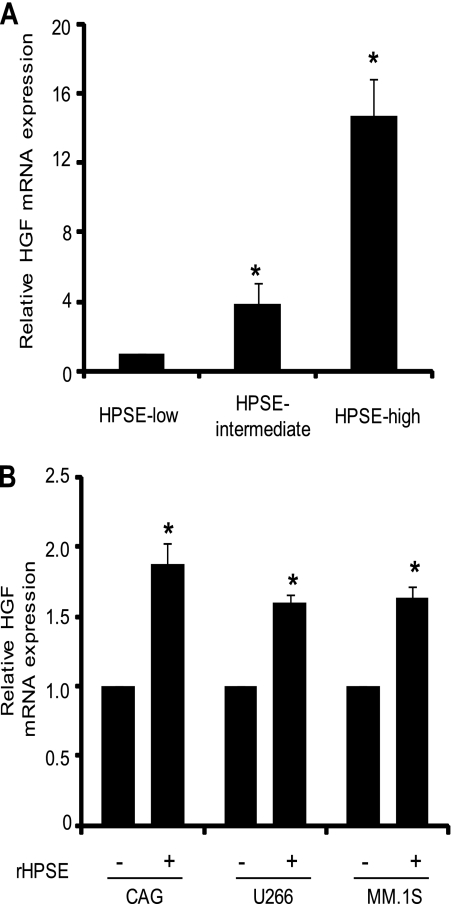
Analysis by real time PCR demonstrated that the increase in HGF protein expression seen following up-regulation of heparanase expression in CAG myeloma cells is paralleled by an increase in total HGF mRNA (Fig. 2A). To extend this finding to other human myeloma cell lines, we investigated the effects of recombinant HPSE on HGF transcript levels. When added to cells, recombinant heparanase influences cell behavior in a manner similar to endogenously produced heparanase and therefore is a useful model to study heparanase-mediated changes in cellular functions (25). Within 12 h after addition of recombinant heparanase (100 ng), there was a significant increase in the level of HGF mRNA in wild-type CAG, U266, and MM.1S human myeloma cell lines (Fig. 2B). These results demonstrate that the change in HGF levels driven by heparanase and seen in the transfected CAG cells is not cell type-specific and is not an artifact related to the transfection of the CAG cells.
FIGURE 2.
Heparanase increases HGF transcript levels in myeloma cell lines. A, stable expression of heparanase up-regulates HGF transcript levels in vitro. Equal numbers of HPSE-high, HPSE-intermediate, and HPSE-low cells were grown for 12 h in serum-free media prior to extraction of RNA. HGF transcript levels were determined by real time PCR and normalized against 28 S rRNA levels in the corresponding samples. Data are mean ± S.D. of three independent experiments. *, p < 0.01 versus HPSE-low. B, 0.5 × 106 cells from CAG, U266, or MM.1S human myeloma cell lines were incubated for 12 h with 100 ng of enzymatically active recombinant HPSE. The level of HGF mRNA transcript from each sample was determined by real time PCR and normalized against a standard housekeeping gene (28 S rRNA). For each cell line tested, HGF transcript levels in untreated cells served as the control. Data are mean ± S.D. of three independent experiments. *, p < 0.01.
HGF Expressed Following Up-regulation by Heparanase Is Functional and Signals via c-met
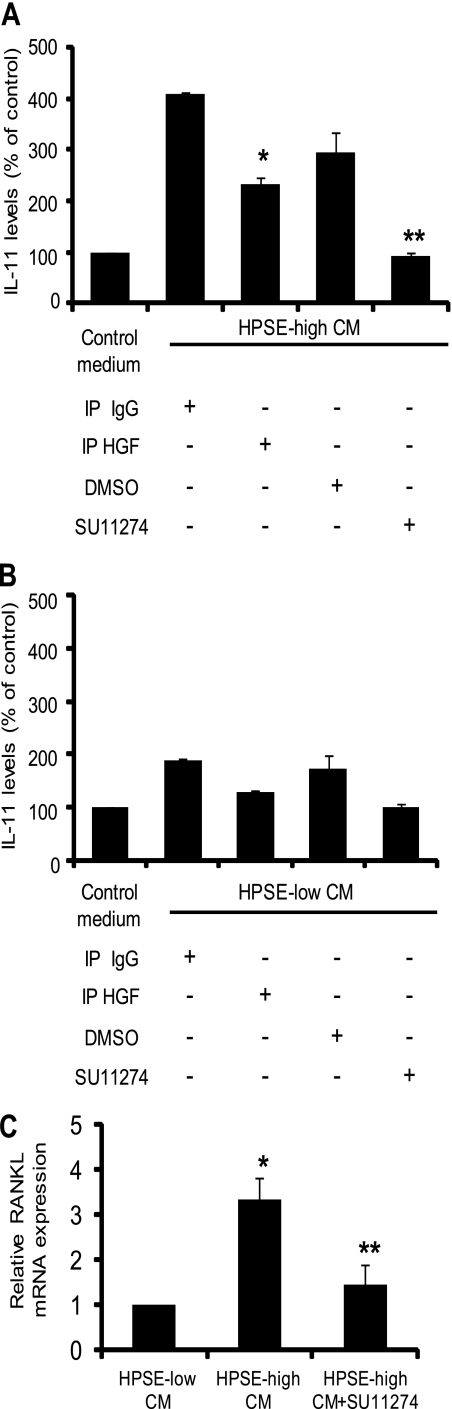
HGF is secreted normally as a 90-kDa single-chain precursor form; however, proteolytic conversion of precursor HGF to its active form is essential for its activity (26). Proteases such as uPA (27) and HGF activator (28) have been implicated in activating HGF. To determine whether the HGF secreted by CAG cells becomes active, we used an established bioassay that measures HGF-induced secretion of IL-11 in osteoblast-like Saos-2 cells (18). Media conditioned for 48 h by HPSE-high or HPSE-low CAG cells were added to a monolayer of Saos-2 cells and incubated for 24 h. The conditioned medium induced a 4-fold increase in IL-11 levels as compared with control medium that was not conditioned by cells (Fig. 3A). In contrast, medium conditioned by HPSE-low cells had a much less dramatic effect on IL-11 production (Fig. 3B). This is reflective of the lower levels of HGF produced by HPSE-low cells as compared with HPSE-high cells. Immunodepletion of HGF from HPSE-high medium (which resulted in an ∼35–40% decrease in total HGF levels4) resulted in a significant decrease in IL-11 production compared with control IgG-treated medium. HGF signals via the c-met receptor, the only known receptor for this cytokine (29). Targeting c-met has been shown to block signaling and the downstream effects induced by active HGF (30). Therefore, as a further confirmation that IL-11 production was induced by active HGF, HPSE-high conditioned medium was incubated in the presence SU11274, a highly specific inhibitor that targets c-met (31). Medium treated in this fashion failed to stimulate IL-11 production as compared with DMSO-treated controls (Fig. 3A). This inhibition of IL-11 production was not due to cytotoxic effects of SU11274 because the low concentration of inhibitor (1 μm) used in the assay did not affect the viability of Saos-2 cells.4 Also, analysis by real time PCR and by ELISA demonstrated that the myeloma cells themselves did not express detectable levels of IL-11.4
FIGURE 3.
HGF expressed by CAG cells is functional and activates c-met signaling. Levels of HGF in the conditioned media from HPSE-high (A) and HPSE-low cells (B) were reduced by immunoprecipitation with anti-HGF antibodies (IP HGF) or isotype-matched control antibody (IP IgG). Treated conditioned media were then incubated with osteoblast-like Saos-2 cells for 24 h, and the levels of secreted IL-11 were determined by ELISA. Saos-2 cells incubated with control medium (medium not conditioned by cells) served as an additional control. The media conditioned by HPSE-high (A) or HPSE-low (B) cells were incubated with Saos-2 cells in the presence of the c-met inhibitor SU11274 (1 μm) or DMSO as a control for 24 h, and the level of IL-11 was determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.01 versus Saos-2 incubated with isotype antibody-treated HPSE-high conditioned medium. **, p < 0.05 versus Saos-2 cells treated with HPSE-high conditioned medium containing DMSO. A and B, raw values for levels of IL-11 induced in Saos-2 cells by untreated conditioned media from HPSE-high and HPSE-low cells were 315.30 ± 26.64 and 158.92 pg/ml, respectively. C, conditioned medium from HPSE-high or HPSE-low transfectants was incubated with Saos-2 cells for 30 h, and the level of RANKL was determined by real time PCR. Control included Saos-2 cells incubated with HPSE-high conditioned medium along with SU11274 (2.5 μm). RANKL transcript levels were normalized against 28S rRNA levels. Data are mean ± S.D. of three independent experiments. *, p < 0.01 versus HPSE-low; **, p < 0.05 versus HPSE-high.
To further examine the downstream effects of HGF signaling, we assessed Saos-2 expression of RANKL for two reasons. First, it has been shown that IL-11 induces expression of RANKL (32); second, RANKL drives osteolytic bone disease in myeloma. Therefore, determining the mechanisms that regulate RANKL production is of great interest in understanding mechanisms that promote myeloma bone disease. Results show that medium conditioned by HPSE-high cells induced a significantly higher expression of RANKL transcript than HPSE-low cells. This effect was significantly lowered in the presence of c-met inhibitor (Fig. 3C).
Heparanase-induced Expression of HGF Is Not Dependent on Heparanase Enzymatic Activity
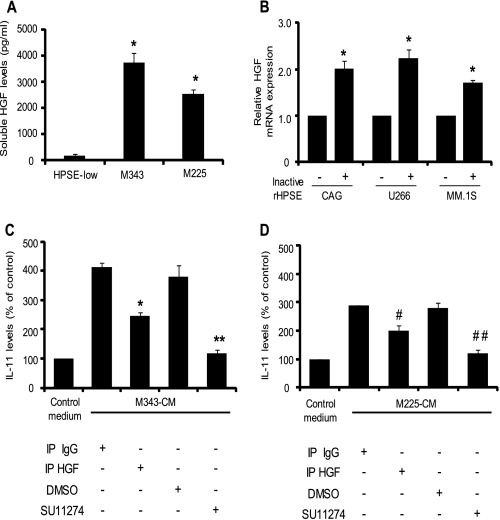
Mutated heparanase lacking heparan sulfate degrading activity has been shown to promote certain cellular functions in several cancers (25, 33). However, it has been demonstrated that heparanase-regulated gene expression in myeloma, which includes changes in levels of MMP-9, uPA/uPA receptor (7), VEGF (6), and shed syndecan-1 (4), all depends on the catalytic endoglucuronidase activity of heparanase. To determine whether the enhanced level of HGF in HPSE-high cells is dependent on the enzymatic activity of heparanase, we tested the levels of soluble HGF in CAG cells expressing mutated, inactive heparanase (mutations at amino acid positions 343 or 225 within the enzyme active site). Results reveal a significantly higher level of HGF in the conditioned media of both of these transfectants when compared with the HPSE-low cells (Fig. 4A). Similar to HPSE-high cells, there was a concomitant increase in HGF transcript levels accompanying the high HGF protein expression.4 Addition of 250 ng of recombinant inactive heparanase enzyme bearing mutations at both 343 and 225 amino acid positions (a double mutant) also induced a significant increase in HGF transcript in CAG (wild type), U266, and MM.1S myeloma cell lines (Fig. 4B).
FIGURE 4.
Up-regulation of HGF expression by heparanase is independent of heparanase enzyme activity. A, CAG cells expressing enzymatically inactive heparanase, mutated at amino acids 343 (M343) or 225 (M225), were seeded at equal density and incubated for 48 h. HGF levels in the conditioned media were determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.01 versus HPSE-low cells. B, human myeloma cell lines CAG, U266, or MM.1S were incubated for 12 h with 250 ng of inactive recombinant heparanase (a double mutant that bears mutation at amino acids 343 and 225). After incubation, RNA was extracted, and the change in the levels of HGF transcript from each sample was determined by real time PCR and normalized against 28 S RNA. For individual cell lines, HGF transcript levels in untreated cells served as a control. Data are mean ± S.D. of three independent experiments. *, p < 0.05 versus untreated controls for each cell line. C and D, HGF induced by inactive heparanase is active. High HGF in the conditioned media from M343 cells (C) or M225 cells (D) was immunodepleted using specific antibodies (IP HGF) or treated with isotype-matched control antibody (IP IgG). Conditioned media after treatments were incubated with Saos-2 cells for 24 h, and the level of soluble IL-11 secreted by Saos-2 cells was determined by ELISA. Additionally, conditioned media from M343 cells (C) or M225 cells (D) were further incubated with Saos-2 cells in the presence of the c-met inhibitor SU11274 (1 μm) or DMSO (as a control) separately for 24 h, and the levels of IL-11 were determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.05 versus Saos-2 incubated with isotype antibody-treated M343 conditioned media. **, p < 0.05 versus Saos-2 cells treated with M343 conditioned medium containing DMSO. #, p < 0.05 versus Saos-2 cells incubated with isotype antibody-treated M225 conditioned media. ##, p < 0.001 versus Saos-2 cells treated with M225 conditioned medium containing DMSO. The raw values for levels of IL-11 induced in Saos-2 cells by untreated conditioned media from M343 and M225 cells were 415.12 ± 15.10 and 320.49 ± 1.22 pg/ml, respectively.
HGF produced by M343 and M225 transfectants was active and was able to induce IL-11 production in Saos-2 cells. Depletion of HGF from the conditioned media of M343 (Fig. 4C) and M225 (Fig. 4D) cells significantly decreased IL-11 production compared with their respective controls. As observed with HPSE-high cells, the presence of c-met inhibitor (SU11274) strongly blocked HGF-induced IL-11 production by conditioned media from M343 (Fig. 4C) and M225 (Fig. 4D) cells in comparison with DMSO-treated controls. Together these results indicate that the ability of heparanase to stimulate HGF expression and activity in myeloma cells is independent of heparanase enzyme activity.
Heparanase-induced HGF Forms an Active Complex with Shed Syndecan-1
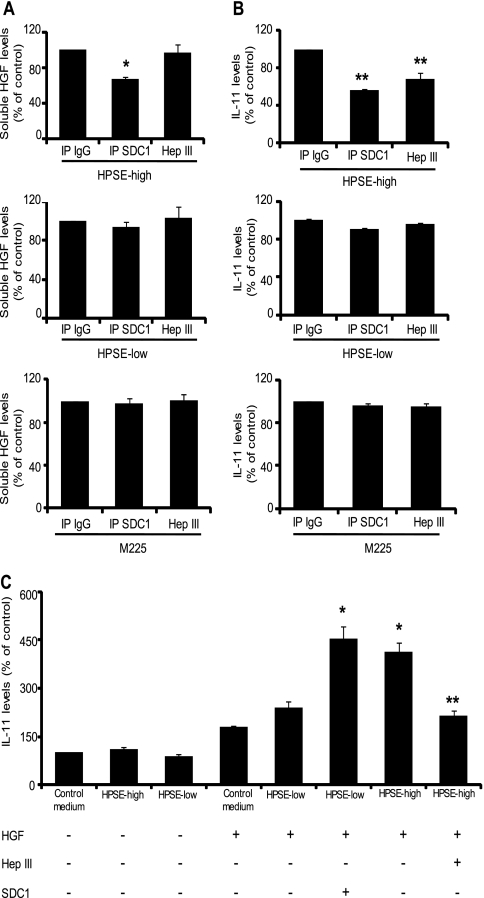
The cells expressing enzymatically inactive forms of heparanase (M225 and M343) secreted 2–3 times more HGF than did the cells expressing the enzymatically active form of heparanase (HPSE-high cells) (see Figs. 1A and 4A). This is not surprising as M225 and M343 express considerably higher levels of heparanase protein compared with HPSE-high cells (4). However, the levels of IL-11 induced in Saos-2 cells by the three cell lines were similar (320.49 ± 1.22, 415.12 ± 15.10, and 315.30 ± 26.64 pg/ml, respectively). Importantly, the level of HGF produced by these cells is well below the maximal amount of HGF to which Saos-2 cells can respond (18). This suggests the presence of an additional “soluble factor” in the conditioned medium of HPSE-high cells that is capable of promoting HGF-induced IL-11 production. Syndecan-1 is known to bind HGF and promote c-met signaling in myeloma cells (19, 20), and because enzymatically active heparanase is known to enhance syndecan-1 expression and shedding (4), we investigated whether syndecan-1 was the soluble factor in the HPSE-high conditioned medium that was partnering with HGF to enhance HGF signaling. Media conditioned by either HPSE-high cells (which contains high levels of both HGF and shed syndecan-1), HPSE-low cells (which contains low levels of both HGF and shed syndecan-1), or M225 cells (which contains high levels of HGF and low levels of shed syndecan-1) were immunodepleted of syndecan-1, and the amount of HGF remaining in the supernatant was determined (Fig. 5A). Only in medium from HPSE-high cells, which contains high levels of HGF and shed syndecan-1, did immunodepletion of syndecan-1 result in a significant decrease in soluble HGF levels (Fig. 5A). These results indicate that soluble syndecan-1 in HPSE-high conditioned medium forms a complex with HGF. The absence of any significant change in HGF levels following immunodepletion of syndecan-1 from HPSE-low or M225 medium is not surprising given that these cells produce low levels of shed syndecan-1 (Fig. 5A). Importantly, immunodepletion of soluble syndecan-1 significantly diminished the capacity of the HPSE-high conditioned medium to stimulate IL-11 production by Saos-2 cells (Fig. 5B). To test whether HGF is present in HPSE-high conditioned medium as a complex with syndecan-1 and is more active than HGF alone, the conditioned media were treated with heparitinase (HepIII), a bacterial enzyme that extensively degrades the heparan sulfate chains of proteoglycans like syndecan-1. The HepIII treatment did not alter total HGF levels in HPSE-high conditioned medium (Fig. 5A) but significantly attenuated HGF-induced production of IL-11 in Saos-2 cells (Fig. 5B). These data demonstrate that the shed syndecan-1 induced by heparanase expression enhances HGF activity.
FIGURE 5.
Heparanase-enhanced shed syndecan-1 and HGF form an active signaling complex. Media conditioned for 48 h by HPSE-high, HPSE-low, and M225 cells were immunodepleted of syndecan-1 using specific antibody (IP SDC1) or treated with isotype control antibody (IP IgG). A separate aliquot of conditioned media from the cell lines was treated with heparitinase (Hep III), a bacterial enzyme that extensively degrades heparan sulfate chains. A, levels of HGF in conditioned media were determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.05 versus IP IgG. B, conditioned media from the three cell groups were added to Saos-2 cells in culture and incubated for 24 h. At the end of incubation, media were removed, and the levels of IL-11 secreted by the Saos-2 cells were determined by ELISA. **, p < 0.05 versus IP IgG. C, heparanase-enhanced shed syndecan-1 is a key regulator of HGF activity. Conditioned media from HPSE-high and HPSE-low cells were treated with heparin-agarose beads overnight to deplete heparin-binding growth factors, including HGF. Saos-2 cells were then incubated with control media or with heparin-agarose-treated HPSE-high or HPSE-low conditioned media alone or in the presence of 1 ng of HGF. After incubation for 24 h, the supernatant was collected, and the levels of IL-11 in the supernatant were measured by ELISA. Treatment groups included either conditioned medium from HPSE-high cells treated with heparitinase (Hep III) and mixed with HGF or HPSE-low conditioned medium mixed with HGF and partially purified syndecan-1 (250 ng), incubated with a monolayer of Saos-2 cells for 24 h. At the end of incubation IL-11 levels secreted by Saos-2 cells was determined by ELISA. Data are mean ± S.D. of three independent experiments. *, p < 0.01 versus conditioned media from HPSE-low cells with HGF alone. **, p < 0.01 versus conditioned media from HPSE-high cells mixed with HGF.
Although purified syndecan-1 along with recombinant HGF is more efficient than HGF alone in inducing IL-11 expression (20), this potentiating activity of HGF and syndecan-1 has never been tested using conditions that resemble cellular events occurring in vivo. Heparanase expression in our myeloma cell lines, which results in enhanced HGF expression and syndecan-1 shedding, presents such a system for testing this biological phenomenon. To examine this, conditioned media from HPSE-high and HPSE-low cells were treated with heparin immobilized on agarose beads to remove heparin binding growth factors, including HGF. The levels of HGF present in these media after heparin-agarose treatment were negligible as reflected by their poor induction of IL-11 expression in Saos-2 cells (Fig. 5C). A fixed amount of HGF (1 ng) was added to the HGF-depleted medium from HPSE-high and HPSE-low cells. The same amount of HGF when added to HPSE-high conditioned medium induced 2-fold higher IL-11 production than when added to HPSE-low conditioned medium (Fig. 5C). Complete degradation of heparan sulfate chains by HepIII treatment of HPSE-high conditioned media decreased IL-11 induction by HGF to almost the same levels seen when HGF was added to medium not conditioned by cells (Fig. 5C, control medium + HGF). Levels of soluble shed syndecan-1 in HPSE-high conditioned medium were 350 ng/ml, whereas HPSE-low conditioned medium had around 100 ng/ml. To further establish that soluble syndecan-1 in HPSE-high conditioned medium was the potentiating factor for HGF activity, 250 ng of partially purified syndecan-1 and 1 ng of HGF were mixed with HPSE-low conditioned medium and then incubated with Saos-2 cells overnight. This enhanced IL-11 production to levels similar to those observed with HPSE-high conditioned medium (Fig. 5C). These data indicate that the syndecan-1 shed by the HPSE-high cells is a potentiating factor for HGF activity.
DISCUSSION
In this study, we demonstrate a novel and dynamic role for heparanase in enhancing HGF signaling by stimulating myeloma tumor cells to secrete high levels of HGF and by enhancing shedding of syndecan-1. Within the tumor microenvironment, soluble HGF and shed syndecan-1 bind together to form a complex that activates c-met signaling much better than HGF alone. Importantly, we show that the increase in HGF expression is independent of heparanase enzyme activity, although in contrast, as shown previously, the enhanced shedding of syndecan-1 is dependent on heparanase enzyme activity. Thus, heparanase is likely driving enhanced HGF signaling via two distinct mechanisms that merge when the HGF binds to syndecan-1. Given the established role of HGF signaling in driving growth and behavior of both myeloma cells and host cells within tumors, these findings underscore the importance of heparanase in myeloma progression and further validate it as a therapeutic target.
High expression of HGF and its effects in myeloma are well established (34). However, the underlying mechanisms triggering enhanced HGF levels and activity are largely unexplored. Our finding that heparanase enhances HGF expression is consistent with our previous work showing that heparanase up-regulates transcription of a number of genes that drive the aggressive tumor phenotype. For example, heparanase enhances myeloma cell expression of VEGF and proteases (MMP-9 and uPA). Enhanced expression of these molecules results from heparanase-stimulated signaling via ERK (6, 7). However, this is not likely the mechanism driving enhanced HGF expression because high levels of active ERK in HPSE-high cells are dependent upon the enzymatic activity of heparanase (7). In contrast, in the present study up-regulation of HGF was stimulated by enzymatically inactive forms of heparanase. This finding strongly suggests that stimulation of HGF expression is not dependent on the enzyme active site of the heparanase molecule but resides in a different domain. Predictions of heparanase structure point to a TIM-barrel domain that bears the enzyme active site and to a C-terminal domain (C-domain) responsible for nonenzymatic functions of heparanase (2). Functionally, the C-domain of heparanase can mediate enhanced Akt phosphorylation (35). Interestingly, it was recently demonstrated that induction of Akt phosphorylation leads to enhanced HGF expression and secretion in human mesenchymal stem cells (36). However, heparanase did not alter levels of Akt phosphorylation in the myeloma cells studied here4 indicating that the C-domain likely has functions that have not yet been uncovered.
Our findings demonstrate that myeloma cells produce levels of HGF and syndecan-1 that are appropriate for forming an effective complex that stimulates signaling. This is important because previous studies have shown that when levels of syndecan-1 are too high relative to levels of HGF, signaling activity is actually inhibited. Conversely, when concentrations of syndecan-1 are too low, they have no effect on HGF signaling (20). The level of heparanase-induced syndecan-1 is therefore crucial in determining HGF activity and c-met signaling in vivo thus providing a sensitive mechanism to modulate tumor behavior.
The positive impact of heparanase expression on HGF activity in myeloma could have multiple effects on progression of this cancer. It has been shown that recombinant HGF enhances myeloma tumor cell proliferation and inhibits apoptosis in vitro (37). However, it is important to note that the effects of HGF on myeloma proliferation are evident only upon treatment with relatively high levels of recombinant HGF (37). In contrast, cells within the myeloma tumor microenvironment, like osteoblasts (18), respond strongly to low levels of HGF resulting in increased IL-11 production. Therefore, it is likely that a major role for heparanase-stimulated HGF in vivo is largely mediated via paracrine c-met signaling. In addition to stimulation of osteoblasts, another HGF/c-met paracrine signaling pathway active in cancer is present in microvascular endothelial cells where HGF signaling enhances tumor-associated angiogenesis (38). Recognition that HGF/c-met signaling is an important stimulator of angiogenesis has led to attempts to therapeutically inhibit this pathway (39–41).
We also found that in the system being studied here, HGF and syndecan-1 acting together stimulated IL-11 production much higher than HGF alone and that this led to an increase in RANKL expression in Saos-2 cells. IL-11, a key regulator of skeletal biology, is a downstream target of c-met signaling (18). IL-11 inhibits bone formation in vitro (42) and supports bone resorption (43) via multiple independent pathways, including up-regulation of RANKL (44–48). Taken together, these data indicate that heparanase might contribute to myeloma bone disease by activating the HGF/c-met/IL-11/RANKL axis (Fig. 6). In addition to these indirect effects on osteoblasts, heparanase also directly stimulates expression of RANKL by myeloma tumor cells (52). Yet another downstream effect of heparanase-mediated HGF/c-met signaling could be in driving mechanisms that support minimal residual disease, a persistent problem that leads to relapse and eventual death of many myeloma patients (49). We have previously demonstrated that high levels of syndecan-1 are present in fibrotic bone marrow of myeloma patients post-treatment (49). This syndecan-1 could facilitate establishment of reservoirs of HGF making it available for nourishing remaining tumor cells that escape chemotherapy, thereby helping to maintain bone disease and stimulating angiogenesis, all of which contribute to emergence of full blown relapse. Numerous studies have highlighted the importance of shed syndecan-1 in regulating the pathophysiology of diseases, including different types of cancers (50). The ability of heparanase to enhance syndecan-1 shedding and the downstream effect of shed syndecan-1 in regulating the activity of various growth factors may be crucial in the pathogenesis of diseases where there are high levels of shed syndecan-1 present (51).
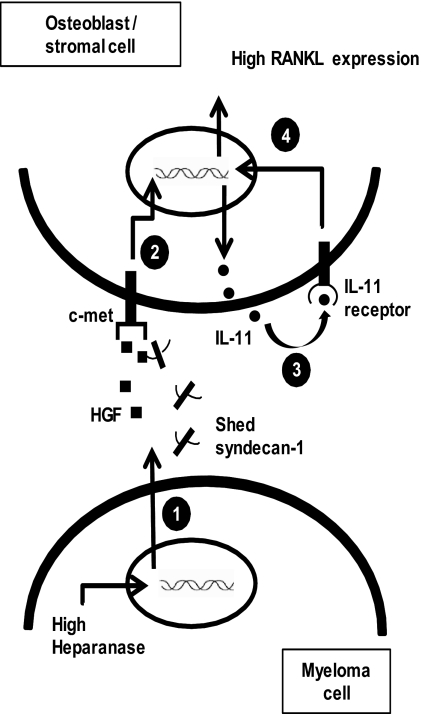
FIGURE 6.
Model of heparanase regulation of the HGF/c-met/IL-11/RANKL axis in myeloma. 1, high heparanase expression in myeloma cells up-regulates the levels of soluble HGF and shed syndecan-1 (4) in the tumor milieu. 2, tumor-derived HGF either alone or as a complex with shed syndecan-1 signals via the c-met receptor in osteoblast or stromal cells present in the tumor microenvironment, thereby resulting in enhanced IL-11 expression. 3, IL-11 secreted by the stromal or osteoblast cells acts in an autocrine manner and activates the IL-11 receptor present in these cells. 4, IL-11 signaling can drive the expression of RANKL (48). Heparanase via the HGF/c-met/IL-11 axis could possibly increase the expression of RANKL in myeloma thereby leading to enhanced osteolytic bone disease.
In summary, the findings presented here provide novel insights into how heparanase regulates HGF activity in myeloma. By controlling the expression and activity of HGF, heparanase very likely impacts tumor angiogenesis, osteolytic bone disease, and perhaps the overall response to therapy in myeloma. Because heparanase can strongly drive the function of molecules that promote osteolysis, targeting heparanase offers a unique window of opportunity for alleviating cancer-associated bone disease. Therefore, inhibitors of heparanase hold great potential as therapeutic tools for targeting the HGF/IL-11/RANKL axis in osteolytic cancers. Together these results further establish heparanase as a master regulator of myeloma pathobiology and may extend to other cancers where HGF plays a pivotal role in tumor progression.
Supplementary Material
Acknowledgments
We thank Dr. Israel Vlodavsky (Technion, Haifa, Israel) for providing recombinant heparanase and antibody to heparanase; Drs. Nancy Krett and Steven Rosen (Northwestern University) for MM.1S cells; Dr. Majd Zayzafoon (University of Alabama at Birmingham) for Saos-2 cells; the Histomorphometry and Molecular Analysis Core (University of Alabama at Birmingham Center for Metabolic Bone Disease) for tissue sections; and Dr. Renee Desmond (University of Alabama at Birmingham Comprehensive Cancer Center) for assistance with statistical analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants CA135075 and CA138340 (to R. D. S.). This work was also supported by Multiple Myeloma Research Foundation research fellow award (to V. C. R.) and Multiple Myeloma Research Foundation senior research award (to Y. Y.).
This article was selected as a Paper of the Week.
V. C. Ramani, unpublished observations.
- uPA
- urokinase-type plasminogen activator
- HGF
- hepatocyte growth factor
- RANKL
- receptor activator for nuclear factor κB ligand
- IP
- immunoprecipitation
- HPSE
- heparanase.
REFERENCES
- 1. Vlodavsky I., Goldshmidt O., Zcharia E., Atzmon R., Rangini-Guatta Z., Elkin M., Peretz T., Friedmann Y. (2002) Semin. Cancer Biol. 12, 121–129 [DOI] [PubMed] [Google Scholar]
- 2. Fux L., Ilan N., Sanderson R. D., Vlodavsky I. (2009) Trends Biochem. Sci. 34, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kelly T., Miao H. Q., Yang Y., Navarro E., Kussie P., Huang Y., MacLeod V., Casciano J., Joseph L., Zhan F., Zangari M., Barlogie B., Shaughnessy J., Sanderson R. D. (2003) Cancer Res. 63, 8749–8756 [PubMed] [Google Scholar]
- 4. Yang Y., Macleod V., Miao H. Q., Theus A., Zhan F., Shaughnessy J. D., Jr., Sawyer J., Li J. P., Zcharia E., Vlodavsky I., Sanderson R. D. (2007) J. Biol. Chem. 282, 13326–13333 [DOI] [PubMed] [Google Scholar]
- 5. Yang Y., Macleod V., Bendre M., Huang Y., Theus A. M., Miao H. Q., Kussie P., Yaccoby S., Epstein J., Suva L. J., Kelly T., Sanderson R. D. (2005) Blood 105, 1303–1309 [DOI] [PubMed] [Google Scholar]
- 6. Purushothaman A., Uyama T., Kobayashi F., Yamada S., Sugahara K., Rapraeger A. C., Sanderson R. D. (2010) Blood 115, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Purushothaman A., Chen L., Yang Y., Sanderson R. D. (2008) J. Biol. Chem. 283, 32628–32636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khotskaya Y. B., Dai Y., Ritchie J. P., MacLeod V., Yang Y., Zinn K., Sanderson R. D. (2009) J. Biol. Chem. 284, 26085–26095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sanderson R. D., Yang Y. (2008) Clin. Exp. Metastasis 25, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neuss S., Becher E., Wöltje M., Tietze L., Jahnen-Dechent W. (2004) Stem Cells 22, 405–414 [DOI] [PubMed] [Google Scholar]
- 11. Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. (2003) Nat. Rev. Mol. Cell Biol. 4, 915–925 [DOI] [PubMed] [Google Scholar]
- 12. Ma P. C., Maulik G., Christensen J., Salgia R. (2003) Cancer Metastasis Rev. 22, 309–325 [DOI] [PubMed] [Google Scholar]
- 13. Börset M., Hjorth-Hansen H., Seidel C., Sundan A., Waage A. (1996) Blood 88, 3998–4004 [PubMed] [Google Scholar]
- 14. Zhan F., Hardin J., Kordsmeier B., Bumm K., Zheng M., Tian E., Sanderson R., Yang Y., Wilson C., Zangari M., Anaissie E., Morris C., Muwalla F., van Rhee F., Fassas A., Crowley J., Tricot G., Barlogie B., Shaughnessy J., Jr. (2002) Blood 99, 1745–1757 [DOI] [PubMed] [Google Scholar]
- 15. Seidel C., Børset M., Turesson I., Abildgaard N., Sundan A., Waage A. (1998) Blood 91, 806–812 [PubMed] [Google Scholar]
- 16. You W. K., McDonald D. M. (2008) BMB Rep. 41, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lesko E., Majka M. (2008) Front. Biosci. 13, 1271–1280 [DOI] [PubMed] [Google Scholar]
- 18. Hjertner O., Torgersen M. L., Seidel C., Hjorth-Hansen H., Waage A., Børset M., Sundan A. (1999) Blood 94, 3883–3888 [PubMed] [Google Scholar]
- 19. Derksen P. W., Keehnen R. M., Evers L. M., van Oers M. H., Spaargaren M., Pals S. T. (2002) Blood 99, 1405–1410 [DOI] [PubMed] [Google Scholar]
- 20. Seidel C., Børset M., Hjertner O., Cao D., Abildgaard N., Hjorth-Hansen H., Sanderson R. D., Waage A., Sundan A. (2000) Blood 96, 3139–3146 [PubMed] [Google Scholar]
- 21. Børset M., Hjertner O., Yaccoby S., Epstein J., Sanderson R. D. (2000) Blood 96, 2528–2536 [PubMed] [Google Scholar]
- 22. Schmittgen T. D., Livak K. J. (2008) Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 23. Reiland J., Sanderson R. D., Waguespack M., Barker S. A., Long R., Carson D. D., Marchetti D. (2004) J. Biol. Chem. 279, 8047–8055 [DOI] [PubMed] [Google Scholar]
- 24. Abdallah B. M., Stilgren L. S., Nissen N., Kassem M., Jørgensen H. R., Abrahamsen B. (2005) Calcif. Tissue Int. 76, 90–97 [DOI] [PubMed] [Google Scholar]
- 25. Zetser A., Bashenko Y., Edovitsky E., Levy-Adam F., Vlodavsky I., Ilan N. (2006) Cancer Res. 66, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 26. Naka D., Ishii T., Yoshiyama Y., Miyazawa K., Hara H., Hishida T., Kidamura N. (1992) J. Biol. Chem. 267, 20114–20119 [PubMed] [Google Scholar]
- 27. Naldini L., Vigna E., Bardelli A., Follenzi A., Galimi F., Comoglio P. M. (1995) J. Biol. Chem. 270, 603–611 [DOI] [PubMed] [Google Scholar]
- 28. Kataoka H., Kawaguchi M. (2010) FEBS J. 277, 2230–2237 [DOI] [PubMed] [Google Scholar]
- 29. Cooper C. S. (1992) Oncogene 7, 3–7 [PubMed] [Google Scholar]
- 30. Yap T. A., de Bono J. S. (2010) Mol. Cancer Ther. 9, 1077–1079 [DOI] [PubMed] [Google Scholar]
- 31. Berthou S., Aebersold D. M., Schmidt L. S., Stroka D., Heigl C., Streit B., Stalder D., Gruber G., Liang C., Howlett A. R., Candinas D., Greiner R. H., Lipson K. E., Zimmer Y. (2004) Oncogene 23, 5387–5393 [DOI] [PubMed] [Google Scholar]
- 32. Ahlen J., Andersson S., Mukohyama H., Roth C., Bäckman A., Conaway H. H., Lerner U. H. (2002) Bone 31, 242–251 [DOI] [PubMed] [Google Scholar]
- 33. Goldshmidt O., Zcharia E., Cohen M., Aingorn H., Cohen I., Nadav L., Katz B. Z., Geiger B., Vlodavsky I. (2003) FASEB J. 17, 1015–1025 [DOI] [PubMed] [Google Scholar]
- 34. Børset M., Seidel C., Hjorth-Hansen H., Waage A., Sundan A. (1999) Leuk. Lymphoma 32, 249–256 [DOI] [PubMed] [Google Scholar]
- 35. Fux L., Feibish N., Cohen-Kaplan V., Gingis-Velitski S., Feld S., Geffen C., Vlodavsky I., Ilan N. (2009) Cancer Res. 69, 1758–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang A., Wang Y., Ye Z., Xie H., Zhou L., Zheng S. (2010) J. Cell. Biochem. 111, 469–475 [DOI] [PubMed] [Google Scholar]
- 37. Derksen P. W., de Gorter D. J., Meijer H. P., Bende R. J., van Dijk M., Lokhorst H. M., Bloem A. C., Spaargaren M., Pals S. T. (2003) Leukemia 17, 764–774 [DOI] [PubMed] [Google Scholar]
- 38. Bussolino F., Di Renzo M. F., Ziche M., Bocchietto E., Olivero M., Naldini L., Gaudino G., Tamagnone L., Coffer A., Comoglio P. M. (1992) J. Cell Biol. 119, 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsumoto K., Nakamura T. (2003) Cancer Sci. 94, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuba K., Matsumoto K., Date K., Shimura H., Tanaka M., Nakamura T. (2000) Cancer Res. 60, 6737–6743 [PubMed] [Google Scholar]
- 41. Date K., Matsumoto K., Kuba K., Shimura H., Tanaka M., Nakamura T. (1998) Oncogene 17, 3045–3054 [DOI] [PubMed] [Google Scholar]
- 42. Hughes F. J., Howells G. L. (1993) Calcif. Tissue Int. 53, 362–364 [DOI] [PubMed] [Google Scholar]
- 43. Shaughnessy S. G., Walton K. J., Deschamps P., Butcher M., Beaudin S. M. (2002) Cytokine 20, 78–85 [DOI] [PubMed] [Google Scholar]
- 44. Morrissey C., Kostenuik P. L., Brown L. G., Vessella R. L., Corey E. (2007) BMC Cancer 7, 148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajgopal R., Butcher M., Weitz J. I., Shaughnessy S. G. (2006) J. Biol. Chem. 281, 20780–20787 [DOI] [PubMed] [Google Scholar]
- 46. Romas E., Udagawa N., Zhou H., Tamura T., Saito M., Taga T., Hilton D. J., Suda T., Ng K. W., Martin T. J. (1996) J. Exp. Med. 183, 2581–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kudo O., Sabokbar A., Pocock A., Itonaga I., Fujikawa Y., Athanasou N. A. (2003) Bone 32, 1–7 [DOI] [PubMed] [Google Scholar]
- 48. Hill P. A., Tumber A., Papaioannou S., Meikle M. C. (1998) Endocrinology 139, 1564–1572 [DOI] [PubMed] [Google Scholar]
- 49. Bayer-Garner I. B., Sanderson R. D., Dhodapkar M. V., Owens R. B., Wilson C. S. (2001) Mod. Pathol. 14, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 50. Manon-Jensen T., Itoh Y., Couchman J. R. (2010) FEBS J. 277, 3876–3889 [DOI] [PubMed] [Google Scholar]
- 51. Barash U., Cohen-Kaplan V., Dowek I., Sanderson R. D., Ilan N., Vlodavsky I. (2010) FEBS J. 277, 3890–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Y., Ren Y., Ramani V. C., Nan L., Suva L. J., Sanderson R. D. (2010) Cancer Res. 70, 8329–8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.