Abstract
The maintenance of rapid and efficient actin dynamics in vivo requires coordination of filament assembly and disassembly. This regulation requires temporal and spatial integration of signaling pathways by protein complexes. However, it remains unclear how these complexes form and then regulate the actin cytoskeleton. Here, we identify a srGAP2 and formin-like 1 (FMNL1, also known as FRL1 or FRLα) complex whose assembly is regulated by Rac signaling. Our data suggest srGAP2 regulates FMNL1 in two ways; 1) Rac-mediated activation of FMNL1 leads to the recruitment of srGAP2, which contains a Rac-specific GAP domain; 2) the SH3 domain of srGAP2 binds the formin homology 1 domain of FMNL1 to inhibit FMNL1-mediated actin severing. Thus, srGAP2 can efficiently terminate the upstream activating Rac signal while also opposing an important functional output of FMNL1, namely actin severing. We also show that FMNL1 and srGAP2 localize to the actin-rich phagocytic cup of macrophage-derived cells, suggesting the complex may regulate this Rac- and actin-driven process in vivo. We propose that after Rac-dependent activation of FMNL1, srGAP2 mediates a potent mechanism to limit the duration of Rac action and inhibit formin activity during rapid actin dynamics.
Keywords: Actin, Cdc42, G Proteins, Rho, Signal Transduction, FMNL1, GAP, Rac, Formin, srGAP2
Introduction
Remodeling of the actin cytoskeleton is a tightly controlled process that directs cellular functions including cell migration, adhesion, polarity, and membrane trafficking. Many studies suggest that the regulation of actin filament assembly and disassembly must be coordinated by a complex interplay between multiple cellular signaling pathways (1–4). One signaling pathway that plays a prominent role in regulating actin is the Rho-family GTPase pathway, which is typified by Rho, Rac, and Cdc42 (5, 6). Rho-family GTPase signaling pathways are inactivated by Rho GTPase-activating proteins (GAPs)2 and activated by Rho guanine nucleotide exchange factors. Once activated by guanine nucleotide exchange factors, Rho-family GTPases bind to and modulate the action of actin binding proteins such as the formins. The mammalian formin family is composed of 15 different members, which suggests that they have widespread roles for regulating distinct actin processes (7–9). However, there is some commonality to the molecular mechanisms regulating the diaphanous-related formins (8, 10, 11). Diaphanous-related formins are autoinhibited by an intramolecular interaction between an N-terminal Dia inhibitory domain (DID) and a C-terminal Dia autoregulatory domain (DAD). Activated Rho-family GTPases disrupt this autoinhibition by binding to the GTPase binding domain (GBD) and DID region (12, 13). This induces the release of the DID-DAD interaction, opening the formin so that the formin homology 2 (FH2) domain can associate with the barbed end of actin filaments to protect them from capping proteins and allow processive elongation. In addition to actin filament elongation, some formins, such as formin-like 1 (FMNL1, also called FRL1 and FRLα), have unique activities, including severing and bundling actin filaments by associating with filament sides (14, 15). Thus, members of the formin family have multiple actin remodeling activities.
In vitro, formins polymerize actin filaments that are much longer than those found in vivo or sever filaments into very small fragments. Because formin activity appears to be exaggerated in vitro compared with their activity in vivo, it is clear that formin activation must be tightly regulated. Additionally, it is estimated that some formin activity cycles last less than 5 s in vivo (16). In vitro, however, formin activity can persist much longer (17). This suggests that cellular mechanisms must exist to turn “off” formin activity, to counterbalance Rho-family GTPase-induced activation of formins. Along these lines, several factors have been identified that appear to inhibit formin activity. These include Bud14p in yeast, the Drosophila Spire, and the mammalian DIP/WISH; however, none of these regulates formin activity at the level of the Rho-family GTPases (16, 18, 19).
Here we report an interaction between the formin FMNL1 and the RhoGAP family member srGAP2 (Slit-Robo GAP family member 2). This complex forms via binding between the FH1 domain of FMNL1 and the SH3 domain of srGAP2. This binding is temporally regulated by the Rac-mediated activation of FMNL1. Additionally, srGAP2 functions as a selective Rac GAP when compared with Cdc42 or RhoA. Finally, actin filament severing assays show that the srGAP2 SH3 domain also directly inhibits FMNL1 actin severing activity. Together, our data suggest two novel mechanisms for srGAP2-mediated regulation of FMNL1, including GAP domain-mediated regulation of local Rac signaling to FMNL1 and steric/allosteric inhibition of actin severing by FMNL1.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Assay
The SH3 domain of srGAP2 was cloned into pLexNA vector, and this vector was transformed into the L40 yeast strain. A mouse embryonic (days 9.5–10.5) cDNA library was screened for SH3 binding partners using the yeast two-hybrid assay as described (20). Positive colonies were cured of the pLexNA-SH3 vector and retransformed with various baits described below to determine the specificity of interaction using 3-aminotriazole and β-galactosidase activity. The clones with the strongest and most specific activity for srGAP2 were then sequenced.
Cell Culture and Transfections
HEK293T cells were cultured in DMEM supplemented with 10% FBS. HeLa cells were cultured in MEM supplemented with 10% FBS, nonessential amino acids, and sodium pyruvate. RAW264.7 cells were cultured in DMEM supplemented with 10% FBS, penicillin, and streptomycin. Transfections were performed using calcium phosphate for HEK293T cells and Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol for HeLa cells.
Plasmid Constructs
A Formin-like 1 (Homo sapiens, Uniprot O95466) construct was kindly provided by the Daniel Billadeau laboratory (Mayo Clinic). A portion of the gene was cloned from I.M.A.G.E. clone 5729432 into the FMNL1 construct. This, resulting in full-length cDNA (amino acids 1–1100) product, was subsequently cloned into pcDNA3.1D-TOPO-V5His (Invitrogen). N-terminal GFP fusion was made by subcloning GFP into pcDNA3.1D-FMNL1-V5His. Mus musculus (Uniprot Q9JL26) FMNL1 WT (amino acids 1–1094)-GFP, FMNL1 L1062D-GFP, and FMNL1 N terminus (amino acids 1–450)-GFP in pAS were generously given by the Michael Rosen laboratory (UT Southwestern). Mutations in srGAP2 (H. sapiens, Uniprot O75044) were made using QuikChange II XL site-directed mutagenesis kit (Stratagene) as per the manufacturer's protocol.
Antibodies and Stains
For Western blots, immunoprecipitations, and immunostaining, antibodies were used against FLAGM2 (Sigma; F3165), GST-conjugated horseradish peroxidase (HRP) (Bethyl; A190-122P), V5 (Invitrogen; 46-0705), GFP (Invitrogen; A11122), actin (Sigma; A5441), Rac1 (BD Transduction; R56220), and FMNL1 D14 (Santa Cruz). FMNL1 and formin-like 3 (FMNL3) antibodies were generously provided by Dr. Daniel Billadeau (Mayo Clinic). A rabbit srGAP2 antibody was raised and purified against an SH3 domain fusion protein. HRP-conjugated secondary was purchased from GE Healthcare, and fluorescent secondary antibodies were purchased from Invitrogen. All Western blots are representative images from at least three different experiments.
Immunoprecipitations and Pulldown Assays
HEK293T were transfected with constructs described above using calcium phosphate for 12–48 h. Cells were rinsed with PBS and lysed with ice-cold lysis buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100) with 4-(2-aminoethyl)benzenesulfonyl fluoride, leupeptin, and pepstatin (RPI Corp.). Lysate was precleared by centrifugation, and antibodies were added to supernatant with protein A- or G-agarose beads (Millipore) or V5-antibody-agarose (Sigma) at 4 °C. For GST pull down assays, cells were lysed and precleared, and GST proteins were added with glutathione-Sepharose (GE Healthcare). After incubation, beads and associated proteins were washed 3 times with lysis buffer with 1 m NaCl and 1 additional wash with lysis buffer at 4 °C. Sample buffer was added to beads, and Western blots were performed. For PDGF-stimulated cells, HEK293T cells were transfected with calcium phosphate overnight. Cells were rinsed with PBS and placed in serum-free DMEM overnight. Cells were rinsed with PBS, and PDGF-BB (Chemicon) in PBS was added at 10 ng/ml for 15 min at 37 °C. Cells that were unstimulated remained in PBS. Cells were lysed, and immunoprecipitations were performed as described above.
Cellular Microscopy
Cells were prepared for microscopy by fixation in 4% paraformaldehyde in PBS for 10 min at room temperature. Coverslips were mounted with FluorSave Reagent (Calbiochem). Images were taken on a Zeiss LSM 710. All images were acquired using a 63×/1.4 NA oil immersion objective. Maximum image projections were also made in ImageJ from confocal z-series images. Additional image preparation was done using Adobe Photoshop.
Protein Expression and Purification
GST and His6-tagged proteins were purified from BL21 Escherichia coli as previously described (21). Full-length srGAP2-V5 for in vitro GAP assays was expressed in HEK293T. Cells were lysed with lysis buffer, lysate was precleared, and srGAP2-V5 was purified using anti-V5-conjugated-agarose beads (Sigma) as previously described (22). FMNL1-C (M. musculus amino acids 449–1094) was purified as previously described (14). Briefly, FMNL1-C in pGEX-KT was expressed in BL21-DE3 E. coli, and after induction with isopropyl-1-thio-β-d-galactopyranoside, protein was extracted via sonication. Supernatant after ultracentrifugation was loaded onto glutathione-Sepharose 4B (Amersham Biosciences) column, washed, and then cleaved with thrombin (Sigma) for 1 h. The eluted protein was further purified via FPLC using a SourceS15 chromatography column (Amersham Biosciences). FMNL1-C-containing fractions were pooled and dialyzed into 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole pH 7.0, 1 mm DTT, and 0.01% sodium azide. The protein was snap-frozen using liquid nitrogen and stored at −80 °C. Actin was purified from rabbit skeletal muscle (23) and gel-filtered on S200 gel filtration column (Amersham Biosciences) (14). AlexaFluor 488-labeled actin was purchased from Invitrogen.
Actin-severing Assays
Recombinant His6-srGAP2 SH3 was purified from E. coli (BL21) using nickel nitrilotriacetic acid-agarose (Qiagen, Valencia, CA) at 4 °C. Purified protein was dialyzed into 1× KMEI (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0) with 1 mm DTT. Assays were performed as previously described (14). Briefly, actin was polymerized for 1 h in 1× KMEI in G-Mg buffer (2 mm Tris, pH 8.0, 0.5 mm DTT, 0.2 mm ATP, 0.1 mm MgCl2, 0.01% sodium azide). FMNL1-C and srGAP2 SH3 were diluted in 1× KMEI in G-Mg buffer with 0.2 mm nonaethylene glycol monododecyl ether (Thesit) (Sigma). Actin filaments were incubated with FMNL1-C and srGAP2 SH3. The reaction was stopped by adding rhodamine phalloidin (Invitrogen) and diluted into dilution buffer (25 mm imidazole, pH 7.0, 25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 0.5% methylcellulose) supplemented with 250 mm NaCl, 100 mm DTT, 3 mg/ml glucose, 100 mg/ml glucose oxidase, and 18 mg/ml catalase. The solution was placed onto an 18-mm coverslip coated with poly-l-lysine and imaged on a Leica DMAR2 microscope. 10–15 images per coverslip were taken of random fields. Filament lengths were quantified using MetaMorph software (Molecular Devices). Percent severing was calculated for each experiment from fractions of filaments greater than 9 μm ((1 − (F − FFMNL1)/(Factin − FFMNL1)) × 100) and from fractions of filaments less than 3 μm (((F − Factin)/(FFMNL1 −Factin)) × 100), where F = fraction of filaments, FFMNL1 = average fraction of filaments with FMNL1 alone, and Factin = fraction of filaments with actin alone. Data were plotted using Prism software (GraphPad).
For TIRF imaging of actin severing, assays were performed as previously described (24). Briefly, imaging chambers were made using glass coverslips attached to glass slides with Parafilm strips. N-Ethylmaleimide-inactivated myosin is wicked through the chamber followed by Superblock (Pierce), then 1× KMEI. 30% AlexaFluor 488-labeled actin was polymerized in 1× KMEI/G-Mg for 30 min at room temperature and placed on ice. FMNL1-C was diluted to working concentrations in imaging buffer (16 mm imidazole, pH 7.0, 40 mm KCl, 0.8 mm EGTA, 1.6 mm MgCl2, 200 mm DTT, 6 mg/ml dextrose, 5 mg/ml methylcellulose, 40 μg/ml glucose oxidase, 20 μg/ml catalase, 0.4 mm ATP). Actin filaments were diluted in imaging buffer to concentrations described below and loaded into imaging chamber using a cut pipette tip. After 5 min, FMNL1-C was added, and images were acquired on Leica AM TIRF MC. Image were processed using ImageJ, Version 1.42q.
Actin Binding Assay
Actin was polymerized in 1× NaMEI buffer (50 mm NaCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0, 0.2 mm Thesit) in G-Mg buffer at room temperature for 1 h and stabilized with unlabeled phalloidin (Invitrogen). FMNL1-C, srGAP2 SH3, and actin filaments were added to concentrations described below in 1× NaMEI/G-Mg and spun at 80,000 rpm in TLA100 rotor for 20 min at 4 °C. Supernatant was removed, concentrated using SpeedVac, and resuspended in sample buffer. Pelleted proteins were resuspended in sample buffer. Samples were subjected to SDS-PAGE, and gels were stained using Flamingo Stain (Bio-Rad). Fluorescent gels were scanned using Typhoon 9400 PhosphorImager (GE Healthcare).
GAP Assays
In vitro GAP assays were performed as previously published (22). Briefly, 300 ng of purified recombinant GST-RhoA, -Rac1, or -Cdc42 was loaded with 10 mCi of [γ-32P]GTP in 20 mm Tris-HCl, pH 7.6, 0.1 mm DTT, 25 mm NaCl, and 4 mm EDTA. Rac and [γ-32P]GTP were incubated at 30 °C for 15 min. [γ-32P]GTP-loaded Rac was stabilized with 17 mm MgCl2 and placed on ice. 3 μl of [γ-32P]GTP-loaded Rac mixture was added to sample containing partially purified srGAP2-V5 protein from HEK293T cells on V5-agarose beads in 20 mm Tris-HCl, pH 7.6, 0.1 mm DTT, 1 mm GTP, and 1 mg/ml BSA and incubated at 30 °C. After 2 min, the reaction was stopped with 1 ml of ice-cold stop buffer containing 50 mm Tris-HCl, pH 7.6, 5 mm NaCl, and 5 mm MgCl2. The solution was filtered through a 0.45-μm nitrocellulose filter (VWR International, West Chester, PA) and washed 3 times with 10 ml of ice-cold stop buffer. Samples were read in a Tri-Carb 2100TR liquid scintillation analyzer (Canberra, Meriden, CT).
Cellular Rac activity assays in HEK293T cells were performed as previously published (22). Briefly, cells were transfected with pcDNA3.1D-srGAP2-V5His using calcium phosphate. After 48 h, cells were rinsed with PBS and lysed with MLB (25 mm HEPES, pH 7.4, 150 mm NaCl, 1% Nonidet P-40 substitute, 10 mm MgCl2, 1 mm EDTA, 2% glycerol). Lysate was incubated with recombinant GST-p21-activated kinase and glutathione beads for 1 h at 4 °C. Pulldown was washed twice with MLB. Sample buffer was added to protein beads, and Western blots were preformed.
RESULTS
srGAP2 Forms a Complex with FMNL1
The srGAP family consists of srGAP1, srGAP2, WRP, and ArhGAP4, all of which contain a conserved N-terminal Inverse F-BAR domain, a central Rho-family GAP domain, and a C-terminal SH3 domain (22, 25, 26). The srGAP1 SH3 domain binds the Robo1 receptor in response to Slit binding and down-regulates Cdc42 activity (26). The WRP SH3 domain binds the proline-rich region of WAVE-1. WAVE-1 is activated by Rac and binds Arp2/3 to induce branched actin polymerization. The activation of WAVE-1 by Rac can be terminated by WRP, a Rac-selective GAP (22). This interaction is important for the development of dendritic spines, synaptic plasticity, and memory retention (27). Together, these findings suggest that the srGAPs coordinately regulate Rho-family GTPase complexes and pathways using a common mechanism; that is, SH3 domain binding to proline-rich ligands in downstream effectors. To identify ligands for SH3 domains of other srGAP family members, we performed a yeast two-hybrid screen of a mouse embryonic day 9.5–10.5 cDNA library using the SH3 domain of srGAP2. This initial screen identified 58 clones that were sequenced for identification. Of these, one clone represented the intracellular tail of Robo-2, confirming previous findings that srGAP2 can bind members of the Robo receptor family (26). Interestingly, another clone corresponded to a portion of the proline-rich FH1 domain of FMNL3 (Fig. 1A). FMNL3 belongs to a subset of three highly homologous diaphanous-related formins, FMNL1, -2, and -3, that are activated downstream of Rho-family GTPases (9). This clone was re-tested for interactions with empty vector (negative control), the srGAP2 SH3 domain, the closely related WRP SH3 domain, and the unrelated SH3 domain of Nck (Fig. 1B). Profilin was used as a positive control, as it binds the proline-rich FH1 domain of many formins. Of the SH3 domains tested, only srGAP2 interacted with FMNL3. To further analyze this interaction and to assess specificity among other formins, we overlaid radiolabeled srGAP2 SH3 domain onto a peptide array of 20-mer peptides spanning the FH1 domains of FMNL1, -2, and -3 and mDia1, -2, and -3, which are variable in length (Fig. 2A). The results showed that, although the srGAP2 SH3 domain could bind to FMNL3, it preferentially bound peptides within the FH1 domain of FMNL1. No binding was detected between srGAP2 and FMNL2 or members of the mDia family. Western blots were used to determine the tissue distribution of FMNL1, FMNL3, and srGAP2. All three are expressed in similar tissues, but srGAP2 (which often migrates as two distinct molecular masses on SDS-PAGE, presumably due to proteolysis) and FMNL1 are preferentially co-expressed within the brain and thymus (Fig. 2B). To confirm the initial two-hybrid results, co-immunoprecipitations were performed between recombinant full-length proteins expressed in HEK293T cells (Fig. 2C). Compared with a negative control (Fig. 2C, lane 1), srGAP2 readily co-immunoprecipitated with FMNL1 (Fig. 2C, lane 2), confirming the interaction between the full-length proteins. This interaction depended on a conserved tryptophan within the SH3 domain required for polyproline peptide recognition (28). Mutation of this tryptophan to alanine completely abolished the interaction (Fig. 2C, lane 3). The binding between srGAP2 and FMNL1 was further confirmed by co-immunoprecipitation of the endogenous proteins (Fig. 2D). Immunoprecipitation of FMNL1 using an FMNL1-specific peptide antibody co-precipitated srGAP2 from HeLa cell extracts (Fig. 2D, lane 1), whereas negative control rabbit IgG did not (Fig. 2D, lane 2). Collectively, these results show that srGAP2, via its SH3 domain, forms a complex with FMNL1, binding specific proline-rich peptides within the formin FH1 domain.
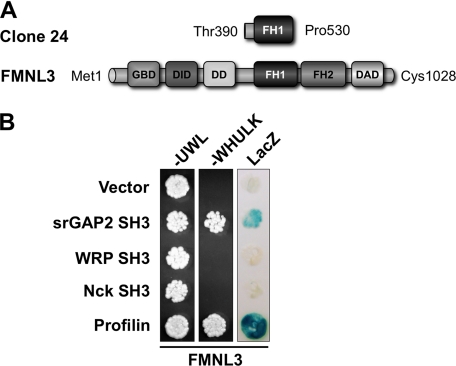
FIGURE 1.
Two-hybrid analysis of srGAP2 SH3 domain interactions. A, a schematic shows the fragment of FMNL3 identified in the library screen using the srGAP2 SH3 domain bait. Amino acid positions are indicated for each. B, specificity of the srGAP2 and FMNL3 interaction was verified in the two-hybrid assay. Yeast containing clone 24 and either empty vector (negative control), srGAP2 SH3, WRP SH3, Nck SH3, or profilin (positive control) were plated onto plates lacking uracil, tryptophan, and leucine (−UWL) or also histidine and lysine (−WHULK). Although all yeast could grow on the −UWL plate, indicating the yeast contain both bait and prey vectors, only the srGAP2 and profilin containing yeast grew on the −WHULK plates, confirming an interaction. Yeast were also assayed for β-galactosidase activity (LacZ), which further confirmed srGAP2 SH3 is specific for FMNL3. DD stands for dimerization domain.
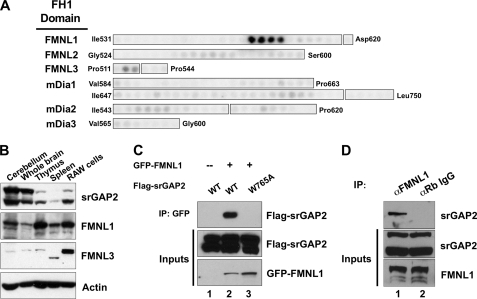
FIGURE 2.
srGAP2 forms a complex with FMNL1. A, shown is a peptide array analysis of the binding specificity of the srGAP2 SH3 domain for the FH1 domains of several formins. The beginning and ending amino acid positions indicate regions synthesized as 20-mer peptides offset every 3 amino acids. Binding was detected by overlay of γ-32P-radiolabeled GST-srGAP2 SH3. Results suggested the srGAP2 SH3 domain prefers peptides within the FMNL1 FH1 domain. B, expression distribution of srGAP2, FMNL1, and FMNL3 were determined by Western blot analysis using specific antibodies. Actin was used as a control for loading. Each panel is labeled to the right, and each tissue or cell line is labeled above each panel. RAW cells are a macrophage-derived cell line. C, full-lengths rGAP2 and FMNL1 interact by co-immunoprecipitation (IP). Cells were transfected with GFP-FMNL1 alone (lane 1) or with FLAG-srGAP2 (lane 2) or FLAG-srGAP2 with a point mutation in the SH3 domain (tryptophan 765 to alanine; lane 3). Immunoprecipitation of FMNL1 co-precipitated wild type srGAP2 (lane 2, top panel), but GFP alone or the SH3 mutant of srGAP2 did not (lanes 1 and 2, top panel). Immunoblot analysis of extracts indicated equal levels of srGAP2 in all three lysates (middle panel) and similar levels of FMNL1 (bottom panel). D, endogenous FMNL1 and srGAP2 form a complex in cells. Immunoprecipitation of FMNL1 co-precipitates srGAP2 from HeLa cells (lane 1), whereas α-rabbit IgG (negative control) does not (lane 2).
srGAP2 Regulates Signaling between Rac and FMNL1
Because srGAP2 is a GTPase-activating protein, we tested the GAP specificity of full-length srGAP2 toward the prototypic Rho-family GTPases (Fig. 3, A–C). We tested the binding preference of srGAP2 with all three GTPases in a GST-pulldown assay (Fig. 3A). Lysates from cells expressing recombinant srGAP2 were incubated with constitutively active GST-Rho, -Rac, or -Cdc42 bound to beads. Western blot analysis of pelleted proteins showed that srGAP2 preferentially bound to Rac, with some interaction also evident for Cdc42 but not RhoA or GST alone (Fig. 3A, top panel). Coomassie staining of each pulldown verified equivalent amounts of each GTPase were used in the assay (Fig. 3A, bottom panel). In vitro GTPase activity measurements were performed using partially purified full-length recombinant srGAP2 and purified wild-type GTPases (Fig. 3B). Recombinant srGAP2 preferentially stimulated the intrinsic GTPase activity of Rac rather than RhoA or Cdc42. Together with the pulldown assays, this indicated srGAP2 is a Rac-GAP. To test the Rac-GAP activity of srGAP2 in cells, a PAK1 pulldown assay for cellular Rac-GTP was performed (Fig. 3C). Lysates from HEK293T cells transfected with vector alone (Fig. 3C, lane 1) contained more Rac-GTP than lysates from cells transfected with srGAP2 (Fig. 3C, lane 2), confirming srGAP2 can function as a Rac-GAP in vitro and in situ.
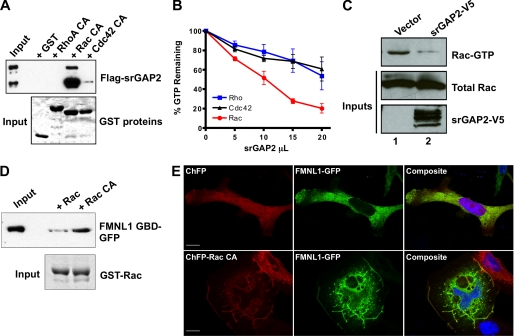
FIGURE 3.
Regulation of the Rac-srGAP2-FMNL1 pathway. A, shown is a GTPase specificity pulldown assay for srGAP2. Lysate from cells expressing srGAP2 (top panel, input) were incubated with GST-fused constitutively active Rho, Rac, or Cdc42 on glutathione beads (bottom panel, Coomassie stain). After centrifugation, bead fractions were assayed for bound srGAP2 by Western blot analysis (top panel). srGAP2 specifically associated with Rac but did not interact with Rho or Cdc42. B, shown is an in vitro GTPase assay for srGAP2 GAP specificity. 300 ng of purified Rho, Rac, and Cdc42 were loaded with radiolabeled GTP and incubated with increasing amounts of full-length srGAP2. srGAP2 exhibited greater GAP activity toward Rac when compared with Rho or Cdc42. C, shown is a cellular assay for srGAP2 Rac-GAP activity. Cells were transfected with empty vector or srGAP2 (bottom panel), and levels of Rac-GTP were analyzed (top panel) compared with total Rac (middle panel) by the p21-activated kinase pulldown assay. Cells expressing srGAP2 had lower levels of Rac-GTP, confirming Rac GAP activity in situ. D, shown is an activity-dependent interaction of FMNL1 with Rac. Lysates (Input) from cells expressing the FMNL1 GTPase binding domain (FMNL1 GBD; amino acids 1–450) were subjected to a pulldown assay using wild-type Rac or constitutively active Rac (RacCA) bound to beads as GST fusion (bottom panel). The FMNL1 GTPase binding domain preferentially interacted with active Rac. E, regulation of membrane targeting of FMNL1 by Rac is shown. Cells were co-transfected with either cherry fluorescent protein (ChFP) and FMNL1-GFP (top panels) or ChFP-Rac CA and FMNL1-GFP (bottom panels). Without active Rac, FMNL1 was predominately cytosolic, whereas with constitutively Rac, FMNL1 was enriched in membrane ruffles where it co-localized with Rac. The scale bar represents 15 μm.
Previous in vitro studies have suggested FMNL1 can bind to Rho-family GTPases, including both Rac and Cdc42, and that endogenous FMNL1 may be associated with Rac (29–31). To confirm the Rac interaction, we performed GTPase pulldown assays with recombinant wild-type or constitutively active Rac (RacQ61L) and an N-terminal fragment containing the GTPase binding domain of FMNL1 expressed in HEK293T cells (Fig. 3D). These results verified previous studies, indicating that FMNL1 preferentially interacts with the activated form of Rac. Prior studies have also shown that Cdc42 translocates FMNL1 to the plasma membrane, suggesting that membrane localization is one feature of FMNL1 activation (29). Therefore, we analyzed the localization of full-length FMNL1-GFP when co-transfected with either soluble cherry fluorescent protein (ChFP) or constitutive active Rac (ChFP-Rac CA). When expressed with ChFP, FMNL1 was predominantly cytosolic (Fig. 3E, top panels). In contrast, co-expression of Rac CA resulted in a marked translocation of FMNL1 to the membrane and dorsal ruffles, suggesting Rac can indeed interact with and activate FMNL1 (Fig. 3E, bottom panels). Together, these results suggest that the srGAP2·FMNL1 complex can be functionally linked to Rac signaling.
Dynamic Formation of the srGAP2·FMNL1 Complex
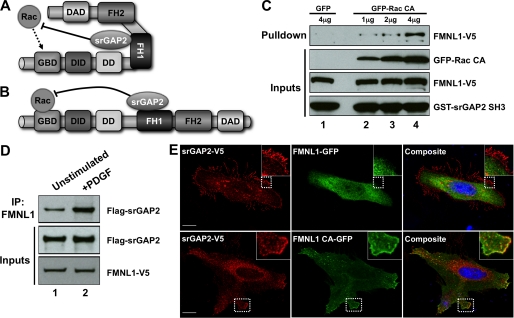
The above results suggest that srGAP2 complexes with FMNL1 for one of two potential reasons; 1) to prevent Rac from activating FMNL1 (Fig. 4A) or 2) to turn off Rac signaling after FMNL1 has been activated (Fig. 4B). If srGAP2 impedes the activation of FMNL1 by Rac, it should constitutively bind the formin or preferentially bind the inactive conformation. Conversely, if srGAP2 functions to terminate Rac-mediated activation of FMNL1, we would predict srGAP2 preferentially binds FMNL1 that has been activated by Rac. To distinguish between these two possibilities, we assayed the interaction between the srGAP2 SH3 domain and FMNL1 in the presence of increasing amounts of constitutively active Rac (Rac CA) (Fig. 4C). Because these experiments utilized the isolated SH3 domain, we could rule out any confounding effects of the GAP domain associating with Rac in the pulldown assay. GST-fused srGAP2 SH3 domain pulled down very little FMNL1 from cell lysates co-expressing GFP alone (Fig. 4C, lane 1). In contrast, the srGAP2 SH3 domain pulled down increasing amounts of FMNL1 from lysates expressing GFP-Rac CA in a dose-dependent manner (Fig. 4C, lanes 2–4). These data suggested the srGAP2·FMNL1 complex is dynamically regulated by the Rac-mediated activation of FMNL1. To confirm this with full-length proteins, HEK293T cells were co-transfected with srGAP2 and FMNL1 and either serum-starved (unstimulated) or treated with PDGF to activate endogenous Rac (32, 33) (Fig. 4D). Western blots of co-immunoprecipitations demonstrated that assembly of the srGAP2·FMNL1 complex was enhanced by PDGF treatment (Fig. 4D, lane 2). To visualize this activation-dependent interaction, HeLa cells expressing srGAP2 were transfected with either wild-type FMNL1 or FMNL1 that is rendered constitutively active (FMNL1 CA) by a point mutation in the DAD (L1062D) (29). When co-expressed, srGAP2 was predominantly at the membrane, whereas wild-type FMNL1 was largely cytosolic (Fig. 4E, top panels). In contrast, both srGAP2 and FMNL1 CA co-localized at the membrane (Fig. 4E, bottom panels), further supporting the idea that srGAP2 preferentially interacts with FMNL1 when it is activated. Together, the results indicate that the srGAP2·FMNL1 complex is formed in response to Rac-mediated activation of FMNL1 in vivo. The cumulative evidence is consistent with a model whereby srGAP2 acts to turn off Rac-mediated activation of FMNL1 (Fig. 4B).
FIGURE 4.
Dynamic assembly of the srGAP2·FMNL1 complex. A and B, shown is a schematic of possible roles for srGAP2 in regulating GTPase signaling to FMNL1. A, in this model srGAP2 is bound to FMNL1 in the inactive state and limits the ability of Rac to activate FMNL1. This could favor the specificity of FMNL1 activation toward other GTPases such as Cdc42. B, in this model srGAP2 only binds to activated FMNL1, where it functions to turn off Rac after activation is achieved. C, GST srGAP2 SH3 (bottom panel) pulldown from cells expressing FMNL1 (second panel from bottom) and increasing amounts of constitutively active Rac (Rac CA; third panel from bottom; lanes 2–4). Whereas very little FMNL1 associated with the srGAP2 SH3 domain in the absence of active Rac (lane 1), increasing amounts of FMNL1 co-associated with the SH3 domain in the presence of increasing levels of active Rac (lanes 2–4). D, association of the srGAP2·FMNL1 complex was analyzed by co-immunoprecipitation without (IP, lane 1) or with (lane 2) PDGF stimulation from cells co-transfected with srGAP2 (middle panel) and FMNL1 (bottom panel). Increased complex association was observed following PDGF stimulation (top panel). E, srGAP2 preferentially associates with the active form of FMNL1 in cells. Cells were co-transfected with srGAP2 and either GFP- tagged wild-type FMNL1 (top panels) or GFP tagged constitutive active FMNL1 (bottom panels; FMNL1 CA). Immunostaining for srGAP2 showed cytosolic and membrane staining that co-localized with FMNL1 CA (bottom panels). The inset shows higher magnification images of the corresponding boxed regions. The scale bar represents 15 μm. DD stands for dimerization domain.
Both srGAP2 and FMNL1 Co-localize with F-actin during Fc-γ Receptor Phagocytosis
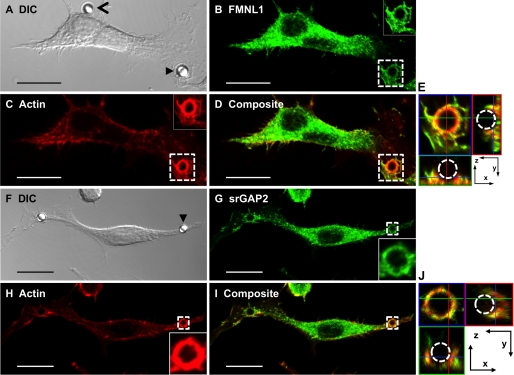
srGAP2 has been characterized primarily as a neuronal GAP, where it may regulate membrane protrusions during cortical neuron migration (25). In contrast, FMNL1 is expressed in cells of hematopoietic lineage, where it can function to regulate either synapse formation in T-cells or phagocytosis in macrophages (29–31). Thus, it was important to examine whether endogenous FMNL1 and srGAP2 co-localize during either of these events. Based on our finding that both proteins are expressed in the macrophage-derived line RAW264.7 (Fig. 2B) and the known role of Rac signaling during Fc-γ receptor-mediated phagocytosis (34), we focused on the potential role of the FMNL1·srGAP2 complex in these cells. One of the earliest events during phagocytosis is the formation of extensive membrane evaginations that engulf Fc-coated particles. Recent work on the membrane binding Inverse F-BAR (IF-BAR) domain has shown that the srGAP family of GAPs can be efficiently recruited to outward membrane protrusions (25). Thus, the possibility that endogenous srGAP2 may co-localize with FMNL1 and actin within membrane extensions during phagocytosis was tested (Fig. 5). In cellular regions where phagocytosis did not occur, FMNL1 and srGAP2 were mostly cytosolic, and neither co-localized with actin very well (Fig. 5, A–D and F–I). However, upon stimulation of Fc-γ receptor phagocytosis using Fc-coated beads, FMNL1 and srGAP2 co-localized with actin at beads undergoing phagocytosis (Fig. 5, A and F, closed arrowheads; B–D and G–I, boxed regions and insets). This was clearly evident in z-projections (Fig. 5, E and J) of the periphagosomal regions, where both FMNL1 and srGAP2 co-localized with actin in the phagocytic membranes as they surrounded the bead (dashed circle). These data indicate that one function of the FMNL1·srGAP2 complex may be to regulate actin dynamics during phagocytic cup formation, which involves both membrane evagination and actin-driven internalization downstream of Rac signaling.
FIGURE 5.
Localization of FMNL1 and srGAP2 to the actin-rich phagosome during phagocytosis. Co-localization of FMNL1 (A–E) or srGAP2 (F–J) with F-actin during phagocytosis is shown. A and F, differential contrast image (DIC) of a RAW cell incubated with Fc-coated beads is shown. Closed arrowheads indicate a bead undergoing phagocytosis, and an open arrowhead indicates a nearby bead that is not being phagocytosed. Maximum projection images depicting immunolocalization of endogenous FMNL1 (B), srGAP2 (G), phalloidin-stained actin (C and H), or composite images (D and I) are shown. Insets correspond to boxed regions. E and J, images depict composite boxed regions from panels D and I. The central image, surrounded by blue in E and J is a 0.25-μm section from the z-stack. The images below and to the side of this section are orthogonal projections of the phagocytosis cup. The images to the bottom (surrounded by green) represent the image projected where the green line bisects the stack. The images to the right (surrounded by red) indicate the image projected where the red line bisects the stack. The blue lines seen in the images to the bottom and right are the z-position of the central image section. The scale bar represents 15 μm.
Direct Regulation of FMNL1 Actin Severing by the srGAP2 SH3 Domain
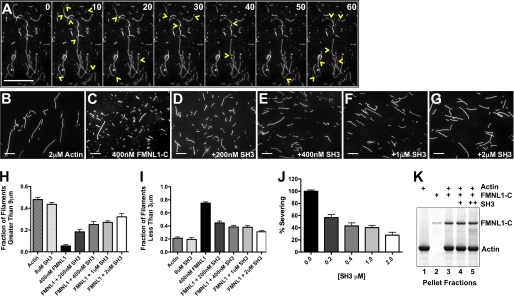
Our data demonstrate that upon FMNL1 activation, srGAP2 binds the FH1 domain. The C terminus of FMNL1, including the FH1 and FH2 domains, is tightly associated with actin filaments and possesses actin severing activity (14). The binding of srGAP2 might modulate this FMNL1 activity. To test this hypothesis, we assayed the ability of the srGAP2 SH3 domain to inhibit severing by the FMNL1 C terminus (FMNL1-C, amino acids 449–1094). First, we visualized FMNL1-C severing actin in a live, single filament assay of using TIRF microscopy (Fig. 6A). In this assay the severing of filaments by FMNL1-C was directly observed in time-lapse every 10 s. Within 1 min, most filaments were severed at multiple sites (Fig. 6A, yellow arrowheads indicate new breaks within each panel), demonstrating that FMNL1 possessed potent severing activity in this assay (14). We next tested whether the binding of the srGAP2 SH3 domain modulated this FMNL1 activity in a quantitative actin severing assay (14) (Fig. 6, B–G). In this assay actin was polymerized and incubated for 5 min with FMNL1-C without and with increasing concentrations of the srGAP2 SH3. The reaction was stopped using rhodamine-labeled phalloidin, which inhibits FMNL1-mediated actin severing (14). The resulting filaments were imaged, and lengths were quantified. F-actin alone showed filaments of varied lengths (Fig. 6B). In contrast, FMNL1-C severed actin into predominantly shorter filaments (Fig. 6C). This potent severing activity was inhibited by the srGAP2 SH3 domain (Fig. 6, D–G). Furthermore, the srGAP2 SH3 domain did not affect actin filaments in the absence of FMNL1, suggesting that the effects were mediated by inhibition of FMNL1 and not a direct effect on actin. To quantify severing, we calculated the fraction of filaments that were either long (greater than 9 μm, which decrease with severing) or short (less than 3 μm, which increase with severing). These data are graphically represented in Fig. 6, H–J. Percent of actin severing at each concentration of the srGAP2 SH3 domain was calculated by normalizing to the effect of FMNL1 alone. This demonstrated a dose-dependent inhibition by srGAP2 SH3 on FMNL1 (Fig. 6J).
FIGURE 6.
Regulation of FMNL1-mediated actin filament severing by the srGAP2 SH3 domain. A, shown are time-lapse images of polymerized actin filaments every 10 s in the presence of purified active FMNL1-C (amino acids 449–1094). Numbers in the upper right corner of each frame indicate time. Extensive severing is observed during the 1-min time-lapse period. Yellow arrowheads mark the locations of new filament severing events in each frame. B–G, shown are images of actin filaments (2 μm) either alone (B), with FMNL1-C (400 nm) (C), or with FMNL1-C and increasing concentrations of the purified srGAP2 SH3 domain (D–G). Scale bar represents 15 μm. H–J, quantification of filament lengths from three independent severing assays show the fraction of filaments longer than 9 μm (H) or shorter than 3 μm for each condition (I). J, shown is percent severing as calculated from H and I for increasing concentrations of the srGAP2 SH3 domain. Scale bars represent 15 μm. Error is ±S.E. K, polymerized actin filaments (250 nm, lane 1), stabilized by phalloidin, are pelleted by ultracentrifugation in the presence of FMNL1-C (100 nm) without and with srGAP2 SH3 (200 and 400 nm, lanes 4 and 5, respectively). In the absence of actin, FMNL1-C does not co-pellet with actin filaments (lane 2). With actin filaments, FMNL1-C does co-pellet (lane 3), and the srGAP2 SH3 domain does not disrupt this interaction (lane 4 and 5).
The effect of the srGAP2 SH3 domain could be mediated by either dissociating FMNL1 from the sides of actin filaments or by an allosteric or steric hindrance effect on severing. The srGAP2 SH3 domain did not alter the ability of the FMNL1-C to bind actin filaments in an actin filament co-sedimentation assay (Fig. 6K). Thus, these results show that whereas FMNL1 can sever actin filaments, the binding of srGAP2 interferes with this activity, most likely by an allosteric or steric hindrance mechanism.
DISCUSSION
In this study we have identified a complex between srGAP2 and FMNL1 that is temporally regulated by Rac signaling and appears to regulate FMNL1 activity by two mechanisms (Fig. 7). This complex forms by a direct interaction between the SH3 domain of srGAP2 and the FH1 domain of FMNL1. Interestingly, our data suggest the accessibility of the FMNL1 binding site is dependent on activation by Rac. Although we found that srGAP2 is selective for Rac-GTPase activity and that FMNL1 could also interact with Rac, it should be noted that FMNL1 might also be regulated by Cdc42 (29). Our data show that srGAP2 likely regulates Rac signaling to FMNL1 by stimulating the cycling of Rac to its inactive state. To our knowledge this is the first description of a mechanism to terminate Rho-family GTPase-dependent activation of a formin, which has been hypothesized to be a key step in regulating formins (11). Furthermore, it is quite likely that GAP-mediated regulation of formins is a conserved mechanism.3
FIGURE 7.
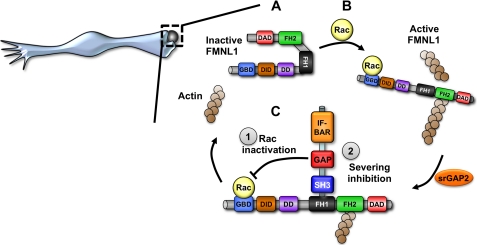
Model of srGAP2 bi-modal regulation of FMNL1-actin signaling during phagocytosis. A, FMNL1 is basally inhibited and cytosolic. Upon activation by Rac (B) FMNL1 translocates to the membrane, and the active form of FMNL1 can bind and sever actin filaments. This potentially regulates Fc-γ receptor-mediated phagocytosis in macrophages. C, in this active state srGAP2 can bind FMNL1 at the membrane to inactivate the Rac signal (1) and inhibit the severing activity of FMNL1 (2). This would return FMNL1 to its quiescent, inactive state. This recycling mechanism could be responsible for rapid turnover of actin filaments and Rac signaling during phagocytosis. DD stands for dimerization domain.
A second mechanism by which srGAP2 regulates FMNL1 is through direct inhibition of its actin severing activity. An active form of FMNL1 (FMNL1-C) containing the C-terminal FH1-FH2 domains severs and bundles preformed actin filaments in addition to enhancing polymerization at barbed ends using profilin and actin (Fig. 6 and Ref. 14). These diverse activities elicit many questions about the function of FMNL1 in vivo and how these activities might be regulated. For example, our work and the work of others show that actin filaments in vitro can be elongated to lengths well over 40 μm, and FMNL1-C can sever these filaments into submicron lengths (14). Neither of these scenarios is overtly physiological, so there must be a balance established between these two; a regulatory factor must exist in vivo. Few inhibitors (DIP/WISH, SPIRE, and Bud14p) and even fewer mechanisms have been identified to inhibit formins (16, 18, 19). In one recent example, Bud14p has been shown to displace a yeast formin, Bnr1p, from the barbed end of actin filaments (16). Bnr1p is a potent elongator of actin filaments, so displacing this protein from the barbed end provides an elegant and practical mechanism for inhibiting formins; that is, physically displacing them from the filaments they polymerize. Interestingly, yeast genetic data suggest Bud14p does not simply oppose Bnr1p activity but, instead, regulates the duration of Bnr1p polymerization.
srGAP2 is the fourth biochemical inhibitor of formin activity identified. Our data show that srGAP2 functions differently from Bud14p in that it does not displace FMNL1 from the sides of actin filaments (Fig. 6K). Interestingly, it seems the FH1 domain might play an important role in regulating the adjacent FH2 domain, acting as a gas or brake pedal to the FH2 engine. For example, profilin-actin complexes accelerate elongation of filaments by formins (17, 35). This effect is directly mediated by binding to the FH1 domain, acting as a gas pedal to elevate polymerization rates at the barbed end. We hypothesize that the srGAP2 SH3 domain acts as a brake, binding the FH1 domain of FMNL1 to oppose severing through an allosteric or steric mechanism. To our knowledge, this is also the first example of a GTPase-activating protein that may also modify the actin cytoskeleton architecture in a GAP-independent manner, utilizing only the SH3 domain. These data suggest that factors that bind the FH1 domain may play a critical role in regulating formin biochemistry and signaling in vivo, warranting future work to identify and characterize their functions.
The cellular mechanisms and functions for many of the mammalian formins are still unknown. FMNL1 is critical for T-cell polarization and subsequent activation as well as Fc-γ receptor-mediated phagocytosis in macrophages, a Rac-dependent process (29, 31, 36, 37). Our data show that both FMNL1 and srGAP2 localize to the actin-rich Fc-γ receptor phagocytic cup in macrophages. Phagocytosis of foreign particles can occur in less than 1 min, indicating that rapid polymerization and turnover of actin is essential for driving the Fc-γ receptor-dependent process (38). Because rapid actin polymerization is necessary, a large pool of free barbed ends of actin filaments is likely required. Elongation of these filaments may also be coordinated with Arp2/3-dependent branching, an essential driving force during the phagocytic process (39, 40). Actin severing mediated by FMNL1 could provide a mechanism for generating new barbed ends for abundant filament growth. Essential to this amplification of new filaments may also be a mechanism for the coordinated regulation of Rac activity and FMNL1 actin severing by srGAP2. Analogous to observations on the Bud14p regulation of Bnr1p in yeast, where Bud14p may promote Bnr1p activity cycling (16), the Rac·FMNL1·srGAP2 complex may provide a succinct mechanism to coordinately recycle FMNL1 and Rac for rapid actin turnover in the phagocytic cup.
Acknowledgments
We thank Dr. Michael Rosen for the FMNL1-GFP constructs, Dr. Ryohei Yasuda for RacQ61L-Cherry, Dr. Daniel Billadeau for FMNL1 and FMNL3 antibodies as well as the FMNL1 cDNA, Dr. Tim Oliver and the Duke Light Microscopy Core Facility for expert imaging and analysis advice, and Dr. James Bear for actin filament TIRF imaging protocol assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant NS059957 (to S. H. S.). This work was also supported by March of Dimes Grant 5-FY07-671 (to S. H. S.) and GM069818 (to H. N. H.).
F. M. Mason and S. H. Soderling, unpublished observations.
- GAP
- GTPase-activating protein
- FMNL1
- formin-like 1
- TIRF
- total internal reflection fluorescence
- ChFP
- cherry fluorescent protein
- CA
- constitutively active
- DID
- Dia inhibitory domain
- DAD
- Dia autoregulatory domain
- GBD
- GTPase binding domain
- FH
- formin homology.
REFERENCES
- 1. Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 2. Weaver A. M., Young M. E., Lee W. L., Cooper J. A. (2003) Curr. Opin. Cell Biol. 15, 23–30 [DOI] [PubMed] [Google Scholar]
- 3. Chesarone M. A., Goode B. L. (2009) Curr. Opin. Cell Biol. 21, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soderling S. H. (2009) Sci. Signal. 2, pe5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 6. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 7. Chesarone M. A., DuPage A. G., Goode B. L. (2010) Nat. Rev. Mol. Cell. Biol. 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 8. Kovar D. R. (2006) Curr. Opin. Cell Biol. 18, 11–17 [DOI] [PubMed] [Google Scholar]
- 9. Higgs H. N., Peterson K. J. (2005) Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higgs H. N. (2005) Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 11. Goode B. L., Eck M. J. (2007) Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 12. Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., Wittinghofer A. (2005) Nature 435, 513–518 [DOI] [PubMed] [Google Scholar]
- 13. Otomo T., Otomo C., Tomchick D. R., Machius M., Rosen M. K. (2005) Mol. Cell 18, 273–281 [DOI] [PubMed] [Google Scholar]
- 14. Harris E. S., Li F., Higgs H. N. (2004) J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 15. Esue O., Harris E. S., Higgs H. N., Wirtz D. (2008) J. Mol. Biol. 384, 324–334 [DOI] [PubMed] [Google Scholar]
- 16. Chesarone M., Gould C. J., Moseley J. B., Goode B. L. (2009) Dev. Cell 16, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 18. Eisenmann K. M., Harris E. S., Kitchen S. M., Holman H. A., Higgs H. N., Alberts A. S. (2007) Curr. Biol. 17, 579–591 [DOI] [PubMed] [Google Scholar]
- 19. Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. (1995) Mol. Cell. Biol. 15, 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Westphal R. S., Soderling S. H., Alto N. M., Langeberg L. K., Scott J. D. (2000) EMBO J. 19, 4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soderling S. H., Binns K. L., Wayman G. A., Davee S. M., Ong S. H., Pawson T., Scott J. D. (2002) Nat. Cell Biol. 4, 970–975 [DOI] [PubMed] [Google Scholar]
- 23. Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 24. Cai L., Makhov A. M., Schafer D. A., Bear J. E. (2008) Cell 134, 828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W. L., Frost A., Polleux F. (2009) Cell 138, 990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong K., Ren X. R., Huang Y. Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L., Wu J. Y., Xiong W. C., Rao Y. (2001) Cell 107, 209–221 [DOI] [PubMed] [Google Scholar]
- 27. Soderling S. H., Guire E. S., Kaech S., White J., Zhang F., Schutz K., Langeberg L. K., Banker G., Raber J., Scott J. D. (2007) J. Neurosci. 27, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erpel T., Superti-Furga G., Courtneidge S. A. (1995) EMBO J. 14, 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seth A., Otomo C., Rosen M. K. (2006) J. Cell Biol. 174, 701–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yayoshi-Yamamoto S., Taniuchi I., Watanabe T. (2000) Mol. Cell. Biol. 20, 6872–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gomez T. S., Kumar K., Medeiros R. B., Shimizu Y., Leibson P. J., Billadeau D. D. (2007) Immunity 26, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takahashi M., Rikitake Y., Nagamatsu Y., Hara T., Ikeda W., Hirata K., Takai Y. (2008) Genes Cells 13, 549–569 [DOI] [PubMed] [Google Scholar]
- 33. Ouyang M., Sun J., Chien S., Wang Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14353–14358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caron E., Hall A. (1998) Science 282, 1717–1721 [DOI] [PubMed] [Google Scholar]
- 35. Paul A. S., Pollard T. D. (2009) Cell Motil. Cytoskeleton 66, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall A. B., Gakidis M. A., Glogauer M., Wilsbacher J. L., Gao S., Swat W., Brugge J. S. (2006) Immunity 24, 305–316 [DOI] [PubMed] [Google Scholar]
- 37. Wang Q. Q., Li H., Oliver T., Glogauer M., Guo J., He Y. W. (2008) J. Immunol. 180, 2419–2428 [DOI] [PubMed] [Google Scholar]
- 38. Diakonova M., Bokoch G., Swanson J. A. (2002) Mol. Biol. Cell 13, 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. May R. C., Caron E., Hall A., Machesky L. M. (2000) Nat. Cell Biol. 2, 246–248 [DOI] [PubMed] [Google Scholar]
- 40. Lorenzi R., Brickell P. M., Katz D. R., Kinnon C., Thrasher A. J. (2000) Blood 95, 2943–2946 [PubMed] [Google Scholar]