Abstract
Ski was originally identified as an oncogene based on the fact that Ski overexpression transformed chicken and quail embryo fibroblasts. Consistent with these proposed oncogenic roles, Ski is overexpressed in various human tumors. However, whether and how Ski functions in mammalian tumorigenesis has not been fully investigated. Here, we show that Ski interacts with p53 and attenuates the biological functions of p53. Ski overexpression attenuated p53-dependent transactivation, whereas Ski knockdown enhanced the transcriptional activity of p53. Interestingly, Ski bound to the histone deacetylase SIRT1 and stabilized p53-SIRT1 interaction to promote p53 deacetylation, which subsequently decreased the DNA binding activity of p53. Consistent with the ability of Ski to inactivate p53, overexpressing Ski desensitized cells to genotoxic drugs and Nutlin-3, a small-molecule antagonist of Mdm2 that stabilizes p53 and activates the p53 pathway, whereas knocking down Ski increased the cellular sensitivity to these agents. These results indicate that Ski negatively regulates p53 and suggest that the p53-Ski-SIRT1 axis is an attractive target for cancer therapy.
Keywords: Cancer Therapy, Oncogene, p53, Signal Transduction, SIRT, Ski
Introduction
Ski was originally identified as the transforming protein (v-Ski) of avian Sloan-Kettering virus (1). Ski was initially characterized as a structurally and functionally related nuclear proto-oncoprotein on the basis of its ability to transform chicken and quail embryo fibroblasts when overexpressed (2). Ski is an important negative regulator of transforming growth factor-β (TGF-β) signaling (3). Because TGF-β potently inhibits the growth of most cell types, loss of the TGF-β anti-proliferative response is thought to enhance the progression of various tumors (4). Ski is thought to suppress TGF-β signaling primarily through transcriptional repression by recruiting the nuclear corepressor (N-CoR) and histone deacetylases to Smad complexes as well as inhibiting the recruitment of the transcriptional coactivator p300/CBP2 (5). In addition, Ski regulates distinct cellular signaling pathways, including nuclear hormone receptors, Wnt/β-catenin signaling, GATA1, PU.1, and retinoblastoma (RB) (2). Consistent with its oncogenic activity, Ski is overexpressed in many tumors, including melanoma (6), colorectal cancer (7), pancreatic cancer (8) esophageal cancer (9), gastric cancer (10), and leukemia (11). In addition, RNA interference (RNAi)-mediated silencing of Ski expression inhibits the tumorigenic properties of cancer cells (12). On the other hand, Ski also has been reported to function as a tumor suppressor. Heterozygous Ski+/− mice are more susceptible to carcinogen-induced tumors than wild-type mice (13) and down-regulating Ski expression enhances tumor metastasis in vivo (14). Therefore, the role of Ski in mammalian tumorigenesis is highly complex, and the impact of Ski on cells is context-dependent and likely influenced by interactions with multiple cellular proteins.
The tumor suppressor p53 plays an important role in regulating cell proliferation during cellular stress (15–18). In addition, p53 functions are lost in most human tumors. Approximately 50% of all malignancies contain a p53 mutation, and a large proportion of tumors without p53 mutations have inactivated the function of p53 by various mechanisms (e.g. overexpression of Mdm2, loss of p14Arf, and expression of viral oncogenes including E1A and SV40 large T antigen) (19). p53 plays an essential role in controlling the transactivation of target genes in each stress response pathway, although some p53 effects may be independent of transcription. The function of p53 is tightly regulated, and p53 expression is maintained at low levels by a mechanism that involves ubiquitin-proteasome-mediated degradation. However, when cells experience a variety of stresses, p53 becomes activated. The mechanisms that activate p53 are generally thought to involve posttranslational modifications of the p53 protein, such as phosphorylation and acetylation (20, 21). For example, phosphorylation of the N-terminal domain of p53 has been shown to regulate its transactivation properties, whereas acetylation of the C-terminal domain activates sequence-specific DNA binding and stabilizes of the p53 protein. p53 is acetylated by histone acetyltransferases, including p300, CBP, PCAF, and Tip60. Recently, Tang et al. (22) showed that p53 acetylation is indispensable for the biological functions of p53.
Conversely, p53 can be deacetylated by distinct histone deacetylase complexes containing HDAC1 or SIRT1 (23). Deacetylation of p53 by these complexes represses p53-dependent transcriptional activation, apoptosis, and growth arrest. In particular, SIRT1-deficient mice have hyperacetylated p53 and increased DNA damage-induced apoptosis (24), and short interfering RNA (siRNA)-mediated knockdown of SIRT1 reduces drug resistance and induces the growth arrest in cancer cells in vitro (25). Because SIRT1 is up-regulated in various tumors (26), it has been suggested that SIRT1 contributes to tumorigenesis through its deacetylase activity.
We performed a screen to identify Ski-interacting proteins and determined that Ski physically interacts with p53. Ski suppresses the transactivation ability of p53 by reducing p53 acetylation and decreasing the ability of p53 to bind to DNA upon DNA damage. Although Ski forms a complex with histone deacetylases, we determined that Ski also interacts with SIRT1. Ski interacts with SIRT1 and cooperates to repress p53 activity by deacetylating p53. Moreover, Ski overexpression confers resistance to genotoxic agents and Nutlin-3, a small-molecule inhibitor that blocks the Mdm2-p53 interaction and activates the p53 pathway. These results indicate that Ski is a negative regulator of p53 and suggest that Ski is a new molecular target for cancer therapy.
EXPERIMENTAL PROCEDURES
Cell Lines, Plasmids, and Transfections
H1299 cells were cultured in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum (FBS) and penicillin/streptomycin. MCF7 cells, 293 cells and HCT116 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% FBS and penicillin/streptomycin. HepG2 cells were cultured in minimum essential medium (Invitrogen) supplemented with nonessential amino acids, sodium pyruvate, penicillin/streptomycin, and 10% FBS.
The original constructs encoding the human Ski and p300 cDNAs were described previously (5, 10, 27). pcDEF3/FLAG-p53 and pcDNA3/FLAG-SIRT1 were PCR-amplified using mRNA derived from MCF7 cells. All constructs were verified by sequencing. H1299 cells and 293 cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Antibodies
Anti-p53 (DO-1) and anti-p21 (Ab-1) antibodies were purchased from Calbiochem. anti-Ski (G-8), anti-Ski (H-329), anti-SIRT1 (H-300), and anti-mSin3A (K-20) antibodies as well as horseradish peroxidase-conjugated anti-p53 antibody (DO-1) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) Anti-FLAG (M2) and anti-β-actin (AC-15) antibodies were obtained from Sigma, and anti-acetyl-p53 (Lys-382), anti-phospho-p53 (Ser-15), anti-phospho-p53 (Ser-20), anti-phospho-p53 (Ser-392) antibodies as well as horseradish peroxidase-conjugated secondary antibodies were purchased from Cell Signaling Technology (Beverly, MA). Anti-phospho-p53 (Ser-46) antibody was obtained from Medical & Biological Laboratories Co. (Nagoya, Japan). Anti-Myc (9E10) and anti-Smad2/3 antibodies were obtained from BD Biosciences.
Luciferase Assay
Cells were transiently transfected with pp53-TA-Luc (Stratagene, La Jolla, CA) or pTA-Luc (Stratagene), pGL4.75 (hRuc/CMV) (Promega, Madison, WI), and the empty vector. The total amount of transfected DNA was the same in each experiment. Luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega), and the values were normalized to the Renilla luciferase activity. All experiments were performed at least three times and are presented as means ± S.D.
Immunoprecipitation and Western Blot Analyses
Cells were lysed in TNTE buffer (20 mm Tris-HCl, pH 7.5, 120 mm NaCl, 1 mm EDTA, and 0.5% Triton-X 100) supplemented with protease and phosphatase inhibitors as described previously (28). The lysates were immunoprecipitated with the appropriate antibodies in the presence of protein G-Sepharose (GE Healthcare). Endogenous Ski proteins were immunoprecipitated with an anti-Ski (G-8) antibody that had been preincubated for 6 h with Dynabeads M-280 sheep anti-mouse IgG (Invitrogen). The immunoprecipitates were washed four times with TNTE buffer and then subjected to SDS-polyacrylamide gel electrophoresis. The gels were transferred onto Fluoro Trans W membranes (Pall, East Hills, NY) and then probed with the indicated antibodies. The Western blot membranes were developed using ECL Western blotting detection reagents (GE Healthcare).
RNA Extraction and Quantitative Real-time PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions as described previously (29). First-strand cDNA was synthesized with PrimeScript reverse transcriptase (TaKaRa Bio Inc., Shiga, Japan) and oligo(dT)20 primers. Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Invitrogen) and the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). The following primer sequences were used: human p21, 5′-GATTTCTACCACTCCAAACGCC-3′ (forward) and 5′-AGAAGATGTAGAGCGGGC-3′ (reverse); human PIG3, 5′-CAGCTGCTGGATTCAATTAC-3′ (forward) and 5′-TGACGTTCTTCTCCCAGTAG-3′ (reverse); human mdm2, 5′-TGTTGGTGCACAAAAAGACA-3′ (forward) and 5′-CACGCCAAACAAATCTCCTA-3′ (reverse); and human HPRT1, 5′-TTTGCTTTCCTTGGTCAGGC-3′ (forward) and 5′-GCTTGCGACCTTGACCATCT-3′ (reverse). The specificity of the detected signals was confirmed by a dissociation curve, which consisted of a single peak.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed as described previously (10). Briefly, the cells were fixed in 1% formaldehyde with gentle shaking for 10 min at room temperature, and the cross-linking reaction was stopped by adding 2.5 m glycine to a final concentration of 0.125 m. After two washes with cold phosphate-buffered saline, the cells were harvested by scraping, pelleted, and then resuspended in SDS lysis buffer (50 mm Tris-HCl, pH 8.0, 1% SDS, 10 mm EDTA, and protease inhibitors). The samples were sonicated three times for 15 s each with 30-s intervals using a UH-50 sonicator (SMT Co., Tokyo, Japan), and centrifuged at 14,000 rpm at 8 °C for 10 min. After removal of a control aliquot (whole cell extract), the supernatants were diluted 10-fold in ChIP dilution buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, and protease inhibitors). The samples were immunoprecipitated with Dynabeads M-280 sheep anti-mouse IgG that had been preincubated with 4 μg of the appropriate antibodies in phosphate-buffered saline containing 0.5% bovine serum albumin. After washing five times with ChIP wash buffer (50 mm Hepes-KOH, pH 7.0, 0.5 m LiCl, 1 mm EDTA, 0.7% deoxycholate, and 1% Nonidet P-40) and once with TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA) the proteins were eluted from the beads with 0.2 ml of elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, and 1% SDS). After reversing the cross-linking, the DNA samples were extracted with a PCR purification kit (Qiagen, Valencia, CA). The purified DNA was analyzed by quantitative real-time PCR using the following specific primers: 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′ for the p21 promoter; 5′-GGTTGACTCAGCTTTTCCTCTTG-3′ and 5′-GGAAAATGCATGGTTTAAATAGCC-3′ for the mdm2 promoter; and 5′-TGTTTGGGCTATTTACTAG-TTG-3′ and 5′-ATAAAATGACTTAAGCCCAGAG-3′ for the HPRT1 first intron.
RNA Interference
siRNAs were obtained from Invitrogen. The siRNA duplexes were as follows: Ski siRNA sense strand, 5′-UGACUCGUUGGCCUCUUUCAUCUUC-3′; Ski siRNA sense strand 2, 5′-UUGUGCGAGUGCACCACGAACUUGU-3′; Ski siRNA sense strand 3, 5′-AUAGUCGAAUUUCUCCUUCACGUCG-3′; p53 siRNA sense strand, 5′-CCAGUGGUAAUCUACUGGGACGGAA-3′; and SIRT1 siRNA sense strand, 5′-UCAUAGAGCCAUGAAGUAUGACAAA-3′. The stealth control siRNA (Invitrogen; StealthTM RNAi negative control medium GC duplex) was used as a control. Cells were transfected with the noted siRNAs using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol.
Generation and Infection of Lentiviruses
FLAG-Ski cDNA was cloned into the pENTR vector (Invitrogen). To create the lentiviral constructs, this cDNA was subcloned into lentivirus vector CSII-EF-RfA (a gift from Dr. H. Miyoshi, RIKEN) using LR clonase (Invitrogen).
To produce replication-defective lentivirus vectors, 293FT cells were co-transfected with the expression plasmids, a VSV-G and Rev expression plasmid (pCMV-VSV-G-RSV-Rev), and the packaging plasmid (pCAG-HIVgp) using Lipofectamine 2000. The viral supernatants were collected at 24 and 48 h after transfection. For the lentiviral infections, 1.0 × 105 cells/well in 6-well plates were infected with the viral particles according to standard protocols.
BrdUrd Incorporation Assay and Cell Viability Assay
BrdUrd incorporation was measured using the BrdU Labeling and Detection Kit I (Roche Molecular Biochemicals). Briefly, the cells were incubated with 10 mm BrdUrd for 1 h and then fixed with 70% ethanol in glycine buffer, pH 2.0, for 30 min. After washing with PBS, the labeled cells were visualized by incubating with an anti-BrdUrd antibody and fluorescein-conjugated anti-mouse IgG antibody. At least 200 cells from each sample were counted for BrdUrd incorporation. Cell viability was measured using the CellTiter-Glo luminescent cell viability assay, which quantifies ATP levels as a measure of metabolically active cells (Promega).
RESULTS
Ski Interacts with p53
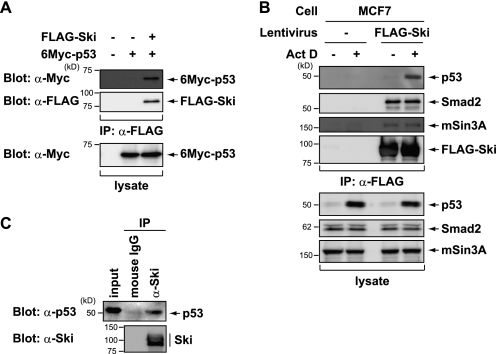
We conducted a screen to identify Ski-interacting proteins and determined that Ski interacts physically with the tumor suppressor p53. As shown in Fig. 1A, ectopically expressed Ski co-immunoprecipitated with p53 in p53-null H1299 cells. To investigate this interaction under more physiological conditions, we next examined MCF7 cells that stably express FLAG-Ski. The interaction between p53 and FLAG-Ski was minimally detected in unstressed cells because of the extremely low p53 levels. However, this interaction was strongly enhanced by treatment with low concentrations of actinomycin D (ActD), which activate a ribosomal stress response and stabilize the p53 protein (Fig. 1B) (30–32). Several previously identified Ski-associated proteins (e.g. Smad2 and mSin3A) also co-immunoprecipitated in the presence and absence of ActD (Fig. 1B). Moreover, endogenous Ski and p53 interacted in HCT116 cells (Fig. 1C). These results indicate that Ski interacts with p53 in vivo and that Ski may regulate the function of p53 during DNA damage.
FIGURE 1.
Ski interacts with p53. A, H1299 cells were transiently transfected with the indicated constructs. After 24 h, the cell lysates were immunoprecipitated (IP) with an anti-FLAG antibody and then immunoblotted with an anti-Myc antibody. B, the cells were treated with or without ActD for 8 h. The FLAG-purified Ski complexes were analyzed by immunoblotting. C, HCT116 cells were treated with 10 μm MG132, a proteasome inhibitor, for 12 h. The cell lysates were immunoprecipitated with an anti-Ski antibody and then immunoblotted with an anti-p53 antibody.
Mapping the Interacting Regions in Ski and p53
We next identified p53-binding domain of Ski using various Ski deletion mutants (supplemental Fig. S1A). Deleting residues 309–728 (Ski D and Ski E) did not affect the Ski-p53 interaction, whereas further truncating Ski to residue 210 (Ski F) diminished the p53 interaction (supplemental Fig. S1B). These results indicate that the region in Ski from residues 210 to 309, which contains the SAND-like domain (2), is necessary to bind to p53. Previous studies have shown that the SAND-like domain of Ski is involved in binding to Smad4 and that a W274E mutation in Ski abolishes the interaction between Ski and Smad4 (33). As shown in supplemental Fig. S1C, Ski W274E failed to interact with p53, indicating that the p53-binding domain in Ski overlaps the binding site for Smad4. Because Smads are known to interact with p53 (34), we examined whether Smad4 was required for the complex formation between Ski and p53 (supplemental Fig. S1D). When endogenous Smad4 was depleted with siRNA, the association between p53 and Ski was hardly affected, suggesting that Smad4 is not required for the interaction between Ski and p53.
The SAND-like domain of Ski also has a high degree of homology with v-Ski (35). Consistent with this, v-Ski was also able to bind to p53 (supplemental Fig. S1C). As the Ski fragment containing residues 76–304 is responsible for the ability of Ski to transform cells (36), we hypothesized that the ability of Ski to bind to p53 is important for the oncogenic activity of Ski.
To map the Ski-binding domain in p53, we also prepared various p53 deletion mutants (supplemental Fig. S1E) and examined the interaction between Ski and these p53 deletion mutants by immunoprecipitation/Western blot analyses. As shown in supplemental Fig. S1F, Ski bound to p53-(1–290), p53-(90–290), and p53-(90–393) but not p53-(290–393). These results suggest that the DNA-binding domain of p53 is important for the interaction with Ski.
Ski Suppresses the Transactivation Ability of p53
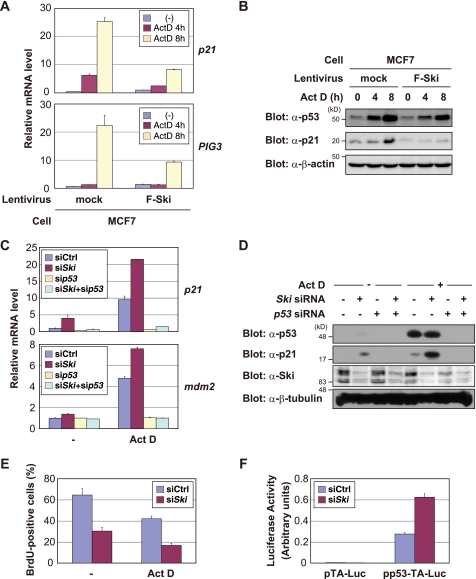
Because Ski has been shown to regulate the activity of transcription factors (2), we examined whether Ski affects the transcriptional activity of p53. Overexpressing Ski in MCF7 cells markedly inhibited the ActD-induced up-regulation of p21 and PIG3 mRNA, which are well characterized p53 target genes (Fig. 2A), and p21 protein (Fig. 2B). Similar results were also obtained with other cell lines (supplemental Fig. S2A). Ski expression inhibited the induction of p21 when p53 was expressed in H1299 cells (supplemental Fig. S2B). Mdm2, a ubiquitin ligase that promotes p53 degradation, is the best characterized negative regulator of p53 to date (26). Mdm2 overexpression blocked the p53-mediated induction of p21 with a dramatic reduction in the p53 protein levels, whereas Ski overexpression only modestly affected the p53 protein levels compared with Mdm2 (supplemental Fig. S2B). These results suggest that Ski and Mdm2 inhibit p53 through different mechanisms.
FIGURE 2.
Ski suppresses the transactivation ability of p53. A and B, Ski inhibits the induction of p21 and PIG3 transcription (A) and p21 protein expression (B) after ActD treatment. A, cells were treated with 1 nm ActD for the indicated time periods, and RNA was purified from each sample and analyzed by quantitative real-time PCR. Each value was normalized to the HPRT1 expression levels. B, cells were treated with 1 nm ActD for the indicated time periods, and the cell lysates were analyzed by immunoblotting with the indicated antibodies. C and D, Ski knockdown augments p21 and mdm2 mRNA (C) and p21 protein (D) expression levels after ActD treatment. C, MCF7 cells were transfected with the indicated siRNAs and treated with 10 nm ActD for 4 h. Quantitative real-time PCR analyses of the p21 and mdm2 mRNA levels in MCF7 cells as in A are shown. D, MCF7 cells were transfected with the indicated siRNAs and treated with 10 nm ActD for 4 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies. E, Ski knockdown enhances cell cycle arrest after ActD treatment. MCF7 cells were transfected with the indicated siRNAs and treated with 10 nm ActD for 8 h. The cells were labeled with 10 mm BrdUrd for 1 h and then immunostained with an anti-BrdUrd antibody. The average percentage of BrdUrd-positive cells is shown. F, transactivation activity of p53 is increased upon Ski knockdown. Reporter gene constructs containing p53-responsive elements (pp53-TA-Luc) or the control vector alone (pTA-Luc) were co-transfected into MCF7 cells with the indicated siRNAs. After 24 h, the luciferase activity was measured. The experiment was performed in triplicate, and the data are represented as mean -fold activation ± S.D.
Conversely, compared with the control siRNA, Ski siRNA increased p21 and mdm2 mRNA levels (Fig. 2C) and p21 protein expression (Fig. 2D), which were induced by ActD. To exclude any off-target effects associated with the siRNAs, two additional Ski siRNAs were used, with similar results obtained (supplemental Fig. S2C). Simultaneous knockdown of both p53 and Ski inhibited this ActD-mediated increase in p21 and Mdm2 (Fig. 2, C and D), indicating that these Ski-mediated effects are p53-dependent. When induced by p53, p21 primarily mediates cell cycle arrest at the G1 phase. Because Ski knockdown results in robust p21 expression after ActD treatment (Fig. 2D), we next examined cell cycle progression in control or Ski-knocked down cells after ActD treatment. As expected, knocking down Ski reduced entry into S phase before and after ActD treatment as measured by BrdUrd incorporation (Fig. 2E). Moreover, down-regulating Ski up-regulated the transcriptional activity of p53 compared with the control, as determined by a reporter assay using a p53-responsive luciferase reporter construct pp53-TA-Luc (Fig. 2F). Taken together, these results suggest that Ski suppresses the transactivation ability of p53.
Ski Suppresses the Levels of Acetylated p53 and Reduces the DNA Binding Activity of p53
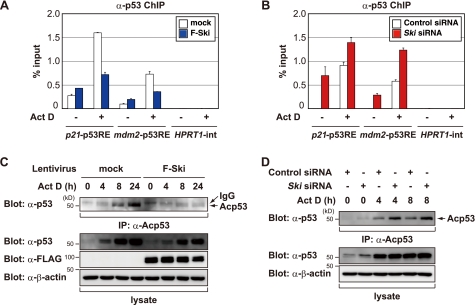
p53 activity is controlled largely by the cellular levels, DNA binding activity, and subcellular localization of p53 as well as by the recruitment of transcriptional coactivators or corepressors (17). Because Ski minimally affected the p53 protein levels (Fig. 2, B and D), we next used a ChIP assay to measure the ability of p53 to bind to the promoters of p53 target genes in response to ActD treatment. ActD treatment increased the ability of p53 to bind to the p21 promoter and the mdm2 promoter but not to a control region (HPRT1 intron), whereas Ski overexpression reduced p53 DNA binding (Fig. 3A). Conversely, siRNA-mediated knockdown of Ski resulted in significantly higher levels of p53 at the p21 and mdm2 promoters in MCF7 cells than in control cells (Fig. 3B). These results indicate that Ski represses the activity of p53, at least in part by decreasing the DNA binding of p53.
FIGURE 3.
Ski suppresses the levels of acetylated p53 and reduces the DNA binding activity of p53. A, Ski expression reduces the amount of p53 on the promoters of p53 target genes. MCF7 cells and MCF7 cells stably expressing Ski were treated with or without 5 nm ActD for 8 h and then analyzed by ChIP to determine the amount of p53 that was bound to the indicated promoters. B, knockdown of endogenous Ski augments p53 occupancy on the promoters of p53 target genes. MCF7 cells were transfected with a control siRNA or Ski siRNA for 48 h and then treated with 5 nm ActD for 4 h. ChIP was performed using an anti-p53 antibody, and quantitative real-time PCR was performed for the indicated promoters. C, Ski suppresses the levels of acetylated p53 after DNA damage. MCF7 cells and MCF7 cells stably expressing Ski were treated with 2 nm ActD for the indicated periods. The levels of acetylated p53 were determined by immunoprecipitation with an anti-acetylated p53 antibody (IP: α-Acp53) and then Western blotting with an anti-p53 antibody. D, knockdown of endogenous Ski increases p53 acetylation after DNA damage. MCF7 cells were transfected with a control siRNA or Ski siRNA. Forty-eight hours after transfection, the cells were treated with 2 nm ActD for the indicated time periods. The levels of acetylated p53 were determined as described in C.
During various types of stress, p53 is phosphorylated and acetylated, suggesting that p53 activation plays a critical role in the stress response (20, 21). Phosphorylation of the p53 protein has been shown to regulate the transactivation properties of p53, whereas acetylation activates the ability of p53 to bind to specific DNA sequences and stabilizes the p53 protein. Given that Ski decreases the DNA binding activity of p53, we next examined the effects of Ski on p53 acetylation in response to ActD treatment. Consistent with previous reports (22, 37), the levels of Lys-382-acetylated p53 were significantly enhanced after ActD treatment. Although Ski overexpression did not measurably alter the total p53 levels, there were decreased levels of Lys-382-acetylated p53 (Fig. 3C). In contrast, Ski knockdown in MCF7 cells increased the acetylation levels after treatment with ActD (Fig. 3D) or etoposide (VP16) (supplemental Fig. S3A). Similar results were obtained with other cell lines (supplemental Fig. S3B). On the other hand, Ski knockdown in MCF7 cells had no significant effect or a somewhat less inhibitory effect on phosphorylation of p53 after treatment with ActD or VP16 (supplemental Fig. S3, C and D). Skp2 is known to suppress p300-mediated acetylation of p53, and siRNA-mediated down-regulation of Skp2 increases the acetylated p53 levels after DNA damage (38). We also observed that the increased levels of acetylated p53 were comparable in both Ski siRNA- and Skp2 siRNA-transfected cells (supplemental Fig. S3B). These results demonstrated that Ski suppressed the levels of acetylated p53 upon DNA damage.
Ski Interacts with SIRT1 and Enhances SIRT1-promoted p53 Deacetylation
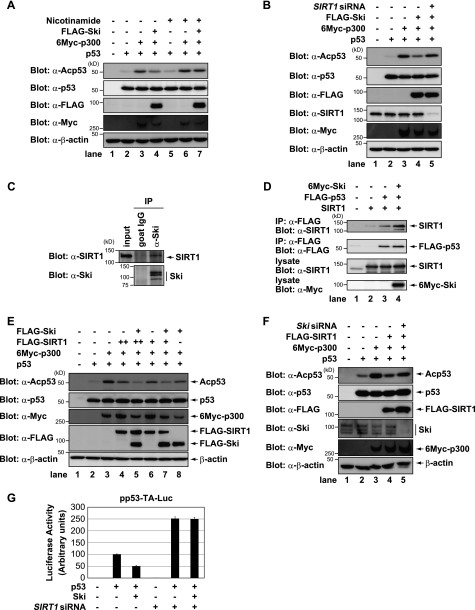
p53 acetylation is reversible in the presence of deacetylases, and class III deacetylase SIRT1 negatively regulates p53 through deacetylation (26). To determine the role of SIRT1 in the function of Ski, we monitored the effects of a SIRT inhibitor on Ski-mediated p53 deacetylation. p300-mediated p53 acetylation was moderately inhibited in the presence of Ski (Fig. 4A). Interestingly, nicotinamide, a specific SIRT inhibitor, reversed the effects of Ski. We also tested the effects of trichostatin A, which inhibits both class I and II deacetylases, on Ski-mediated p53 deacetylation (supplemental Fig. S4A). Remarkably, trichostatin A enhanced p300-mediated p53 acetylation, but Ski-mediated deacetylation of p53 could not be blocked by trichostatin A. In addition, SIRT1 knockdown attenuated Ski-mediated deacetylation of p53 (Fig. 4B). Although Ski and SIRT1 have not been shown to interact functionally, these results indicate that SIRT1 is clearly involved in Ski-mediated deacetylation of p53. Therefore, we examined whether Ski interacts with SIRT1. H1299 cells were transiently transfected with FLAG-Ski and Myc-SIRT1, and co-immunoprecipitation analyses showed that SIRT1 co-precipitated with Ski (supplemental Fig. S4B). Furthermore, an endogenous Ski-SIRT1 complex was detected in MCF7 cells (Fig. 4C). Next, we determined whether Ski promotes the formation of the p53 and SIRT1 complex. To test this possibility, FLAG-p53 and SIRT1 were transfected into H1299 cells together with or without 6Myc-Ski. Although SIRT1 interacted weakly with p53, this interaction was dramatically enhanced by co-expressing Ski (Fig. 4D). This result indicated that Ski recruits SIRT1 into a complex that contains p53. To further investigate this finding, the ability of SIRT1 to associate with the p53-responsive element in the mdm2 promoter was examined by ChIP (supplemental Fig. S4C). The recruitment of SIRT1 to the mdm2 promoter was enhanced upon ActD treatment in control cells, whereas SIRT1 recruitment to the mdm2 promoter was minimally affected in Ski knockdown cells. These results suggest that Ski stabilizes p53-SIRT1 complexes on the promoters of p53-responsive genes.
FIGURE 4.
Ski interacts with SIRT1 and enhances SIRT1-promoted p53 deacetylation. A, Ski-mediated p53 deacetylation is blocked by treatment with nicotinamide. H1299 cells were transfected with the indicated constructs and treated with 10 mm nicotinamide for 12 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies. B, SIRT1 knockdown abrogates Ski-mediated p53 deacetylation. H1299 cells were transfected with the indicated constructs and siRNAs. The cell lysates were analyzed by immunoblotting with the indicated antibodies. C, endogenous Ski and SIRT1 interact in MCF7 cells. The cell lysates were immunoprecipitated (IP) with normal goat IgG or an anti-Ski antibody and then blotted for SIRT1 or Ski. D, Ski promotes p53-SIRT1 binding. H1299 cells were transfected with the indicated constructs, and FLAG-p53 was immunoprecipitated with anti-FLAG antibodies. The co-precipitated SIRT1 was detected by immunoblotting with an anti-SIRT1 antibody. E, Ski and SIRT1 cooperate to inhibit p53 acetylation. H1299 cells were transfected with the indicated constructs, and the levels of acetylated p53 were determined by immunoblotting with an anti-acetylated p53 antibody. F, Ski knockdown abrogates SIRT1-mediated p53 deacetylation. H1299 cells were transfected with the indicated constructs and siRNAs. The cell lysates were analyzed by immunoblotting with the indicated antibodies. G, SIRT1 knockdown abolishes Ski-mediated repression of p53 transcriptional activity. H1299 cells were transfected with pp53-TA-Luc in combination with the indicated constructs and siRNAs. After 24 h, luciferase activity was measured. The experiment was performed in triplicate, and the data are represented as mean -fold activation ± S.D.
p300/CBP participates in p53-mediated transcription and acetylates Lys-373 and Lys-382 (20, 21). Furthermore, p53 acetylation is important to efficiently recruit the p300/CBP complexes to promoter regions in vivo and to activate p53-targeted gene expression (39). As shown in supplemental Fig. S4D, the ability of CBP to bind to the p53-responsive elements in the p21 and mdm2 promoters was enhanced upon DNA damage. Upon Ski knockdown, CBP recruitment to the p21 and mdm2 promoters increased in the absence or presence of DNA damage. Moreover, RNAi-mediated knockdown of both Ski and p53 reduced the extent that CBP associated with the p53-responsive elements in the p21 promoter, suggesting that Ski blocks the recruitment of CBP and p53 to p53-responsive promoters (supplemental Fig. S4E). These results suggest that Ski changes the function of p53 from a coactivator-binding protein to a corepressor-binding protein, which inhibits transcriptional activation of p53.
We next investigated whether Ski and SIRT1 cooperate to inhibit the acetylation of p53. As shown in Fig. 4E, p300 was co-transfected to stimulate p53 acetylation (lane 3). SIRT1 expression modestly deacetylated p53 (Fig. 4E, lanes 4 and 6), but co-expression of both SIRT1 and Ski dramatically reduced the levels of acetylated p53 (lanes 5 and 7). Moreover, Ski knockdown attenuated the SIRT1-mediated deacetylation of p53 (Fig. 4F). Finally, we examined whether SIRT1 was required for Ski-mediated transcriptional inactivation of p53. As shown in Fig. 4G, Ski repressed p53-mediated transcription measured with a p53-responsive luciferase reporter. On the other hand, the silencing of SIRT1 enhanced the transcriptional activity of p53, which supports a role for SIRT1 as a negative regulator of p53. Importantly, when endogenous SIRT1 was suppressed by the siRNA treatment, the negative effect of Ski on p53 transcriptional activity was diminished, indicating that SIRT1 is necessary for the suppression of p53 transcriptional activity by Ski. Taken together, these results suggest that Ski and SIRT1 cooperate to inhibit p53 acetylation, leading to the inactivation of p53.
Ski Overexpression Confers Resistance to Chemotherapy Agents
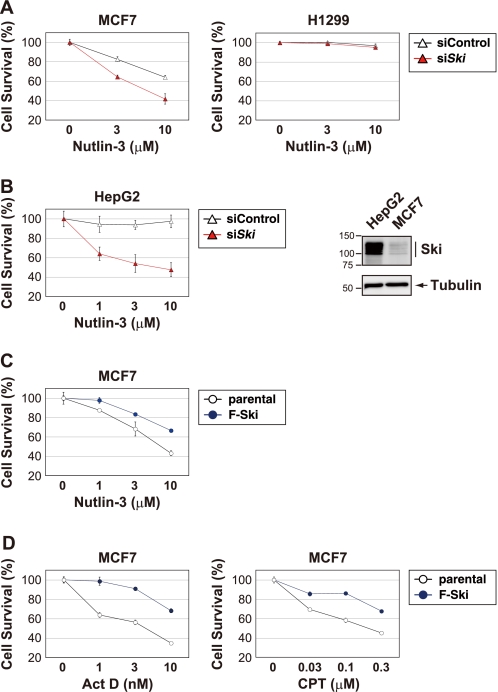
Inactivating p53 inhibits the ability of p53 to induce apoptosis, which promotes tumor development and drug resistance. Mdm2 is overexpressed in many human tumors that retain wild-type p53, and therefore, Mdm2 is an attractive target for novel anti-cancer agents. Nutlin-3 has been shown to bind Mdm2, block the Mdm2-p53 interaction, and activate wild-type p53 without inducing genotoxic stress (40). The finding that Ski negatively regulates p53 prompted us to investigate whether down-regulating Ski in tumor cells could increase their sensitivity to Nutlin-3. As shown in Fig. 5A and supplemental Fig. S5A, siRNA-mediated knockdown of Ski increased the sensitivity of MCF7 cells to Nutlin-3 but had little effect on p53-null H1299 cells. We also found that HepG2 cells had high levels of endogenous Ski and were less sensitive to Nutlin-3 (Fig. 5B and Ref. 41). However, Ski siRNA-treated HepG2 cells had decreased survival after Nutlin-3 treatment (Fig. 5B and supplemental Fig. S5A). Moreover, overexpressing Ski attenuated the cytotoxic effects of Nutlin-3 on MCF7 cells (Fig. 5C). These results suggest that Ski expression levels in tumor cells determine their sensitivity to Nutlin-3.
FIGURE 5.
Ski overexpression confers resistance to chemotherapy agents. A, MCF7 cells and H1299 cells were transfected with a control siRNA or Ski siRNA. Forty-eight hours after transfection, the cells were treated with Nutlin-3 for 48 h and then examined for cell viability using the CellTiter-Glo assay. B, left panel, 48 h after Ski or control siRNA transfection, HepG2 cells were treated with Nutlin-3 for 48 h and analyzed for cell viability as described in A. Right panel, immunoblot analysis of Ski levels in HepG2 cells and MCF7 cells. β-Tubulin expression was examined as an internal control. C, MCF7 cells and MCF7 cells stably expressing Ski were treated with Nutlin-3 for 48 h and then analyzed for cell viability as described in A. D, MCF7 cells and MCF7 cells stably expressing Ski were treated with ActD or camptothecin (CPT) for 48 h and then analyzed for cell viability as described in A.
p53 also plays a pivotal role in maintaining the cellular response to DNA damage-induced genotoxic stress. To investigate whether Ski overexpression confers resistance to other anti-cancer drugs, we treated MCF7 cells and MCF7 cells that were stably expressing Ski with various chemotherapy agents, including ActD, camptothecin, VP16, and 5FU. As shown in Fig. 5D and supplemental Fig. S5B, compared with the parental cells, overexpressing Ski enhanced cellular survival after treatment of cells with these chemotherapy agents. Similar results were obtained with HCT116 cells and HepG2 cells (data not shown). These results suggest that Ski represses the function of p53 and mediates cell survival in response to genotoxic agents and Nutlin-3. In addition, these findings indicate that targeting Ski to sensitize tumor cells is an attractive strategy for cancer therapeutics.
DISCUSSION
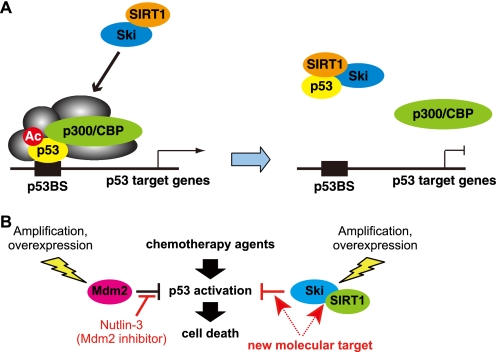
In this article, we have shown that Ski, which is often overexpressed in many types of cancers, performs the novel role of suppressing the function of p53 (Fig. 6A). Ectopically expressing Ski suppresses p53 transactivation activity, whereas knocking down endogenous Ski augments the activity of p53. Although Ski does not have deacetylase activity, Ski binds to the histone deacetylase SIRT1 and stabilizes the p53-SIRT1 interaction to promote p53 deacetylation, which subsequently decreases the DNA binding activity of p53. Furthermore, overexpressing Ski desensitizes cells to genotoxic drugs and Nutlin-3, a small-molecule antagonist of Mdm2 that stabilizes p53 and activates the p53 pathway, whereas knocking down Ski increases the sensitivity of cells to these agents. Our findings indicate that Ski negatively regulates p53 and confers a survival and growth advantage on cells that are treated with DNA-damaging agents (Fig. 6B).
FIGURE 6.
Ski facilitates repression of p53 activity by recruiting SIRT1 and mediates cell survival in response to chemotherapy agents. A, schematic representation of a mechanistic model for Ski-mediated repression of p53. In response to genotoxic stress, activated p53 binds to the target promoters and recruits various transcriptional coactivators to initiate transcription. However, Ski overexpression, which is often observed in many human cancers, renders p53 transcriptionally inactive. Ski interacts with SIRT1 and cooperates to repress p53 activity through p53 deacetylation. B, chemotherapeutic targeting of the p53-Ski-SIRT1 axis. Small molecules that inhibit the p53-Ski interaction in tumor cells that overexpress Ski proteins may increase the cellular sensitivity to chemotherapeutic agents, inducing a stronger p53 response and promoting the apoptosis of tumor cells.
Some of the oncogenic functions of Ski have been attributed to its ability to relieve TGF-β-mediated cytostatic effects (2). Ski interacts with Smads and prevents Smad complexes from activating the transcription of TGF-β target genes. High levels of Ski may promote cell proliferation by inhibiting Smad proteins. Indeed, many cancer cell lines express high levels of Ski and are refractory to TGF-β-induced growth arrest. Here, we show that a major tumor suppressor protein, p53, is also a target of Ski. Recent evidence indicates that Ski and the closely related SnoN protein may inhibit p53-induced p21 expression (12, 42, 43). Zhu et al. (42) have shown that p21 expression is elevated in a TGF-β signal-independent manner in SnoN knockdown A549 cells that express wild-type p53. We also observed that knocking down Ski or SnoN increased p21 expression in several cell lines and that knocking down both p53 and Ski or SnoN canceled these effects (Fig. 2, C and D, and data not shown). These findings suggest that Ski negatively regulates two major pathways, the TGF-β-signaling and p53 pathway, that control cellular proliferation.
SIRT1 is an NAD-dependent deacetylase that regulates cell survival, energy homeostasis, and life span extension (26). Several SIRT1 substrates, including p53, have been well characterized, but relatively little is known about the factors that regulate SIRT1 activity. Active regulator of SIRT1 (AROS) activates the deacetylase activity of SIRT1, thereby increasing SIRT1-mediated p53 deacetylation (44). DBC1 acts as a native inhibitor of SIRT1 by directly binding to the catalytic domain of the SIRT1 protein (45, 46). Unexpectedly, we found that Ski interacted with SIRT1 and that Ski potentiated SIRT1-mediated deacetylation of p53 by facilitating their association. On the basis of these results, we propose that Ski is a key component of the complex that contains SIRT1 and p53 and that Ski may modulate the substrate specificity of SIRT1. In future studies, it will be interesting to examine whether Ski also regulates other SIRT1 substrates, including Smad7, Foxo, and PGC-1a, in cooperation with SIRT1.
Although Ski and SnoN are considered oncogenes, emerging evidence suggests that both proteins also act as tumor suppressors in a context-dependent manner, similar to the bidirectional functions of TGF-β in tumorigenesis. For example, Ski and SnoN heterozygous mice have unexplained paradoxes in their functions. Heterozygous Ski+/− and SnoN+/− mice are more susceptible to carcinogen-induced tumors than wild-type mice (13, 47). Very recently, it was reported that SnoN can act as a tumor suppressor by inducing cellular senescence (48). In addition, it has been shown that knocking down Ski or SnoN suppresses tumor growth but enhances tumor metastasis in vivo (14, 42, 49). These anti-metastatic activities of Ski and SnoN may be related to their ability to repress TGF-β signaling, which promotes tumor invasiveness and metastasis (4). Alternatively, Ski and SnoN may exert different effects on different aspects or stages of tumorigenesis.
Finally, Ski may prove to be an especially valuable therapeutic target. We demonstrated that forced expression of Ski desensitized cells to genotoxic agents and the Mdm2 inhibitor Nutlin-3 and that RNAi-mediated Ski down-regulation increased the cellular sensitivity to these agents. Ski is expressed at low levels in most tissues but is overexpressed in many tumors. Therefore, Ski overexpression likely attenuates p53 activation and triggers uncontrolled cell proliferation, and the combination of these events promotes tumorigenesis. Considering our findings, we may be able to design effective chemotherapeutic interventions by inhibiting the interaction between p53 and Ski and repairing the p53 response in tumor cells with minimal side effects (Fig. 6B). In particular, it is known that activating p53 with Mdm2 inhibitors like Nutlin-3 is not toxic to normal cells (50). Therefore, targeting Ski may be a useful chemotherapeutic intervention in combination with Nutlin-3-like compounds.
In conclusion, we have identified a new molecular mechanism in which Ski promotes oncogenesis by inactivating the tumor suppressor p53. Thus, our study provides a rationale for using Ski as a diagnostic marker and for targeting Ski as a potential tumor therapy.
Supplementary Material
Acknowledgments
We thank A. Hikita and A. Hanyu for discussions and suggestions and N. Kaneniwa and Y. Yuuki for technical assistance.
This work was supported by Grant-in-aid for Scientific Research on Innovative Areas 22113004, “Fluorescence Live Imaging,” and Grants-in-aid for Scientific Research on Priority Areas 21025036 and 20058038 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and by Grant-in-aid for Scientific Research (B) 20390407 and Grant-in-aid for Young Scientists (B) 22700890 from the Japan Society for the Promotion of Science (JSPS).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- CBP
- CREB-binding protein
- CREB
- cAMP-responsive element-binding protein
- ActD
- actinomycin D
- BrdUrd
- bromodeoxyuridine.
REFERENCES
- 1. Li Y., Turck C. M., Teumer J. K., Stavnezer E. (1986) J. Virol. 57, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deheuninck J., Luo K. (2009) Cell Res. 19, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyazono K., Suzuki H., Imamura T. (2003) Cancer Sci. 94, 230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akiyoshi S., Inoue H., Hanai J., Kusanagi K., Nemoto N., Miyazono K., Kawabata M. (1999) J. Biol. Chem. 274, 35269–35277 [DOI] [PubMed] [Google Scholar]
- 6. Medrano E. E. (2003) Oncogene 22, 3123–3129 [DOI] [PubMed] [Google Scholar]
- 7. Buess M., Terracciano L., Reuter J., Ballabeni P., Boulay J. L., Laffer U., Metzger U., Herrmann R., Rochlitz C. (2004) Neoplasia 6, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heider T. R., Lyman S., Schoonhoven R., Behrns K. E. (2007) Ann. Surg. 246, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukuchi M., Nakajima M., Fukai Y., Miyazaki T., Masuda N., Sohda M., Manda R., Tsukada K., Kato H., Kuwano H. (2004) Int. J. Cancer 108, 818–824 [DOI] [PubMed] [Google Scholar]
- 10. Takahata M., Inoue Y., Tsuda H., Imoto I., Koinuma D., Hayashi M., Ichikura T., Yamori T., Nagasaki K., Yoshida M., Matsuoka M., Morishita K., Yuki K., Hanyu A., Miyazawa K., Inazawa J., Miyazono K., Imamura T. (2009) J. Biol. Chem. 284, 3334–3344 [DOI] [PubMed] [Google Scholar]
- 11. Ritter M., Kattmann D., Teichler S., Hartmann O., Samuelsson M. K., Burchert A., Bach J. P., Kim T. D., Berwanger B., Thiede C., Jäger R., Ehninger G., Schäfer H., Ueki N., Hayman M. J., Eilers M., Neubauer A. (2006) Leukemia 20, 437–443 [DOI] [PubMed] [Google Scholar]
- 12. Chen D., Lin Q., Box N., Roop D., Ishii S., Matsuzaki K., Fan T., Hornyal T. J., Reed J. A., Stavnezer E., Timchenko N. A., Medrano E. E. (2009) Pigment Cell Melanoma Res. 6, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shinagawa T., Nomura T., Colmenares C., Ohira M., Nakagawara A., Ishii S. (2001) Oncogene 20, 8100–8108 [DOI] [PubMed] [Google Scholar]
- 14. Le Scolan E., Zhu Q., Wang L., Bandyopadhyay A., Javelaud D., Mauviel A., Sun L., Luo K. (2008) Cancer Res. 68, 3277–3285 [DOI] [PubMed] [Google Scholar]
- 15. Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 16. Oren M. (2003) Cell Death Differ. 10, 431–442 [DOI] [PubMed] [Google Scholar]
- 17. Prives C., Hall P. A. (1999) J. Pathol. 187, 112–126 [DOI] [PubMed] [Google Scholar]
- 18. Vousden K. H., Lane D. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 275–283 [DOI] [PubMed] [Google Scholar]
- 19. Hainaut P., Hollstein M. (2000) Adv. Cancer Res. 77, 81–137 [DOI] [PubMed] [Google Scholar]
- 20. Brooks C. L., Gu W. (2003) Curr. Opin. Cell Biol. 15, 164–171 [DOI] [PubMed] [Google Scholar]
- 21. Olsson A., Manzl C., Strasser A., Villunger A. (2007) Cell Death Differ. 14, 1561–1575 [DOI] [PubMed] [Google Scholar]
- 22. Tang Y., Zhao W., Chen Y., Zhao Y., Gu W. (2008) Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ford J., Jiang M., Milner J. (2005) Cancer Res. 65, 10457–10463 [DOI] [PubMed] [Google Scholar]
- 26. Brooks C. L., Gu W. (2009) Nat. Rev. Cancer 9, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagano Y., Mavrakis K. J., Lee K. L., Fujii T., Koinuma D., Sase H., Yuki K., Isogaya K., Saitoh M., Imamura T., Episkopou V., Miyazono K., Miyazawa K. (2007) J. Biol. Chem. 282, 20492–20501 [DOI] [PubMed] [Google Scholar]
- 28. Inoue Y., Kitagawa M., Taya Y. (2007) EMBO J. 26, 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fukunaga E., Inoue Y., Komiya S., Horiguchi K., Goto K., Saitoh M., Miyazawa K., Koinuma D., Hanyu A., Imamura T. (2008) J. Biol. Chem. 283, 35660–35667 [DOI] [PubMed] [Google Scholar]
- 30. Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gilkes D. M., Chen L., Chen J. (2006) EMBO J. 25, 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu J. W., Krawitz A. R., Chai J., Li W., Zhang F., Luo K., Shi Y. (2002) Cell 111, 357–367 [DOI] [PubMed] [Google Scholar]
- 34. Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. (2003) Cell 113, 301–314 [DOI] [PubMed] [Google Scholar]
- 35. Stavnezer E., Brodeur D., Brennan L. A. (1989) Mol. Cell. Biol. 9, 4038–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng G., Teumer J., Colmenares C., Richmond C., Stavnezer E. (1997) Oncogene 15, 459–471 [DOI] [PubMed] [Google Scholar]
- 37. Ito A., Lai C. H., Zhao X., Saito S., Hamilton M. H., Appella E., Yao T. P. (2001) EMBO J. 20, 1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kitagawa M., Lee S. H., McCormick F. (2008) Mol. Cell 29, 217–231 [DOI] [PubMed] [Google Scholar]
- 39. Barlev N. A., Liu L., Chehab N. H., Mansfield K., Harris K. G., Halazonetis T. D., Berger S. L. (2001) Mol. Cell 8, 1243–1254 [DOI] [PubMed] [Google Scholar]
- 40. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 41. Nagano Y., Koinuma D., Miyazawa K., Miyazono K. (2010) J. Biochem. 147, 545–554 [DOI] [PubMed] [Google Scholar]
- 42. Zhu Q., Krakowski A. R., Dunham E. E., Wang L., Bandyopadhyay A., Berdeaux R., Martin G. S., Sun L., Luo K. (2007) Mol. Cell. Biol. 27, 324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nanjundan M., Cheng K. W., Zhang F., Lahad J., Kuo W. L., Schmandt R., Smith-McCune K., Fishman D., Gray J. W., Mills G. B. (2008) Mol. Oncol. 2, 164–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim E. J., Kho J. H., Kang M. R., Um S. J. (2007) Mol. Cell 28, 277–290 [DOI] [PubMed] [Google Scholar]
- 45. Kim J. E., Chen J., Lou Z. (2008) Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 46. Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008) Nature 451, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shinagawa T., Dong H. D., Xu M., Maekawa T., Ishii S. (2000) EMBO J. 19, 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pan D., Zhu Q., Luo K. (2009) EMBO J. 28, 3500–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang P., Chen Z., Meng Z. Q., Fan J., Luo J. M., Liang W., Lin J. H., Zhou Z. H., Chen H., Wang K., Shen Y. H., Xu Z. D., Liu L. M. (2009) Carcinogenesis 30, 1497–1506 [DOI] [PubMed] [Google Scholar]
- 50. Shangary S., Wang S. (2009) Annu. Rev. Pharmacol. Toxicol. 49, 223–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.