Abstract
The human Eg5 (HsEg5) protein is unique in its sensitivity to allosteric agents even among phylogenetic kin. For example, S-trityl-l-cysteine (STC) and monastrol are HsEg5 inhibitors that bind to a surface pocket created by the L5 loop, but neither compound inhibits the Drosophila Kinesin-5 homologue (Klp61F). Herein we ask whether or not drug sensitivity can be designed into Klp61F. Two chimeric Klp61F motor domains were engineered, bacterially expressed, and purified to test this idea. We report that effector binding can elicit a robust allosteric response comparable with HsEg5 in both motor domain chimeras. Furthermore, isothermal titration calorimetry confirms that the Klp61F chimeras have de novo binding affinities for both STC and monastrol. These data show that the mechanism of intramolecular communication between the three ligand binding sites is conserved in the Kinesin-5 family, and reconstitution of a drug binding cassette within the L5 pocket is sufficient to restore allosteric inhibition. However, the two compounds were not equivalent in their allosteric inhibition. This surprising disparity in the response between the chimeras to monastrol and STC suggests that there is more than one allosteric communication network for these effectors.
Keywords: Allosteric Regulation, ATPases, Calorimetry, Drug Resistance, Enzyme Catalysis, Enzyme Inactivation, Enzyme Inhibitors, Enzyme Kinetics, Enzyme Mutation, Kinesin
Introduction
The Kinesin-5 family of motor proteins plays a conserved role in the morphogenesis of the mitotic spindle. In particular, human Eg5 kinesin (HsEg5)2 forms a homotetramer that is capable of cross-linking adjacent microtubule bundles during spindle formation and is essential for mitotic progression. High-throughput screens for small chemical inhibitors of mitosis have often yielded compounds that target this kinesin in particular. At the time of their discovery, these compounds represented the only known antimitotic compounds that did not directly affect cellular microtubule assembly or function and constituted an entirely new class of potential anticancer compounds. Important for our purposes, they also serve as tools to explore fundamental questions about motor protein function.
To date, small chemical inhibitors have been discovered that bind to at least three different sites within the motor domain of HsEg5 (1–4). The most highly explored allosteric site is a single pocket whose absolute location was defined by x-ray crystallography (for examples, see Ref. 5–11). This site is formed by the α2 and α3 helices and capped by the L5 loop. This L5 pocket is on the surface of the motor domain and is ∼12 Å and 22 Å from the nucleotide binding site and microtubule (MT)-binding site, respectively.
Interactions of the L5 loop are at the crux of long distance, allosteric communication with the active site and the MT-binding site. The biochemical role of the L5 loop has been confirmed by kinetic measurements and mutagenesis efforts. Mutations throughout the loop result in varying degrees of inhibition of basal rates and MT-stimulated ATPase rates (3, 7, 12–15). Not limited in its effects upon the orthosteric site, a dihydropyrimidine derivative, monastrol, has been shown to affect how the motor domain interacts with microtubules. At the cellular level, monastrol-induced inhibition results in catastrophic disruption of the mitotic spindle (16, 17). More detailed in situ experiments, however, reveal frictionless motion along microtubules by monastrol-HsEg5 in gliding assays/tug-of-war experiments (18).
There are still outstanding unanswered questions regarding this allosteric site in Kinesin-5 proteins. First, it remains unclear how the L5 loop transmits the inhibitory signal or conformational change that affects the orthosteric and MT-binding sites. Formally, three events in series are required for allosteric inhibition in this model system; these include, binding of the small compound, allosteric changes in the L5 loop, and propagation of allosteric signals to the distal sites. In crystallographic studies of HsEg5-ADP complexed with monastrol (7, 10) and other L5 pocket inhibitors (for example, Ref. 19), the wild type Kinesin-5 motor domain displays a similar conformer irrespective of the chemical nature of the allosteric drug. Regardless, examination of co-crystal structures of drugs complexed with HsEg5 do not reveal any pronounced perturbation of the active site residues, such as those of the P-loop, or Mg2+ cofactor that might explain the aborted catalytic cycle.
Second, no biochemical or biological action has been ascribed or accepted for the L5 loop in the motor domain. Current models suggest that these inhibitors somehow drive the loop to trap HsEg5 predominantly in an inactive ADP-bound state (20, 21) with atomic contacts that vary by inhibitor and prevent the displacement of ADP with ATP for a subsequent round of hydrolysis. Multivariate analysis of HsEg5 solution structures defined that the L5 loop may act in concert with the core β-sheet to serve as a transducer between the nucleotide site and the force generator (13). A similar definition was proposed for myosin motor proteins, for which this analogous protein sector (22) is responsible for releasing ADP from myosin (23).
Third, it is not understood why there is such exquisite drug sensitivity for only some of the Kinesin-5 proteins and not others. Although both human and Xenopus Eg5 kinesin are sensitive to L5 pocket inhibitors, other homologs, such as Klp61F in Drosophila, are not sensitive to drug inhibition. Furthermore, drug-mediated inhibition is abolished in HsEg5 chimeras formed by replacing the endogenous L5 loop with roughly cognate segments from either human kinesin heavy chain (7) or Neurospora kinesin heavy chain (1). This loss-of-function strategy does not provide insight as to whether sequence alteration in L5 alone is responsible for loss of small-molecule binding or results in sequence incompatibility within an allosteric network between the surface loop and the motor domain core.
To distinguish between these explanations, we chose a gain-of-function mutational approach. The motor domain of Drosophila Klp61F is 59% identical to HsEg5, contains a similar size L5 loop, and is not measurably inhibited by either monastrol or STC. Here we report that, consistent with the lack of inhibition, Klp61F does not bind the allosteric effectors with any measurable affinity. We surmise that, if the pathway for allosteric inhibition is conserved, then reconstitution of effector binding to the L5 pocket of modified Klp61F should confer druggability and result in long-distance allosteric inhibition. On the other hand, if drug binding to the L5 pocket of Klp61F can be reconstituted without accompanying allosteric inhibition, then it is probable that the mechanism of allosteric inhibition is a unique attribute to an ensemble of residues or contacts found only within HsEg5.
MATERIALS AND METHODS
Motor Protein Expression and Purification
The motor domain of HsEg5 as well as the Klp61F, Klp61F-L5, and Klp61F-L5-α3 motor domain constructs were expressed in BL21 (DE3) cell lines (Invitrogen) and purified by cation exchange chromatography as described in Wojcik et al. (24). The motor protein samples were estimated to be >90% pure based on SDS-PAGE analysis.
The initial plasmid construct of the Klp61F motor domain with C-terminal His tag was a gift from Dr. Richard A. Walker (Virginia Tech). The C-terminal His6 tag was removed from the wild type Klp61F motor domain expression construct by inserting a TAG stop codon before the tag sequence. We synthesized the Klp61F-L5 variant in which the native L5 loop (Met113–Ile133) was replaced with the homologous HsEg5 L5 loop (Met113–Ile133). We also synthesized the Klp61F-L5-α3 variant further containing the K214A substitution.
ATPase Activity Assays
Basal and MT-stimulated ATPase activities of the motor proteins were measured using a coupled pyruvate kinase/lactate dehydrogenase assay (2, 25) in a 96-well plate using a SpectraMax 2E spectrophotometer. Basal ATPase reactions contained 2.5 μm motor, whereas MT-stimulated ATPase reaction mixtures contained 50 nm motor and 4 μm tubulin stabilized with 20 μm paclitaxel (Calbiochem). To establish a threshold for the sensitivity of both the basal and MT-stimulated ATPase assay, 2.5 μm BSA was substituted for enzyme in mock time-course reactions. These control reactions established a noise base line and never recorded a value in excess of 0.009 ADP/protein/s (average = 0.004 ± 0.002 ADP/protein/s (n = 6)). Inhibitor concentrations were 0–200 μm for the basal and MT-stimulated reactions. The potency of the inhibitors was calculated in Igor Pro (Wavemetrics Inc.). Different curve-fitting algorithms were tested, and the quality of the fits was assessed by χ2 analysis. The best curve fits typically resulted from application of the Hill equation algorithm. However, in the case of STC binding to HsEg5, a classic “tight binding” situation (12), the best fit arose from a curve generated by the Morrison equation (26), from which we calculated kinetic parameters for this data.
Isothermal Titration Calorimetry
The binding affinities of STC (Sigma #164739) and monastrol (Sigma #M8515) for the motor proteins were determined by isothermal titration calorimetry (ITC) using a VP-ITC instrument (MicroCal). Protein samples were dialyzed against sample buffer (50 mm Hepes (pH 7), 150 mm NaCl, 2 mm MgCl2, 2 mm β-mercaptoethanol, and 5% glycerol) before the experiment. STC and monastrol were prepared as stock solutions of 50 mm in DMSO and thereafter diluted to desired concentrations in the sample buffer. The final percentage of DMSO was kept below 3% for all the experiments. The concentration of DMSO in the syringe and the cell was kept constant by adding an equal percentage to the protein solution before injection of the drug. All solutions were degassed before loading to reduce the noise. For a typical experiment, 10 μl of STC (300 μm) or monastrol (1 mm) was injected into 1.45 ml of the protein solution (25–50 μm) at 20 °C. A total of 29 injections were carried out at 3-min intervals for each experiment. The heat generated by the dilution of STC or monastrol into 1.45 ml of the buffer was subtracted for base-line correction. The binding affinities of the drugs for the motor proteins were determined by plotting the best-fit curve for the experimental binding isotherms using Origin 5.0 software. For all our measurements, the single-site binding model resulted in the best fit compared with alternatives, including the two-binding-site models.
RESULTS
Engineering an Effector Binding Pocket in Klp61F
The only Kinesin-5 family member that has been structurally analyzed by x-ray crystallography is HsEg5. Therefore, we used a homology-model approach to predict the homologous L5 pocket residues for Klp61F and design a chimeric binding pocket. The extensive identity of pocket residues permitted good alignment of Klp61F L5 pocket residues to those of HsEg5 (Fig. 1A). This alignment formed the template for the construction of our Klp61F chimeras.
FIGURE 1.
Alignment and spatial location of residues composing the L5 binding pocket of HsEg5 to cognate sequences from Klp61F. A, sequence alignment of the L5 region of HsEg5 to Klp61F is shown. Identical residues are shaded in gray. Residues shaded in yellow mark the L5 loop. Open triangles mark residues within HsEg5 in close contact to STC, and filled black triangles mark residues within HsEg5 in close contact to monastrol. B, shown is an overview of the location of the STC-binding site within HsEg5. STC (magenta) is surrounded by a schematic view of HsEg5 with the L5 loop in yellow and helices α2 and α3 in green. Contact side chains for STC are shown in stick view, with Ala218 of helix α3 in orange. This PyMol-generated image is derived from PDB 3KEN (13).
To design the L5 pocket Klp61F chimeras, we examined the residues in the L5 pocket of HsEg5 co-crystallized with either STC or monastrol. Our criteria were that the residues must reside within 3–4 Å of either compound and must be predicted (27) to participate in chemical interactions.
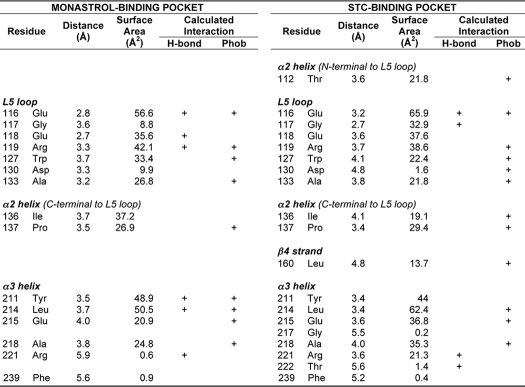
STC binding to HsEg5 appears to be driven primarily by hydrophobic interactions, with 12 of 19 contact residues oriented for hydrophobic interactions to aromatic and nonpolar moieties of STC and 4 remaining contact residues in position for predicted H-bond formation to the cysteinyl moiety (Fig. 1 and Table 1). In contrast, the monastrol binding pocket is composed of a subset of 15 residues of the 19 that form STC contacts. In this case, nine residues are predicted to contribute to hydrophobic interactions with monastrol, and six are in favorable orientation for H-bonding to the polar components of this effector. Perhaps due to its mobility, all but two residues of HsEg5 (Tyr211 and Ala218) that form the closest contacts or appear to contribute most to the effector binding environment in the co-crystal structures are resident within the L5 loop. Seven residues within the loop (Fig. 1A and Table 1) each share key contacts with both monastrol and STC.
TABLE 1.
Summary of the monastrol and STC binding pocket residues in HsEg5; distance from effector and bond predictions
Data were derived with LPC software (27). Two crystal structure datasets were utilized for this analysis. PDB code IX88 (58) was used to assess the monastrol binding pocket, and PDB code 3KEN (13) was used to assess the STC binding pocket interactions.
Based on our analysis, we first opted to swap the entire Klp61F L5 loop with that of HsEg5 (Met113–Ile133) to create chimera Klp61F-L5 (Fig. 1A, box). With the exception of Ala218 in HsEg5, contact residues outside of loop L5 are conserved between HsEg5 and Klp61F. Thus, this chimera preserves the closest contacts and a majority of the predicted pocket contacts.
Second, we opted to bring Klp61F-L5 pocket residues into closer agreement with the predicted HsEg5 drug binding contacts by substituting Lys214 with the cognate residue, Ala218, in HsEg5. In HsEg5, Ala218 contributes significantly to the hydrophobic surface area of the pocket and approaches to within nearly 4 Å of STC and 3.8 Å of monastrol (Table 1). Therefore, we generated the substitution K214A in helix α3 of Klp61F-L5, hereafter termed Klp61F-L5-α3 (Fig. 1A, asterisk). This mutation assumes that there is conserved rotational orientation of helix α3 in Klp61F, with the same side chains facing the L5 pocket as HsEg5 (Fig. 1B).
Monastrol and STC Do Not Bind to the Wild Type Klp61F Motor Domain
Under equivalent conditions, we expressed and purified the wild type and two chimeric Klp61F motor domains with a C-terminal breakpoint in an identical location as our HsEg5 construct. Similar to other reported rates (2), our wild type Klp61F motor domain had lower basal and MT-stimulated ATPase rates (Table 2) than achievable with HsEg5 motor domain. We confirmed, as had previously been shown (2, 7), that wild type Klp61F motor domain basal and MT-stimulated ATPase activity is not detectably inhibited by STC or monastrol (Figs. 2A and 3A, respectively).
TABLE 2.
Basal and MT-stimulated ATPase rates for wild type and modified kinesin-5 motor domains
Data are the average values for three replicates obtained from three independent enzyme preparations. S.E. are also reported. Klp61F-WT exhibited a higher basal and MT-stimulated ATPase rate than the chimeras (unpaired t test, p < 0.001), whereas the -fold increase upon MT stimulation was indistinguishable among the Klp61F constructs (15-fold, unpaired t test, p < 0.05).
| Motor domain | Rate of ATP hydrolysis |
|
|---|---|---|
| Basal | MT-stimulated | |
| ADP motor−1s−1 | ||
| HsEg5 | 0.17 ± 0.05 | 7.69 ± 0.17 |
| Klp61F-WT | 0.05 ± 0.01 | 0.66 ± 0.02 |
| Klp61F-L5 | 0.04 ± 0.01 | 0.51 ± 0.02 |
| Klp61F-L5-α3 | 0.04 ± 0.01 | 0.53 ± 0.02 |
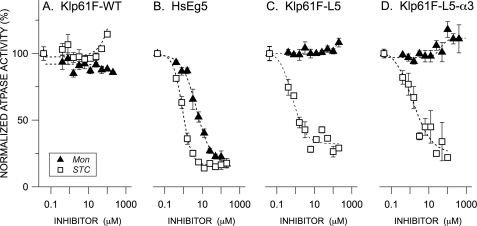
FIGURE 2.
Normalized rates of ATP hydrolysis for wild type HsEg5 and Drosophila melanogaster homolog Klp61F as a function of allosteric effector concentration. Basal ATPase rates (ADP/motor/sec) for wild type Drosophila Klp61F (A), HsEg5 (B), Klp61F-L5 (C), and Klp61F-L5-α3 (D) were measured in the presence of either STC (open squares) or monastrol (filled black triangles). The averages from 2–10 measurements and S.E. are normalized against the parent wild type kinesin motors. The monastrol inhibition curves exhibited by Klp61F-WT and both chimeras are indistinguishable (Wilcoxon signed rank test (α= 0.05)). In contrast, the STC inhibition curves exhibited by Klp61F-L5 and Klp61F-L5-α3 are indistinguishable from one another but are significantly different from that of Klp61F-WT (Wilcoxon signed rank test (α = 0.05).
FIGURE 3.
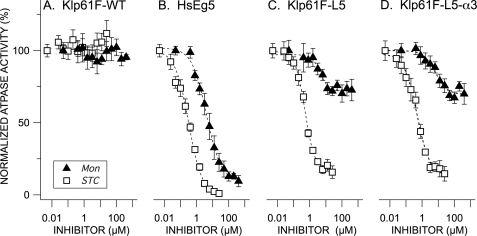
Normalized MT-stimulated rates of ATP hydrolysis for wild type HsEg5 and D. melanogaster homologue Klp61F as a function of allosteric effector concentration. Steady-state, MT-ATPase rates (ADP/motor/s) for wild type Drosophila Klp61F (A), HsEg5 (B), Klp61F-L5 (C), and Klp61F-L5-α3 (D) were measured in the presence of either STC (open squares) or monastrol (filled black triangles). 4 μm taxol-stabilized microtubules were present in each assay. The averages of 3–10 measurements and S.E. are normalized against the parent kinesin motor, which has a rate of 100%. All the inhibition curves exhibited by the Klp61F chimeras are significantly different from the corresponding traces exhibited by Klp61F-WT (Wilcoxon signed rank test (α = 0.05).
To determine whether Klp61F is simply refractory to allosteric inhibition due to limited effector binding activity, we explored whether these effectors exhibit measurable affinity to wild type Klp61F by ITC. We find that there is no detectable binding activity of either effector to wild type Klp61F motor domain over a wide range of molar ratios and concentrations (see Figs 4 and supplemental Fig S1). The lack of any detectable monastrol binding to wild type Klp61F by ITC is consistent with our earlier work that tested the binding activity of radiolabeled monastrol (2).
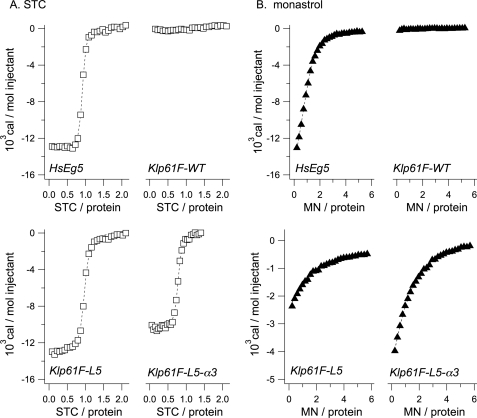
FIGURE 4.
ITC analyses of STC and monastrol binding to Kinesin-5 motor domain in the presence of ATP. Shown is the heat evolved upon STC (A) or monastrol (B) binding to HsEg5, Klp61F-WT, Klp61F-L5, and Klp61F-L5-α3. Data are plotted versus molar ratio of effector/motor. Shown are representative traces from experiments performed in triplicate of three independent enzyme preparations.
As a positive control for effector inhibition and binding, we examined wild type HsEg5 kinetics and equilibrium dissociation constants. In our hands, this motor exhibits basal and MT-stimulated ATPase activity of 0.17 and 7.7 ADP/motor/s, respectively (Table 2), in a range that is typical for this enzyme. As expected (4, 12, 28), monastrol and STC both inhibit HsEg5 motor domain basal ATPase activity (Fig. 2B) with IC50 values of 4.9 and 0.9 μm, respectively, and MT-stimulated ATPase activity (Fig. 3B) with IC50 values of 4.1 and 0.3 μm, respectively (Table 3).
TABLE 3.
Basal and MT-stimulated ATPase assay IC50 values for kinesin-5 motor domains treated with either monastrol or STC
These median inhibitory concentration values were obtained from the data curve fits to the Hill equation as shown in Figs. 2 and 3. We find no significant differences in the response of the chimeras to either monastrol or STC (Wilcoxon signed rank test (α = 0.05)). S.E. are reported. ND, not detectable.
| Motor domain | Monastrol IC50 |
STC IC50 |
||
|---|---|---|---|---|
| Basal | MT-stimulated | Basal | MT-stimulated | |
| μm | μm | |||
| HsEg5 | 4.9 ± 0.6 | 4.1 ± 0.4 | 0.9 ± 0.05 | 0.3 ± 0.02a |
| Klp61F-WT | ND | ND | ND | ND |
| Klp61F-L5 | ND | 4.4 ± 0.9 | 0.7 ± 0.2 | 0.5 ± 0.03 |
| Klp61F-L5-α3 | ND | 6.7 ± 1.9 | 1.3 ± 0.7 | 0.5 ± 0.06 |
a The Morrison equation was used to obtain IC50 values for HsEg5 treated with STC in the presence of saturating MT.
Our ITC measurements detected a change in enthalpy caused by the binding of either effector to the L5 pocket of wild type HsEg5 (Fig. 4, A and B). Based on this data, we calculated the Kd values of monastrol and STC to wild type HsEg5 in the presence of ATP to be 8.1 μm and 81 nm, respectively (Tables 4 and 5). Note that our monastrol binding data and ATPase activity measurements were performed with a racemic mixture of monastrol. Although it is not known whether both enantiomers of monastrol have differing affinity for the wild type HsEg5 allosteric site, it has been shown that both can inhibit the motor, whereas the (S)-enantiomer is a more effective inhibitor than the (R)-enantiomer (4).
TABLE 4.
Isothermal titration calorimetry data of STC binding different kinesin-5 motor domains in the presence of ATP
Data include the calculated number of binding sites (N), enthalpy (ΔH) and entropy (ΔS) values, and Kd. Averaged data are shown and were obtained from 3–5 replicates per kinesin-5 protein. Furthermore, a set of replicates was obtained for three independent enzyme preparations. In all cases except Klp61F-WT, the overall ΔG for the binding reaction was approximately −9 kcal/mol. ND, not detectable.
| Motor domain | N | ΔH° | ΔS° | Kd |
|---|---|---|---|---|
| kcal/mol | cal mol−1K−1 | μm | ||
| HsEg5 | 0.93 ± 0.05 | −14.8 ± 1.3 | −17.9 ± 4.7 | 0.08 ± 0.01 |
| Klp61F-WT | ND | ND | ND | ND |
| Klp61F-L5 | 0.92 ± 0.04 | −12.7 ± 0.6 | −12.2 ± 2.1 | 0.17 ± 0.01 |
| Klp61F-L5-α3 | 0.89 ± 0.11 | −11.3 ± 0 | −7.2 ± 0.1 | 0.15 ± 0.01 |
TABLE 5.
Isothermal titration calorimetry data of monastrol binding different kinesin-5 motor domains in the presence of ATP
Data include the calculated number of binding sites (N), enthalpy and entropy values, and Kd. Averaged data from three replicates, each from three independent enzyme preparations, are shown. The ΔG values for the monastrol binding reaction of HsEg5 and Klp61F-L5 are both approximately −7 kcal/mol, whereas ΔG for monastrol binding to Klp61F-L5-α3 is approximately −5 kcal/mol. ND, not detectable.
| Motor domain | N | ΔH° | ΔS° | Kd |
|---|---|---|---|---|
| kcal/mol | cal mol−1K−1 | μm | ||
| HsEg5 | 0.89 ± 0.02 | −21.4 ± 1.8 | −49.5 ± 6.2 | 8.1 ± 0.5 |
| Klp61F-WT | ND | ND | ND | ND |
| Klp61F-L5 | 0.9a | −12.9 ± 2.3 | −20.3 ± 10.2 | 142.3 ± 11.7 |
| Klp61F-L5-α3 | 0.9a | −13.5 ± 1.9 | −29.7 ± 7.8 | 48.6 ± 1.2 |
a Although the ITC algorithms found the best fit to the single-site model in these cases, the weak binding curve necessitated fixing n = 0.9 to calculate the associated thermodynamic parameters.
Our ITC measurements are on a par with other reports in the literature. Sheth et al. (15) also directly measured the Kd of racemic monastrol with HsEg5-ADP (1.4 μm) and HsEg5-ATPγS (2.0 μm) by isothermal titration calorimetry; although the N-terminal portion of the human Kinesin-5 motor domain was absent in these experiments, the Kd values are in reasonable agreement with our data derived from HsEg5 in the presence of ATP. The Kd values reported by Cochran and co-workers (29) are in closer accord with this work, most likely due to examination of similar protein construct. In contrast, there is no detectable stereospecificity in the binding or inhibition caused by S-trityl-l-cysteine or S-trityl-d-cysteine (30). Overall, for HsEg5, the binding reactions of both STC and monastrol have comparable free energy changes, with ΔG values of approximately −9 and −7 kcal/mol, respectively (Tables 4 and 5).
The Klp61F Chimeric Enzymes Retain ATPase Activity Similar to the Parent Protein
We examined the basal and MT-stimulated ATPase rates achievable by both Klp61F chimera constructs using an NADH-coupled assay and found that the L5 loop substitution caused no decrease in basal ATPase rates of the chimeras, whereas the MT-stimulated ATPase rates decreased ∼20% (Table 2). The additional substitution present in Klp61F-L5-α3 resulted in no further statistically significant change in activity.
These data are consistent with efforts (7, 12) that introduced elements of the Klp61F and Neurospora KHC L5 loops into that of HsEg5. Both of these constructs (7, 12) resulted in a ∼50% reduction in basal ATPase rates. These modifications also significantly ablated drug-mediated inhibition of HsEg5 (7, 12), which is in contrast to our current results below.
The Basal ATPase Rate of Klp61F Chimera Is Inhibited by STC but Not Monastrol
Unlike wild type Klp61F, the basal ATPase rates of both Klp61F chimeras (Klp61F-L5 and Klp61F-L5-α3) were inhibited by STC (Fig. 2, C and D, open boxes). With IC50 values of 0.7 ± 0.2 and 1.3 ± 0.7 μm, respectively, the degree of STC inhibition is similar to that of HsEg5 under the same conditions (Table 3). Of note, the Klp61F-L5-α3 variant showed no significant change in sensitivity to STC over Klp61F-L5, indicating only minor involvement of residue 214 in affecting the inhibition of the chimeras.
On the other hand, monastrol was unable to measurably inhibit the basal ATPase rate of either Klp61F chimera (Fig. 2, C and D, filled black triangles; Table 3) even at the highest inhibitor concentrations. Alteration of the residue 214 side chain did not affect these results. Note that both effector compounds recorded a small increase in ATPase activity at the highest inhibitor concentrations, an effect that could also be observed in control experiments performed in the absence of enzyme and attributable to solubility limits of the drug (data not shown).
STC and Monastrol Inhibit MT-stimulated ATPase Activity of the Drosophila Chimeras
As expected, similar to wild type HsEg5, in the presence of saturating levels of MTs (supplemental Fig. S2), ATPase activities of both Klp61F chimeras were similarly well inhibited by STC (Fig. 3), with an IC50 value of 0.5 μm for both Klp61F-L5 and Klp61F-L5-α3 (Table 3). Again, the presence of the K214A substitution resulted in no change in sensitivity to STC inhibition.
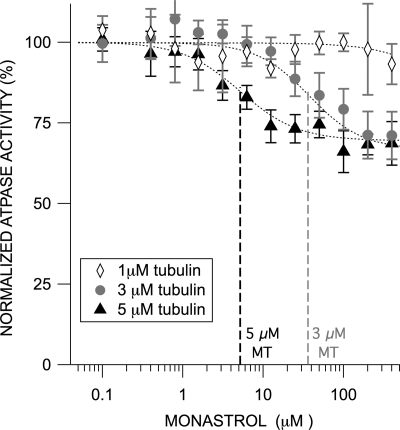
Although monastrol failed to inhibit basal ATPase rates of the Klp61F chimeras, we observed clear inhibition of MT-stimulated rates (Fig. 3, C and D, filled black triangles). At higher concentrations, monastrol elicited a maximum of 30% inhibition of ATPase activity of both chimeras in the presence of saturating levels of MTs. In addition, the IC50 of this reaction was dependent on the concentration of microtubules (Fig. 5); increasing MTs in these reactions decreases the IC50 for monastrol.
FIGURE 5.
Fractional occupancy of the microtubule-binding site impacts the efficacy of allosteric monastrol-based inhibition of Klp61F-L5. In the presence of saturating ATP, the monastrol IC50 for Klp61F-L5 decreases with increasing microtubule concentration. In the presence of 1 μm taxol-stabilized microtubules, Klp61F-L5 motor domains are not inhibited by monastrol. However, the monastrol IC50 is 39 ± 10 and 5 ± 2 μm in the presence of 3 and 5 μm tubulin, respectively. The Wilcoxon signed rank test confirmed that all three datasets are significantly different from one another (α = 0.05).
STC Binding Is Reconstituted in Both Klp61F Chimeras
The disparity between the STC and monastrol sensitivity of chimeric Klp61F ATPase activity is unexpected given the overlapping binding site contacts between the two inhibitors. To clarify the nature of this disparity, we probed the binding activity of both effectors by isothermal titration calorimetry. Remarkably, STC exhibited tight binding kinetics with both Klp61F-L5 and Klp61F-L5-α3 (Fig. 4A) with Kd values of 165 and 149 nm, which are in good agreement with the Kd of HsEg5 for STC (Table 4). In this case, the presence of the K214A substitution did not markedly affect the overall binding affinity of STC to the chimera. We observe that the chimeras effectively reconstitute STC binding activity and thereby modulate a heretofore unobserved mechanism for allosteric inhibition of Klp61F ATPase activity.
Monastrol Binds to the Chimeras but Only Inhibits MT-stimulated ATPase Activity
Surprisingly, although neither chimera showed any measurable inhibition by monastrol in basal ATPase assays, our ITC analysis found unambiguous binding affinity to this compound. With a monastrol binding curve displaying its expected weak binding curve shape (Fig. 4B), the Kd values for Klp61F-L5 and Klp61F-L5-α3 were measured at 142 and 48.6 μm, respectively (Table 5). Although substantially weaker, these Kd values approach the wild type HsEg5 values for monastrol of 8 μm. However, in contrast to STC binding, the K214A substitution has a major impact on monastrol binding, with almost 3-fold greater binding affinity.
Based on the Kd values measured in the ITC experiments, the molar ratios of drug to enzyme utilized in the basal and MT-stimulated assays are expected to bind a majority of the enzyme in the activity assay. For example, at 2.5 μm enzyme with a Kd of 48.6 μm (Table 5), 100–200 μm monastrol would occupy 67–80% of Klp61F-L5-α3, whereas these concentrations of monastrol would occupy 92–96% of HsEg5. The Kd of STC binding to the Klp61F chimeras are in the nanomolar range (Table 4) and, therefore, would be saturated at very low concentrations of STC. Despite binding to a majority of the motor at equilibrium, monastrol causes little or no measurable inhibition of ATPase activity. Therefore, we are led to conclude that basal ATPase rates of the chimeric Klp61F motor domains are not inhibited by monastrol despite significant occupancy of the L5 pocket by monastrol. Yet in the presence of microtubules, ATPase rates of the chimeric Klp61F motor domains are inhibited by monastrol.
Overall, both Klp61F chimeras show similar effector binding thermodynamics to HsEg5. The ITC data found both effectors bound in primarily enthalpy-driven reactions that overcome small entropically unfavorable components, with good agreement in the overall ΔG for the reaction in each case (−5 to −7 kcal/mol).
DISCUSSION
The exquisite specificity of allosteric inhibitors to the human Kinesin-5 protein has been established for the last decade (17), a finding remarkable given the overall sequence identity of the kinesin motor domain and the large number of protein members in the kinesin superfamily. The study of allosteric regulation in other protein families has yielded two developing schools of thought. The first argument is that allosteric sites are idiosyncratic features in individual family members, and thus, homologous proteins can have different allosteric mechanisms (31). The second idea is that allosteric networks and control are conserved features in protein families (32). The fundamental question of whether site-to-site communication is conserved is one of therapeutic relevance, as allostery is a major consideration in drug design not only for the human Kinesin-5 proteins but for other protein families as well (33–35).
Although the characterization of many different small molecule inhibitors of HsEg5 has generated a plethora of kinetic and structural data, to date little is known about the detailed mechanism of inhibition of this enzyme. On one hand, structure-activity relationship approaches to drug design have produced a growing list of tight-binding effectors to the L5 pocket but without concomitant insight for modeling the chemistry of inhibition (for examples, see Refs. 9, 36, and 37). Despite recent progress in resolving key catalytic intermediate steps including a prehydrolytic state for HsEg5 (8), structural and/or biochemical analyses have yet to elucidate how the L5 loop participates in native ATP hydrolysis or explain why L5 pocket effectors halt catalytic progression (4, 7, 8, 12, 14, 15, 20, 21, 26, 30, 36, 38–41).
To attack the question of the mechanism of inhibition, we chose a gain-of-function approach with a related Kinesin-5, Klp61F, which is not inhibited by STC or monastrol. This study has similarities with other reports in the literature that engineer new allosteric sites within proteins (35, 42). However, our work distinguishes itself by testing whether the effectors fail to inhibit Klp61F due to a lack of site-specific connections at the L5 pocket or due to a pattern of intraprotein interactions that are unique to HsEg5. A structural comparison is difficult as there are no reported crystal structures of Drosophila Klp61F motor domain, and the network of atomic-level contacts that mediate allosteric inhibition of HsEg5 have not been identified.
We predict that if Klp61F retains a conserved, intrinsic, allosteric network, then it should be uncovered through simple reconstitution of the effector binding pocket. Hence, we opted to substitute the entire L5 loop of HsEg5 into Klp61F. We argue that this would minimize disruption of native biochemical features of the human L5 loop and preserve the noted overall flexibility or mobility of the loop (7, 28, 43) that would likely be affected by the alternative point mutation strategy. Our results show that the human L5 loop can, even without directed optimization, produce coupled long distance activities in the designed chimeric proteins.
We employed isothermal titration calorimetry to directly measure the interactions of drug and protein. Stoichiometric binding ratios, equilibrium dissociation constants, and thermodynamic parameters were obtained for the effectors in solution. These data were then correlated to the effector-enzyme activity data to distinguish any allosteric effects. These ITC data provide unambiguous determination of the number of binding sites of the effector to the Kinesin-5 macromolecule. The N values of HsEg5 and the Klp61F proteins range from 0.89 to 0.93 (Tables 4 and 5), which indicates that the examined Kinesin-5 proteins possess only one binding site for these effectors. The correspondence between our experimentally determined binding stoichiometries in solution and the available crystal structures is a valuable control for our analysis; it supports the conclusion that the kinesin motor protein preparations were of high and consistent quality, and it eliminates the possibility that we are observing spurious avidity effects. Moreover, these consistent binding stoichiometries bolster our argument that the kinetic rates are a clear phenotypic measure of a “gain-of-function” in the chimeras.
In the determination of equilibrium dissociation constants, both Klp61F chimeras exhibited tight binding to STC with Kd values in the nanomolar range irrespective of residue 214. These ITC data, which closely parallel those of STC binding to HsEg5, indicate robust reconstitution of all critical STC binding elements in the chimeras and classify the chemical nature of the side chain of residue 214 as a relatively unimportant player in this regard. In the measurement of the equilibrium binding strength of monastrol, reconstitution of the HsEg5 effector binding sites in Klp61F-L5 and Klp61F-L5-α3 also restores affinity for this compound with Kd values of 142 and 48 μm, respectively, albeit weaker when compared with our Kd value of 8 μm for HsEg5.
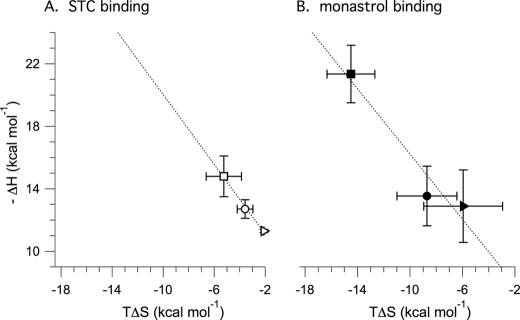
By measuring the thermodynamics of effector-enzyme complex formation, insight is gained toward understanding the physiochemical forces that modulate complex formation. Our data indicate that both STC and monastrol exhibit similar binding thermodynamics across all the motor domains tested (Tables 4 and 5) despite variability in the absolute values of ΔH and ΔS that are influenced by the sequence composition of the protein and chemistry of the effector. Enthalpy-entropy compensation plots reflect the changes between these two thermodynamic parameters; our plot of −ΔH versus TΔS for STC binding (Fig. 6A) show a linear relationship with a slope of 1.12. A comparable plot for monastrol binding to Kinesin-5 motor domains (Fig. 6B) shows a slope slightly smaller than that for STC but still greater than 1. A slope greater than unity in enthalpy-entropy compensation plots is associated with the free energy of binding being predominantly driven by enthalpy, whereas slope values less than unity are linked with dominant entropy contributions (44). Although the significance of such analysis is debated (see Refs. 45–48), we conclude from these often-used analyses (discussed in Refs. 44, 47, and 49–52) that the free energy of L5-directed inhibitor binding is primarily enthalpic. Therefore, it is probable that the increase in enthalpy and small decrease in entropy that is characteristic for both effectors causes the formation of a number of hydrogen bonds and/or restricts the motional freedom in the Kinesin-5-inhibitior complexes.
FIGURE 6.
Enthalpy-entropy compensation plots for the binding of allosteric effectors to Kinesin-5 motor domains. The plots for STC (A) and monastrol (B) binding have slopes of 1.12 and 1.04, respectively. Motors shown include HsEg5 (square), Klp61F-L5-α3 (circle), and Klp61F-L5 (triangle).
We find that these thermodynamic terms are correlated with the level of disorder within the protein matrix. For HsEg5-STC and HsEg5-monastrol, both crystal structures (e.g. PDB code 3KEN (13) and 1X88 (7)) and FT-IR measurements (13) detect an overall increase in ordered secondary structure in the enzyme upon drug binding that correlates well with our measured net decrease in entropy. Based on their thermodynamic similarities, we suspect that effector binding elicits parallel structural changes in the Drosophila Kinesin-5 chimeras as those that occur in the human isoform.
A major conclusion of our work is that the long distance allosteric network detected originally in HsEg5 is conserved in Klp61F. We surmise that the network(s) of amino acid residues involved in allosteric communication between the L5 loop, the active site, and the MT-binding site are, therefore, well conserved across Kinesin-5 family members. Given that the Klp61F and HsEg5 motor domains are 59% identical, it is unclear whether the conserved allosteric network is based on a shared network of identical residues. Alternatively, it is possible that higher order structural elements are conserved that are not predicated on specific amino acid identity but that retain allosteric signaling capability (13). Distinguishing these models awaits detailed elucidation of the nature of the Kinesin-5 allosteric networks.
A second major conclusion is that simple reconstitution of effector affinity to the L5 pocket is not sufficient to cause inhibition of mechanochemistry. In both chimeric motor domains, the gain of STC binding capability correlates with robust inhibition of basal and MT-stimulated ATPase activities. In contrast, binding of monastrol to either Klp61F chimera elicits no measurable inhibition of basal ATPase rates and yet exhibits moderate inhibition of MT-stimulated ATPase rates.
Why is there a difference in the allosteric responses of the chimeras to monastrol and STC? There are two possible explanations that are not mutually exclusive; these compounds are not synonymous, and the residue network for allosteric communication may not be the same for monastrol and STC. Our working model is that monastrol and STC elicit inhibitory responses through different pathways that are contingent on their different modes of contact to the L5 pocket.
Without an available Klp61F crystal structure, we can only surmise that chimeras suffer from a missing or broken element that is critical for the transmission of the monastrol inhibitory effect to the basal ATPase engine. As there are differences in the overall secondary structure of HsEg5 bound to monastrol and HsEg5 bound to STC (13), it is possible that the chimeras can access the analogous latter state upon STC binding but not the former. This would imply that the two effectors operate through at least partially different allosteric communication networks to affect inhibition.
Consistent with this hypothesis, adding a third allosteric effector, microtubules, to the chimera-monastrol complex permits some stimulation of monastrol-bound motor ATPase activity over basal albeit at clearly inhibited levels. We conclude that, although the mechanism of inhibition of the chimera-monastrol basal ATPase activity is broken, the complex nonetheless remains competent to interfere with MT-stimulated cooperativity in elevating ATPase activity levels. Supporting this view, our data show that the two inhibitors result in different long distance inhibitory effects on our chimeras.
A third key observation, which is related to the prior point, is the existence of a conserved communication linkage between the L5 loop and the MT binding pocket. Reconstitution of drug-mediated inhibition in Klp61F chimeras provides direct support for this model. In addition, we observe that mutations in the L5 loop of both chimeras mediate changes in the Km of the distal microtubule-binding site (supplemental Fig. S2). This behavior correlates with our finding that the fractional occupancy of the microtubule-binding site affects the IC50 of monastrol-mediated inhibition of the Drosophila chimeras (Fig. 5). Together, these data provide the first direct support for interdependence and cooperativity of the L5 loop with the microtubule-binding site of kinesins.
Our formal kinetic evidence has support in the literature. It has recently been shown that the closure of the HsEg5 L5 loop is correlated with flexing of the central β sheet in the motor domain (13) and with neck linker positioning (53). Therefore, it is possible that the linkage between the L5 loop and neck linker is driven by the cooperativity we observe between the L5 loop and microtubule-binding site. The cooperativity across this 22 Å distance may be mediated or communicated through the central β strands of the motor domain.
The fourth significant observation in this work is deciphering the initial atomic-level step of Kinesin-5 inhibition; residues in helix α3 are key to the disparate effects of STC and monastrol. This idea is supported by data herein on Klp61F residue 214 and data from other laboratories on Val210 of HsEg5. Homology models predict Lys214 of Klp61F should occupy the same spatial position within helix α3 as Ala218 in HsEg5 (see Fig. 1A). Our Klp61F-L5-α3 ITC experiments register a large positive effect by the K214A substitution on monastrol affinity but no detectable effect on STC affinity (Fig. 4). Because HsEg5 Ala218 is positioned 3.8 and 4.0 Å away from the monastrol methyl side chain and STC, respectively (Table 1), it is unlikely that the K214A substitution introduces a side chain capable of significant direct contributions to the affinity for either inhibitor in the chimera. However, in light of the large contribution by HsEg5 Ala218 to the hydrophobic surface within the L5 pocket (Table 1), it is possible that this residue position acts as a hydrophobic “gatekeeper” for monastrol in both HsEg5 and Klp61F-L5-α3 and facilitates entry of the nonpolar effector into the binding cavity while excluding water (see Fig. 1B).
Interestingly, we note that the V210A point mutation within helix α3 in the HsEg5 allosteric site (54) mimics the negative response of our Drosophila chimeras to monastrol while maintaining sensitivity to STC. Therefore it is possible that Val210 is an element of the inhibitory network of HsEg5 that is unique to monastrol. In addition, various inhibitory molecules, such as gossypol (55), biaryl compounds (3), and benzimidazole derivatives (56), have been identified that only inhibit HsEg5 MT-stimulated ATPase activity similar to chimera-monastrol. Together, these effectors may target the same element of the allosteric network linking the microtubule-binding site to the L5 loop.
Last, we conclude that, as the mechanism for allosteric inhibition in HsEg5 is conserved in Klp61F, it may also be conserved in other kinesins. For example, recent cross-linking and mutagenesis analysis suggest that allosteric inhibition of human kinesin-7 family member CENP-E by GSK923295 also occurs within the corresponding L5 pocket (57). Furthermore, allosteric inhibition may be reconstituted in all kinesins either through targeting the corresponding L5 pocket by small chemical effectors or modeling the corresponding L5 loop after that of HsEg5. Although more work remains to fully understand and harness these principles, this approach may open the door to the design of kinesin-based molecular machines that can be selectively regulated in the cell by synthetic small molecule effectors.
Supplementary Material
Acknowledgments
We thank Lindsey Ryals for dedicated technical support. Rich Walker and Sarah Learman (Virginia Tech) provided Klp61F cDNA and valuable discussions that initiated this work. We also thank the Worthylake laboratory (LSU Health Sciences Center, New Orleans) for assistance with the VP-ITC instrument. We are indebted to Tim Mitchison, Christine Field, and Rebecca Ward (Harvard Medical School, Boston) for support and discussions related to this project.
This work was supported, in whole or in part, by National Institutes of Health Grant GM066328 (to E. J. W.). This work was also supported by a Louisiana Board of Regents grant (to S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HsEg5
- human Eg5 kinesin
- MT
- microtubule
- ITC
- isothermal titration calorimetry
- STC
- S-trityl-l-cysteine
- FT-IR
- Fourier transform infrared spectroscopy.
REFERENCES
- 1. Brier S., Lemaire D., Debonis S., Forest E., Kozielski F. (2004) Biochemistry 43, 13072–13082 [DOI] [PubMed] [Google Scholar]
- 2. Learman S. S., Kim C. D., Stevens N. S., Kim S., Wojcik E. J., Walker R. A. (2009) Biochemistry 48, 1754–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo L., Parrish C. A., Nevins N., McNulty D. E., Chaudhari A. M., Carson J. D., Sudakin V., Shaw A. N., Lehr R., Zhao H., Sweitzer S., Lad L., Wood K. W., Sakowicz R., Annan R. S., Huang P. S., Jackson J. R., Dhanak D., Copeland R. A., Auger K. R. (2007) Nat. Chem. Biol. 3, 722–726 [DOI] [PubMed] [Google Scholar]
- 4. Maliga Z., Kapoor T. M., Mitchison T. J. (2002) Chem. Biol. 9, 989–996 [DOI] [PubMed] [Google Scholar]
- 5. Cox C. D., Coleman P. J., Breslin M. J., Whitman D. B., Garbaccio R. M., Fraley M. E., Buser C. A., Walsh E. S., Hamilton K., Schaber M. D., Lobell R. B., Tao W., Davide J. P., Diehl R. E., Abrams M. T., South V. J., Huber H. E., Torrent M., Prueksaritanont T., Li C., Slaughter D. E., Mahan E., Fernandez-Metzler C., Yan Y., Kuo L. C., Kohl N. E., Hartman G. D. (2008) J. Med. Chem. 51, 4239–4252 [DOI] [PubMed] [Google Scholar]
- 6. Kaan H. Y., Ulaganathan V., Hackney D. D., Kozielski F. (2010) Biochem. J. 425, 55–60 [DOI] [PubMed] [Google Scholar]
- 7. Maliga Z., Mitchison T. J. (2006) BMC Chem. Biol. 6, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parke C. L., Wojcik E. J., Kim S., Worthylake D. K. (2010) J. Biol. Chem. 285, 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schiemann K., Finsinger D., Zenke F., Amendt C., Knöchel T., Bruge D., Buchstaller H. P., Emde U., Stähle W., Anzali S. (2010) Bioorg. Med. Chem. Lett. 20, 1491–1495 [DOI] [PubMed] [Google Scholar]
- 10. Yan Y., Sardana V., Xu B., Homnick C., Halczenko W., Buser C. A., Schaber M., Hartman G. D., Huber H. E., Kuo L. C. (2004) J. Mol. Biol. 335, 547–554 [DOI] [PubMed] [Google Scholar]
- 11. Zhang B., Liu J. F., Xu Y., Ng S. C. (2008) Biochem. Biophys. Res. Commun. 372, 565–570 [DOI] [PubMed] [Google Scholar]
- 12. Brier S., Lemaire D., DeBonis S., Forest E., Kozielski F. (2006) J. Mol. Biol. 360, 360–376 [DOI] [PubMed] [Google Scholar]
- 13. Kim E. D., Buckley R., Learman S., Richard J., Parke C., Worthylake D. K., Wojcik E. J., Walker R. A., Kim S. (2010) J. Biol. Chem. 285, 18650–18661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marshall C. G., Torrent M., Williams O., Hamilton K. A., Buser C. A. (2009) Arch. Biochem. Biophys. 484, 1–7 [DOI] [PubMed] [Google Scholar]
- 15. Sheth P. R., Basso A., Duca J. S., Lesburg C. A., Ogas P., Gray K., Nale L., Mannarino A. F., Prongay A. J., Le H. V. (2009) Biochemistry 48, 11045–11055 [DOI] [PubMed] [Google Scholar]
- 16. Kapoor T. M., Mayer T. U., Coughlin M. L., Mitchison T. J. (2000) J. Cell Biol. 150, 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999) Science 286, 971–974 [DOI] [PubMed] [Google Scholar]
- 18. Crevel I. M., Alonso M. C., Cross R. A. (2004) Curr. Biol. 14, R411–R412 [DOI] [PubMed] [Google Scholar]
- 19. Garcia-Saez I., DeBonis S., Lopez R., Trucco F., Rousseau B., Thuéry P., Kozielski F. (2007) J. Biol. Chem. 282, 9740–9747 [DOI] [PubMed] [Google Scholar]
- 20. Cochran J. C., Gilbert S. P. (2005) Biochemistry 44, 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krzysiak T. C., Wendt T., Sproul L. R., Tittmann P., Gross H., Gilbert S. P., Hoenger A. (2006) EMBO J. 25, 2263–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Halabi N., Rivoire O., Leibler S., Ranganathan R. (2009) Cell 138, 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coureux P. D., Sweeney H. L., Houdusse A. (2004) EMBO J. 23, 4527–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wojcik E. J., Dalrymple N. A., Alford S. R., Walker R. A., Kim S. (2004) Biochemistry 43, 9939–9949 [DOI] [PubMed] [Google Scholar]
- 25. Deavours B. E., Reddy A. S., Walker R. A. (1998) Cell Motil. Cytoskeleton 40, 408–416 [DOI] [PubMed] [Google Scholar]
- 26. Luo L., Carson J. D., Molnar K. S., Tuske S. J., Coales S. J., Hamuro Y., Sung C. M., Sudakin V., Auger K. R., Dhanak D., Jackson J. R., Huang P. S., Tummino P. J., Copeland R. A. (2008) J. Am. Chem. Soc. 130, 7584–7591 [DOI] [PubMed] [Google Scholar]
- 27. Sobolev V., Sorokine A., Prilusky J., Abola E. E., Edelman M. (1999) Bioinformatics 15, 327–332 [DOI] [PubMed] [Google Scholar]
- 28. DeBonis S., Simorre J. P., Crevel I., Lebeau L., Skoufias D. A., Blangy A., Ebel C., Gans P., Cross R., Hackney D. D., Wade R. H., Kozielski F. (2003) Biochemistry 42, 338–349 [DOI] [PubMed] [Google Scholar]
- 29. Zhao Y. C., Kull F. J., Cochran J. C. (2010) J. Biol. Chem. 285, 25213–25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skoufias D. A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R. H., Hackney D., Kozielski F. (2006) J. Biol. Chem. 281, 17559–17569 [DOI] [PubMed] [Google Scholar]
- 31. Kuriyan J., Eisenberg D. (2007) Nature 450, 983–990 [DOI] [PubMed] [Google Scholar]
- 32. Goodey N. M., Benkovic S. J. (2008) Nat. Chem. Biol. 4, 474–482 [DOI] [PubMed] [Google Scholar]
- 33. Bharatham K., Bharatham N., Kwon Y. J., Lee K. W. (2008) J. Comput. Aided Mol. Des. 22, 925–933 [DOI] [PubMed] [Google Scholar]
- 34. Lewis J. A., Lebois E. P., Lindsley C. W. (2008) Curr. Opin. Chem. Biol. 12, 269–280 [DOI] [PubMed] [Google Scholar]
- 35. Zhang X. Y., Bishop A. C. (2007) J. Am. Chem. Soc. 129, 3812–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Debonis S., Skoufias D. A., Indorato R. L., Liger F., Marquet B., Laggner C., Joseph B., Kozielski F. (2008) J. Med. Chem. 51, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 37. Roecker A. J., Coleman P. J., Mercer S. P., Schreier J. D., Buser C. A., Walsh E. S., Hamilton K., Lobell R. B., Tao W., Diehl R. E., South V. J., Davide J. P., Kohl N. E., Yan Y., Kuo L. C., Li C., Fernandez-Metzler C., Mahan E. A., Prueksaritanont T., Hartman G. D. (2007) Bioorg. Med. Chem. Lett. 17, 5677–5682 [DOI] [PubMed] [Google Scholar]
- 38. Cochran J. C., Krzysiak T. C., Gilbert S. P. (2006) Biochemistry 45, 12334–12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lad L., Luo L., Carson J. D., Wood K. W., Hartman J. J., Copeland R. A., Sakowicz R. (2008) Biochemistry 47, 3576–3585 [DOI] [PubMed] [Google Scholar]
- 40. Lakämper S., Thiede C., Düselder A., Reiter S., Korneev M. J., Kapitein L. C., Peterman E. J., Schmidt C. F. (2010) J. Mol. Biol. 399, 1–8 [DOI] [PubMed] [Google Scholar]
- 41. Luo L., Carson J. D., Dhanak D., Jackson J. R., Huang P. S., Lee Y., Sakowicz R., Copeland R. A. (2004) Biochemistry 43, 15258–15266 [DOI] [PubMed] [Google Scholar]
- 42. Lee J., Natarajan M., Nashine V. C., Socolich M., Vo T., Russ W. P., Benkovic S. J., Ranganathan R. (2008) Science 322, 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bodey A. J., Kikkawa M., Moores C. A. (2009) J. Mol. Biol. 388, 218–224 [DOI] [PubMed] [Google Scholar]
- 44. Dam T. K., Torres M., Brewer C. F., Casadevall A. (2008) J. Biol. Chem. 283, 31366–31370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cooper A., Johnson C. M., Lakey J. H., Nöllmann M. (2001) Biophys. Chem. 93, 215–230 [DOI] [PubMed] [Google Scholar]
- 46. Dunitz J. D. (1995) Chem. Biol. 2, 709–712 [DOI] [PubMed] [Google Scholar]
- 47. Gallicchio E., Kubo M., Levy R. (1998) J. Am. Chem. Soc. 120, 4526–4527 [Google Scholar]
- 48. Sharp K. (2001) Protein Sci. 10, 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fisicaro E., Compari C., Braibanti A. (2004) Phys. Chem. Chem. Phys. 6, 4156–4166 [Google Scholar]
- 50. Freire E. (2008) Drug Discov. Today 13, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whitesides G. M., Krishnamurthy V. M. (2005) Q. Rev. Biophys. 38, 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams D. H., Stephens E., O'Brien D. P., Zhou M. (2004) Angew. Chem. Int. Ed. Engl. 43, 6596–6616 [DOI] [PubMed] [Google Scholar]
- 53. Larson A. G., Naber N., Cooke R., Pate E., Rice S. E. (2010) Biophys. J. 98, 2619–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tcherniuk S., van Lis R., Kozielski F., Skoufias D. A. (2010) Biochem. Pharmacol. 79, 864–872 [DOI] [PubMed] [Google Scholar]
- 55. DeBonis S., Skoufias D. A., Lebeau L., Lopez R., Robin G., Margolis R. L., Wade R. H., Kozielski F. (2004) Mol. Cancer Ther. 3, 1079–1090 [PubMed] [Google Scholar]
- 56. Sheth P. R., Shipps G. W., Jr., Seghezzi W., Smith C. K., Chuang C. C., Sanden D., Basso A. D., Vilenchik L., Gray K., Annis D. A., Nickbarg E., Ma Y., Lahue B., Herbst R., Le H. V. (2010) Biochemistry 49, 8350–8358 [DOI] [PubMed] [Google Scholar]
- 57. Wood K. W., Lad L., Luo L., Qian X., Knight S. D., Nevins N., Brejc K., Sutton D., Gilmartin A. G., Chua P. R., Desai R., Schauer S. P., McNulty D. E., Annan R. S., Belmont L. D., Garcia C., Lee Y., Diamond M. A., Faucette L. F., Giardiniere M., Zhang S., Sun C. M., Vidal J. D., Lichtsteiner S., Cornwell W. D., Greshock J. D., Wooster R. F., Finer J. T., Copeland R. A., Huang P. S., Morgans D. J., Jr., Dhanak D., Bergnes G., Sakowicz R., Jackson J. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5839–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maliga Z., Xing J., Cheung H., Juszczak L. J., Friedman J. M., Rosenfeld S. S. (2006) J. Biol. Chem. 281, 7977–7982 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.