Abstract
NafY participates in the final steps of nitrogenase maturation, having a dual role as iron-molybdenum cofactor (FeMo-co) carrier and as chaperone to the FeMo-co-deficient apo-NifDK (apo-dinitrogenase). NafY contains an N-terminal domain of unknown function (n-NafY) and a C-terminal domain (core-NafY) necessary for FeMo-co binding. We show here that n-NafY and core-NafY have very weak interactions in intact NafY. The NMR structure of n-NafY reveals that it belongs to the sterile α-motif (SAM) family of domains, which are frequently involved in protein-protein interactions. The presence of a SAM domain in NafY was unexpected and could not be inferred from its amino acid sequence. Although SAM domains are very commonly found in eukaryotic proteins, they have rarely been identified in prokaryotes. The n-NafY SAM domain binds apo-NifDK. As opposed to full-length NafY, n-NafY impaired FeMo-co insertion when present in molar excess relative to FeMo-co and apo-NifDK. The implications of these observations are discussed to offer a plausible mechanism of FeMo-co insertion. NafY domain structure, molecular tumbling, and interdomain motion, as well as NafY interaction with apo-NifDK are consistent with the function of NafY in FeMo-co delivery to apo-NifDK.
Keywords: Iron-Sulfur Protein, Metalloproteins, Nitrogen Metabolism, Nitrogenase, NMR, Protein Structure
Introduction
The versatile properties of iron-sulfur clusters make them essential to a variety of biological processes (1). The most common [Fe-S] clusters have [2Fe-2S], [3Fe-4S], or [4Fe-4S] composition and are referred to as simple [Fe-S] clusters. There are, however, a few examples of [Fe-S] clusters with higher structural complexity, which often contain additional metals. The iron-molybdenum cofactor (FeMo-co4; [Mo-7Fe-9S-X-homocitrate]) located at the active site of the nitrogenase5 enzyme has been extensively studied and serves as a paradigm to study complex [Fe-S] cluster assembly and incorporation into apoproteins.
Complex metalloclusters are generated from simple [Fe-S] clusters by incorporating additional metals and organic/inorganic ligands. In the case of the FeMo-co of nitrogenase, simple [Fe-S] clusters are the substrates of the NifB protein, which uses them to generate NifB-co, a complex [Fe-S] cluster with a proposed [6Fe-9S-X] structure (2–4). Molybdenum, homocitrate, and an additional iron are subsequently incorporated into NifB-co to generate FeMo-co in a series of biosynthetic steps involving the activities of the NifX, NifEN, NifH, NifQ, and NifV proteins (5–7). Finally, the completed FeMo-co is incorporated into its target apoprotein, a FeMo-co-deficient form of dinitrogenase (apo-NifDK), yielding an enzyme that is competent for N2 reduction.
NafY (nitrogenase accessory factor Y), which is able to independently bind FeMo-co and apo-NifDK, has been proposed to have a role as an FeMo-co insertase (8, 9). Consistent with this role, the presence of NafY enhances the in vitro conversion of apo-NifDK into active NifDK by FeMo-co (10). In addition, NafY has been shown to stabilize apo-NifDK prior to FeMo-co insertion, although deletion of the nafY gene does not abolish active nitrogenase biosynthesis (9).
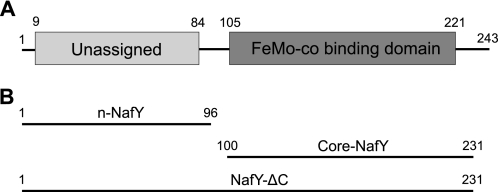
A NifX-like domain6 (termed core-NafY) that spans residues 105–231 has been previously identified in NafY (Fig. 1). Core-NafY binds one molecule of FeMo-co with high affinity through contacts that include a surface-exposed histidine residue (11). X-ray crystallography showed that core-NafY has a different structure than the FeMo-co-binding region of NifDK, precluding the identification of a FeMo-co-binding region in NafY based on structural similarity (12). Core-NafY was shown not to bind apo-NifDK tightly (12). There have been no reports on the structure or function of the 12-kDa N-terminal domain (residues 1–96, which we will refer to as n-NafY) or the C-terminal region of NafY (residues 231–243).
FIGURE 1.
NafY domain architecture. A, diagram illustrating the domain organization of NafY. B, truncated NafY variants used in this study. Amino acid residues comprising the constructs studied are indicated.
In this work, we show that n-NafY is sufficient to bind apo-NifDK with substantial affinity. In addition, we present the solution structure of n-NafY and characterize domain interactions in NafY by NMR. Surprisingly, n-NafY has a sterile α-motif (SAM) fold, a protein-protein interaction domain that could not be inferred from its amino acid sequence. These findings strengthen the model that NafY targets apo-NifDK and delivers the cofactor using two essentially independent specialized domains.
EXPERIMENTAL PROCEDURES
Purification of NafY, NafY-ΔC, n-NafY, and Core-NafY
Escherichia coli cells induced for the overexpression of the corresponding polypeptide were resuspended in buffer A (10 mm sodium phosphate, 1.8 mm potassium phosphate (pH 7.3), 140 mm NaCl, and 2.7 mm KCl) and disrupted at 12,000 p.s.i. with a French press. Cell debris was removed by two centrifugation steps at 25,000 × g for 40 min. The cell-free extracts were loaded onto a 25-ml GSH-Sepharose Fast Flow affinity column (GE Healthcare). The column was washed with 250 ml of buffer A supplemented with 1% Triton X-100, and the GST-tagged fusion protein was eluted in 75 ml of buffer B (50 mm Tris-HCl (pH 8.0) and 10 mm GSH). The GST tag was cleaved by the addition of 10 μg of pure tobacco etch virus protease/mg of GST-tagged protein, followed by 2 h of incubation at 30 °C. The protein mixture was then subjected to a series of chromatographic steps: gel filtration on Sephadex G-25 (to remove GSH), affinity to GSH-Sepharose (to remove the GST tag), and nickel affinity chromatography (to remove the tobacco etch virus protease). A typical purification procedure yielded 1 mg of protein from 6 g of E. coli cell paste. NafY, n-NafY, core-NafY, and NafY-ΔC were estimated to be >95% pure based on SDS-PAGE analysis (supplemental Fig. S1). The resulting purified proteins were concentrated by ultrafiltration using an Amicon cell equipped with a YM10 membrane or an Ultrafree-0.5 device equipped with a PBCC Biomax 5-kDa membrane (Millipore) to ∼1 mm.
In Vitro FeMo-co Insertion and Nitrogenase Activity Assays
The in vitro FeMo-co insertion reactions were performed in 3-ml serum vials sealed with rubber stoppers. The vials were repeatedly evacuated and flushed with argon gas and finally rinsed with 0.3 ml of anaerobic 25 mm Tris-HCl (pH 7.5) and 1 mm sodium dithionite. The reactions shown in Fig. 5C contained 25 mm Tris-HCl (pH 7.5), 1 mm sodium dithionite, 5 mg/ml BSA, 0.22 nmol of apo-NifDK, 6.28 nmol of NifH, and variable amounts of FeMo-co. The reactions shown in Fig. 5D contained 25 mm Tris-HCl (pH 7.5), 1 mm sodium dithionite, 5 mg/ml BSA, 0.22 nmol of apo-NifDK, 6.28 nmol of NifH, 0.215 nmol of FeMo-co, and variable amounts of purified n-NafY. The reactions (total volume of 325 μl) were incubated at 30 °C for 30 min to allow for FeMo-co insertion. The resulting activation of apo-NifDK present in the reaction mixture was analyzed by the acetylene reduction assay after the addition of 0.4 mg of NifH and 0.8 ml of ATP-regenerating mixture (13).
FIGURE 5.
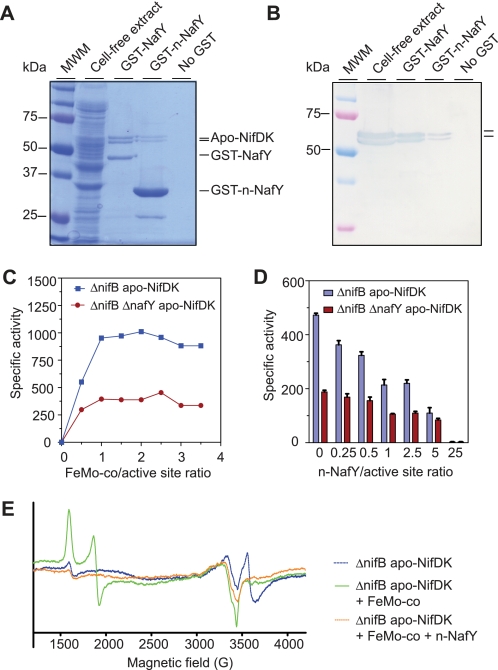
Binding and effect of n-NafY on apo-NifDK reconstitution. A, SDS-PAGE analysis of the interaction between n-NafY and apo-NifDK. An UW146 (nifB ΔnafY) cell-free extract containing apo-NifDK (second lane) was applied to a GSH-Sepharose resin carrying bound GST-NafY, bound GST-n-NafY, or no protein. After thorough washing, proteins bound to the resin were eluted by the addition of GSH and analyzed by SDS-PAGE. Third lane, protein eluted from GST-NafY resin; fourth lane, protein eluted from GST-n-NafY resin; fifth lane, protein eluted from GSH-Sepharose resin. Molecular mass markers (first lane) are indicated to the left. B, immunoblot analysis of SDS gel from A developed with antibodies to NifDK. The position of apo-NifDK is indicated to the right. C, activation by FeMo-co of ΔnifB apo-NifDK and ΔnifB ΔnafY apo-NifDK. Specific activity is given in nanomoles of C2H4 produced per min/mg of apo-NifDK added to the reaction mixture. Data are the average of two independent determinations. D, effect of n-NafY on the activation of ΔnifB apo-NifDK and ΔnifB ΔnafY apo-NifDK by suboptimal amounts of FeMo-co. Specific activity is given in nanomoles of C2H4 produced per min/mg of apo-NifDK added to the reaction mixture. Data are the average of two to four independent determinations. E, continuous wave-EPR spectra from apo-NifDK (blue line) and apo-NifDK samples incubated with FeMo-co (green line) and n-NafY plus FeMo-co (orange line).
NMR Spectroscopy and n-NafY Structure Determination
NMR samples of n-NafY, core-NafY, and NafY-ΔC were obtained by concentrating purified protein in buffer (pH 7.3) containing 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 2 mm EDTA, and 0.02% (w/v) NaN3 in either 100% 2H2O or 10% (v/v) 2H2O to a protein concentration of ∼1 mm. All n-NafY NMR data were collected on a 98-amino acid polypeptide containing the first 96 residues (positions 1–96) of NafY plus two N-terminal residues (Gly-His) introduced during cloning. All core-NafY NMR data sets were collected on a 135-amino acid polypeptide containing 132 residues of NafY (Arg-100–Phe-231) plus three N-terminal residues (Gly-His-Met) introduced during the cloning procedure. For assignment of NafY-ΔC (residues 1–231), protein was made with a high level of deuteration as well as uniform 13C and 15N labeling but using [2H,13C]glucose and 15NH4Cl with growth in 98% 2H2O. NMR experiments were recorded at 298 K on a Bruker Avance 800, a Bruker Avance DMX 600 equipped with a cryoprobe, or a Bruker Avance II 900 equipped with a cryoprobe. Transverse relaxation optimized spectroscopy versions of the triple resonance experiments were used for NafY-ΔC. T1, T2, and heteronuclear NOE relaxation data were collected using 1H/13C/15N-labeled NafY-ΔC. NMR data were processed using NMRPipe or rNMRtk, and spectra were analyzed with NMRDraw (14) and CARA.
RESULTS
Solution Structure of n-NafY
The three-dimensional structure of n-NafY in solution was determined by NMR spectroscopy. Chemical shift assignment of both backbone and side chain resonances n-NafY was obtained using triple resonance coherence transfer and NOESY experiments (15), as described under “Experimental Procedures.” Supplemental Fig. S2 shows the assigned 1H-15N heteronuclear single quantum coherence spectrum of n-NafY. Twelve amide resonances, including those from residues 2–8, which include two extra N-terminal residues introduced during cloning and most residues in the loop region between residues 35 and 42, remain unassigned. The Glu-39 and Gly-40 resonances were identified by their coupling patterns and distinctive chemical shifts. The Glu-39 and Gly-40 amide peaks and several of the unassigned resonances were significantly broadened. In addition, residues 34–43 showed few NOE cross-peaks, consistent with this region of the protein undergoing conformational exchange on the microsecond-to-millisecond timescale.
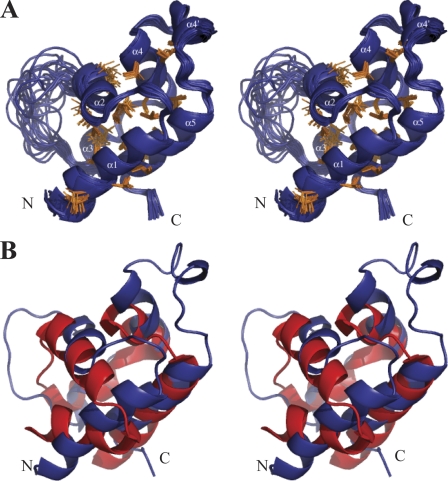
Structures of n-NafY were calculated using NOE-derived distances, chemical shift-derived dihedral angle restraints, J coupling-derived dihedral angle restraints, and global orientation restraints from residual dipolar couplings with the CYANA program (16). The 20 structures (from 200 random starting structures) most consistent with the NMR data were chosen to represent the solution structure ensemble of n-NafY (Fig. 2A). The statistics for these structures are summarized in supplemental Table S1. The structure with the minimum violations is presented in Fig. 2B, with the calculated electrostatic potential shown in supplemental Fig. S3. Within the well structured region of the protein (residues 9–33 and 44–85), the average root mean square difference from the mean structure of this group of 20 lowest energy structures is 0.45 Å for backbone atoms and 0.96 Å for all heavy atoms. The domain adopts an all α-helical fold with two long helices, α1 and α5, which are separated by four short helices, α2, α3, α4, and α4′, and a flexible loop composed of residues 34–43 between α2 and α3.
FIGURE 2.
NMR structure of n-NafY. A, stereo drawing of the 20 best structures. Hydrophobic side chains within the well structured region of the protein (residues 9–33 and 44–85) are shown in orange. B, structural overlay of n-NafY and the SAM domain that is the closest structural homolog (Ste11 from S. cerevisiae, Protein Data Bank code 1OW5).
A BLAST search for n-NafY homologs in protein databases did not reveal any protein of known structure with significant amino acid sequence similarity to n-NafY. A structure homology search of the Protein Data Bank using the Secondary Structure Matching algorithm (17) with the well defined region of n-NafY (residues 8–88) as a query was performed. The results (supplemental Table S2) identified structural similarity between n-NafY and the SAM domain family. The SAM domain motif is frequently involved in mediating protein-protein interactions, although interactions with RNA have also been reported (18). The fold of typical SAM domains consists of a five-helix bundle, with a helical hairpin formed by the first two helices and the three additional helices packed on one side of the hairpin (18, 19). SAM domains have been generalized to include proteins with four helices (generally two helical hairpins), with somewhat different packing than the typical versions. n-NafY also has some differences relative to the typical SAM domains: 1) α1 and α2 are at nearly a right angle, rather than being packed close to antiparallel in a hairpin; 2) α3 has been reduced to a single helical turn; 3) there is an extra helix, α4′, inserted between α4 and α5; and 4) the loop linking α2 and α3 is poorly ordered. Fig. 2B shows an overlay of the n-NafY structure nearest to the mean and the SAM domain with closest structural similarity, that of the Saccharomyces cerevisiae protein Ste11 (Protein Data Bank code 1OW5) (20). Despite the differences, the basic fold of n-NafY is relatively close to typical SAM domains.
Domain Interactions in NafY
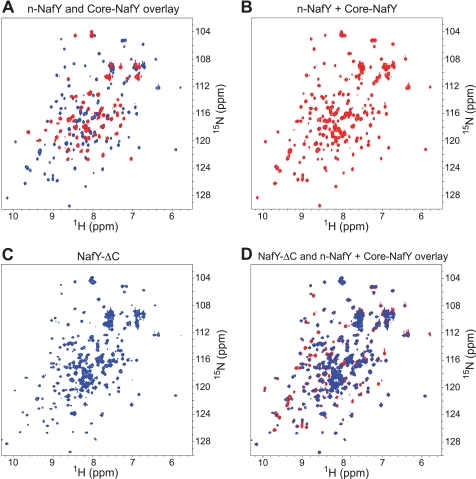
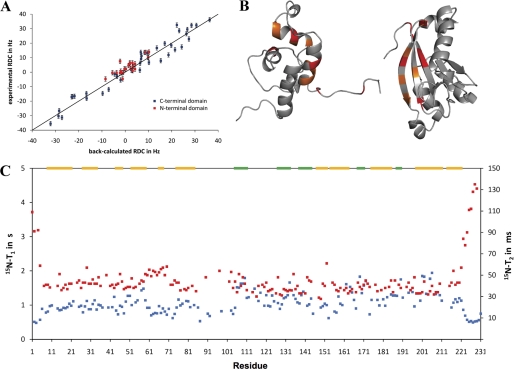
The structures determined for n-NafY and core-NafY show clearly that these are independently folded domains. However, it was not clear whether there is any interaction between them in the intact protein. To address this issue, mixtures of n-NafY and core-NafY were studied and compared with spectra of the naturally linked version NafY-ΔC. Both core-NafY and NafY-ΔC have a C-terminal deletion of NafY amino acid residues 232–243, which are not conserved among NafY-like proteins and were not observed in the crystal structure of core-NafY reported previously (12). 1H-15N heteronuclear single quantum coherence spectra of n-NafY, core-NafY, and NafY-ΔC are shown in Fig. 3. No significant chemical shift changes were observed in either domain when n-NafY and core-NafY were mixed 1:1 in solution (Fig. 3B), consistent with the conclusion that the two domains fold independently and do not interact to form a defined quaternary structure. The spectrum of NafY-ΔC (Fig. 3C, with assignments shown in supplemental Fig. S4A) was also compared with those from the individual domains, n-NafY and core-NafY (Fig. 3D). Small shift differences were detected, which are plotted as a function of residue in supplemental Fig. S4. There are relatively few residues affected, with shift changes that are modest in magnitude. The residues with the largest shifts are indicated on the structure in Fig. 4B. These do not define a contiguous surface, suggesting that only transient interactions occur. 1H-15N residual dipolar coupling measurements on NafY-ΔC ordered in a phage medium showed that the two domains have very different order tensors and hence are not well ordered with respect to one another (Fig. 4A). From the much different residual dipolar coupling ranges found, we conclude that the two domains tumble independently. 15N T1/T2 ratios for NafY-ΔC clearly indicated different effective rotational correlations times for the two domains, again supporting flexible linkage of n-NafY and core-NafY (Fig. 4C). However, the derived rotational correlation times for the individual domains are substantially longer than expected if there was no interaction at all between them. This behavior is analogous to the WW and prolyl-isomerase domains of Pin1, also disordered but with some interaction (21).
FIGURE 3.
1H-15N heteronuclear single quantum coherence spectra of NafY domains and domain combinations. A, overlay of n-NafY (red) and core-NafY (blue). B, spectrum of an equimolar mixture of n-NafY and core-NafY. C, NafY-ΔC spectrum. D, overlaid spectra of NafY-ΔC (blue) and the equimolar mixture of n-NafY and core-NafY (red).
FIGURE 4.
Domain interactions in NafY. A, residual dipolar coupling (RDC) values of NafY-ΔC. NafY-ΔC was made to tumble anisotropically by the addition of Pf1 phage, which aligns in strong magnetic fields and interferes with the isotopic tumbling of other solutes. Residual dipolar coupling sizes are dependent on both the orientation of the bond vector and the degree of anisotropy in the tensor that describes molecular tumbling (34). A larger range of residual dipolar couplings denotes a higher degree of anisotropy in the molecular motion. B, diagram showing the residues in n-NafY and core-NafY that exhibit chemical shift differences when compared with NafY-ΔC. Residues that are shifted by >1 S.D. above the mean are shown in orange; those shifted by >2 S.D. are shown in red. C, 15N T1 (blue) and T2 (red) values of NafY-ΔC. The secondary structure is depicted above. α-Helices are shown in yellow; β-strands are shown in green.
n-NafY Binds Apo-NifDK
NafY, NafY-ΔC, core-NafY, and n-NafY were expressed in E. coli as GST fusion proteins and purified as described under “Experimental Procedures.” Purified GST-NafY, GST-NafY-ΔC, GST-core-NafY, and GST-n-NafY were individually applied to columns containing GSH-Sepharose to generate resins with potential affinity to apo-NifDK. Cell-free extracts from Azotobacter vinelandii UW146 (nifB ΔnafY) containing NafY-deficient apo-NifDK were then applied to each affinity column. After extensive washing, apo-NifDK remained bound to the GST-NafY affinity resin, as expected (Fig. 5, A and B). Removal of the C-terminal 232–243 amino acid residues in the NafY-ΔC variant did not affect binding to either apo-NifDK or FeMo-co (supplemental Fig. S5).
The GST-n-NafY affinity resin pulled down apo-NifDK protein from UW146 extracts (Fig. 5, A and B), demonstrating that n-NafY binds apo-NifDK in the absence of core-NafY. The efficiency of apo-NifDK retention by n-NafY was lower than that of NafY or NafY-ΔC, suggesting that, although not essential for binding, the presence of core-NafY increases the stability of the NafY·apo-NifDK complex. Consistently, weaker interaction between core-NafY and apo-NifDK was observed in a similar assay (supplemental Fig. S5A). No binding of apo-NifDK to the resin was observed when purified GST protein was used as affinity ligand, ruling out any interaction of apo-NifDK with the GST moiety or the resin (data not shown).
The same results were observed when using purified apo-NifDK instead of UW146 extracts in all NafY affinity pulldown assays (supplemental Fig. S5A), demonstrating that the interaction of NafY with apo-NifDK requires no additional factors. This interaction was specific to apo-NifDK. NafY exhibited no affinity for purified holo-NifDK in the pulldown assays (supplemental Fig. S5C).
Excess of n-NafY Impairs Reconstitution of NifDK Activity
The effect of n-NafY on apo-NifDK activation was studied by comparing the specific activity of NifDK reconstituted by FeMo-co in the absence and presence of increasing amounts of n-NafY. Two purified apo-NifDK variants were used for these studies, the ΔnifB apo-NifDK from strain DJ1143 (22) and the ΔnifB ΔnafY apo-NifDK from strain UW269 (supplemental Table S3). Both variants were purified as tetramers free from bound NafY to facilitate comparison of results obtained from FeMo-co insertion experiments. Extensive washing with imidazole-containing buffer (which weakens the NafY-apo-NifDK interaction) was required to completely remove NafY from the ΔnifB apo-NifDK.
Activation of apo-NifDK by the addition of FeMo-co was essentially complete at a 1:1 molar ratio of FeMo-co- to apo-NifDK-binding sites, independent of the genetic background (Fig. 5C). Specific activity was ∼2-fold higher in activated ΔnifB apo-NifDK than in ΔnifB ΔnafY apo-NifDK.
Fig. 5D shows the effect of n-NafY on reconstitution of NafY-free apo-NifDK with FeMo-co. The reaction mixtures contained limiting amounts of FeMo-co (0.5:1 molar ratio of FeMo-co to apo-NifDK active sites) with variable amounts of n-NafY. The presence of n-NafY had a negative effect on apo-NifDK activation; an n-NafY:apo-NifDK active site ratio of 1:1 decreased activation by 50%. The level of inhibition increased with the amount of n-NafY added to the reaction and was completely inhibited at a 25:1 ratio of n-NafY to apo-NifDK active sites. The negative effect of n-NafY was largely ameliorated by doubling the amount of FeMo-co in the reaction (supplemental Fig. S6).
To understand the nature of n-NafY-dependent inhibition of NifDK reconstitution, larger scale experiments were carried out with reaction mixtures containing a 0.5:1 molar ratio of FeMo-co to apo-NifDK active sites in the absence or presence of a 25-fold excess of n-NafY. The products were analyzed by EPR spectroscopy. Fig. 5E shows the EPR signatures of apo-NifDK (blue line), FeMo-co-activated apo-NifDK (green line), and apo-NifDK incubated with FeMo-co and an excess of n-NafY (orange line). The EPR signatures of apo-NifDK and holo-NifDK proteins have been reported previously (22). Purified apo-NifDK (blue line) exhibited the EPR signal of the P-clusters in the P1+ state but no FeMo-co EPR signal, as expected. Activated apo-NifDK (green line) exhibited the typical EPR signals arising from FeMo-co, indicating that cofactor insertion took place normally. In contrast, apo-NifDK incubated with FeMo-co in the presence of excess n-NafY (orange line) exhibited an EPR spectrum lacking signals attributable to FeMo-co. This result indicates that excess n-NafY impairs insertion of FeMo-co into apo-NifDK.
DISCUSSION
Three proteins have been shown to interact with apo-NifDK: NifH, NifY, and NafY. NifH interacts with apo-NifDK during the formation of the P-cluster (23). In Klebsiella pneumoniae, NifY was shown to associate with apo-NifDK and dissociate upon FeMo-co insertion (24, 25). Likewise, the A. vinelandii NafY protein interacts with apo-NifDK carrying mature P-clusters but lacking FeMo-co (8). The pulldown experiments described above show that n-NafY binds to apo-NifDK with significant affinity, although the core-NafY domain appears to contribute to the stability of the NafY·apo-NifDK complex. Measurements of chemical shifts, relaxation rates, and residual dipolar couplings indicate that n-NafY and core-NafY are connected by a flexible tether (residues 85–104) and are not significantly ordered with respect to one another but do have some transient interactions. It remains to be determined whether the interaction and ordering of these domains increase in the presence of FeMo-co or apo-NifDK, and how such interactions might affect function.
The structure of n-NafY shows that it belongs to the SAM family of protein-protein interaction domains (18), which is completely in line with the functional role we have defined for it. However, this finding was unexpected because amino acid sequence comparisons did not identify this fold. Furthermore, not even conserved key residues were evident by aligning n-NafY with structurally similar SAM domains (data not shown). Because there are very few structures of complexes of SAM domains available, it is not possible to identify by analogy the likely surface of NifDK that interacts with the SAM domain.
Although the SMART and Pfam Databases identify >2500 SAM domains, only 11 of them are from prokaryotic proteins, and at present, only the AR1 domain of the NusA protein from E. coli (26) and a four-helix SAM domain, part of TipαN34 from Helicobacter pylori (27), have been structurally characterized. SAM domains are frequently responsible for interaction with other proteins, sometimes with other SAM domains but with many other folds as well. Interactions appear to be diverse in character, with electrostatics and/or exposed hydrophobic surfaces implicated, and involve different surfaces within the SAM domain.
In A. vinelandii cell-free extracts, the presence of NafY was shown to be important for complete apo-NifDK activation by FeMo-co, which suggested a role for NafY in apo-NifDK stabilization (9). Similar results had been observed in cell-free extracts of K. pneumoniae nifY mutants (24, 25), leading to the hypothesis that NifY acts transiently as a molecular prop to keep apo-NifDK in a conformation that can be activated by FeMo-co (24). Because K. pneumoniae NifY and A. vinelandii NafY exhibit primary structure similarity throughout their sequences (9), it is likely that they would share a mechanism of action. Interestingly, in addition to nafY, A. vinelandii carries a nifY gene in a genetic context similar to that of the K. pneumoniae nifY. The function of the A. vinelandii nifY gene is unknown.
NafY-free apo-NifDK variants purified from both nafY-lacking and nafY-containing genetic backgrounds were also differentially activated by FeMo-co (Fig. 5C), demonstrating that these variants are inherently different from each other. Thus, it can be concluded that the presence or absence of interactions between apo-NifDK and NafY in vivo will define the activation properties of apo-NifDK by FeMo-co. The exact mechanism by which NafY protects or “marks” apo-NifDK to allow complete reconstitution by FeMo-co insertion is not known, but we hypothesize that preventing oxidation of the α-Cys275 residue could be part of this mechanism (see below).
There are at least four potential explanations for the observed inhibition of apo-NifDK reconstitution by an excess of n-NafY in vitro. First, n-NafY could exhibit higher affinity for FeMo-co than apo-NifDK and would sequester the cofactor. This is not the case because n-NafY did not show FeMo-co-binding activity when analyzed by anaerobic native gel electrophoresis (supplemental Fig. S5). Second, because NafY stabilizes the “open” conformation of apo-NifDK (28) and dissociates from NifDK upon FeMo-co insertion (8), an excess of n-NafY might lock the FeMo-co-bound enzyme into the open conformation by preventing the conformational change that generates active NifDK. To reconcile this possibility with the observed lack of a FeMo-co EPR signal, we would have to assume that FeMo-co bound to the open conformation of NifDK is in an EPR-silent state or that the conformational change must occur to keep FeMo-co in place. Third, the binding of n-NafY could sterically block the access of FeMo-co to the apo-NifDK active site. Because the surface of n-NafY displays a large negatively charged region (supplemental Fig. S3), it is possible that n-NafY would bind between the positively charged insertion funnel and the disordered region of apo-NifDK (NifD residues 381–407) that would act as a lid (29). Selective alkylation studies have shown that NafY-stabilized apo-NifDK has the side chain of α-Cys275 exposed to solvent (α-Cys275 serves as ligand to FeMo-co) (28). Fourth, it could also be possible that n-NafY increases the KD of the FeMo-co·apo-NifDK complex. The negative effect of n-NafY can be largely prevented by increasing the amount of FeMo-co in the reconstitution assay to match that of n-NafY (supplemental Fig. S6), which is consistent with the last two possibilities.
The relief of the inhibitory effect of n-NafY by FeMo-co might be physiologically relevant because it could represent a fine-tuning scheme to link successful FeMo-co synthesis to apo-NifDK activation. Consistently, whereas equimolar amounts of apo-NifDK and full-length NafY enhanced activation by FeMo-co by 2-fold (10), the same amount of n-NafY (which cannot bind FeMo-co) decreased apo-NifDK activation by 50%. Additionally, NafY could prevent insertion of clusters different from FeMo-co, such as NifB-co or other biosynthetic intermediates, into the apo-NifDK active site. Consistent with this, NafY exhibits much higher affinity for FeMo-co (KD = 60 nm) than NifB-co (KD in the low micromolar range) (11).
Our combined structural and biochemical study indicates that NafY relies on its N-terminal domain (n-NafY), of previously unknown function, to bind tightly to apo-NifDK. Through this interaction, the FeMo-co-binding module of NafY, core-NafY, is recruited to apo-NifDK. Colocalization of core-NafY and apo-NifDK increases their relative local concentrations, which could be a potential mechanism to facilitate the transfer of FeMo-co from NafY to apo-NifDK.
Supplementary Material
Acknowledgments
We thank Paul Ludden (Southern Methodist University) for discussions and support; Dennis Dean (Virginia Tech) for providing strains DJ and DJ1143; Professor David Britt and Constantino Aznar (Chemistry and Biochemistry Department, University of California, Davis) for use of the EPR instrument; Leonardo Curatti, Basem Soboh, and Robert Igarashi for helpful discussions; and Gerhard Wagner for providing pulse sequences.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-35332 (to Paul Ludden) and Grant 62163 from NIGMS (to D. E. W.). This work was also supported by European Research Council Starting Grant 205442 and Ministerio de Ciencia e Innovación de España Grant BIO2009-12661 (to L. M. R.) and by Midwestern University intramural funds (to J. A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S6, Tables S1–S3, and references.
Nitrogenase is composed of two oxygen-sensitive metalloproteins termed dinitrogenase and dinitrogenase reductase (13). Dinitrogenase reductase is a homodimer of the nifH gene product that carries one [4Fe-4S] cluster (30). Dinitrogenase is an α2β2-tetramer of the nifD and nifK gene products that carries two pairs of complex metallocenters, the P-cluster ([8Fe-7S] cluster) and FeMo-co (31). The FeMo-co-deficient form of dinitrogenase, referred to as apo-dinitrogenase or apo-NifDK, co-purifies with NafY, a protein that appears to act as stabilizing chaperone (9, 32).
The NifX-like domain of NafY is found in a family of proteins that comprises NifX, NafY, NifY, and NifB, and has been proposed to bind either FeMo-co, NifB-co, or the VK-cluster (9). In particular, NifX has been proposed to serve as a transient reservoir of FeMo-co precursors and thus help control their flux during FeMo-co synthesis (33).
- FeMo-co
- iron-molybdenum cofactor
- SAM
- sterile α-motif.
REFERENCES
- 1. Beinert H., Holm R. H., Münck E. (1997) Science 277, 653–659 [DOI] [PubMed] [Google Scholar]
- 2. Curatti L., Ludden P. W., Rubio L. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5297–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. George S. J., Igarashi R. Y., Xiao Y., Hernandez J. A., Demuez M., Zhao D., Yoda Y., Ludden P. W., Rubio L. M., Cramer S. P. (2008) J. Am. Chem. Soc. 130, 5673–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao D., Curatti L., Rubio L. M. (2007) J. Biol. Chem. 282, 37016–37025 [DOI] [PubMed] [Google Scholar]
- 5. Hu Y., Fay A. W., Lee C. C., Yoshizawa J., Ribbe M. W. (2008) Biochemistry 47, 3973–3981 [DOI] [PubMed] [Google Scholar]
- 6. Rubio L. M., Ludden P. W. (2008) Annu. Rev. Microbiol. 62, 93–111 [DOI] [PubMed] [Google Scholar]
- 7. Schwarz G., Mendel R. R., Ribbe M. W. (2009) Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 8. Homer M. J., Dean D. R., Roberts G. P. (1995) J. Biol. Chem. 270, 24745–24752 [DOI] [PubMed] [Google Scholar]
- 9. Rubio L. M., Rangaraj P., Homer M. J., Roberts G. P., Ludden P. W. (2002) J. Biol. Chem. 277, 14299–14305 [DOI] [PubMed] [Google Scholar]
- 10. Curatti L., Hernandez J. A., Igarashi R. Y., Soboh B., Zhao D., Rubio L. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17626–17631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rubio L. M., Singer S. W., Ludden P. W. (2004) J. Biol. Chem. 279, 19739–19746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dyer D. H., Rubio L. M., Thoden J. B., Holden H. M., Ludden P. W., Rayment I. (2003) J. Biol. Chem. 278, 32150–32156 [DOI] [PubMed] [Google Scholar]
- 13. Shah V. K., Brill W. J. (1973) Biochim. Biophys. Acta 305, 445–454 [DOI] [PubMed] [Google Scholar]
- 14. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 15. Ferentz A. E., Wagner G. (2000) Q. Rev. Biophys. 33, 29–65 [DOI] [PubMed] [Google Scholar]
- 16. Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 17. Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 18. Qiao F., Bowie J. U. (2005) Sci. STKE 2005, re7 [DOI] [PubMed] [Google Scholar]
- 19. Ponting C. P. (1995) Protein Sci. 4, 1928–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kwan J. J., Warner N., Pawson T., Donaldson L. W. (2004) J. Mol. Biol. 342, 681–693 [DOI] [PubMed] [Google Scholar]
- 21. Bernadó P., Fernandes M. X., Jacobs D. M., Fiebig K., García de la Torre J., Pons M. (2004) J. Biomol. NMR 29, 21–35 [DOI] [PubMed] [Google Scholar]
- 22. Christiansen J., Goodwin P. J., Lanzilotta W. N., Seefeldt L. C., Dean D. R. (1998) Biochemistry 37, 12611–12623 [DOI] [PubMed] [Google Scholar]
- 23. Hu Y., Fay A. W., Lee C. C., Ribbe M. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10424–10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. White T. C., Harris G. S., Orme-Johnson W. H. (1992) J. Biol. Chem. 267, 24007–24016 [PubMed] [Google Scholar]
- 25. Homer M. J., Paustian T. D., Shah V. K., Roberts G. P. (1993) J. Bacteriol. 175, 4907–4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonin I., Mühlberger R., Bourenkov G. P., Huber R., Bacher A., Richter G., Wahl M. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13762–13767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tosi T., Cioci G., Jouravleva K., Dian C., Terradot L. (2009) FEBS Lett. 583, 1581–1585 [DOI] [PubMed] [Google Scholar]
- 28. Magnuson J. K., Paustian T. D., Shah V. K., Dean D. R., Roberts G. P., Rees D. C., Howard J. B. (1997) Tetrahedron 53, 11971–11984 [Google Scholar]
- 29. Schmid B., Ribbe M. W., Einsle O., Yoshida M., Thomas L. M., Dean D. R., Rees D. C., Burgess B. K. (2002) Science 296, 352–356 [DOI] [PubMed] [Google Scholar]
- 30. Georgiadis M. M., Komiya H., Chakrabarti P., Woo D., Kornuc J. J., Rees D. C. (1992) Science 257, 1653–1659 [DOI] [PubMed] [Google Scholar]
- 31. Chan M. K., Kim J., Rees D. C. (1993) Science 260, 792–794 [DOI] [PubMed] [Google Scholar]
- 32. Paustian T. D., Shah V. K., Roberts G. P. (1990) Biochemistry 29, 3515–3522 [DOI] [PubMed] [Google Scholar]
- 33. Hernandez J. A., Igarashi R. Y., Soboh B., Curatti L., Dean D. R., Ludden P. W., Rubio L. M. (2007) Mol. Microbiol. 63, 177–192 [DOI] [PubMed] [Google Scholar]
- 34. Fischer M. W., Losonczi J. A., Weaver J. L., Prestegard J. H. (1999) Biochemistry 38, 9013–9022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.