Abstract
Galectins are β-galactoside-binding lectins that regulate diverse cell behaviors, including adhesion, migration, proliferation, and apoptosis. Galectins can be expressed both intracellularly and extracellularly, and extracellular galectins mediate their effects by associating with cell-surface oligosaccharides. Despite intensive current interest in galectins, strikingly few studies have focused on a key enzyme that acts to inhibit galectin signaling, namely β-galactoside α2,6-sialyltransferase (ST6Gal-I). ST6Gal-I adds an α2,6-linked sialic acid to the terminal galactose of N-linked glycans, and this modification blocks galectin binding to β-galactosides. This minireview summarizes the evidence suggesting that ST6Gal-I activity serves as an “off switch” for galectin function.
Keywords: Apoptosis, Carbohydrate-binding Protein, Cell Death, Glycosylation, Lectin, ST6Gal-I, Galectin, Sialic Acid
Introduction
Sialic acids comprise a family of nine-carbon sugars added to the termini of oligosaccharides found on secreted or cell-surface glycoproteins and glycolipids (1). Because of their negative charge and relatively large size, sialic acids can mask important functional domains on surface glycoproteins and also serve more generally to protect the cell from various types of assault (2). However, evidence is emerging that sialic acids mediate specific cellular and molecular recognition by regulating association with glycan-binding proteins such as lectins. For example, sialic acids bind specifically to the siglec2 family of lectins (3), whereas other types of glycan/lectin interactions are conversely inhibited by sialylation. Thus, sialic acids are positioned to play a pivotal role in regulating lectin-dependent cell/cell and cell/matrix interactions. Sialic acids are added to glycans via α2,3-, α2,6-, or α2,8-linkages, and these linkages are directed by distinct sialyltransferases. β-Galactoside α2,6-sialyltransferase (ST6Gal-I) is one of the principal enzymes responsible for the addition of α2,6-linked sialic acids to the Galβ1,4GlcNAc disaccharide (4), which is found mainly on N-glycans and, to a lesser extent, on O-glycans. In this minireview, we summarize the evidence suggesting that ST6Gal-I-mediated α2,6-sialylation inhibits binding of N-glycans to galectin-type lectins, thereby serving as a negative regulator of galectin-dependent cell responses (of note, α2,6-sialic acid/siglec interactions, although of equal biologic importance, will be not be discussed herein due to the availability of other reviews on this topic (3, 5)).
Galectins
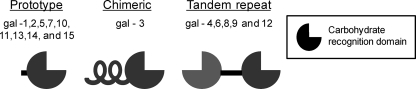
Galectins are animal lectins that bind β-galactosides through their conserved carbohydrate recognition domains (CRDs) (6, 7). At least 15 mammalian galectins have been identified, and these are subdivided into three different groups based on their biochemical structure (Fig. 1). The prototype group (galectin (Gal)-1, -2, -5, -7, -10, -11, -13, -14, and -15) contains one CRD and a short N-terminal sequence. Members of this group typically assemble into noncovalent homodimers. The chimeric group, of which Gal-3 is the only member, contains one CRD and an extended N-terminal domain with a repeated collagen-like sequence. The tandem repeat group (Gal-4, -6, -8, -9, and -12) is composed of a single polypeptide chain that forms two distinct but homologous CRDS, separated by a short linker. Galectins are found intracellularly in the nucleus and cytoplasm (6) but are also secreted through a nonclassical mechanism that is not well understood (8). Extracellular galectins bind glycoproteins on the cell surface and in the extracellular matrix (9, 10), whereas intracellular galectins can associate with cytoplasmic and nuclear proteins through carbohydrate-independent interactions (11). Galectins have been implicated in numerous biologic processes, including cell adhesion, migration, proliferation, differentiation, transformation, apoptosis, angiogenesis, and immune responses (9–15).
FIGURE 1.
Galectins are categorized into three distinct groups. The prototype group contains one CRD and a short N-terminal sequence. The chimeric group, of which Gal-3 is the only member, contains one CRD and an extended N-terminal domain with a repeated collagen-like sequence. The tandem repeat group is composed of a single polypeptide chain that forms two distinct but homologous CRDs, separated by a short linker domain.
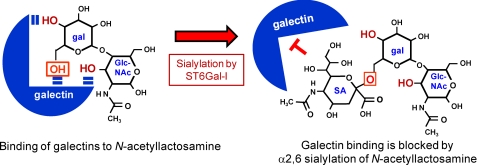
Although all galectins bind to β-galactosides, each galectin subtype has selectivity for certain galactose-containing oligosaccharides, which occurs as a consequence of variability in the CRD sequence (7). In general, galectins have particular affinity for poly-N-acetyllactosamine (N-acetyllactosamine is defined as the GlcNAc-Gal disaccharide) (Fig. 2); however, finer specificity is conferred by compositional features of the glycan, including the number of N-acetyllactosamine units within a poly-N-acetyllactosamine chain, the presence of terminal sugars on the chain such as sialic acid and fucose, and the degree of N-glycan branching (7, 16, 17). Many galectins exhibit stronger binding to β1,6-branched glycans (7), a structure generated by β1,6-N-acetylglucosaminyltransferase V (designated GnTV or Mgat5). Because the expression of Mgat5 changes significantly during pathologic conditions such as carcinogenesis (18, 19), this enzyme can serve as a central regulator of cell responses to galectins. Another important characteristic of galectins is that the binding affinity between an individual galectin and its minimal glycan ligand is typically low; however, the propensity of galectins to oligomerize enhances avidity. In turn, this facilitates cross-linking of cell-surface glycans, leading to the formation of stabilized lattices. These lattices have multiple functions, but one critical activity is to control the retention of selected glycoproteins on the cell surface (17, 20, 21).
FIGURE 2.
Structural studies suggest that three free OH groups are necessary for galectin binding to N-acetyllactosamine: 4-OH and 6-OH on galactose (gal) and 3-OH on GlcNAc (note that N-acetyllactosamine is defined as the GlcNAc-Gal disaccharide). The ST6Gal-I enzyme adds a sialic acid (SA; depicted as Neu-5-Ac) to 6-OH of galactose (boxed OH shown in red). The addition of sialic acid at this position blocks galectin binding.
Inhibition of Galectin Binding by α2,6-Sialylation
Much of what is known regarding structural determinants for galectin binding has been gleaned from studies of synthetic oligosaccharides. Results from such studies suggest that most (if not all) galectins exhibit diminished binding to β-galactosides capped with α2,6-sialic acid. Hirabayashi et al. (7) used frontal affinity chromatography to show that α2,3-sialylation of β-galactosides was tolerated by some galectins, but none of the 13 galectins studied, including Gal-1, -3, -8, or -9, could bind to β-galactosides that were α2,6-sialylated. It was concluded in this investigation that there was a strict requirement for the 6-OH of galactose (the site for addition of α2,6-linked sialic acid) to remain unmodified in order for galectins to associate with N-acetyllactosamine (Fig. 2). Similarly, fluorescence-based solid-phase assays were used to determine that dimeric Gal-1 could bind unsialylated and α2,3-sialylated poly-N-acetyllactosamines with approximately equal affinity, whereas binding was completed inhibited by α2,6-sialylation (22). It was also reported that α2,6-sialylation blocked the interaction of Gal-1, -2, and -3 with N-acetyllactosamine in glycan microarrays (23).
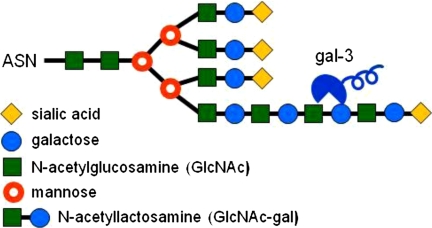
Despite these extensive results implicating α2,6-sialylation as a generic inhibitor of galectin binding, it is becoming apparent that the effects of α2,6-sialylation on the binding of Gal-3, compared with other galectins, may be more complex than initially appreciated. Chammas and co-workers (24) detected some binding of Gal-3 to α2,6-sialylated poly-N-acetyllactosamine, although the binding was lower than that observed with unsialylated or α2,3-sialylated poly-N-acetyllactosamine. Likewise, Cummings and co-workers (23) reported that α2,6-sialylation was less effective at blocking the association of Gal-3 with poly-N-acetyllactosamine compared with Gal-1 and Gal-2. These findings are in striking contrast to the strong inhibitory effect of α2,6-sialylation on Gal-3 binding to a single N-acetyllactosamine unit. One plausible explanation for this incongruity is that Gal-3 (unlike Gal-1) may bind laterally to the internal N-acetyllactosamines within an extended poly-N-acetyllactosamine chain (23, 25), thus weakening the inhibitory effect of the sialic acid on the terminal N-acetyllactosamine (Fig. 3).
FIGURE 3.
Gal-3 may bind internal N-acetyllactosamine units on an extended poly-N-acetyllactosamine chain, thus attenuating the inhibitory effect of the terminal α2,6-sialic acid.
ST6Gal-I-mediated α2,6-Sialylation of N-Glycoproteins
Synthetic oligosaccharides have been invaluable for characterizing determinants of galectin binding. However, cell-surface glycans have much greater structural complexity, and many of the biologic glycan structures cannot currently be synthesized (23, 26). In addition, the mode of glycan presentation, either in solid phase or in solution, can alter the binding specificity of galectins (23), and the glycan/lectin interaction might be conformation-specific. Glycan/lectin interactions may also be altered through lateral association with other membrane glycoproteins and glycolipids. For all of these reasons, it is important to study glycan/galectin interactions within the context of the cell, and moreover, the biologic relevance of these interactions and the potential significance of α2,6-sialylation in controlling them need further elucidation. Within the cell, variant α2,6-sialylation of N-linked glycans occurs primarily as a consequence of differential ST6Gal-I activity, secondary to changes in ST6Gal-I expression.
Given the putative role of ST6Gal-I as a negative regulator of galectins, it is surprising that so few studies have focused on this enzyme. In particular, there is still very little known regarding factors such as 1) signaling mechanisms controlling ST6Gal-I expression, 2) extracellular stimuli that might initiate such signaling mechanisms, 3) the biologic relevance of variant ST6Gal-I mRNA isoforms, 4) the specific substrates for the enzyme, and 5) the functional consequences associated with variant α2,6-sialylation of specific substrates. Much of the ST6Gal-I-related research has centered on correlating cell responses with global changes in cell-surface α2,6-sialylation; however, our understanding of the biologic importance of this enzyme can be complete only when we have defined the role of α2,6-sialylation in regulating the activity of specific target molecules.
ST6Gal-I-mediated α2,6-sialylation of glycoproteins likely influences cell behavior through several molecular mechanisms, including modulation of glycoprotein conformation, alterations in receptor clustering or retention at the cell surface, and regulation of protein/protein interactions. For example, studies from our group suggest that α2,6-sialylation alters the conformation and function of the β1 integrin, thereby regulating cell adhesion and migration (27–30). Baum and co-workers (31) have shown that α2,6-sialylation inhibits clustering of the CD45 tyrosine phosphatase on T cells, leading to diminished signaling, whereas Kitazume et al. (32) conversely reported that α2,6-sialylation is necessary for clustering and cell-surface retention of PECAM (platelet endothelial cell adhesion molecule) on endothelial cells. In another noteworthy study, α2,6-sialylation of the core glycan in the IgG Fc domain was shown to regulate IgG binding to Fc receptors, and coordinately, the loss of sialylation switched IgG from having anti-inflammatory effects at steady state to having pro-inflammatory activity after antigen challenge (33). Finally, the hemagglutinin of human (but not avian) influenza viruses predominantly binds α2,6-sialic acid structures on the nonciliated cells of human trachea (34). These examples highlight the many ways in which α2,6-sialylation can alter the activity of specific molecules or molecular interactions. However, it is emerging that one of the major physiologic roles for α2,6-sialylation may be to block galectin-dependent responses. This important function of ST6Gal-I-mediated α2,6-sialylation has been most extensively studied in immunology and cancer biology.
Role of ST6Gal-I-mediated α2,6-Sialylation in Regulating Galectin-dependent Immune Cell Responses
ST6Gal-I expression is tightly regulated in many immune cell types and varies as a consequence of cell activation or differentiation status. Glycan profiling studies reveal that α2,6-sialylated structures comprise the predominant type of complex N-glycans in freshly isolated CD4 and CD8 T cells, whereas activated T cells exhibit a dramatic decrease in α2,6-sialylated glycans due to down-regulated expression of ST6Gal-I (35, 36). ST6Gal-I expression and activity are similarly down-regulated during dendritic cell maturation (37, 38) and differentiation of primary monocytes and promonocytic cell lines along the macrophage lineage (27, 30, 39). Collectively, these results hint that decreased α2,6-sialylation may be necessary for some aspect of immune cell maturation or activation. Indeed, ST6Gal-I-deficient mice exhibit alterations in thymopoiesis and granulopoiesis (40, 41); disruptions in eosinophil and dendritic cell profiles (42, 43); and finally, deficits in B cell proliferation and antibody production (44). Undoubtedly, some of these phenotypes are due, at least in part, to elimination of the ligand for α2,6-sialic acid-specific siglecs. For instance, it is well established that impaired B cell responses observed in ST6Gal-I deficient mice occur as consequence of diminished signaling from the B cell siglec, CD22, due to loss of its α2,6-sialylated ligand (45, 46). Nevertheless, one anticipates that deletion of ST6Gal-I also contributes to immunopathology through effects on galectin signaling. One very important function of extracellular galectins is to induce apoptosis (15). It is tempting to speculate that diminished ST6Gal-I-mediated α2,6-sialylation, resulting in exposure of galectin-binding galactosyl-type glycans, provides a mechanism for limiting the life span of activated and/or differentiated immune cells.
Some of the earliest evidence supporting α2,6-sialylation as a negative regulator of galectin-mediated immune cell apoptosis was provided by Baum and co-workers (31). In this study, ST6Gal-I expression was forced in a murine T cell line, and it was found that α2,6-sialylation blocked Gal-1 binding as well as Gal-1-induced cell death. These effects were mediated by α2,6-sialylation of CD45, which was shown to be a selective target for ST6Gal-I. ST6Gal-I-dependent resistance to Gal-1 may have particular relevance in the positive selection of maturing thymocytes; α2,6-sialylation is highly enriched in mature medullary thymocytes (47), which in turn exhibit resistance to Gal-1-induced apoptosis (48, 49). Interestingly, there appears to be selectivity not only in the glycoproteins bound by various galectins but also in the cell-surface receptors responsible for translating galectin-initiated signals into specific cell responses. For example, Gal-3 binds a different (although overlapping) complement of receptors than Gal-1, and of these Gal-3-binding partners (including β1 integrin, CD43, CD45, and CD71), only CD45 appeared to be required for Gal-3-induced apoptosis of several T cell lines (50). Fukumori et al. (51) alternately suggested that the β1 integrin and CD7 receptors were involved in Gal-3-directed apoptosis of the MOLT-4 T cell line. The important implication emerging from these studies is that there is an apparent dependence on specific receptors to direct galectin-induced responses, although this feature of galectin signaling is not well understood at this time.
More recently, it has been reported that α2,6-sialylation is a critical factor controlling the expansion of selected CD4 T cell subtypes. Effector CD4 T cells (TH1, TH2, and TH17) orchestrate the functional activity of both the innate and adaptive immune systems, and the homeostatic process is often accompanied by a shift toward a TH2 profile. In an elegant study, Toscano et al. (52) found that TH2 cells have higher ST6Gal-I protein expression, ST6Gal-I enzyme activity, and α2,6-sialic acid compared with TH1 and TH17 cells. This elevated surface α2,6-sialylation was associated with protection of TH2 cells from Gal-1-induced cell death. Similarly, Gal-1-deficient mice developed hyper-TH1 and hyper-TH17 responses after antigenic challenge, reflecting better survival of TH1 and TH17 cells in the absence of Gal-1, whereas no disruption was observed in the levels of TH2 cells. These combined results suggest that Gal-1 may function to preferentially eliminate antigen-specific TH1 and TH17 cells (due to low levels of surface α2,6-sialylation) (52), and they may also explain the prior observation that administration of exogenous Gal-1 suppresses chronic inflammation and skews the immune response toward a TH2 cytokine profile (53). Intriguingly, TH1 and TH2 cells exhibit equivalent levels of cell death when exposed to Gal-3 (52), which mirrors the results from synthetic oligosaccharide studies indicating that α2,6-sialylation does not always block the activity of Gal-3 as it does for other galectins. In support of this concept, the enzymatic removal of α2,6-sialic acids from the surface of HL-60 promyelocytic cells sensitizes cells to Gal-1- but not Gal-3-directed apoptosis (23). It remains to be determined whether the persistence of Gal-3 activity observed in various models results from binding of Gal-3 to internal N-acetyllactosamines or is alternatively due to other mechanisms. Recently, it was shown that Gal-3 can bind to extended type 1 glycans, which contain the Galβ1,3GlcNAc linkage (lacto-N-biose) rather than Galβ1,4GlcNAc (54). Given that ST6Gal-I has preferential activity toward the Galβ1,4GlcNAc disaccharide, alterations in ST6Gal-I-directed sialylation may have little effect on Gal-3 binding to cells presenting extended type 1 surface glycans. It is also possible that Gal-3 binding to certain O-linked glycans would be independent of ST6Gal-I-mediated α2,6-sialylation. Clearly, further studies are needed to dissect the complex relationship between ST6Gal-I activity and Gal-3.
In addition to effects on T cell responses, protection from galectin-mediated apoptosis through α2,6-sialylation has been reported in human B cells. Suzuki et al. (55) determined, in several B lymphoma cell lines, that α2,6-sialylation prevents the binding and apoptotic activity of Gal-1. Cell-surface sialylation also inhibits Gal-3-induced apoptosis of HBL-2 B lymphoma cells, although the specific type of sialyl linkage was not determined in this study (56). Finally, sialylation-dependent blockade of galectin signaling may contribute to the worse prognosis known to be associated with diffuse large B cell lymphoma patients harboring tumors that express sialylated oligosaccharides (57).
ST6Gal-I-dependent Inhibition of Galectin Function May Promote Tumor Cell Survival
Another example of variant ST6Gal-I expression is found in tumor cells. ST6Gal-I is overexpressed in many types of human cancers, including colon (58–62), breast (63), ovarian (64), gastric (65), oral (66), cervical (67), choriocarcinoma (68), leukemia (69), and brain tumors (70), and high expression positively correlates with tumor metastasis and poor prognosis (61, 63, 66). Furthermore, both in vitro cell culture and animal studies have implicated ST6Gal-I in regulating tumor cell invasiveness and differentiation state, as well as metastasis (71–79). Although mechanisms regulating ST6Gal-I expression have not been widely investigated, it is known that ST6Gal-I is up-regulated by oncogenic Ras (reviewed in Ref. 28) signaling through a Ral guanine exchange factor-dependent mechanism (80). The functional consequences of ST6Gal-I up-regulation are not well defined but are likely mediated through multiple molecular pathways impacting tumor cell behaviors such as adhesion to matrix and cell migration and survival. Recent studies suggest that, as with immune cells, epithelial tumor cells are protected against galectin-mediated apoptosis via α2,6-sialylation of surface receptors. Notably, like ST6Gal-I, Gal-3 is commonly up-regulated in several types of cancers (81–83), raising the paradox of why a tumor cell would up-regulate a sugar structure that blocks Gal-3 binding. To address this issue, our group forced expression of ST6Gal-I in SW48 cells, a colon epithelial cell line that lacks both α2,3- and α2,6-sialyltransferases (84), and then evaluated apoptosis induced by recombinant Gal-3 (added extracellularly). These studies showed that parental cells lacking sialylation had significantly greater binding to exogenous Gal-3 than ST6Gal-I expressors (85). Using a blot overlay approach, it was shown that Gal-3 binds directly to the β1 integrin but not when this integrin carries α2,6-sialic acids (85). Moreover, α2,6-sialylation of the β1 integrin was found to protect cells against Gal-3-mediated cell apoptosis (85). Thus, increased ST6Gal-I-mediated receptor sialylation protects cancer cells from the pro-apoptotic function of secreted Gal-3. However, intracellular Gal-3 is known to have many protumorigenic functions, including enhancement of Ras signaling and inhibition of pro-apoptotic mitochondrial proteins (11, 12, 86). These carbohydrate-independent functions would not be affected by ST6Gal-I activity; therefore, on balance, simultaneous up-regulation of ST6Gal-I and Gal-3 should provide a survival advantage for tumor cells. It is also noteworthy that, in this cell model system (unlike HL-60 myelocytic cells), α2,6-sialylation by ST6Gal-I served as a strong inhibitor of Gal-3-induced apoptosis. These results point to a role for cell type-specific glycans in the regulation of Gal-3 efficacy. Factors such as N-glycan branching and chain length, expression of type 1 versus type 2 glycans, and/or the presence of certain O-linked oligosaccharides are likely important, and all of these structures are correspondingly controlled by the unique complement of glycosylating enzymes expressed by each distinct cell type.
In contrast to reports of simultaneous up-regulation of ST6Gal-I and Gal-3 in tumor cells, Gabius and co-workers (87) suggested that there was an inverse relationship between the expression of Gal-1 and the levels of α2,6-sialylation. This group forced expression of the p16INK4a tumor suppressor in pancreatic epithelial cells (87). It is well known that abrogation of the Rb/p16INK4a pathway is found in virtually all pancreatic carcinomas (88), although the mechanism is still not fully elucidated. It was found that pancreatic carcinoma cell lines stably transfected with p16INK4a had increased Gal-1 protein expression but decreased α2,6-sialylation on N-glycans (although ST6Gal-I expression and activity were not directly evaluated in this study). Nonetheless, the effects of α2,6-sialylation on Gal-1 function observed by Gabius and co-workers were consistent with the larger literature; reduced α2,6-sialylation was associated with greater Gal-1 binding, leading to p16INK4a-mediated anoikis in pancreatic cell lines (87).
Regulation of Surface α2,6-Sialylation by Extracellular Sialic Acid-modifying Enzymes
Variant α2,6-sialylation of N-glycosylated proteins typically occurs as a consequence of changes in the levels of ST6Gal-I within the trans-Golgi, resulting from either transcriptional or post-transcriptional mechanisms. The gene encoding ST6Gal-I (siat1) displays multiple promoter sequences, and several alternatively spliced mRNAs have been identified (4, 89–93). In addition, glycoprotein sialylation can be down-regulated following shedding of ST6Gal-I from cells after cleavage in the Golgi by the BACE1 β-secretase (30, 94). ST6Gal-I activity may also be altered through oligomerization of the enzyme within the Golgi (95). Regardless of these various modes of regulation, it has generally been thought that α2,6-sialylation has a relatively long-lived effect on glycoprotein function. Because α2,6-sialic acids are added during biosynthesis of N-glycosylated proteins, this modification is expected, at least in theory, to be retained for the lifetime of a protein targeted to the plasma membrane. However, exciting new evidence suggests a potential mechanism for inducing rapid loss of α2,6-sialic acid from receptors already translocated to the cell surface, which hints at a complexity in sialic acid signaling not previously appreciated. More specifically, Cha et al. (96) reported that the TRPV5 Ca2+ channel is retained at the cell surface through an interaction with extracellular Gal-1 and that α2,6-sialylation by ST6Gal-I can block this interaction, leading to receptor internalization. The seminal finding by this group is that cells secrete an α2,6-specific sialidase enzyme known as Klotho, which cleaves the α2,6-sialic acids from TRPV5 and restores galectin binding and galectin-mediated receptor retention. The Klotho enzyme appears to have a restricted specificity for TRPV5 and related ion channels, which prompts speculation regarding the possibility of other receptor-specific sialidases. The identification of a surface-acting α2,6 sialidase suggests a putative mechanism for directing rapid glycoform switching and an exquisite level of control over glycan/galectin interactions.
Conclusions and Future Directions
There is currently intensive interest in characterizing galectin structure and function, which is not surprising given the many important cell responses regulated by this class of lectins, as well as accumulating evidence implicating galectins in human disease. In contrast, there is a marked dearth of research centered on ST6Gal-I, despite the strong inhibitory effect of ST6Gal-I-mediated sialylation on glycan/galectin interactions. As with galectins, ST6Gal-I expression is dynamically regulated in many cell types, and thus, the degree of receptor α2,6-sialylation can change as a consequence of cell status or in response to microenvironmental cues. Accordingly, defining ST6Gal-I regulatory mechanisms and specific ST6Gal-I substrates will be necessary for a complete understanding of the physiologic function of galectins and may also have translational relevance. Recombinant galectins and galectin inhibitors are currently being developed for use in cancer (and other) treatments (97–100). However, there is a good likelihood that the elevated ST6Gal-I expression known to occur during carcinogenesis may alter the efficacy of interventions targeting galectin pathways. As an alternative (or possibly complementary) approach, it may be fruitful to directly target ST6Gal-I expression as a mechanism to modulate glycan/galectin associations. In sum, the emerging role for ST6Gal-I as one of the principal negative regulators of galectin-mediated events highlights the need for future studies aimed at defining molecular pathways regulating this critical glycosyltransferase.
Supplementary Material
Acknowledgment
We gratefully acknowledge Dr. Linda Baum (University of California, Los Angeles) for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA-84248 from NCI. This work was also supported by a grant-in-aid from the American Heart Association. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- siglec
- sialic acid-binding Ig-like lectin
- ST6Gal-I
- β-galactoside α2,6-sialyltransferase
- CRD
- carbohydrate recognition domain
- Gal
- galectin.
REFERENCES
- 1. Chen X., Varki A. (2010) ACS Chem. Biol. 5, 163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schauer R. (2009) Curr. Opin. Struct. Biol. 19, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varki A., Angata T. (2006) Glycobiology 16, 1R–27R [DOI] [PubMed] [Google Scholar]
- 4. Dall'Olio F. (2000) Glycoconj. J. 17, 669–676 [DOI] [PubMed] [Google Scholar]
- 5. Crocker P. R., Paulson J. C., Varki A. (2007) Nat. Rev. Immunol. 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 6. Yang R. Y., Rabinovich G. A., Liu F. T. (2008) Expert Rev. Mol. Med. 10, e17 [DOI] [PubMed] [Google Scholar]
- 7. Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 8. Hughes R. C. (1999) Biochim. Biophys. Acta 1473, 172–185 [DOI] [PubMed] [Google Scholar]
- 9. Elola M. T., Wolfenstein-Todel C., Troncoso M. F., Vasta G. R., Rabinovich G. A. (2007) Cell. Mol. Life Sci. 64, 1679–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He J., Baum L. G. (2006) Methods Enzymol. 417, 247–256 [DOI] [PubMed] [Google Scholar]
- 11. Liu F. T., Patterson R. J., Wang J. L. (2002) Biochim. Biophys. Acta 1572, 263–273 [DOI] [PubMed] [Google Scholar]
- 12. Liu F. T., Rabinovich G. A. (2005) Nat. Rev. Cancer 5, 29–41 [DOI] [PubMed] [Google Scholar]
- 13. Liu F. T., Rabinovich G. A. (2010) Ann. N.Y. Acad. Sci. 1183, 158–182 [DOI] [PubMed] [Google Scholar]
- 14. Nakahara S., Raz A. (2006) Methods Enzymol. 417, 273–289 [DOI] [PubMed] [Google Scholar]
- 15. Hsu D. K., Yang R. Y., Liu F. T. (2006) Methods Enzymol. 417, 256–273 [DOI] [PubMed] [Google Scholar]
- 16. Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Cell 129, 123–134 [DOI] [PubMed] [Google Scholar]
- 17. Grigorian A., Torossian S., Demetriou M. (2009) Immunol. Rev. 230, 232–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dennis J. W., Granovsky M., Warren C. E. (1999) Biochim. Biophys. Acta 1473, 21–34 [DOI] [PubMed] [Google Scholar]
- 19. Guo H. B., Randolph M., Pierce M. (2007) J. Biol. Chem. 282, 22150–22162 [DOI] [PubMed] [Google Scholar]
- 20. Garner O. B., Baum L. G. (2008) Biochem. Soc. Trans. 36, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabinovich G. A., Toscano M. A., Jackson S. S., Vasta G. R. (2007) Curr. Opin. Struct. Biol. 17, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leppänen A., Stowell S., Blixt O., Cummings R. D. (2005) J. Biol. Chem. 280, 5549–5562 [DOI] [PubMed] [Google Scholar]
- 23. Stowell S. R., Arthur C. M., Mehta P., Slanina K. A., Blixt O., Leffler H., Smith D. F., Cummings R. D. (2008) J. Biol. Chem. 283, 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Melo F. H., Butera D., Medeiros R. S., Andrade L. N., Nonogaki S., Soares F. A., Alvarez R. A., Moura da Silva A. M., Chammas R. (2007) J. Histochem. Cytochem. 55, 1015–1026 [DOI] [PubMed] [Google Scholar]
- 25. Brewer C. F. (2004) Glycoconj. J. 19, 459–465 [DOI] [PubMed] [Google Scholar]
- 26. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Semel A. C., Seales E. C., Singhal A., Eklund E. A., Colley K. J., Bellis S. L. (2002) J. Biol. Chem. 277, 32830–32836 [DOI] [PubMed] [Google Scholar]
- 28. Bellis S. L. (2004) Biochim. Biophys. Acta 1663, 52–60 [DOI] [PubMed] [Google Scholar]
- 29. Seales E. C., Shaikh F. M., Woodard-Grice A. V., Aggarwal P., McBrayer A. C., Hennessy K. M., Bellis S. L. (2005) J. Biol. Chem. 280, 37610–37615 [DOI] [PubMed] [Google Scholar]
- 30. Woodard-Grice A. V., McBrayer A. C., Wakefield J. K., Zhuo Y., Bellis S. L. (2008) J. Biol. Chem. 283, 26364–26373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amano M., Galvan M., He J., Baum L. G. (2003) J. Biol. Chem. 278, 7469–7475 [DOI] [PubMed] [Google Scholar]
- 32. Kitazume S., Imamaki R., Ogawa K., Komi Y., Futakawa S., Kojima S., Hashimoto Y., Marth J. D., Paulson J. C., Taniguchi N. (2010) J. Biol. Chem. 285, 6515–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaneko Y., Nimmerjahn F., Ravetch J. V. (2006) Science 313, 670–673 [DOI] [PubMed] [Google Scholar]
- 34. Gagneux P., Cheriyan M., Hurtado-Ziola N., van der Linden E. C., Anderson D., McClure H., Varki A., Varki N. M. (2003) J. Biol. Chem. 278, 48245–48250 [DOI] [PubMed] [Google Scholar]
- 35. Kaech S. M., Hemby S., Kersh E., Ahmed R. (2002) Cell 111, 837–851 [DOI] [PubMed] [Google Scholar]
- 36. Comelli E. M., Sutton-Smith M., Yan Q., Amado M., Panico M., Gilmartin T., Whisenant T., Lanigan C. M., Head S. R., Goldberg D., Morris H. R., Dell A., Paulson J. C. (2006) J. Immunol. 177, 2431–2440 [DOI] [PubMed] [Google Scholar]
- 37. Jenner J., Kerst G., Handgretinger R., Müller I. (2006) Exp. Hematol. 34, 1212–1218 [DOI] [PubMed] [Google Scholar]
- 38. Videira P. A., Amado I. F., Crespo H. J., Algueró M. C., Dall'Olio F., Cabral M. G., Trindade H. (2008) Glycoconj. J. 25, 259–268 [DOI] [PubMed] [Google Scholar]
- 39. Taniguchi A., Higai K., Hasegawa Y., Utsumi K., Matsumoto K. (1998) FEBS Lett. 441, 191–194 [DOI] [PubMed] [Google Scholar]
- 40. Marino J. H., Tan C., Davis B., Han E. S., Hickey M., Naukam R., Taylor A., Miller K. S., Van De Wiele C. J., Teague T. K. (2008) Glycobiology 18, 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nasirikenari M., Segal B. H., Ostberg J. R., Urbasic A., Lau J. T. (2006) Blood 108, 3397–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nasirikenari M., Chandrasekaran E. V., Matta K. L., Segal B. H., Bogner P. N., Lugade A. A., Thanavala Y., Lee J. J., Lau J. T. (2010) J. Leukocyte Biol. 87, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crespo H. J., Cabral M. G., Teixeira A. V., Lau J. T., Trindade H., Videira P. A. (2009) Immunology 128, e621–e631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hennet T., Chui D., Paulson J. C., Marth J. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4504–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Collins B. E., Smith B. A., Bengtson P., Paulson J. C. (2006) Nat. Immunol. 7, 199–206 [DOI] [PubMed] [Google Scholar]
- 46. Ghosh S., Bandulet C., Nitschke L. (2006) Int. Immunol. 18, 603–611 [DOI] [PubMed] [Google Scholar]
- 47. Baum L. G., Derbin K., Perillo N. L., Wu T., Pang M., Uittenbogaart C. (1996) J. Biol. Chem. 271, 10793–10799 [DOI] [PubMed] [Google Scholar]
- 48. Perillo N. L., Uittenbogaart C. H., Nguyen J. T., Baum L. G. (1997) J. Exp. Med. 185, 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vespa G. N., Lewis L. A., Kozak K. R., Moran M., Nguyen J. T., Baum L. G., Miceli M. C. (1999) J. Immunol. 162, 799–806 [PubMed] [Google Scholar]
- 50. Stillman B. N., Hsu D. K., Pang M., Brewer C. F., Johnson P., Liu F. T., Baum L. G. (2006) J. Immunol. 176, 778–789 [DOI] [PubMed] [Google Scholar]
- 51. Fukumori T., Takenaka Y., Yoshii T., Kim H. R., Hogan V., Inohara H., Kagawa S., Raz A. (2003) Cancer Res. 63, 8302–8311 [PubMed] [Google Scholar]
- 52. Toscano M. A., Bianco G. A., Ilarregui J. M., Croci D. O., Correale J., Hernandez J. D., Zwirner N. W., Poirier F., Riley E. M., Baum L. G., Rabinovich G. A. (2007) Nat. Immunol. 8, 825–834 [DOI] [PubMed] [Google Scholar]
- 53. Rabinovich G. A., Daly G., Dreja H., Tailor H., Riera C. M., Hirabayashi J., Chernajovsky Y. (1999) J. Exp. Med. 190, 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song X., Xia B., Stowell S. R., Lasanajak Y., Smith D. F., Cummings R. D. (2009) Chem. Biol. 16, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suzuki O., Nozawa Y., Abe M. (2006) Int. J. Oncol. 28, 155–160 [PubMed] [Google Scholar]
- 56. Suzuki O., Abe M. (2008) Oncol. Rep. 19, 743–748 [PubMed] [Google Scholar]
- 57. Suzuki O., Nozawa Y., Kawaguchi T., Abe M. (1999) Pathol. Int. 49, 874–880 [DOI] [PubMed] [Google Scholar]
- 58. Dall'Olio F., Malagolini N., di Stefano G., Minni F., Marrano D., Serafini-Cessi F. (1989) Int. J. Cancer 44, 434–439 [DOI] [PubMed] [Google Scholar]
- 59. Sata T., Roth J., Zuber C., Stamm B., Heitz P. U. (1991) Am. J. Pathol. 139, 1435–1448 [PMC free article] [PubMed] [Google Scholar]
- 60. Gessner P., Riedl S., Quentmaier A., Kemmner W. (1993) Cancer Lett. 75, 143–149 [DOI] [PubMed] [Google Scholar]
- 61. Lise M., Belluco C., Perera S. P., Patel R., Thomas P., Ganguly A. (2000) Hybridoma 19, 281–286 [DOI] [PubMed] [Google Scholar]
- 62. Petretti T., Kemmner W., Schulze B., Schlag P. M. (2000) Gut 46, 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Recchi M. A., Harduin-Lepers A., Boilly-Marer Y., Verbert A., Delannoy P. (1998) Glycoconj. J. 15, 19–27 [DOI] [PubMed] [Google Scholar]
- 64. Wang P. H., Lee W. L., Juang C. M., Yang Y. H., Lo W. H., Lai C. R., Hsieh S. L., Yuan C. C. (2005) Gynecol. Oncol. 99, 631–639 [DOI] [PubMed] [Google Scholar]
- 65. Gretschel S., Haensch W., Schlag P. M., Kemmner W. (2003) Oncology 65, 139–145 [DOI] [PubMed] [Google Scholar]
- 66. Shah M. H., Telang S. D., Shah P. M., Patel P. S. (2008) Glycoconj. J. 25, 279–290 [DOI] [PubMed] [Google Scholar]
- 67. López-Morales D., Velázquez-Márquez N., Valenzuela O., Santos-López G., Reyes-Leyva J., Vallejo-Ruiz V. (2009) Invest. Clin. 50, 45–53 [PubMed] [Google Scholar]
- 68. Fukushima K., Hara-Kuge S., Seko A., Ikehara Y., Yamashita K. (1998) Cancer Res. 58, 4301–4306 [PubMed] [Google Scholar]
- 69. Mondal S., Chandra S., Mandal C. (2010) Leukocyte Res. 34, 463–470 [DOI] [PubMed] [Google Scholar]
- 70. Kaneko Y., Yamamoto H., Kersey D. S., Colley K. J., Leestma J. E., Moskal J. R. (1996) Acta Neuropathol. 91, 284–292 [DOI] [PubMed] [Google Scholar]
- 71. Seales E. C., Jurado G. A., Brunson B. A., Wakefield J. K., Frost A. R., Bellis S. L. (2005) Cancer Res. 65, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 72. Christie D. R., Shaikh F. M., Lucas J. A., 4th, Lucas J. A., 3rd, Bellis S. L. (2008) J. Ovarian Res. 1, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shaikh F. M., Seales E. C., Clem W. C., Hennessy K. M., Zhuo Y., Bellis S. L. (2008) Exp. Cell Res. 314, 2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Le Marer N., Stéhelin D. (1995) Glycobiology 5, 219–226 [DOI] [PubMed] [Google Scholar]
- 75. Lin S., Kemmner W., Grigull S., Schlag P. M. (2002) Exp. Cell Res. 276, 101–110 [DOI] [PubMed] [Google Scholar]
- 76. Zhu Y., Srivatana U., Ullah A., Gagneja H., Berenson C. S., Lance P. (2001) Biochim. Biophys. Acta 1536, 148–160 [DOI] [PubMed] [Google Scholar]
- 77. Hedlund M., Ng E., Varki A., Varki N. M. (2008) Cancer Res. 68, 388–394 [DOI] [PubMed] [Google Scholar]
- 78. Harvey B. E., Toth C. A., Wagner H. E., Steele G. D., Jr., Thomas P. (1992) Cancer Res. 52, 1775–1779 [PubMed] [Google Scholar]
- 79. Bresalier R. S., Rockwell R. W., Dahiya R., Duh Q. Y., Kim Y. S. (1990) Cancer Res. 50, 1299–1307 [PubMed] [Google Scholar]
- 80. Dalziel M., Dall'Olio F., Mungul A., Piller V., Piller F. (2004) Eur. J. Biochem. 271, 3623–3634 [DOI] [PubMed] [Google Scholar]
- 81. Sakaki M., Fukumori T., Fukawa T., Elsamman E., Shiirevnyamba A., Nakatsuji H., Kanayama H. O. (2010) J. Med. Invest. 57, 152–157 [DOI] [PubMed] [Google Scholar]
- 82. Prieto V. G., Mourad-Zeidan A. A., Melnikova V., Johnson M. M., Lopez A., Diwan A. H., Lazar A. J., Shen S. S., Zhang P. S., Reed J. A., Gershenwald J. E., Raz A., Bar-Eli M. (2006) Clin. Cancer Res. 12, 6709–6715 [DOI] [PubMed] [Google Scholar]
- 83. Zaia Povegliano L., Oshima C. T., de Oliveira Lima F., Andrade Scherholz P. L., Manoukian Forones N. (2011) J. Gastrointest. Cancer, in press [DOI] [PubMed] [Google Scholar]
- 84. Dall'Olio F., Chiricolo M., Lollini P., Lau J. T. (1995) Biochem. Biophys. Res. Commun. 211, 554–561 [DOI] [PubMed] [Google Scholar]
- 85. Zhuo Y., Chammas R., Bellis S. L. (2008) J. Biol. Chem. 283, 22177–22185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nangia-Makker P., Nakahara S., Hogan V., Raz A. (2007) J. Bioenerg. Biomembr. 39, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. André S., Sanchez-Ruderisch H., Nakagawa H., Buchholz M., Kopitz J., Forberich P., Kemmner W., Böck C., Deguchi K., Detjen K. M., Wiedenmann B., von Knebel Doeberitz M., Gress T. M., Nishimura S., Rosewicz S., Gabius H. J. (2007) FEBS J. 274, 3233–3256 [DOI] [PubMed] [Google Scholar]
- 88. Schutte M., Hruban R. H., Geradts J., Maynard R., Hilgers W., Rabindran S. K., Moskaluk C. A., Hahn S. A., Schwarte-Waldhoff I., Schmiegel W., Baylin S. B., Kern S. E., Herman J. G. (1997) Cancer Res. 57, 3126–3130 [PubMed] [Google Scholar]
- 89. Dalziel M., Huang R. Y., Dall'Olio F., Morris J. R., Taylor-Papadimitriou J., Lau J. T. (2001) Glycobiology 11, 407–412 [DOI] [PubMed] [Google Scholar]
- 90. Wuensch S. A., Huang R. Y., Ewing J., Liang X., Lau J. T. (2000) Glycobiology 10, 67–75 [DOI] [PubMed] [Google Scholar]
- 91. Taniguchi A., Hasegawa Y., Higai K., Matsumoto K. (2000) Glycobiology 10, 623–628 [DOI] [PubMed] [Google Scholar]
- 92. Aas-Eng D. A., Asheim H. C., Deggerdal A., Smeland E., Funderud S. (1995) Biochim. Biophys. Acta 1261, 166–169 [DOI] [PubMed] [Google Scholar]
- 93. Wen D. X., Svensson E. C., Paulson J. C. (1992) J. Biol. Chem. 267, 2512–2518 [PubMed] [Google Scholar]
- 94. Kitazume S., Tachida Y., Oka R., Shirotani K., Saido T. C., Hashimoto Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fenteany F. H., Colley K. J. (2005) J. Biol. Chem. 280, 5423–5429 [DOI] [PubMed] [Google Scholar]
- 96. Cha S. K., Ortega B., Kurosu H., Rosenblatt K. P., Kuro-O M., Huang C. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9805–9810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lin C. I., Whang E. E., Donner D. B., Jiang X., Price B. D., Carothers A. M., Delaine T., Leffler H., Nilsson U. J., Nose V., Moore F. D., Jr., Ruan D. T. (2009) Mol. Cancer Res. 7, 1655–1662 [DOI] [PubMed] [Google Scholar]
- 98. Glinsky V. V., Raz A. (2009) Carbohydr. Res. 344, 1788–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Salatino M., Croci D. O., Bianco G. A., Ilarregui J. M., Toscano M. A., Rabinovich G. A. (2008) Expert Opin. Biol. Ther. 8, 45–57 [DOI] [PubMed] [Google Scholar]
- 100. Thijssen V. L., Poirier F., Baum L. G., Griffioen A. W. (2007) Blood 110, 2819–2827 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.