Abstract
The antisense RNA, CopA, regulates the replication frequency of plasmid R1 through inhibition of RepA translation by rapid and specific binding to its target RNA (CopT). The stable CopA–CopT complex is characterized by a four-way junction structure and a side-by-side alignment of two long intramolecular helices. The significance of this structure for binding in vitro and control in vivo was tested by mutations in both CopA and CopT. High rates of stable complex formation in vitro and efficient inhibition in vivo required initial loop–loop complexes to be rapidly converted to extended interactions. These interactions involve asymmetric helix progression and melting of the upper stems of both RNAs to promote the formation of two intermolecular helices. Data presented here delineate the boundaries of these helices and emphasize the need for unimpeded helix propagation. This process is directional, i.e. one of the two intermolecular helices (B) must form first to allow formation of the other (B′). A binding pathway, characterized by a hierarchy of intermediates leading to an irreversible and inhibitory RNA–RNA complex, is proposed.
Keywords: antisense RNA/loop–loop complex/plasmid/replication control/RNA–RNA interaction
Introduction
Naturally occurring antisense RNAs in prokaryotes are generally short, highly structured and untranslated (Wagner and Simons, 1994; Zeiler and Simons, 1998). Many of these antisense RNAs have been identified as plasmid copy-number regulators and are extremely efficient inhibitors; efficiency of in vivo control correlates with high in vitro binding rates to their respective target RNAs. Several in vitro studies indicated that binding rates are maximal with association rate constants in the range of 106 M–1 s–1 (reviewed in Wagner and Simons, 1994; Zeiler and Simons, 1998), and that the reaction is determined by the on-rate (e.g. Nordström and Wagner, 1994). By contrast, antisense RNAs used in artificial gene silencing strategies generally display much lower binding rates, often ∼100-fold lower (e.g. Patzel and Sczakiel, 1998; E.G.H.Wagner, unpublished data). These results emphasize the important role of antisense and target RNA sequence and structure in promoting high pairing rates and high specificity of regulation. They also suggest that the structures of these RNAs have evolved to optimize their regulatory functions. Kinetic studies performed on antisense–target RNA pairs of plasmids ColE1 (Tomizawa, 1984), R1 (Persson et al., 1988), pMU720 (Siemering et al., 1994) and ColIb-P9 (Asano et al., 1998; Asano and Mizobuchi, 2000) demonstrated that the initial step of binding involves a loop–loop interaction mediated by Watson–Crick base pairs, the so-called kissing complex, which is subsequently converted into more stable complexes. Loop–loop interactions play diverse roles in various biological systems, e.g. in dimerization of HIV-1 genomic RNA (for a review see Paillart et al., 1996), in the interaction between human tRNALys and HIV-1 genomic RNA (Isel et al., 1993), and in the localization of Drosophila bicoid mRNA (Ferrandon et al., 1997). Furthermore, in phi29 RNA, a dimer was shown to be formed by a loop–loop interaction that constitutes a building block for the assembly of the hexamer that gears the DNA-translocating machinery of this bacteriophage (Chen et al., 2000). Loop–loop interactions also participate in the architecture of large RNAs (for reviews see Pyle and Green, 1995; Brion and Westhof, 1997). However, the specific properties of antisense–target RNA systems are distinct from these latter cases: rapid kissing complex formation is a prerequisite for proper control, but rapid conversion to sufficiently stable complexes is essential to obtain maximal inhibition rates. The precise mechanism of formation of stable and inhibitory RNA–RNA complexes is still not understood in detail. Hence, it is important to delineate steps in the binding pathway as well as to identify structural features that permit unimpeded progression to the inhibitory complex.
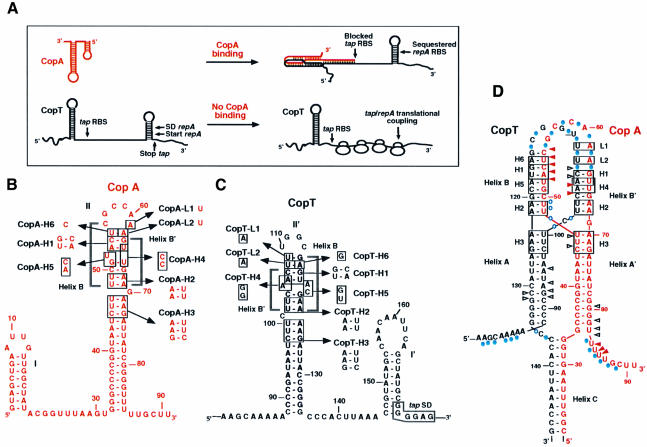
Replication of plasmid R1 is controlled at the translational level by binding of the antisense RNA (CopA) to its target site (CopT) in the leader region of repA mRNA (Nordström et al., 1984). Synthesis of the replication initiator protein RepA requires translation of a short leader peptide (tap), located upstream of repA. CopA binding prevents tap translation and thereby repA expression (Figure 1A; Blomberg et al., 1992, 1994; Malmgren et al., 1996). CopA and CopT are fully complementary, and both RNAs contain a major stem–loop structure as the main element responsible for high binding rate and control (Wagner and Nordström, 1986; Öhman and Wagner, 1989; Hjalt and Wagner, 1992, 1995). Binding initiates by an interaction between these complementary hairpin loops (Persson et al., 1990a,b). Structure mapping of the CopA–CopT complex revealed that the initial loop–loop interaction (kissing complex) is rapidly followed by more extended interactions, and that full duplex formation is a slow process in vitro (Malmgren et al., 1997; Kolb et al., 2000). The major product of this binding reaction is an extended kissing complex that is characterized by a four-helix junction whose formation involves extensive breakage of intramolecular base pairs and the formation of two intermolecular helices (Figure 1; Kolb et al., 2000). This bulky structure is further stabilized by the formation of a third long intermolecular helix involving the 5′ tail of CopA and the complementary region of CopT (Kolb et al., 2000). In the absence of the 5′-most 30 nucleotides of CopA (truncated RNA variant denoted CopI; Persson et al., 1990b; Malmgren et al., 1997), the stabilizer helix cannot form, yet the complex is inhibitory (Wagner et al., 1992; Malmgren et al., 1996).
Fig. 1. Schematic model for antisense RNA control of RepA synthesis (A) and secondary structures of the antisense RNA CopA (B), its target site CopT (C) and the stable CopA–CopT complex (D). (A) Binding of CopA prevents ribosome binding at the tap ribosomal binding site (RBS), and the presence of a stem–loop structure sequesters the repA RBS so that translation of RepA is inhibited. In the absence of CopA binding, translation of tap permits ribosome entry at the repA loading site (translational coupling). SD, Shine–Dalgarno sequence. (B–D) The mutated nucleotides (H1–H6, L1 and L2) are boxed. The secondary structure model for the stable CopA–CopT complex (D) was derived from chemical and enzymatic probing (Kolb et al., 2000). RNase V1 cleavages in CopA and CopT that occur in the CopA–CopT complex are shown by arrows: increased cleavages are shown by red arrows, unchanged cleavages are shown by black arrows; Pb2+-induced cleavages in the CopA–CopT complex are shown by blue circles.
Previous experiments had shown that bulged residues located in the stems of both RNAs were required for rapid binding in vitro and control in vivo (Hjalt and Wagner, 1995). We speculated that these bulged residues would destabilize the upper stem to facilitate inter-strand helix propagation. Therefore, we have defined here the requirements for inter-strand base pairing and its importance for formation of the functional inhibitory complex in vivo and in vitro. By mutational analysis, we have identified a new binding intermediate and have demonstrated that the two intermolecular helices B and B′ (Figure 1D) are not of equivalent importance. This leads us to propose a binding pathway characterized by a hierarchy of distinguishable steps, ultimately resulting in the formation of an irreversible, inhibitory antisense–target RNA complex.
Results
Conversion of the initial kissing complex to extended interactions is required for rapid pairing in vitro
Structure probing of the stable CopA–CopT complex indicated that the primary loop–loop interaction is converted to extended interactions: intra-strand base pairs are melted and two intermolecular helices, B and B′, are formed (Kolb et al., 2000). This extended kissing complex is further stabilized by the formation of an additional intermolecular helix C (shown schematically in Figure 1D). Base pair inversions were introduced at three positions within stem II of CopA and stem II′ of CopT to create mismatches upon the formation of heterologous CopA– CopT complexes (mutations H1, H2 and H3; Figure 1). These mismatches should affect the binding rate if the formation of the helical segments is important. The rate of stable complex formation between homologous or heterologous CopA–CopT pairs was analysed by gel shift assays using denaturing polyacrylamide–urea gel (Persson et al., 1988; Hjalt and Wagner, 1995). Second-order binding rate constants (kapp) were calculated according to Persson et al. (1988) and are summarized in Table I.
Table I. Binding rate constants of wild-type and mutant CopA–CopT pairs.
| CopA–CopT RNAs useda | kapp (M–1 s–1)b | Impairment relative to wt RNA pairc |
|---|---|---|
| wt–wt | 1.6 × 106 [± 43%] | 1.0× |
| H1–H1 | 4.0 × 105 [± 23%] | 4× |
| H1–wt | 2.3 × 104 [± 37%] | 69.5× |
| wt–H1 | 1.2 × 104 [± 0%] | 133.3× |
| H2–H2 | 2.2 × 105 [± 20%] | 7.2× |
| H2–wt | 3.3 × 104 [± 0%] | 48.5× |
| wt–H2 | 2.9 × 104 [± 0%] | 55.2× |
| H3–H3 | 3.8 × 105 [± 73%] | 4.2× |
| H3–wt | 8.7 × 105 [± 47%] | 1.8× |
| wt–H3 | 2.3 × 105 [± 0%] | 6.9× |
| H4–H4 | 2.5 × 105 [± 38%] | 6.4× |
| H4–wt | 5.9 × 105 [± 59%] | 2.7× |
| wt–H4 | 2.2 × 105 [± 47%] | 7.2× |
| H5–H5 | 6.5 × 105 [± 28%] | 2.5× |
| H5–wt | 7.0 × 104 [± 26%] | 22.8× |
| wt–H5 | 8.6 × 104 [± 27%] | 18.6× |
| H6–H6 | 1.1 × 106 [± 10%] | 1.5× |
| H6–wt | 9.0 × 104 [± 22%] | 17.8× |
| wt–H6 | 2.1 × 105 [± 26%] | 7.6× |
| L1–L1 | 1.4 × 106 [± 52%] | 1.1× |
| L1–wt | 2.1 × 106 [± 79%] | 0.8× |
| wt–L1 | 8.7 × 105 [± 16%] | 1.8× |
| L2–L2 | 2.0 × 106 [± 18%] | 0.8× |
| L2–wt | 1.2 × 106 [± 7%] | 1.3× |
| wt–L2 | 2.1 × 106 [± 43%] | 0.8× |
aThe RNAs produced in vitro are indicated as follows: wt, wild-type CopA or CopT; H1, H2, H3, H4, H5, H6, L1 and L2 mutated RNAs.
bkapp represents the rate constant of stable complex formation as determined from experiments such as in Figure 2. Values are averages of four independent experiments for the heterologous and homologous mutated RNA complexes, and of six for the wild-type pairs. Standard deviations are represented in brackets.
cThe relative impairment is the (wt/wt) kapp value divided by the kapp value for the respective pair tested.
The values obtained for the homologous pairs CopA-H1–CopT-H1 and CopA-H3–CopT-H3 were only 4-fold lower compared with the wild-type (wt) RNA pair, whereas CopA-H2–CopT-H2 was decreased by 7-fold (Table I). Thus, base pair inversions have only relatively minor effects on the rate of formation of stable complexes. In agreement with this, structure mapping of the three homologous complexes revealed a topology of the extended kissing complex similar to that of the wild-type complex (Kolb et al., 2000). By contrast, the heterologous CopA-H1–CopT-wt and CopA-H2–CopT-wt pairs were decreased by 70- and 49-fold, respectively, compared with the wild-type RNA pair. The reverse combinations showed similar decreases. Compared with the homologous mutated CopA-H2–CopT-H2 pairs, the effect of the H2 mutations on the rate constants in the heterologous complexes is reduced but still significant (∼10-fold). Finally, the corresponding value for CopA-H3–CopT-wt was identical to that of the wild-type pair, and its reverse was only 6-fold lower (Table I).
The results in Table I thus show that mismatches in upper stem segments (H1, H2) decreased binding rates, whereas a mismatch below the lower bulge (H3) had no effect. This indicates that inter-strand base pairing throughout the upper two stem regions is required for high binding rates. Restored binding rates of the homologous mutant H1 and H2 RNA pairs additionally suggests that base pairing per se, and not the nucleotide sequence in this region, is the critical determinant for the rate of complex formation. These data are congruent with recent chemical probing results performed on the heterologous and homologous complexes (Kolb et al., 2000).
Mutations that destabilize helix B decrease CopA–CopT complex formation rates
The previous data obtained on mutants H1–H3 cannot assess whether both intermolecular helices (B and B′; Figure 1) are equally important for stable complex formation. In order to test whether there was a preference in the directionality of helix progression subsequent to the first loop–loop interaction, the upper stems of CopA or CopT were mutated to alter only helix B′ (mutation H4) or helix B (H5, H6). Mutations H4 and H5 had also been designed so that the structures of both CopA and CopT were preserved. In addition, two point mutations were introduced: in the major hairpin loops of CopA (A61→U, CopA-L1) and CopT (U109→A, CopT-L1), and at the base pair that closed the hairpin loops of CopA (A62→U, CopA-L2) and CopT (U108→A, CopT-L2). These nucleotides were known to be unpaired in the stable CopA–CopT complex (Figure 1D). The kapp values of the homologous and heterologous wild-type and mutant CopA–CopT pairs are shown in Table I.
The binding rates of heterologous or homologous complexes carrying either mutations H4, L1 or L2 were similar to that of the wild-type pair (Table I). Only a 6-fold decrease in kapp was observed for the CopA-wt–CopT-H4 and CopA-H4–CopT-H4 pairs. This could be due to the formation of an additional G–C pair in the upper stem II′ of CopT-H4, resulting in increased stability of the stem and slight interference with helix propagation. By contrast, the binding rate constants of the heterologous CopA-H5–CopT-wt and CopA-H6–CopT-wt pairs were decreased by 23- and 18-fold, respectively, compared with the wild-type RNA pair or the homologous mutant pairs. A similar decrease in the kapp value was also obtained for the CopA-wt–CopT-H5 complex (18.6-fold), but only an 8-fold decrease was observed for the CopA-wt–CopT-H6 complex (Table I). Mutation H6 in CopT changed the intermolecular base pair U55(CopA)–A115(CopT) in helix B by a U(CopA)–G(CopT-H6) wobble pair, thus explaining the wild-type-like behaviour.
Thus, the effect of the H5 and H6 mutations indicates that inter-strand base pairing through helix B is required for high rates of formation of the stable CopA–CopT complex (which also contains helix C), whereas the formation of helix B′ (H4) does not appear to be essential. The silent phenotype of the L1 and L2 mutations in the in vitro binding assay also supports a directional mechanism for helix propagation.
Formation of helix B is a prerequisite for formation of helix B′
Pb2+-induced hydrolysis occurs preferentially at unpaired residues and is very sensitive to subtle RNA rearrangements. This probe can be used to obtain a signature of the extended kissing complex (Malmgren et al., 1997): strong cleavages occur in CopA at positions C56–A62 and U47–U49, and in CopT at U100–C101, C107–U110 and G112–C113. Furthermore, in the CopA–CopT complex, the formation of the intermolecular helix C, which greatly enhances complex stability, was characterized by strong protections in regions A8–G30 in CopA and A139–A164 in CopT (Kolb et al., 2000). Here, Pb2+ cleavages were used on homologous and heterologous complexes formed with the H4, H5, H6, L1 and L2 RNA variants (Figures 2 and 3).
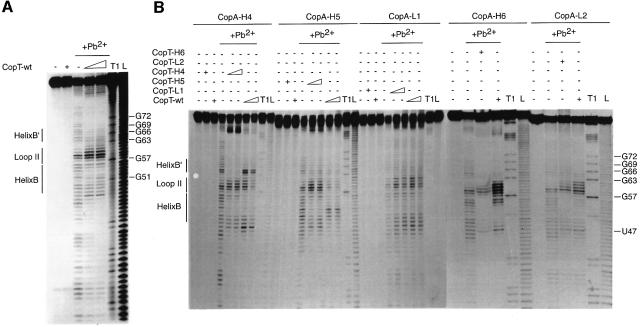
Fig. 2. Pb2+-induced hydrolysis of homologous or heterologous CopA–CopT complexes using end-labelled CopA species. Hydrolysis was performed on 5′-end-labelled wild-type CopA (A) or CopA-H4, CopA-H5, CopA-L1, CopA-H6 and CopA-L2 (B), alone (–) or in the presence of an excess of wild-type or mutant CopT (+). Complex formation was performed at 37°C for 15 min in TMN buffer. Lanes T1 and L, RNases T1 and alkaline ladders, respectively.
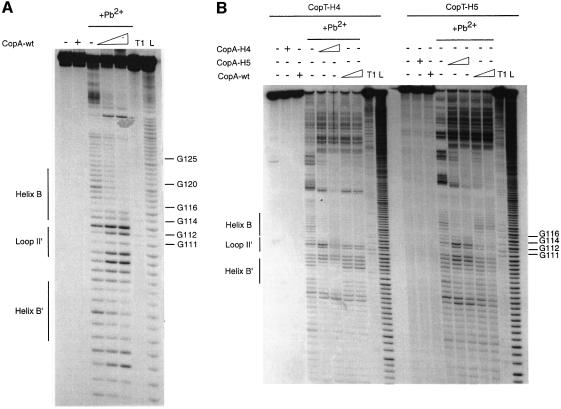
Fig. 3. Pb2+-induced hydrolysis of homologous or heterologous CopA–CopT complexes using end-labelled CopT species. Hydrolysis was performed on 5′-end-labelled wild-type CopT (A) or CopT-H4 and CopT-H5 (B), alone (–) or in the presence of an excess of wild-type or mutant CopA (+). Lanes T1 and L, RNases T1 and alkaline ladders, respectively.
Structure probing of end-labelled CopA and CopT RNAs indicated almost identical cleavage patterns for free mutant and wild-type RNAs (Figures 2 and 3). Hence, no major structural changes were caused by the mutations. The homologous complexes of CopA–CopT (H4, H5, H6, L1 and L2), and the heterologous CopA-L1–CopT-wt and CopA-L2–CopT-wt complexes and their reverse, showed characteristic wild-type-like cleavage patterns (Figures 2 and 3). The heterologous CopA-H4–CopT-wt complex was almost identical to the wild-type complex except that strong cleavages were induced at C64 and C65 of CopA-H4 (Figure 2B). This shows that several inter-strand base pairs in helix B′ were not formed due to the mismatches introduced by mutation H4, whereas—as in the homologous complexes—the intermolecular helices B and C were supported by strong protections in regions C50–U55 and A8–G30, respectively (Figure 2B). Similarly, Pb2+ cleavages at positions U100–U108 in CopT-H4 (Figure 3B) in the reverse complex, CopA-wt–CopT-H4, also supported the finding that helix B′ was not formed. The wild-type-like cleavage pattern, which was restored in complexes formed between the fully complementary H4 mutant RNAs, further argues for the presence of helix B′ (Figures 2B and 3B). Mutation H6 is expected to disrupt the second intermolecular base pair in helix B (Figure 1C). Indeed, structure mapping of CopA-H6–CopT-wt complexes revealed a new Pb2+ cleavage at position C56 and an increased cleavage at U55 (Figure 2B). These data suggest a destabilization/disruption of the two first intermolecular base pairs in helix B. Conversely, structure mapping of CopT-H6–CopA-wt complexes supported a characteristic wild-type-like cleavage pattern. Thus, replacement of the intermolecular base pair U55(CopA)– A115(CopT) by U–G had no significant effect on the four-way junction topology. A strikingly different result was obtained with mutation H5, designed to alter two intermolecular base pairs in helix B. Here, the Pb2+ cleavage pattern of CopA-H5, when in complex with CopT-wt, was significantly different from that of a homologous pair. The only significant protections now occurred at nucleotides A53–C59, whereas protections at U49–C52 were no longer observed (Figure 2B). In the reverse pair (CopA-wt–CopT-H5), significant protections were restricted to residues G112–U117 in CopT-H5 (Figure 3B). This indicates that neither helix B nor B′ could form properly.
Thus, the difference in effects of mutations H4, H5 and H6 in binding experiments (Table I) was paralleled by effects on complex structure. In particular, disruption of two Watson–Crick base pairs in helix B (H5) affected the conversion of the initial kissing complex to the four-way junction structure. Conversely, destabilization of helix B′ (H4) did not interfere with the formation of helices B and C. These data suggest that the formation of the intermolecular helix B′, but also of helix C, depends on prior formation of helix B. The fact that the L1 and L2 mutations did not show any effect on structure and on in vitro binding rate further supports a B→B′ hierarchy of helix formation.
Directionality of helix progression is supported by competition experiments
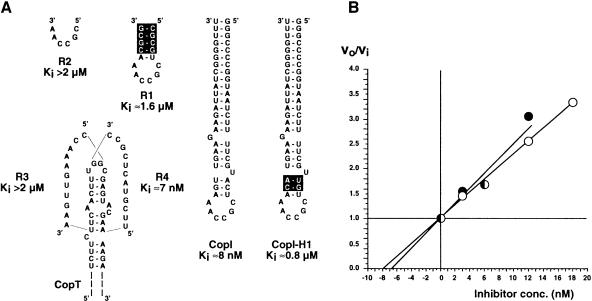
Competition assays can be used to obtain information about the rate of formation and stability of early intermediates in a binding pathway (Tomizawa, 1985; Asano et al., 1998). In the experiments summarized in Figure 4, standard binding rate measurements were performed in the presence or absence of competitor RNAs. Initial rates of stable CopA–CopT complex formation were determined over a range of competitor concentrations, and a Ki value was calculated from graphs of the ratio of initial rates, vo/vi, against the concentration of inhibitor (Tomizawa, 1985). As expected, CopI, unable to form a stable complex but capable of forming the extended kissing complex, inhibited stable complex formation with a Ki value of 8 nM (Figure 4). Two RNA fragments (R3 and R4) were designed to mimic the formation of helices B′ and B, respectively. The 12mer R4 was equally efficient as CopI (Ki = 7 nM), whereas the putative helix B′ former R3 was >>100-fold less effective, as was CopI-H1 (inverted upper stem). The two shorter RNAs, R2, only consisting of the CopA six base loop sequence, and R1, which carries the CopA loop closed by a GC-rich stem (not complementary to target), were both unable to compete at submicromolar concentrations. The high Ki values of R1 and R2 may indicate that the stability of a putative initial loop–loop interaction is low.
Fig. 4. Competitive inhibition between CopA and different RNA variants for the formation of the stable CopA–CopT complex. (A) The inhibitory RNAs used in this study and their corresponding Ki values are shown. Experimental conditions are described in detail in Materials and methods. Nucleotides that are not complementary to CopT are boxed. (B) Determination of the Ki values for CopI and RNA R4. The ratio vo/vi was calculated from the inhibition experiments using different concentrations of CopI (open circles) or of RNA R4 (closed circles) at time intervals between 1 and 6 min at 37°C, and are plotted against the inhibitor concentrations.
From these experiments it is evident that R4 RNA and wild-type CopI were the only two efficient inhibitors of stable complex formation (Figure 4), and that their respective Ki values were practically identical to equilibrium dissociation constants determined previously (Kd = 7.4 nM; Hjalt and Wagner, 1995). The inability of R3 to compete successfully with CopA for binding to CopT, in spite of an expected helix stability similar to R4, clearly indicates that the formation of helix B must precede that of helix B′ (and helix C) to form the stable complex.
Mutations affecting in vitro binding also affect in vivo control
Mutations that affect antisense–target RNA binding rates should have proportionate effects on control in vivo. To assay the effects of the H1–H5 and L1 mutations, we used translational fusions between the repA and lacZ genes, preceded by wild-type or mutant copA/copT control regions. All plasmids of the pGW177-III-L series (Blomberg et al., 1994) carry a copA promoter down-mutation to abolish CopA transcription and to permit supply of heterologous CopA from a second, compatible plasmid. RepA–LacZ fusion protein synthesis was measured in extracts of cells carrying appropriate combinations of plasmids. Assays of unrepressed repA–lacZ expression (no CopA in trans) indicated that neither of the tested mutations significantly affected target RNA stability or translatability [Table II: pGW177III-(H1–H5 or L1)-L].
Table II. β-galactosidase synthesis in cells carrying repA–lacZ translational fusion plasmids, alone or in the presence of CopA supplied in trans.
| Plasmidsa | CopA/CopT pairsb | Specific β-gal activityc | Relative β-gal activityd |
|---|---|---|---|
| pGEM2 | –/– | 9 ± 4 | <0.01 |
| pGW177-L | [wt/wt]e | 459 ± 101 | 0.16 |
| pGW177-III-L | –/wt | 2940 ± 992 | 1.00 |
| pGW177-H1-III-L | –/H1 | 3469 ± 198 | 1.18 |
| pGW177-H2-III-L | –/H2 | 3065 ± 406 | 1.04 |
| pGW177-H3-III-L | –/H3 | 3086 ± 278 | 1.05 |
| pGW177-H4-III-L | –/H4 | 3223 ± 765 | 1.10 |
| pGW177-H5-III-L | –/H5 | 3389 ± 885 | 1.15 |
| pGW177-L1-III-L | –/L1 | 3239 ± 641 | 1.10 |
| pGEM2-H1 + pGW177-H1-III-L | H1/H1 | 10 ± 2 | <0.01 |
| pGEM2-H2 + pGW177-H2-III-L | H2/H2 | 12 ± 3 | <0.01 |
| pGEM2-H3 + pGW177-H3-III-L | H3/H3 | 8 ± 0 | <0.01 |
| pGEM2-H4 + pGW177-H4-III-L | H4/H4 | 15 ± 5 | <0.01 |
| pGEM2-H5 + pGW177-H5-III-L | H5/H5 | 3 ± 2 | <0.01 |
| pGEM2-L1 + pGW177-L1-III-L | L1/L1 | 6 ± 2 | <0.01 |
| pGEM2-H1 + pGW177-III-L | H1/wt | 662 ± 300 | 0.23 |
| pGEM2-H2 + pGW177-III-L | H2/wt | 124 ± 62 | 0.04 |
| pGEM2-H3 + pGW177-III-L | H3/wt | 9 ± 2 | <0.01 |
| pGEM2-H4 + pGW177-III-L | H4/wt | 4 ± 1 | <0.01 |
| pGEM2-H5 + pGW177-III-L | H5/wt | 578 ± 24 | 0.20 |
| pGEM2-L1 + pGW177-III-L | L1/wt | 5 ± 1 | <0.01 |
| pGW643 + pGW177-III-L | wt/wt | 8 ± 3 | <0.01 |
| pGW643 + pGW177-H1-III-L | wt/H1 | 1163 ± 411 | 0.40 |
| pGW643 + pGW177-H2-III-L | wt/H2 | 216 ± 69 | 0.07 |
| pGW643 + pGW177-H3-III-L | wt/H3 | 11 ± 6 | <0.01 |
| pGW643 + pGW177-H4-III-L | wt/H4 | 16 ± 2 | <0.01 |
| pGW643 + pGW177-H5-III-L | wt/H5 | 911 ± 342 | 0.31 |
| pGW643 + pGW177-L1-III-L | wt/L1 | 23 ± 5 | <0.01 |
aFusion plasmids and CopA-donor plasmids: pGW63 carries the copA gene in vector plasmid derivative from pSP64 (Persson et al., 1990); pGW177-L plasmid carries the control region of plasmid R1 (Blomberg et al., 1992); pGW177-III-L derivative of pGW177-L contains the mutation III in copA promoter (Blomberg et al., 1994). The mutated plasmids (pGW177-III-L and pGEM2 series) were constructed as described in Materials and methods.
bThe RNAs produced in vivo are indicated as follows: wt, wild-type CopA or CopT; H1, H2, H3, H4, H5 and L1 mutated RNAs; –, no RNA.
cβ-galactosidase (β-gal) activity assays were performed as in Materials and methods, and represent averages of four independent determinations. Values are given in Miller units (Miller, 1972). Standard deviations are indicated.
dMiller units were converted to relative activities. The activity obtained with pGW177-III-L was set to unity.
eCopA encoded from fusion plasmid (the CopA concentration is lower than when encoded in trans).
Wild-type CopA supplied from a high-copy-number plasmid in trans decreased RepA–LacZ synthesis to background levels (pGW177-III-L + pGW643; Table II), and all mutant CopAs were equally effective on their cognate mutant CopT targets. When heterologous antisense–target RNA combinations were assayed, the CopA-H1–CopT-wt, CopA-H2–CopT-wt and CopA-H5–CopT-wt pairs, and their reverse, were clearly less inhibitory than any of the homologous combinations (0.23/0.40 relative activities for H1/wt; 0.04/0.07 for H2/wt; 0.2/0.31 for H5/wt; Table II). By contrast, the CopA-H3–CopT-wt, CopA-H4–CopT-wt and CopA-L1–CopT-wt pairs, and their reverse, were as efficient in inhibition as any homologous pair. For comparison, note that cells carrying pGW177-L alone give a value of 0.14. This plasmid supplies CopA from its own, intact gene. When CopA is supplied in trans (pGEM2 plasmids; Table II), it is present at ∼20-fold higher concentration (due to a higher copy number of carrier plasmid and lack of convergent transcription; Stougaard et al., 1982 and data not shown). This emphasizes that, for example, CopA-H1 is >20 times less active on a wild-type target [cf. pGW177-L and (pGEM2-H1 + pGW177-III-L); Table II]. The in vivo data correlate well with the in vitro results. All heterologous combinations of wild type and mutant that impaired inhibitory activity in vivo (H1, H2 and H5) also decreased in vitro binding rates (Tables I and II), whereas mutations H3, H4 and L1 induced no significant effect on stable complex formation in vitro and no phenotype in vivo. It has to be noted that no significant effect in vivo was observed for mutations that induced less than a 7-fold decrease in the binding rate constant.
In conclusion, only base pair inversions located above the lower bulged nucleotide in both RNAs (H1 and H2) were important for in vivo inhibition efficiency. The data also indicated that formation of helix B (H5), but not helix B′ (H4), is necessary and sufficient to initiate the inhibition of repA translation. We infer that the H1, H2 and H5 mutations impair CopA’s inhibitory capacity by impeding the rapid progression through the early intermediates to the stable CopA–CopT complex.
Discussion
Rapid binding between antisense and target RNAs is a prerequisite for efficient control. This is particularly important for plasmids regulating their copy numbers, since antisense RNAs are the principal regulators that have to accomplish inhibition of their target RNAs within a very short time frame. The structural features that favour the rapid formation of inhibitory complexes are not well understood, and the complexity of binding pathways makes general conclusions difficult. Nevertheless, in several systems, the initial recognition involves formation of a reversible loop–loop interaction (kissing complex).
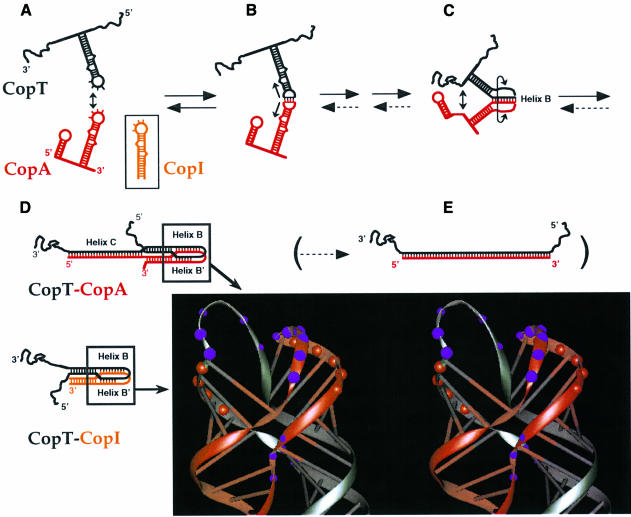
Based on the previous and present data, a tentative binding pathway between CopA (or CopI) and CopT is shown in Figure 5. The initial kissing complex occurs between a subset of loop bases in both RNAs (Figure 5B), and is defined by the location of copy number mutations (5′-GGCG in CopT; e.g. Givskov and Molin, 1984). This interaction is fully reversible and most likely very unstable, as indicated by the inability of loop-only sequences to compete for CopA binding to CopT (R1 and R2; Figure 4). The results presented show that subsequent helix progression proceeds unidirectionally into the upper stem regions. In support of this, changes at residues located at the 3′ side of the CopA loop (L1 and L2) had no effect on antisense efficiency in vivo and in vitro. Furthermore, only the mutations that altered helix B (H5 and H6; Figure 1) but not helix B′ (H4) significantly affected the binding rate (Table I), the structure of heterologous CopA–CopT complexes (Figures 2 and 3) and control in vivo (Table II). Thus, these data suggest that helix B must be formed (structure C in Figure 5) to permit subsequent formation of helices B′ and C. Whether, in a wild-type context, one of the latter two helices (B′ and C) is formed earlier than the other, cannot be determined. Conversely, whether or not helix B′ can form, activity tests in vitro (Table I) and in vivo (Table II), as well as the structure of the complex (except for helix B′ itself; Figures 2 and 3), are wild-type like. Thus, the stable complex is only formed under conditions in which helix B is formed, whereas helix B′ is dispensable. In line with these data, the competition experiments in Figure 4 argue that helix B formation is an early step required to commit the interacting RNAs to stable complex formation, since the RNA fragment R4 gave a Ki value comparable to that obtained with the complete CopI stem–loop, whereas the R3 RNA (helix B′) was >100-fold less effective.
Fig. 5. The binding pathway of CopA–CopT. (A–E) refer to steps in the pathway as explained in the Discussion. The parentheses indicate that full duplex formation is slow and biologically irrelevant (step E). Dotted arrows denote very slow reactions. The structure model of the four-helix junction derived from computer modelling by Kolb et al. (2000) is shown. Red and purple circles represent the RNase V1 and Pb2+-induced cleavages.
The asymmetric strand migration of helix B requires stem disruption above the position of the lower bulge (Figure 1C), and frees the opposing side of the stem for the second intermolecular helix B′ to form. The experiments presented here do not address the reason for the observed directionality of the step from the first loop–loop contact to structure C, and subsequently D (Figure 5). However, an interesting possibility is suggested by the recent finding that CopT, like antisense or target loops in most antisense RNA systems, carries a putative U-turn (π-turn) structure motif (YUNR, Y = pyrimidine, R = purine; Asano et al., 1998; Franch et al., 1999). Such a motif has recently been proposed in the target loop of the rep mRNA of ColIb-P9 (Asano et al., 1998), and is also present in the CopT loop (5′-UUGGCG). In this motif, the sharp turn at the 3′-phosphate of the invariant U positions the bases downstream in a pre-formed A-helical conformation, stabilized by additional hydrogen bonds within the loop. This structural module, in the tRNA context, is crucial for rapid and specific decoding (Grosjean et al., 1998; Ashraf et al., 1999) and it has been suggested that its ubiquitous occurrence in antisense–target RNA systems might reflect the requirement for binding specificity and rate (Franch and Gerdes, 2000). In CopT, the bases immediately following the invariant U are the ones at which binding initiates (5′-GGCG; Givskov and Molin, 1984; Nordström et al., 1984). Extension of this early intermolecular helix (to give B) would not require a rearrangement of the U-turn structure in CopT loop; hence, the orientation of the U-turn might determine the preference for helix B over helix B′ formation.
Once helix B is formed, the interacting RNAs are committed to form the extended kissing complex shown as structure D in Figure 5. This four-helix junction structure is formed as either a stable CopA–CopT complex, or as the less stable CopI–CopT complex lacking helix C. Altogether, the results presented here support the model of the extended kissing complex, which was derived from computer modelling, structure probing data, site-directed mutagenesis and appropriate stereochemistry (Kolb et al., 2000; Figure 5D). This structure shows two long helical domains, each one generated by co-axial stacking of two helices (Figure 1). The base pairs that initiate binding in CopA and CopT are disrupted upon conversion to the extended kissing complex, and serve as connecting loops between helices B and B′. Since mismatches between CopA and CopT below the bulged residues in stems II/II′ (mutation H3) show no phenotype in vitro and in vivo (Tables I and II), this supports the view that intermolecular base pairing proceeds to, and is arrested at, the position of the lower bulged nucleotides (CopA:G69/CopT:C101) to form the four-helix junction (Figure 1C). Progression of the intermolecular helices B and B′ is arrested, probably due to topological stress. This four-way junction adopts an asymmetrical X-shaped conformation as the result of strand exchange (Figure 5). This folding and the constraints imposed by the connecting loops force a side- by-side alignment of the helical segments that, in turn, facilitates the formation of the stabilizer intermolecular helix C in CopA–CopT (Kolb et al., 2000). The stable CopA–CopT complex (Figure 5D) is sufficient to exert inhibition in vitro (Figure 1A; Malmgren et al., 1996) and, under these conditions, full duplexes are neither formed nor required (Malmgren et al., 1996, 1997). In conclusion, the pathway of binding suggested in Figure 5 represents a stepwise progression from free RNAs through a hierarchy of distinguishable intermediates to the inhibitory stable complex.
Analysis of binding kinetics in the CopA–CopT (Persson et al., 1990b) system had shown that overall rates of stable complex formation are very similar to the rate constants of the extended kissing complex formation, at ∼106 M–1 s–1. These data indicate that dissociation rarely occurs once the RNAs have encountered each other in a compatible orientation, and consequently, binding proceeds quasi-irreversibly. This process requires that subsequent steps, such as helix B formation, have to be unimpeded by topology. In accordance with this requirement, CopA and CopT carry upper stem bulges whose destabilizing effects are crucial for rapid binding and inhibition (Hjalt and Wagner, 1995). Thus, the characteristic high rates of stable CopA–CopT complex formation (Figure 5D) imply an almost instantaneous conversion of the reversible kissing complex (Figure 5B) to the extended kissing complex (four-way junction structure; Figure 5D).
Several other plasmids, belonging to incompatibility groups IncIα, B and Z, share a similar genetic organization. Their mechanism of regulation is different from that of plasmid R1; antisense RNAs primarily inhibit activator pseudoknot formation (e.g. Asano et al., 1991; Wilson et al., 1993). Analyses of pMU720 (IncB) and ColIb-P9 (IncIα) have indicated similarities to R1 in that: (i) the structural features of the antisense stem–loops are reminiscent of those of CopA (identical loop sequences, destabilized upper stems, single-stranded region 5′ of stem–loop); (ii) full duplex formation is not required for control; and (iii) a stable binding intermediate involves inter-strand pairing within the upper stems of antisense and target RNAs (Siemering et al., 1993, 1994; Asano et al., 1998). Based on enzymatic probing experiments, Asano and Mizobuchi (2000) recently reported on a late intermediate antisense–target RNA complex in ColIb-P9, of similar overall topology to CopA–CopT, but differing in the lengths of the intermolecular helices (analogous to B and B′). Asano and Mizobuchi (2000) have also postulated that the initial loop–loop propagates in both directions to promote formation of the four-helix junction. However, the results presented here indicate that the two helices in the CopA–CopT complex are formed in a hierarchical order. Whether or not these differences will stand further experimental tests, we suggest similar binding pathways for antisense and target RNAs of plasmids belonging to the incompatibility groups FII, B, Z and Iα.
A binding pathway has also been proposed for RNAI–RNAII of ColE1 (Tomizawa, 1984, 1990; Eguchi and Tomizawa, 1991). Binding of RNAI and RNAII is initiated by at least two loop–loop interactions followed by a series of reactions that progressively lead to the formation of a stable duplex (Tomizawa, 1990; Eguchi and Tomizawa, 1991). NMR studies were performed on a complex between two RNA hairpins carrying seven-membered complementary loops derived from a mutant pair of RNAI–RNAII (Marino et al., 1995; Lee and Crothers, 1998). These studies indicated that all seven loop bases were paired in the loop–loop helix, and continuous stacking of the loop nucleotides on the 3′ side of their respective stems was observed. Thus, both the R1 and ColE1 systems employ recognition loop sequences for the first reversible interaction, but, depending on their particular topological constraints, use radically different pathways to proceed rapidly to more stable intermediates and ultimately to stable inhibitory complexes. In spite of this, these as well as most other naturally occurring antisense systems show comparable binding rate constants.
In conclusion, this study shows that structural features in both CopA and CopT determine the topologically possible and kinetically favoured pathway. The pathway proposed is characterized by a hierarchy of intermediates that leads to a stable and functional RNA–RNA complex (Figure 5D). The results presented, together with previous studies on the effects of loop size and bulge mutations (Hjalt and Wagner, 1992, 1995), also emphasize the requirement for unimpeded helix progression immediately subsequent to initial kissing, in order to sustain the high binding rates characteristic of efficient plasmid copy-number regulators. One can speculate that such a step-wise mechanism for RNA–RNA recognition may also play an important regulatory role in the folding of large RNA molecules and in many other cellular functions involving RNA–RNA interaction.
Materials and methods
Bacterial strains and plasmids
The Escherichia coli strain DH5α [F– endA1, hsdR17, supE44, thi-1, recA1, gyrA96, relA1, Δ(argF-lacZYA)U169, Φ80lacZΔM15 (Hanahan, 1985)] was used for plasmid constructions and in vivo repA–lacZ expression assays. Plasmid pGW177-L carries the control region of plasmid R1 (Blomberg et al., 1992). Plasmids of the pGW177-III-L series used for β-galactosidase measurements are derivatives of pGW177-L (Blomberg et al., 1994). Furthermore, these plasmids carry the mutation III [promoter-down mutation in copA, silent in tap (Öhman and Wagner, 1991)], and thus CopA is not transcribed. To create plasmid pGW177-H1-III-L, two PCRs were performed, using primers HE27-bio/GW110 and GW109-bio/HE40, respectively, on pGW58-III (Blomberg et al., 1994) template DNA. Each biotinylated PCR fragment was immobilized on streptavidin-coated Dynabeads® M (Dynal, Norway), and the complementary strands were separated by denaturation. The biotinylated (+) strand of the HE27-bio/GW110 fragment and the non-biotinylated (–) strand of the GW109-bio/HE40 fragment were annealed, followed by fill-in using the Klenow enzyme in the presence of 100 µM dNTPs. A further PCR using the external primers HE27 and HE40 was performed to obtain a fragment containing the desired mutation. This fragment was cleaved with the restriction enzymes BglII and SalI and ligated to the larger cleavage product of plasmid pGW177-L, cleaved with the same enzymes. pGW177-H2-III-L and pGW177-H3-III-L were constructed using the same protocol, except that the primer combinations differed: HE27-bio/GW112 and GW111-bio/HE40 (mutation H2); HE27-bio/GW114 and GW113-bio/HE40 (mutation H3). Construction of pGW177-H4-III-L, pGW177-H5-III-L, pGW177-H6-III-L, pGW177-L1-III-L and pGW177-L2-III-L was achieved using the Stratagene Quickchange™ Site-Directed Mutagenesis Kit with plasmid pGW58-III and primers H4-FW/H4-Rev, H5-FW/H5-Rev, H6-FW/H6-Rev, L1-FW/L1-Rev and L2-FW/L2-Rev, respectively. The mutated plasmids were digested with SalI and BglII, and fragments were inserted into pGW177-L cleaved with the same two enzymes. pGEM2 cloning vector was purchased from Promega. Plasmids pGEM2-H1, pGEM2-H2 and pGEM2-H3, carrying the mutant copA genes, were constructed by using the above-mentioned primer combinations on pGW58 template DNA (Blomberg et al., 1990) for PCR, yielding H1, H2 and H3 fragments, which were then amplified with primers HE27 and HE40, cleaved with SmaI and SalI, and ligated into SmaI–SalI-cleaved pGEM2 DNA. Plasmids pGEM2-H4, pGEM2-H5 and pGEM2-L1 were obtained using the Stratagene Quickchange™ Site-Directed Mutagenesis Kit with pGW58 plasmid and the same set of primers as described above. The mutated plasmids were digested with XmnI and SalI, and the generated fragments were ligated into SmaI–SalI-cleaved pGEM2 DNA. Purification of plasmid DNA, restriction enzyme cleavages and other DNA techniques were essentially according to Sambrook et al. (1989).
Oligodeoxyribonucleotides
Oligodeoxyribonucleotides were from Amersham Pharmacia Biotech, Interactiva Biotechnologie GmbH and NAPS Goettingen GmbH. For construction of plasmids containing mutations H1–H6, L1 and L2, the following primer pairs were used: for H1, HE27-bio (5′ biotin– GTGATCTTCCGTCACAGGTAT) and GW110 (5′-CGTTGTCGCCAACATGAAGAAGATT), GW109 (5′-CATGTTTGGCGACAACGA AAAGATTA) and HE40 (5′-GAATTTCGACCTCTAGACCAA); for H2, HE27-bio and GW112 (5′-TAACGTACTCGCCAAAGTTGTTGAAGATT), GW111-bio (5′ biotin–CAACAACTTTGGCGAGTACGTTAAGATTA) and HE40; for H3, HE27-bio and GW114 (5′-TGA ATTCGTACTCGCCAAAGTTGAAGTTCATTATCGG), GW113-bio (5′ biotin–TGAACTTCAACTTTGGCGAGTACGAATTCATTACCG) and HE40; for H4, H4-FW (5′-CCGATAATCTTCTTCGGCTTTGGCGAGTAC) and H4-Rev (5′-GTACTCGCCAAAGCCGAAGAAGATTATCGG); for H5, H5-FW (5′-TTCAACTTTGGCGAGTGTGAA AAGATTACCGGG) and H5-Rev (5′-CCCGGTAATCTTTTCACACTCGCCAAAGTTGAA); for H6, H6-FW (5′-CGATAATCTTCTTCAACTTTGGCGGGTACGAAAAGATTACCGG) and H6-Rev (5′-CCG GTAATCTTTTCGTACCCGCCAAAGTTGAAGAAGATTATCG); for L1, L1-FW (5′-TAATCTTCTTCAACTATGGCGAGTACGAAAA) and L1-Rev (5′-TTTTCGTACTCGCCATAGTTGAAGAAGATTA); for L2, L2-FW (5′-CGATAATCTTCTTCAACATTGGCGAGTACGAAAAG) and L2-Rev (5′-CTTTTCGTACTCGCCAATGTTGAAGAAGATTATCG).
For sequence determinations, PCR fragments were generated from the pGW177-III-L-series plasmids using primer pairs: GW58-bio (5′ biotin– CAGGCTCAGTTCGTTGAGAAAA) and GW59 (5′-CACCGCCTTTTCCATCAGTTT). Primer GW60 (5′-GGATTCGGGTTCTTTAC) was used for sequencing. For the pGEM2 plasmid series, inserts were amplified using primer GW120-bio (5′ biotin–CATACGATTTAGGTGACACTAT) and GW45 (5′-GAAATTAATACGACTCACTATA). GW45 was used as sequencing primer.
For in vitro RNA transcription, all CopT mutant PCR fragments were generated from the pGW58 plasmid using a 5′ primer that contains the appropriate mutations (e.g. CopT-H1: 5′-ACGTACTTTAAAGCAAAAACCATGTTTGGCGACAACGAAAAGATTACCG) and the 3′ primer (5′-CGCGGATCCCGGATTCGGGTTCTTTA). The resulting PCR fragments were cloned in the pUT7 vector under the control of T7 promoter (Serganov et al., 1997). The wild-type CopT PCR fragment was synthesized with primers T7-G3 (5′-GAAATTAATACGACTCACTATAGGGTTAAGGAATTTTGTGGCTGG; T7 promoter sequence underlined) and SeqP/II (5′-CGGATTCGGGTTCTTTA). Wild-type CopI template was synthesized with primers T7SI (5′-GAAATTAATACGACTCACTATAGGGCCCCGGTAATCTTTTCGT) and T7EI (5′-AAACCCCGATAATCTTCTTCA), and mutated CopI species were synthesized with 5′ primers containing the corresponding mutations. Finally, the wild-type and mutant (H1, H2, H4, H5, H6, L1 and L2) CopA templates were synthesized with primers T7SA (5′-GAAATTAATACGACTCACTATAGTAGCTGAATTGTTGGCTATACG) and T7EA (5′-AAAGCAAAAACCCCGATAATCTTC), and for CopA-H3 the 3′ primer was T7EA-short (5′-AAAGCAAAAACCCCGATAAT).
RNA preparation and labelling
Mutant CopT RNAs were synthesized with T7 RNA polymerase using BamHI-linearized pUT7 plasmids. Wild-type CopT and wild-type or mutant CopA and CopI were transcribed from PCR-generated DNA fragments (Hjalt and Wagner, 1992). Purification of RNAs was performed either by fast protein liquid chromatography (FPLC; Pharmacia) on a Bio-Sil TSK250 column or by polyacrylamide–urea gel electrophoresis as described previously (Kolb et al., 2000). Transcription of CopT yields a run-off product of 302 nucleotides, initiated with GG instead of the GU sequence of the wild-type repA mRNA. The CopA RNA contains a 5′ terminal G instead of an A residue. Neither of these nucleotide changes affect structure or binding properties (Kolb et al., 2000).
5′-end labelling of dephosphorylated RNA was performed for 30 min at 37°C with T7 polynucleotide kinase and [γ-32P]ATP (Sambrook et al., 1989), and 3′-end labelling of RNA with T4 RNA ligase and [32P]pCp (England and Uhlenbeck, 1978). For competition experiments, CopT RNA was uniformly labelled using [α-32P]UTP in the transcription reaction. Labelled RNAs were purified by polyacrylamide–urea gel electrophoresis, eluted, and precipitated twice with ethanol. Before use, unlabelled or labelled RNAs were dissolved in RNase-free water and renaturated by incubation at 90°C for 2 min, followed by slow cooling at 20°C in TMN buffer (20 mM Tris–acetate pH 7.5, 10 mM magnesium acetate, 100 mM sodium acetate).
Determination of rate constants of stable CopA–CopT complex formation and competitive inhibition with different CopA variants
Binding rate constants of CopA–CopT pairs were measured as described previously (Persson et al., 1988). RNAs R1–R4 used as competitors were purchased from Interactiva (Germany). Binding of 32P-labelled CopT to an excess of unlabelled CopA (1.5 nM) was performed at 37°C in the presence of different concentrations of inhibitors (R1, R2 and R3: from 15 to 1000 nM; R4 and CopI: as indicated in Figure 4; CopI-H1: from 3 to 1000 nM). Samples were withdrawn at various time points (1–6 min), added to gel application buffer and immediately loaded onto a denaturing 5% polyacrylamide–urea gel. The gel was run at constant voltage (500 V) for 3 h and subsequently dried. Bands corresponding to the CopA–CopT complex and free CopT, respectively, were quantified using a PhosphorImager (Molecular Dynamics). The inhibition constant Ki was determined according to Tomizawa (1985), using the equation vo/vi = 1 + io/Ki, where vo and vi are the initial rates of hybridization in the absence and in the presence of inhibitory RNA, and io is the initial concentration of the inhibitory RNA.
Pb2+-induced hydrolysis
Pb2+-induced hydrolysis was carried out on end-labelled CopA or CopT, free or in complex, as described by Kolb et al. (2000). Hydrolysis was conducted in 20 mM HEPES–NaOH pH 7.5, 10 mM magnesium acetate, 100 mM sodium acetate, in the presence of 8 or 16 mM Pb(OAc)2 for 5 min. Reactions were stopped by the addition of 50 mM EDTA, followed by ethanol precipitation. Identification of the cleavage sites was carried out according to Kolb et al. (2000).
In vivo repA–lacZ expression assays
RepA–LacZ fusion protein synthesis was measured in cell extracts from exponentially growing cultures. The protocol used for the experiments reported in Table II was essentially as in Berzal-Herranz et al. (1991), except that cultures were grown in M9 minimal medium supplied with 0.2% glucose, 0.2% casamino acids, ampicillin (100 µg/ml) and/or kanamycin (50 µg/ml), tryptophan (2 µg/ml) and thiamine (1 µg/ml), or in Luria–Bertani medium supplied with ampicillin (100 µg/ml) and/or kanamycin (50 µg/ml).
Acknowledgments
Acknowledgements
We thank Steve Lodmell, Hervé Moine and Christine Brunel for stimulating discussions and critical reading of the manuscript. F.A.K. was supported by a FEBS summer fellowship. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS), the Swedish Natural Science Research Council (NFR; E.G.H.W.) and the Swedish Research Council for Engineering Sciences (TFR; E.G.H.W.).
References
- Asano K. and Mizobuchi,K. (2000) Structural analysis of late intermediate complex formed between plasmid ColIb-P9 Inc RNA and its target RNA. J. Biol. Chem., 275, 1269–1274. [DOI] [PubMed] [Google Scholar]
- Asano K., Kato,A., Morikawi,H., Hama,C., Shiba,K. and Mizobuchi,K. (1991) Positive and negative regulations of plasmid ColIb-P9 repZ gene expression at the translational level. J. Biol. Chem., 266, 3774–3781. [PubMed] [Google Scholar]
- Asano K., Niimi,T., Yokoyama,S. and Mizobuchi,K. (1998) Structural basis for binding of the plasmid ColIb-P9 antisense Inc RNA to its target RNA with the 5′-rUUGGCG-3′ motif in the loop sequence. J. Biol. Chem., 273, 11826–11838. [DOI] [PubMed] [Google Scholar]
- Ashraf S.S., Ansari,G., Guenther,R., Sochacka,E., Malkiewicz,A. and Agris,P.F. (1999) The uridine in ‘U-turn’: Contributions to tRNA–ribosomal binding. RNA, 5, 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzal-Herranz A., Wagner,E.G.H. and Diaz-Orejas,R. (1991) Control of replication of plasmid R1: the intergenic region between copA and repA modulates the level of expression of repA. Mol. Microbiol., 5, 97–108. [PubMed] [Google Scholar]
- Blomberg P., Wagner,E.G.H. and Nordström,K. (1990) Control of replication of plasmid R1: the duplex between the antisense RNA, CopA and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J., 9, 2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P., Nordström,K. and Wagner,E.G.H. (1992) Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J., 11, 2675–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P., Engdahl,H.M., Malmgren,C., Romby,P. and Wagner,E.G.H. (1994) Replication control of plasmid R1: disruption of an inhibitory RNA structure that sequesters the repA ribosome binding site permits tap-independent RepA synthesis. Mol. Microbiol., 12, 49–60. [DOI] [PubMed] [Google Scholar]
- Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- Chen C., Sheng,S. and Guo,P. (2000) A dimer as a building block in assembling RNA. J. Biol. Chem., 275, 17510–17516. [DOI] [PubMed] [Google Scholar]
- Eguchi Y. and Tomizawa,J. (1991) Complexes formed by complementary RNA stem–loops. Their formations, structures and interaction with ColE1 Rom protein. J. Mol. Biol., 220, 831–842. [DOI] [PubMed] [Google Scholar]
- England T.E. and Uhlenbeck,O.C. (1978) 3′-terminal labeling of RNA with T4 RNA ligase. Nature, 275, 560–561. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Koch,I., Westhof,E. and Nüsslein-Volhard,C. (1997) RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR Staufen ribonucleoprotein particles. EMBO J., 16, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch T. and Gerdes,K. (2000) U-turns and regulatory RNAs. Curr. Opin. Microbiol., 3, 159–164. [DOI] [PubMed] [Google Scholar]
- Franch T., Petersen,M., Wagner,E.G.H., Jacobsen,J.P. and Gerdes,K. (1999) Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol., 294, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Givskov M. and Molin,S. (1984) Copy mutants of plasmid R1: effects of base pair substitutions in the copA gene on the replication control system. Mol. Gen. Genet., 194, 286–292. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Houssier,C., Romby,P. and Marquet,R. (1998) Modulatory role of modified nucleotides in RNA loop–loop interaction. In Grosjean,H. and Benne,B. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 113–133. [Google Scholar]
- Hanahan D. (1985) In Glover,D.M. (ed.), DNA Cloning I. IRL Press, Oxford, UK, pp. 109–135. [Google Scholar]
- Hjalt T. and Wagner,E.G.H. (1992) The effect of loop size in antisense and target RNAs on the efficiency of antisense RNA control. Nucleic Acids Res., 20, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalt T. and Wagner,E.G.H. (1995) Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res., 23, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isel C., Marquet,R., Keith,G., Ehresmann,C. and Ehresmann,B. (1993) Modified nucleotides of tRNA(3Lys) modulate primer/template loop–loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem., 268, 25269–25272. [PubMed] [Google Scholar]
- Kolb F.A., Malmgren,C., Westhof,E., Ehresmann,C., Ehresmann,B., Wagner,E.G.H. and Romby,P. (2000) An unusual structure formed by antisense–target RNA binding involves an extended kissing complex with a four-way junction and a side-by-side helical alignment. RNA, 6, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.J. and Crothers,D.M. (1998) The solution structure of an RNA loop–loop complex: the ColE1 inverted loop sequence. Structure, 6, 993–1005. [DOI] [PubMed] [Google Scholar]
- Malmgren C., Engdahl,H.M., Romby,P. and Wagner,E.G.H. (1996) An antisense/target RNA duplex or a strong intramolecular RNA structure 5′ of a translation initiation signal blocks ribosome binding: the case of plasmid R1. RNA, 2, 1022–1032. [PMC free article] [PubMed] [Google Scholar]
- Malmgren C., Wagner,E.G.H., Ehresmann,C., Ehresmann,B. and Romby,P. (1997) Antisense RNA control of plasmid R1 replication: the dominant product of the antisense RNA–mRNA binding is not a full RNA duplex. J. Biol. Chem., 272, 12508–12512. [DOI] [PubMed] [Google Scholar]
- Marino J.P., Gregorian,R.S., Csankovski,G. and Crothers,D.M. (1995) Bent helix formation between RNA hairpins with complementary loops. Science, 268, 1448–1454. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) In Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 352–355. [Google Scholar]
- Nordström K. and Wagner,E.G.H. (1994) Kinetics aspects of control of plasmid replication by antisense RNA. Trends Biochem. Sci., 19, 294–300. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin,S. and Light,J. (1984) Control of replication of bacterial plasmids: genetics, molecular biology and physiology of the plasmid R1 system. Plasmid, 12, 71–90. [DOI] [PubMed] [Google Scholar]
- Öhman M. and Wagner,E.G.H. (1989) Secondary structure analysis of the RepA mRNA leader transcript involved in control of plasmid R1. Nucleic Acids Res., 17, 2557–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhman M. and Wagner,E.G.H. (1991) Regulation of replication of plasmid R1: an analysis of the intergenic region between copA and repA. Mol. Gen. Genet., 230, 321–328. [DOI] [PubMed] [Google Scholar]
- Paillart J.C., Marquet,R., Skripkin,G., Ehresmann,C. and Ehresmann,B. (1996) Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie, 78, 639–653. [DOI] [PubMed] [Google Scholar]
- Patzel V. and Sczakiel,G. (1998) Theoretical design of antisense RNA structures substantially improves annealing kinetics and efficacy in human cells. Nature Biotechnol., 16, 64–68. [DOI] [PubMed] [Google Scholar]
- Persson C., Wagner,E.G.H. and Nordström,K. (1988) Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA and its target, CopT. EMBO J., 7, 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner,E.G.H. and Nordström,K. (1990a) Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J., 9, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner,E.G.H. and Nordström,K. (1990b) Control of replication of plasmid R1: formation of an initial transient complex is rate-limiting for antisense RNA–target RNA pairing. EMBO J., 9, 3777–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A.M. and Green,J.B. (1995) RNA folding. Curr. Opin. Struct. Biol., 5, 303–310. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Serganov A., Rak,A., Garber,M., Reinbolt,J., Ehresmann,B., Ehresmann,C., Grunberg-Manago,M. and Portier,C. (1997) Ribosomal protein S15 from Thermus thermophilus: Cloning, sequencing, overexpression of the gene and RNA-binding properties of the protein. Eur. J. Biochem., 246, 291–300. [DOI] [PubMed] [Google Scholar]
- Siemering KR., Praszkier,J. and Pittard,A.J. (1993) Interaction between the antisense and target RNAs involved in the regulation of IncB plasmid replication. J. Bacteriol., 175, 2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemering K.R., Praszkier,J. and Pittard,A.J. (1994) Mechanism of binding of the antisense and target RNAs involved in the regulation of IncB plasmid replication. J. Bacteriol., 176, 2677–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard P., Light,J. and Molin,S. (1982) Convergent transcription interferes with expression of the copy number control gene, CopA, from plasmid R1. EMBO J., 1, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. (1984) Control of ColE1 plasmid replication: The process of binding of RNA I to the primer transcript. Cell, 38, 861–870. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. (1985) Control of ColE1 plasmid replication: initial interaction of RNAI and the primer transcript is reversible. Cell, 40, 527–535. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. (1990) Control of ColE1 plasmid replication. Interaction of Rom protein with an unstable complex formed by RNA I and RNA II. J. Mol. Biol., 212, 695–708. [DOI] [PubMed] [Google Scholar]
- Wagner E.G.H. and Nordström,K. (1986) Structural analysis of an RNA molecule involved in replication control of plasmid R1. Nucleic Acids Res., 14, 2523–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.G.H. and Simons,R.W. (1994) Antisense RNA control in bacteria, phages and plasmids. Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]
- Wagner E.G.H., Blomberg,P. and Nordström,K. (1992) Replication control in plasmid R1: duplex formation between the antisense RNA, CopA and its target, CopT, is not required for inhibition of RepA synthesis. EMBO J., 11, 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.W., Praszkier,J. and Pittard,A.J. (1993) Mutations affecting pseudoknot control of the replication of B group plasmids. J. Bacteriol., 175, 6476–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler B.N. and Simons,R.W. (1998) Antisense RNA structure and function. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 437–464. [Google Scholar]