Abstract
The 2.2 Å crystal structure of a ternary complex formed by yeast arginyl-tRNA synthetase and its cognate tRNAArg in the presence of the l-arginine substrate highlights new atomic features used for specific substrate recognition. This first example of an active complex formed by a class Ia aminoacyl-tRNA synthetase and its natural cognate tRNA illustrates additional strategies used for specific tRNA selection. The enzyme specifically recognizes the D-loop and the anticodon of the tRNA, and the mutually induced fit produces a conformation of the anticodon loop never seen before. Moreover, the anticodon binding triggers conformational changes in the catalytic center of the protein. The comparison with the 2.9 Å structure of a binary complex formed by yeast arginyl-tRNA synthetase and tRNAArg reveals that l-arginine binding controls the correct positioning of the CCA end of the tRNAArg. Important structural changes induced by substrate binding are observed in the enzyme. Several key residues of the active site play multiple roles in the catalytic pathway and thus highlight the structural dynamics of the aminoacylation reaction.
Keywords: aminoacylation reaction/arginyl-tRNA synthetase/crystal structure/tRNA
Introduction
Aminoacyl-tRNA synthetases (aaRSs) constitute a family of RNA-binding proteins that are responsible for the correct translation of the genetic code by covalently linking the appropriate amino acid to the 3′ end of the correct tRNA. In most organisms, there are 20 distinct aaRSs, each one of them being responsible for aminoacylating its cognate tRNA(s) with a unique amino acid in a two-step catalytic reaction. The first step, which requires ATP and Mg2+ ions, leads to the formation of an enzyme-bound aminoacyl-adenylate and is followed by the transfer of the amino acid to the 3′ end of the tRNA to form an aminoacyl-tRNA. Due to their fundamental importance for cell life, the aaRSs are likely to be one of the most ancient families of enzymes and have therefore been analyzed extensively (Martinis et al., 1999). Determination of the crystal structures of several aaRSs, either in the free state or engaged in complexes with the other partners of the aminoacylation reaction, led to fundamental progress in understanding the structure–function relationship of this heterogeneous family of proteins. However, each new structure reveals unexpected results that illustrate the complexity of this biological process. Moreover, complete sequencing of several archaeal genomes has led to the discovery of novel pathways and enzymes for the synthesis of several aminoacyl-tRNAs (Ibba et al., 2000). Phylogenetic analysis of the 20 aaRSs has also revealed a complex evolutionary picture (Woese et al., 2000). In this context, new structures are essential to gain structural insight from sequence block alignments and therefore to decipher the relationships between function, evolution and sequences. For class I aaRSs, our present understanding of the second step of the aminoacylation reaction, which involves specific tRNA recognition, is still based essentially on the crystal structure of the GlnRS–tRNAGln complex (Rould et al., 1989).
According to sequence analysis and structural information, class I aaRSs can be subdivided into three subgroups. Arginyl-tRNA synthetase (ArgRS) and the five aaRSs specific for hydrophobic amino acids are gathered in subclass Ia. We present here the structure determination and the structural analysis of two complexes involving ArgRS from the yeast Saccharomyces cerevisiae (yArgRS) and one of its cognate isoacceptor tRNAs. The first complex, a ternary complex, which contains yArgRS, tRNAArg and l-arginine (l-Arg) bound to the active site, has been solved and refined at a resolution of 2.2 Å. The second one, a binary complex that only contains yArgRS and tRNAArg, has been solved and refined at 2.9 Å. This is the first example of a complex involving a class I aaRS from a eukaryotic organism. The crystal structure of yArgRS with l-Arg bound to the active site has already been described (Cavarelli et al., 1998).
Results and discussion
Structure determination
yArgRS, a monomeric class Ia aminoacyl-tRNA synthetase of 607 residues, was cloned, expressed and purified as described elsewhere (Cavarelli et al., 1998). In the S.cerevisiae genome, there are 19 genes that encode four different tRNAArg isoacceptors containing 75 or 76 nucleotides. Their D (dihydrouridine)-loop contains seven or eight nucleotides and is characterized by two structural features: (i) the nucleotide in the canonical position 17 is missing in all yeast tRNAArg isoacceptors; and (ii) an extra nucleotide (canonical numbering 20a) is inserted between nucleotides 20 and 21 in two tRNAArg isoacceptors. The second major tRNAArg isoacceptor, tRNAArgICG, where ICG (inosine, cytosine, guanosine) represents the three bases of the anticodon of the tRNA, contains 76 nucleotides and is characterized by eight modified nucleotides, one of them being inosine 34 (Ino34) in the anticodon loop, which allows the reading of three different codons. tRNAArgICG contains a 4 bp D-stem including a Cyt–Cyt mismatch and an eight nucleotide D-loop (see Figure 1A for details). Yeast tRNAArgICG was purified from counter-current fractions and contains all the modified bases as revealed by the crystal structure. The crystallization and preliminary X-ray crystallographic analysis of three different crystal forms of complexes between yArgRS and tRNAArgICG have already been published (Delagoutte et al., 2000).
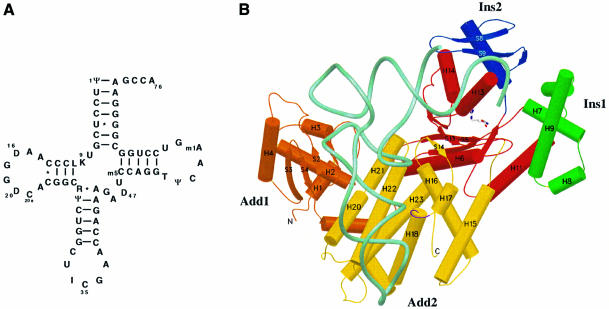
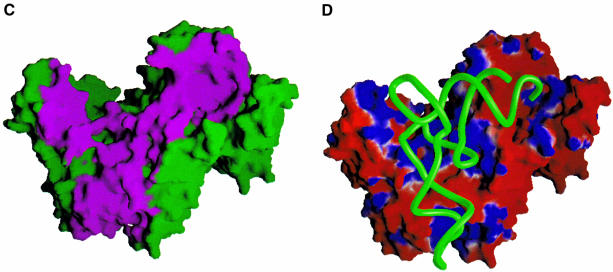
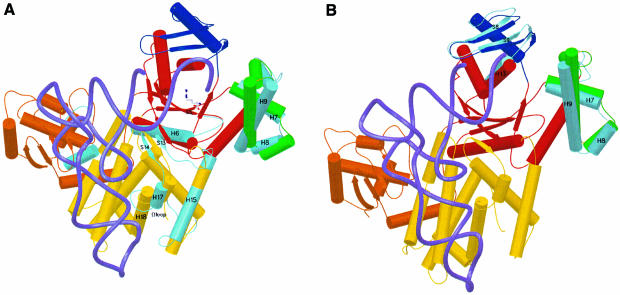
Fig. 1. Overview of yArgRS–tRNAArg interactions. (A) The cloverleaf structure of tRNAArgICG. The one-letter code is used for the nucleotides in all figures. The following code has been used for the modified bases: ψ, pseudouridine; D, dihydrouridine; I, inosine; K, 1-methylguanosine; L, N2-methylguanosine; R, N2,N2-dimethylguanosine; m5C, 5-methylcytidine; m1A, 1-methyladenosine; T, 5-methyluridine. (B) Overview of one monomer of yArgRS interacting with tRNAArgICG (drawn with SETOR; Evans, 1993) showing the modular architecture of yArgRS: Add1 (residues 1–143) is colored in orange; the catalytic domain in red (residues 143–194, 266–293 and 345–410); Ins1 in green (residues 194–266); Ins2 (residues 293–345) in blue; and Add2 (residues 410–607) in yellow. The tRNA backbone is drawn with its phosphate chain traced as a thick cyan line. Numbering of strands and helices is according to the structure of the ‘tRNA-free’ yArgRS (Cavarelli et al., 1998). The water molecules are not shown. (C) A schematic representation showing the footprint of the tRNAArg (in pink) on the surface of yArgRS (in green) (drawn with GRASP; Nicholls and Honig, 1991). (D) The molecular surface of yArgRS showing the electrostatic potential calculated with GRASP (Nicholls and Honig, 1991): negatively charged regions are in red and positively charged areas in blue. The orientation of the yArgRS molecule is similar in all three figures. The tRNA backbone is drawn with its phosphate chain traced as a thick green line.
The final model for the ternary complex (yArgRS, tRNA and l-Arg) has been refined at 2.2 Å resolution to a crystallographic R-factor of 19.0% (Rfree = 23.3%) with good stereochemistry (see Table I for statistics). The crystallographic asymmetric unit contains one molecule of yArgRS, the full tRNA molecule (76 nucleotides), the l-Arg substrate, 588 water molecules and one sulfate ion. The final model for the binary complex (yArgRS and tRNA) has been refined at 2.9 Å resolution to a crystallographic R-factor of 19.4% (Rfree = 24.4%) with good stereochemistry (see Table I for statistics). The crystallographic asymmetric unit contains one molecule of yArgRS, 73 nucleotides of the tRNA molecule and 44 water molecules. The CCA end of the tRNA molecule is not visible in the electron density map of the binary complex. In both structures, the first residue at the N-terminus of yArgRS is not visible in the electron density map. While all crystal forms grow in the presence of ATP, neither ATP nor AMP molecules are visible in the electron density maps. Packing effects cannot explain this absence (see ‘Functional implications’ below).
Table I. Statistics for crystallographic refinement.
| Data sets | Ternary complex | Binary complex |
|---|---|---|
| ArgRS–tRNA–l-Arg | ArgRS–tRNA | |
| Space group | P21212 | I222 |
| Unit cell parameters (a, b, c) (Å) | 129.6, 107.5, 71.4 | 107.7, 129.6, 184.0 |
| Refinement program | CNS | CNS |
| Resolution (Å) | 15–2.2 | 25–2.9 |
| No. of reflections | 51 111 | 28 856 |
| Completeness (%) | 99.8 (99.3)a | 100.0 (100.0)b |
| No. of non-hydrogen atoms: proteinc, tRNAd | 4892, 1629 | 4892, 1567 |
| No. of water molecules | 588 | 44 |
| R-factore, Rfree (%)f | 19.0, 23.3 | 19.4, 24.4 |
| Average B-factors (Å2): overall, protein, tRNA | 33.2, 32.9, 21.6 | 37.9, 40.9, 31.9 |
| Overall G-factorg | 0.41 | 0.35 |
| R.m.s. deviations from ideal geometryh | ||
| Bonds (Å), angles (°) | 0.0044, 1.0521 | 0.0061, 1.1395 |
| Ramachandran plot quality | ||
| Residues in core, allowed, generous, regions (%) | 90.8, 8.6, 0.2 | 89.2, 10.3, 0.2 |
| Coordinate errors from cross-validated Luzzati plots (Å) | 0.28 | 0.41 |
Values in parentheses are statistics for the highest resolution bin: a2.3–2.2 Å, b3.0–2.9 Å.
cThe first residue of the protein has not been located in both complexes.
dIn the binary complex, nucleotides 74, 75 and 76 are disordered.
e,fR = Σhkl|Fobs – Fcalc|/Σhkl Fobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes for reflection hkl, applied to the work (Rfactor) and test (Rfree) sets, respectively. The test set contains, respectively, 2558 reflections (5% of the data) for the ternary complex and 2875 reflections (10% of the data) for the binary complex.
gOverall ‘normality’ as calculated by Procheck (Laskowski et al., 1993).
hR.m.s. deviations were calculated using Engh and Huber parameters (Engh and Huber, 1991).
Overview of the ternary complex
The structure of yArgRS is built around a catalytic domain that contains the class I active site, to which four structurally defined domains are appended. Two of them, called additional domains 1 and 2 (Add1 and Add2), are attached respectively at the N- and C-terminal sides of the active site (Figure 1B). Two domains (Ins1 and Ins2) are inserted into the catalytic core. The α-helical C-terminal domain of yArgRS (Add2) is similar in topology to the C-terminal domains of MetRS [Escherichia coli and Thermus thermophilus (Mechulam et al., 1999)] and IleRS [Thermus thermophilus and Staphylococcus aureus (Nureki et al., 1998; Silvian et al., 1999)] and is therefore the most widespread domain in aaRSs, after the two catalytic domains, characteristic of each class. Add1, a two-layer α/β unit, has now also been found in two other RNA-binding proteins: Bacillus stearothermophilus ribosomal protein S4 (Davies et al., 1998; Markus et al., 1998) and the module N2 of E.coli threonyl-tRNA synthetase (Sankaranarayanan et al., 1999).
The enzyme and the tRNAArg form an extensive interface with a buried surface area of 3000 Å2. Add1 and Add2 of yArgRS cooperate for tRNAArg recognition, and the contact area can be divided schematically into three different parts (Figure 1B and C): (i) the first zone of interaction involves Add2 of the protein and the anticodon loop of the tRNA; (ii) the second zone involves the D-stem and D-loop of the tRNA and Add1 of the protein; and (iii) the third zone of contact involves the end of the acceptor stem and the terminal CCA interacting with the catalytic center of the protein. The distribution of the electrostatic potential on the solvent-accessible surface of yArgRS also displays three predominantly positive regions (Figure 1D) that correspond to the surface binding zone defined above. The overall tRNA binding mode is similar to that described in the GlnRS–tRNAGln complex in E.coli (Rould et al., 1989): (i) the variable loop of the tRNAArg faces the solvent; (ii) the catalytic center of the protein interacts with the minor groove of the acceptor stem of the tRNAArg; and (iii) the terminal CCA of the tRNA adopts a hairpin turn in order to reach the active site of the enzyme (Figure 1B). However, despite a similar conformation of the last two nucleotides (Cyt75 and Ade76) of tRNAArg and tRNAGln when bound to their respective synthetase, the stabilization of the CCA hairpin is achieved by a different molecular mechanism in tRNAArg compared with the tRNAGln (see below for more details).
Anticodon loop recognition
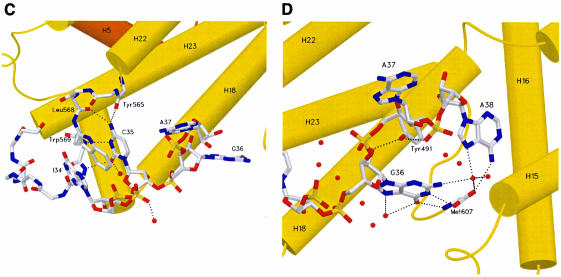
yArgRS approaches the tRNAArg from the minor groove side of the anticodon stem, and the anticodon loop binds in a pocket delimited by five helices of Add2 (H22, H15, H16, H17 and H18). The anticodon loop undergoes a dramatic structural change when compared with the expected canonical structure of a free tRNA, as found in yeast tRNAAsp or yeast tRNAPhe for example. The conformation of this loop is characterized by three structural features: (i) the formation of a bulge at the level of Ade38; (ii) the intercalation of Ade37 between the last base pair of the anticodon stem (Gua31–Cyt39) and nucleotide Cyt32 (see Figure 2A and B for details); and (iii) the splaying out of three bases (Uri33, Ino34 and Cyt35). yArgRS specifically recognizes Cyt35 and interacts with Gua36 and Ade38, but does not interact with Uri33. The complete catalog of typical protein–nucleic acid interactions is used by the enzyme for RNA recognition or binding: (i) exposed aromatic or aliphatic residues that are involved in van der Waals and hydrophobic interactions; (ii) positively charged residues that interact with the sugar–phosphate backbone; and (iii) polar side chains that are involved in direct or water-mediated interactions with the nucleic acid.
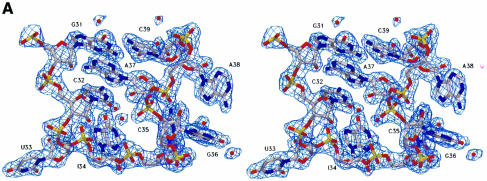
Fig. 2. Recognition of the anticodon loop of tRNAArgICG by yArgRS. (A) Stereo view of a final (2Fobs – Fcalc) cross-validated σA-weighted omit map, contoured at 1.5σ, showing the nucleotides of the anticodon loop (resolution limits 15–2.2 Å, all data used, calculated with CNS; Brünger et al., 1998). The protein residues are not shown for reasons of clarity. (B) Stereo view of the anticodon-binding site. yArgRS approaches the tRNAArg from the minor groove side of the anticodon stem, and the anticodon loop binds in a pocket delimited by five helices of Add2 (shown in yellow). The conformation of the anticodon loop is characterized by: (i) the formation of a bulge at the level of A38; (ii) the intercalation of A37 between the base pair (G31–C39) and nucleotide C32 and; (iii) the splaying out of three bases (U33, I34 and C35). (C) Recognition of the identity determinant C35 by yArgRS. C35, the strongest identity determinant for tRNAArg, is recognized mainly by main chain atoms of the protein belonging to the loop between helices H22 and H23 and by a stacking interaction with Trp569. (D) Interactions of Met607 with A38 and G36. Met607, the last residue of yArgRS, interacts, via its main chain atoms, with G36 and A38, and stabilizes the conformation of the anticodon loop, therefore explaining the strong evolutionary pressure on the C-terminal end of ArgRS. The side chain atoms of Met607 are not shown for reasons of clarity. Figures 2–5 were drawn with SETOR (Evans, 1993). The water molecules are shown as red spheres.
Cyt35, which has been shown to be the strongest identity determinant for tRNAArg (Giegé et al., 1998), is recognized mainly by main chain atoms of the protein belonging to the loop between helices H22 and H23 (Figure 2C). This is quite unusual for a strong determinant. This is reinforced by a stacking interaction, where Trp569 is intercalated between Ino34 and Cyt35. Several amino acids, e.g. Tyr491, Arg495, Arg501, Tyr565 and Met607, which are strictly conserved in all ArgRS sequences known to date (53 sequences, data not shown), govern the conformation of the anticodon loop. A typical example is Tyr491, which forms two hydrogen bonds from its OH group, one with the O2′ atom of Gua36 and the other with the O1P atom of Ade38 (Figure 2D).
Met607, the last residue of the protein, is a key player in the anticodon loop recognition and clearly illustrates the relationships between function, evolution, sequence and structure. Analysis of ArgRS sequences suggests a strong evolutionary pressure on the C-terminal side of the protein as all ArgRS sequences finish at the same residue for all species. Analyses of the sequences of proteins present in several databases show that few proteins present a similar sequence behavior (data not shown). The structure of the ternary complex provides a functional explanation for this strong sequence constraint, as the main chain atoms of Met607 interact with Gua36 and Ade38 and stabilize the conformation of the anticodon loop (Figure 2D).
A structural motif, which has been called the Ω loop (residues 480–485) (Cavarelli et al., 1998), forms part of the recognition interface. This loop, located just after the tRNA-anchoring platform (see below), joins helices H17 and H18 and creates a protruding motif at the surface of the protein. It is located on the major groove side of the anticodon stem and plays a dual functional role. It builds up the roof of the Ade38-binding pocket and stabilizes the conformation of the tRNA by closing the crevice formed on the major groove surface and delimited by the phosphate atoms of Gua25 and Cyt39. Thus, this Ω loop allows an intimate approach of the tRNA by the protein, which is required for the correct positioning of the anticodon. Residue Gly483 plays a crucial role in this Ω loop as any other side chain at this position would interfere with tRNA positioning. In vivo experiments have shown that a mutation of Gly483 to a serine is lethal for cell growth (Geslain et al., 2000), which gives support to the functional role of this residue as seen in the crystal structure.
Comparison of the tRNAArg anticodon loop with the structures of the anticodon loops of five different aaRS–tRNA complexes reveals different important structural changes upon interaction with the corresponding synthetase. Each aaRS induces a unique conformation of the cognate tRNA anticodon loop, illustrating once more the great flexibility and plasticity of single-stranded RNA molecules.
D-loop recognition
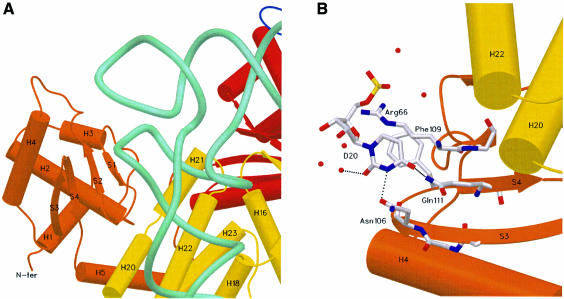
As predicted by model building (Cavarelli et al., 1998), Add1 of yArgRS is strongly involved in tRNA recognition. This structural module recognizes the D-loop region of the tRNA (Figure 3A). This is the first structural example of a tRNA synthetase complex where this loop is involved in synthetase recognition or binding. IleRS also relies on the recognition of the D-loop of the tRNAIle for its editing activity, but the mechanisms of interaction remain completely undefined (Nureki et al., 1998; Silvian et al., 1999). yArgRS recognizes the sugar backbone conformation and interacts specifically with the two nucleotides Dhu16 (Dhu, dihydrouridine) and Dhu20. Dhu16 binds in a pocket formed by strand S1 of Add1 and helices H21–H22 of Add2, while Dhu20 binds on the surface of the small four-stranded antiparallel β-sheet of Add1 and interacts mainly with the β-hairpin S3–S4.
Fig. 3. Interaction of the D-loop of the tRNAArg with yArgRS. (A) Overview. yArgRS recognizes the sugar backbone conformation and interacts specifically with nucleotides D16 and D20. D16 binds in a pocket formed by strand S1 of Add1 and helices H21–H22 of Add2, while D20 interacts mainly with β-hairpin S3–S4. The tRNA backbone is drawn with its phosphate chain traced as a thick light green line. (B) Recognition of D20 by yArgRS, illustrating the co-evolution of aaRS and tRNAs sequences. D20 is recognized mainly by Asn106, Phe109 and Gln111. Phe109, a highly conserved residue in ArgRS sequences, is involved in a stacking-type interaction with D20. The D20–Gln111 interaction is specific for the arginine system in S.cerevisiae. All other ArgRS–tRNAArg complexes from other species use an A20–Asn interaction at this position.
The D-loop of tRNAArg isoacceptors in all species usually has an extra nucleotide (canonical numbering 20a) inserted between nucleotides 20 and 21. However, this nucleotide is not present in tRNAArgUCU, the major tRNAArg isoacceptor in yeast. Our structure shows that this extra nucleotide does not interact with yArgRS and, furthermore, is not involved in the stabilization of the tRNA conformation. It should also be pointed out that the four yeast tRNAArg isoacceptors, like yeast tRNAAsp, do not contain any nucleotide at the canonical position 17 of the D-loop. A relationship between arginine and aspartate systems has already been highlighted in yeast, as yArgRS is able to mischarge the native tRNAAsp with low efficiency, and a transcript of tRNAAsp, deprived of modified bases, is only 30-fold less arginylated that the cognate tRNAArg (Sissler et al., 1996).
The recognition scheme of Dhu20 is another typical illustration of the relationships between function, structure, sequences and co-evolutions of aaRS and tRNAs. Dhu20 is recognized mainly by three residues of the protein: Asn106, Phe109 and Gln111 (Figure 3B). Phe109, a highly conserved residue in ArgRS sequences (it is sometimes replaced by a tyrosine residue), is involved in a stacking-type interaction with Dhu20. Dhu20 is specific to yeast tRNAArg; all other tRNAArg sequences have an adenine in position 20. Gln111 is a residue conserved only in S.cerevisiae ArgRS sequences; all other ArgRS sequences contain an asparagine residue (data not shown) at this position. In all ArgRS sequences, except from S.cerevisiae, position 106 is occupied by a small residue, which is a prerequisite to accommodate an adenosine nucleotide at position 20 of the tRNA. Any large residue at this position would interfere with the adenosine. From the Asn–Dhu20–Gln interaction found in the yeast system, one can easily model and build the Ade20–Asn interaction, which should be present in all other ArgRS–tRNA complexes from other species. Indeed, in vivo and in vitro genetic studies have shown that, while in S.cerevisiae specific arginylation of tRNAArg by ArgRS is strongly linked to the presence of Cyt35, Ade20 is also required in E.coli (Giegé et al., 1998).
One of the four yeast tRNAArg isoacceptors has a cytosine at position 20. Based on the recognition mode of Dhu20 found in the present structure, one can easily imagine how a cytosine can be recognized by Gln111 and Asn106. It only requires a flipping of the two side chain extremities to fulfill the hydrogen bond scheme completely.
Acceptor stem recognition
The binding of the amino acid acceptor stem of tRNAArg involves Ins2, the second half of the Rossmann fold and the so-called tRNA-anchoring platform. The active site of the protein interacts with the minor groove of the acceptor stem helix of the tRNAArg, mainly the N-terminal side of helix H14. The interactions with the first four base pairs of the helical acceptor stem, which are mainly water mediated, are localized on one side of the RNA helix and involve only one strand (Gua69–Ade72). The first nucleotide, Psu1 (pseudouridine), which carries the 5′ phosphate group, is engaged in two hydrogen bonds with Ade72. A different situation was found in the GlnRS–tRNAGln complex, where the first nucleotide was not visible in the electron density map, suggesting that this base pair is broken.
The tRNA-anchoring platform is a structural motif of yArgRS (Cavarelli et al., 1998), made of two strands [S13 (residues 402–406) and S14 (residues 468–473)]. Located after the second half of the Rossmann fold, this motif interacts with the inside L-corner of tRNAArg. It is involved in the anchoring of the tRNA molecule to the synthetase and was first visualized in the GlnRS–tRNAGln complex. At this level, the interface between the two macromolecules is highly hydrated and the protein interacts mainly with the sugar backbone atoms of the tRNA.
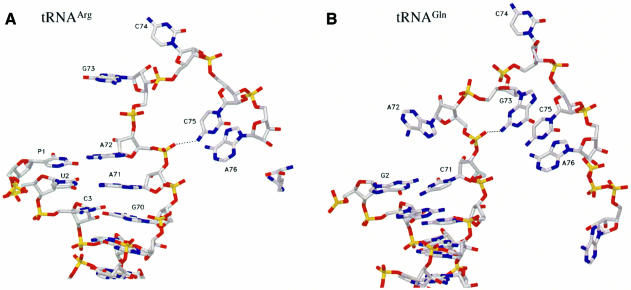
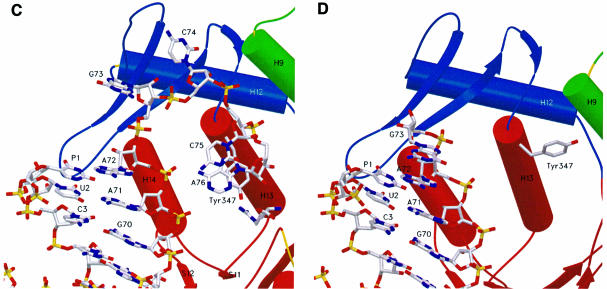
The terminal CCA adopts a hairpin conformation, reminiscent of that observed in the complex formed by GlnRS and tRNAGln. A comparison of the CCA end of tRNAArg and tRNAGln shows that only the last two bases (Cyt75 and Ade76) have a similar conformation. However, this conformation, which is required for the catalytic reaction, is stabilized by two different molecular mechanisms involving a different interaction within the tRNA. In tRNAGln, the Gua73 has been shown to be an important recognition element of this system. This nucleotide is involved in a stacking interaction with Cyt75 and Ade76 and also stabilizes the bending of the 3′-terminal CCA by a hydrogen bond involving its 2-amino group and the phosphate oxygen atom of nucleotide 72 (Figure 4B). In tRNAArg, the stacking interaction involving Gua73– Cyt75–Ade76 is not present, and the bending of the CCA is now stabilized by a hydrogen bond involving the 4-amino group of Cyt75 and the phosphate oxygen atom of nucleotide 72 (Figure 4A). The enzyme also stabilizes the hairpin structure of the CCA end by specific interactions with Cyt74 and Ade76.
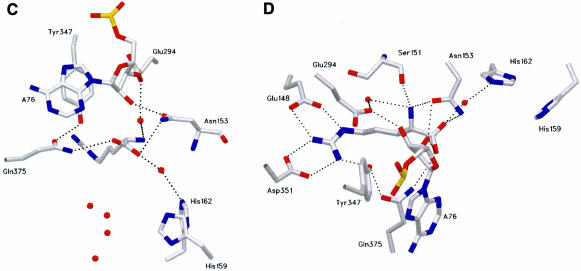
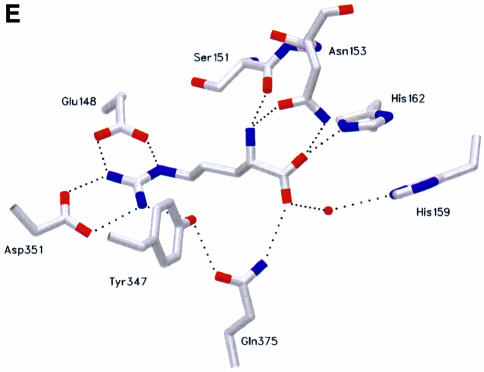
Fig. 4. Conformation of the acceptor arm of tRNAArg and l-Arg recognition. Comparison of the CCA hairpin conformation in tRNAArg and tRNAGln. The similar conformation found for the nucleotides C75 and A76 is stabilized by two different molecular mechanisms involving a different intramolecular interaction within the tRNA. (A) In tRNAArg, the bending of the 3′-terminal CCA is stabilized by a hydrogen bond involving the 4-amino group of C75 and the phosphate oxygen atom of residue 72. The water molecules are not shown. (B) In tRNAGln, nucleotide G73 stabilizes the bending by a hydrogen bond involving its 2-amino group and the phosphate oxygen atom of nucleotide 72, and is also involved in a stacking interaction with C75 and A76. l-Arg recognition. Comparison of the recognition mode of the l-Arg substrate (C and D) in the ternary complex with tRNAArg and (E) in the absence of the tRNAArg molecule. The two structures show a similar scheme of interactions for the guanidinium moiety, involving amino acids strictly conserved in all ArgRS sequences. The recognition of A76 in the ternary complex illustrates the role of Asn153, Glu294, Gln375 and Tyr347. The water molecules that occupy the putative AMP-binding site are shown as red spheres. tRNA binding produces structural changes of the conformation of the two histidines of the first signature motif characteristic of class I aaRSs; moreover, Asn153 and Tyr347 play a multiple role.
Active site of the ternary complex
The active site of yArgRS, which forms the scaffold of the Rossman fold, consists of two halves and binds all the substrates involved in the aminoacylation reaction. In the ternary complex, the l-Arg substrate binds at the C-terminal end of the β-strand in a crevice formed between the two symmetrical halves. This corresponds to the l-Arg-binding site, which was already described in the structure of yArgRS in the absence of the tRNAArg. In the ternary complex, the l-Arg substrate is located just below the last adenosine (Ade76) of the tRNA (Figure 4C). The correct positioning of Ade76 is controlled by three residues, strictly conserved in all ArgRS sequences: Glu294, Tyr347 and Asn153. The side chain atoms of Asn153 interact by a hydrogen bond with the 2′ OH group of Ade76, while the 3′ OH group is locked by a hydrogen bond with Glu294.
Comparison of the recognition mode of the l-Arg substrate in the ternary complex with tRNAArg (Figure 4D) and the absence of the tRNAArg molecule (‘tRNA-free’ yArgRS structure) (Figure 4E) shows a similar scheme of interactions for the guanidinium moiety, involving amino acids strictly conserved in all ArgRS sequences. However, Asn153, which interacts with the α-amino group and the α-carboxylate of the l-Arg molecule, now also interacts with the 2′ oxygen atom of the ribose of Ade76. Tyr347 deserves a special mention since this tyrosine is also strictly conserved in GlnRS, GluRS, TyrRS and TrpRS. Tyr347 cooperates in the recognition of the η-nitrogen atom of the l-Arg substrate, both in the ‘tRNA-free’ yArgRS and in the ternary complex. However, in the latter, it is also in contact with the adenine ring of Ade76 of the tRNA and continues the stacking interaction involving Ade76 and Cyt75.
Three different crystal forms corresponding to three different states of the arginylation reaction have been crystallized and solved for yArgRS (Cavarelli et al., 1998; Delagoutte et al., 2000): the first involves only yArgRS and the l-Arg molecule and the others involve a complex between yArgRS and tRNAArg, with and without the l-Arg substrate. It should be pointed out that all three crystal forms were grown from solutions containing the ATP molecule (Delagoutte et al., 2000). However, neither ATP nor AMP molecules are visible in the electron density maps. For the ‘tRNA-free’ yArgRS structure, this was explained by packing effects that lock the mobile ‘KMSKS’ loop in a non-productive conformation. Several results have indeed been obtained in other systems and have shown that this loop is involved in the stabilization of the first step of the aminoacylation reaction (First, 1998). In the tRNA-bound yArgRS structures, the putative ATP-binding site is accessible and no packing effects can be advocated for the lack of ATP in the active site (see below for discussion).
Water molecules
It is now well accepted that water molecules are extremely important in defining the interactions between biological molecules. The X-ray structures of several DNA-binding proteins complexed with their respective DNA targets have shown that water molecules contribute to the geometric complementarity between the interacting surfaces, as well as chemical complementarity through water-mediated polar interactions (Nadassy et al., 1999). Despite the fact that few RNA–protein complexes are known at high resolution, extensive work has been done on RNA molecules and has shown an extensive hydration of grooves in RNA, compared with that observed in DNA, and a specific hydration pattern correlated with the presence of the 2′ OH group of the ribose (Auffinger and Westhof, 1998; Draper, 1999).
The 2.2 Å electron density map of the ternary complex allows the identification of 588 water molecules, and ∼42% of them make at least one hydrogen bond with an atom of the tRNA molecule and, therefore, either mediate the protein–RNA interactions or stabilize the RNA conformation. However, two schemes of interactions are found. Interactions with the three important recognition signals of the tRNAArg (the nucleotides of the anticodon loop, the Dhu20 of the D-loop and the 3′-terminal CCA) are mainly direct protein–RNA interactions, while the binding of the amino acid acceptor stem is achieved mainly by water-mediated interactions. These different schemes of interactions may be correlated with the high variability in the sequence of the amino acid acceptor end of the four tRNAArg isoacceptors in S.cerevisiae. One may therefore hypothesize that the water-mediated interactions confer a high adaptability to the interface while providing the required specificity and affinity. A similar situation has also been found in the tRNAAsp–AspRS complex in E.coli (Eiler et al., 1999).
Structural changes upon tRNA binding
Comparison of the ‘tRNA-free’ yArgRS structure (yArgRS; l-Arg) with the structure of the ternary complex involving yArgRS, tRNAArg and the l-Arg substrate reveals structural changes due to tRNA binding. Superposition of these two yArgRS structures leads to an r.m.s. deviation between corresponding Cα atoms of 3.6 Å. Conformational changes are located mainly on one side of the enzyme and involve four regions (Figure 5A): the N-terminal helix H1, the Ins1 module, the two catalytic motifs and two peptides of Add2. Structural changes found in helix H1 and in module Ins1 are due mainly to different crystal packing arrangements and may therefore not be biologically relevant.
Fig. 5. Structural changes on yArgRS upon substrate binding. The yArgRS backbone (in orange, red, green, heavy blue and yellow) corresponds to the structure found in the ternary complex. The tRNA backbone is drawn with its phosphate chain traced as a thick purple line. Superpositions were carried out by superimposing the entire protein. (A) Comparison of the structure of the ternary complex with the ‘tRNA-free’ structure of yArgRS shows the structural movements due to the tRNA binding. Structural elements colored in light blue correspond to the ‘tRNA-free’ yArgRS structure. Only large movements are displayed. The conformations of two peptides are particularly altered: the first goes from strand S13 to helix H15 and the second involves strand S14, helix H17 and the Ω loop. Structural changes of the conformation of helix H15 induce the modification of the structure of the two signature motifs characteristic of class I aaRSs; the ‘H159A160G161H162’ loop is located between strand S5 and helix H6, while the ‘M408S409T410R411’ loop is located between strand S13 and helix H15. (B) Comparison of the structure of the ternary complex with the binary complex shows the structural movements due to the l-Arg binding. Structural elements colored in light blue correspond to the conformation found in the binary complex. Conformational changes are located mainly in the two insertion modules (Ins1 and Ins2) and helices H13 and H14 of the second moiety of the Rossmann fold. The overall conformation of the tRNA is the same; however, the absence of l-Arg substrate in the active site strongly affects the conformation of the CCA end (see below). Active site of yArgRS: (C) in the ternary complex and (D) in the binary complex, illustrating the molecular switch control by Tyr347 and l-Arg. In the absence of l-Arg substrate (D), G73 extends the helical conformation of the acceptor stem, and the last three nucleotides C74C75A76 are not visible in the electron density map and are therefore certainly disordered. The water molecules are not shown.
tRNA binding produces structural movements in the Add2 domain that are severe in the region around Met607, the last residue of the protein. The conformations of two peptides are particularly altered: the first one goes from strand S13 to helix H15 and the second one involves strand S14, helix H17 and the Ω loop. Helix H15 (residues 417–435) builds up one side of the pocket recognizing Gua36 and Ade38. Structural changes in the conformation of helix H15 found in the ternary complex induce the modification of the structure of the two signature motifs characteristic of class I aaRSs (H159A160G161H162 and M408S409T410R411 in yArgRS), which are close in space. Helix H15 is located in the sequence just after the ‘MSTR’ loop, and in space just below the ‘HAGH’ motif. The ‘MSTR’ loop flips from a ‘down’ conformation to an ‘up’ conformation in the ternary complex. There are no direct interactions between this loop and the tRNA except for one van der Waals interaction between the side chain atoms of Met404 and the ribose of Gua70, and one hydrogen bond between the ribose of Gua69 and Gln406. Moreover, it should be noted that, even at 2.2 Å, the electron density is not well defined from residues 409 to 413, reflecting a very mobile peptide even in the presence of the tRNA (see below for functional implications).
The conformational changes are also large at the N-terminal side of helix H6 (the ‘HAGH’ motif) and in the loop before it, producing a more open active site crevice. All these movements establish a direct link between the anticodon binding recognition and structural changes in the active site of the enzyme. Therefore, this region of the synthetase structure can be considered as the central knot that controls the communication between the catalytic platform and the anticodon recognition center. Any information related to the anticodon binding can be transferred directly to the active site. This may be related to the peculiar behavior of ArgRS, which requires its cognate tRNA for the first step of the aminoacylation reaction, the amino acid activation (see below for discussion). Full details of this analysis will be published elsewhere.
Structural changes upon l-Arg binding
Comparison of the two ‘tRNA-bound’ yArgRS complexes with and without the l-Arg substrate (ternary and binary complex, respectively) emphasizes the conformational changes due to the binding of the small l-Arg substrate. Superposition of these two yArgRS structures leads to an r.m.s. deviation between corresponding Cα atoms of 1.7 Å. Conformational changes are located mainly in the two insertion modules (Ins1 and Ins2) and the helices H13 and H14 of the second moiety of the Rossmann fold (Figure 5B). Structural changes found in module Ins1 are due mainly to different crystal packing arrangements and may therefore not be biologically relevant.
The overall conformation of the tRNA is the same; however, the absence of the l-Arg substrate in the active site strongly affects the conformation of the CCA end. In the binary complex, the last three nucleotides are not visible in the electron density map and are therefore certainly disordered. Moreover, in the absence of the l-Arg substrate, Gua73 is stacked on the first base pair of the tRNA, therefore extending the helical conformation of the acceptor stem (Figure 5D). Superposition of these two tRNA structures leads to an r.m.s. deviation between all corresponding atoms of 0.68 Å (nucleotides 1–72, excluding nucleotides 20a and 47 whose bases are not well ordered in both structures).
Our results thus show that l-Arg binding is a prerequisite that triggers the correct positioning of the CCA end of the tRNA. Important movements are found at the N-terminal side of helix H13 (residues 346–373). Tyr347, which is a residue strictly conserved in all ArgRS sequences and also in four other class I synthetases (GlnRS, GluRS, TyrRS and TrpRS), is a key player in this molecular switch. Tyr347 adopts two different conformations, which are only controlled by the binding of the l-Arg substrate, regardless of the presence/absence of the tRNA. When the l-Arg substrate is bound to the active site, Tyr347 interacts with the substrate as has been described above and adopts a ‘down’ conformation that stabilizes the conformation of the CCA end (Figure 5C). In the absence of l-Arg, Tyr347 adopts an ‘up’ conformation that is stabilized by a hydrogen bond with the carbonyl atom of Trp192, preventing the correct positioning of Ade76 (Figure 5D). Full details of this analysis will be published elsewhere.
Functional implications
As previously observed, analyses of the evolutionary profile of ArgRS have revealed a complex picture that violates the generally accepted canonical scheme (Woese et al., 2000). The crystal structure of the complexes presented here gives several clues for understanding the relationship between function, structures, sequences and evolution in this system. A full account of this structure-based sequence analysis will be published elsewhere.
We have already mentioned several recognition schemes used for the D-loop and anticodon binding, which may explain the results found from solution studies by others. The yArgRS complex is the first structural example in which the D-loop plays a crucial role in tRNA selectivity. It was not expected that two nucleotides, Dhu16 and Dhu20, were involved in this process in S.cerevisiae. In E.coli, it has been known for a long time that arginyl identity was strongly linked to the presence of Ade20 and Cyt35. The recognition mode of Dhu20 by yArgRS illustrates the co-evolution of synthetase and tRNA sequences and explains the unique scheme used by the yeast enzymes in contrast to all other species. This gives a simple explanation for the observed species-specific arginylation reaction: E.coli ArgRS cannot charge the transcript of yeast tRNAArg but is able to aminoacylate efficiently a mutant of yeast tRNAArg that carries an adenine in position 20 (Liu et al., 1999).
The original scheme used by yArgRS for anticodon binding explains the results obtained in solution with tRNAArg variants, where mutations were made in the anticodon loop (Sissler et al., 1997). The tight recognition of Cyt35 by main chain atoms of the protein reveals an elegant way to exclude any other nucleotide. The guanosine nucleotide in position 36 of the anticodon loop is sometimes replaced by an uridine in tRNAArg sequences, which is the only nucleotide that can fulfill a similar scheme of direct hydrogen bond interactions with Met607.
Among all tRNAs, only four have a cytosine in position 35 of the anticodon loop: tRNACys, tRNATrp, tRNAGly and tRNAArg. In these four tRNAs, Cyt35 has been shown to be an identity determinant. It seems reasonable to think that one position may not be enough to give sufficient selectivity. Involvement of the C-terminal residue of the protein in the recognition of Ade38 and Gua36 emphasizes the contribution of these two positions for the specificity of the reaction, as was already observed in genetic studies (Schulman and Pelka, 1989; Tamura et al., 1992; Sissler et al., 1996). It is also worth pointing out that in vivo selection of mutations lethal for cell growth identified several residues involved in tRNA binding, thus highlighting this method as a useful tool for functional analysis (Geslain et al., 2000).
As described above, the binding of tRNAArg triggers structural changes of the ‘MSTR’ loop, which may be required to build the adequate ATP-binding site. Based on the conformation of the ATP molecule found in the GlnRS–tRNAGln complex, which was crystallized in the presence of ATP, a model of the ATP molecule can be built in the yArgRS active site. This modeling shows that a part of the ‘MSTR’ loop (residues 409–413), which is not ordered in the 2.2 Å density map of the ternary complex, may interact with the β and γ phosphates of the ATP molecule. This is in agreement with the extensive solution studies published by others (First, 1998) that have also shown that the canonical ‘KMSKS’ loop of several class I aaRSs was involved in the stabilization of the first transition state of the aminoacylation reaction and recognizes mainly the β and γ phosphates of the ATP molecule.
As already mentioned, the ternary (yArgRS, tRNAArg and l-Arg) and the binary (yArgRS and tRNAArg) crystal forms correspond to co-crystals grown in the presence of ATP in the crystallization drops. However, neither ATP nor AMP molecules are visible in the electron density maps, and this experimental result cannot be explained by packing effects. Mass spectroscopy experiments on the crystal used for data collection of the ternary complex confirmed that neither ATP nor AMP was present in the crystal. Two main hypotheses may be formulated to explain the apparent low affinity of yArgRS for ATP in those crystal forms. The first postulates that the crystallization medium and, most importantly, the high concentration of ammonium sulfate may inhibit ATP binding. Thus, the picture observed in the ternary complex would correspond to a snapshot taken before the beginning of the aminoacylation reaction. However, the 2.2 Å electron density map of the ternary complex does not show any sulfate ions, which would explain the inhibition; the putative ATP-binding site is only occupied by water molecules. It should be pointed out that a high concentration of ammonium sulfate was used in several other crystal structures (Cavarelli et al., 1994; Poterszman et al., 1994; Belrhali et al., 1995) and that no inhibition of ATP binding was observed. The second hypothesis may be that the arginylation reaction took place in solution before the crystallization process, and therefore we may think that the ternary complex mimics an arginyl-tRNAArg just after deacylation. The possibility remains that the structure of the binary complex shows that at least in the absence of the l-Arg substrate, where such a scenario is excluded, yArgRS exhibits a low affinity for ATP.
A prolonged scientific debate was started more than 30 years ago involving the detailed mechanism of the aminoacylation reaction by ArgRS, GlnRS and GluRS (Mitra and Smith, 1969; Fersht et al., 1978). These enzymes do not catalyze the pyrophosphate exchange reaction in the absence of their cognate tRNA. Our results do not give a clear answer to this controversy but do show that (i) the tRNAArg produces conformational changes of the putative ATP-binding site and (ii) the l-Arg substrate controls the structure of the active conformation of the CCA end.
Crystallographic analysis of GlnRS–tRNAGln complexes (Rath et al., 1998) has shown that GlnRS has a low affinity for glutamine, while our results have shown that yArgRS has a low affinity for ATP, at least in the crystalline states obtained by us. In both cases, it seems impossible to freeze the natural aminoacyl-adenylate in the active site in the presence of the tRNA as was possible in other systems such as the AspRS–tRNAAsp complex (Cavarelli et al., 1994; Eiler et al., 1999). Analysis of the GlnRS (Rath et al., 1998) and yArgRS active sites shows that, while using a similar geometrical platform that governs the stereospecificity of the reaction, each enzyme uses an original protocol for transferring the aminoacid moiety to the 2′ OH group of the 3′-terminal adenosine. This shows again that aaRSs are very complex biological molecules and no single crystal structure can explain all aspects of their function.
Materials and methods
Crystallization and data collection
Gene expression and purification of yArgRS and tRNAArgICG followed protocols already published (Cavarelli et al., 1998; Delagoutte et al., 2000). The different crystal forms of yArgRS–tRNAArg complexes have been crystallized by the hanging drop vapor diffusion method in the presence of ammonium sulfate as previously described (Delagoutte et al., 2000). Crystals of the ternary complex (yArgRS, tRNA and l-Arg), which diffract beyond 2.2 Å resolution at the European Synchrotron Radiation Facility (ESRF) ID14-4 beam line, belong to the orthorhombic space group P21212, with unit cell parameters a = 129.6, b = 107.5, c = 71.4 Å. Crystals of the binary complex (yArgRS and tRNA) belong to the orthorhombic space group I222 with unit cell parameters a = 107.7, b = 129.6, c = 184.0 Å and diffracted beyond 2.9 Å resolution at the ESRF ID14-3 beam line. The two crystal forms were solved by the molecular replacement method using the coordinates of the free yArgRS (Delagoutte et al., 2000).
Structure determination and refinement
For the ternary complex, the refined model contains one yArgRS molecule of 606 residues, the full tRNA molecule (76 nucleotides), the l-Arg substrate, 588 water molecules and one sulfate ion. The crystallographic R-factor is 19.0% using all reflections between 15 and 2.2 Å with no σ cut-off (Rfree = 23.3%). For the binary complex, the current model contains one yArgRS molecule of 606 residues, 73 nucleotides of the tRNA molecule and 44 water molecules. The CCA end of the tRNA molecule is not visible in the electron density map for this second complex. The crystallographic R-factor is 19.4% using all reflections between 25 and 2.9 Å with no σ cut-off (Rfree = 24.4%). For both structures, the first residue at the N-terminus of yArgRS is not visible in the final electron density map. The models have been refined with the program CNS (Brünger et al., 1998), using the Engh and Huber stereochemical parameters (Engh and Huber, 1991). All rebuilding and graphics operations were done with O and related Uppsala programs (Kleywegt and Jones, 1996). All crystallographic calculations were carried out with the CCP4 package (CCP4, 1994). The stereochemistry of the models was inspected by Procheck (Laskowski et al., 1993) (see Table I for detailed analysis) and the quality of the refined structures was assessed using the Biotech validation suite for protein structures (Vriend, 1990; Wodak et al., 1995). Two residues in both structures, Lys131 and Ser150, are in a forbidden region of the Ramachandran plot. However, the electron density is of very high quality in this region and allows unambiguous building.
Acknowledgments
Acknowledgements
We thank Sean McSweeney and the staff of the ESRF beam line ID14-4, and Ed Mitchell and the staff of the ESRF beam line ID14-3, for use of their synchrotron instrumentation and help during data collection. We also thank Gilbert Eriani, Jean Gangloff and Gerard Keith for fruitful discussions, and Bernard Rees and Julie Thompson for careful reading of the manuscript. This work was supported by grants from the CNRS and by EEC contracts. The atomic coordinates and the structure factors have been deposited at the RCSB Protein Data Bank (PDB code 1F7U for the ternary complex and 1F7V for the binary complex).
References
- Auffinger P. and Westhof,E. (1998) Hydration of RNA base pairs. J. Biomol. Struct. Dyn., 16, 693–707. [DOI] [PubMed] [Google Scholar]
- Belrhali H., Yaremchuck,A., Tukalo,M., Berthet-Colominas,C., Rasmussen,B., Bosecke,P., Diat,O. and Cusack,S. (1995) The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure, 3, 341–352. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cavarelli J. et al. (1994) The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J., 13, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Delagoutte,B., Eriani,G., Gangloff,J. and Moras,D. (1998) l-Arginine recognition by yeast arginyl-tRNA synthetase. EMBO J., 17, 5438–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Davies C., Gerstner,R.B., Draper,D.E., Ramakrishnan,V. and White,S.W. (1998) The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J., 17, 4545–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagoutte B., Keith,G., Moras,D. and Cavarelli,J. (2000) Crystallization and preliminary X-ray crystallographic analysis of yeast arginyl-tRNA synthetase–yeast tRNAArg complexes. Acta Crystallogr. D, 56, 492–494. [DOI] [PubMed] [Google Scholar]
- Draper D.E. (1999) Themes in RNA–protein recognition. J. Mol. Biol., 293, 255–270. [DOI] [PubMed] [Google Scholar]
- Eiler S., Dock-Bregeon,A., Moulinier,L., Thierry,J. and Moras,D. (1999) Synthesis of aspartyl-tRNAAsp in Escherichia coli—a snapshot of the second step. EMBO J., 18, 6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Evans S.V. (1993) SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Fersht A.R., Gangloff,J. and Dirheimer,G. (1978) Reaction pathway and rate-determining step in the aminoacylation of tRNAArg catalyzed by the arginyl-tRNA synthetase from yeast. Biochemistry, 17, 3740–3746. [DOI] [PubMed] [Google Scholar]
- First E.A. (1998) Catalysis of tRNA aminoacylation by class I and class II aminoacyl-tRNA synthetases. In Sinnott,M. (ed.), Comprehensive Biological Catalysis. Academic Press, London, UK, pp. 573–607. [Google Scholar]
- Geslain R., Martin,F., Delagoutte,B., Cavarelli,J., Gangloff,J. and Eriani,G. (2000) In vivo selection of lethal mutations reveals two functional domains in arginyl-tRNA synthetase. RNA, 6, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M., Becker,H.D., Stathopoulos,C., Tumbula,D.L. and Soll,D. (2000) The adaptor hypothesis revisited. Trends Biochem. Sci., 25, 311–316. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1996) Efficient rebuilding of protein structures. Acta Crystallogr. D, 52, 829–832. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Liu W., Huang,Y., Eriani,G., Gangloff,J., Wang,E. and Wang,Y. (1999) A single base substitution in the variable pocket of yeast tRNAArg eliminates species-specific aminoacylation. Biochim. Biophys. Acta, 1473, 356–362. [DOI] [PubMed] [Google Scholar]
- Markus M.A., Gerstner,R.B., Draper,D.E. and Torchia,D.A. (1998) The solution structure of ribosomal protein S4 Δ41 reveals two subdomains and a positively charged surface that may interact with RNA. EMBO J., 17, 4559–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999) Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie, 81, 683–700. [DOI] [PubMed] [Google Scholar]
- Mechulam Y., Schmitt,E., Maveyraud,L., Zelwer,C., Nureki,O., Yokoyama,S., Konno,M. and Blanquet,S. (1999) Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol., 294, 1287–1297. [DOI] [PubMed] [Google Scholar]
- Mitra S.K. and Smith,C.J. (1969) Absolute requirement for transfer RNA in the activation of arginine by arginyl transfer RNA synthetase of yeast. Biochim. Biophys. Acta, 190, 222–224. [DOI] [PubMed] [Google Scholar]
- Nadassy K., Wodak,S.J. and Janin,J. (1999) Structural features of protein–nucleic acid recognition sites. Biochemistry, 38, 1999–2017. [DOI] [PubMed] [Google Scholar]
- Nicholls A. and Honig,B. (1991) A rapid finite difference algorithm utilizing successive over-relaxation to solve the Poisson–Boltzmann equation. J. Comp. Chem., 12, 435–445. [Google Scholar]
- Nureki O. et al. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- Poterszman A., Delarue,M., Thierry,J.-C. and Moras,D. (1994) Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase. J. Mol. Biol., 244, 158–167. [DOI] [PubMed] [Google Scholar]
- Rath V.L., Silvian,L.F., Beijer,B., Sproat,B.S. and Steitz,T.A. (1998) How glutaminyl-tRNA synthetase selects glutamine. Structure, 6, 439–449. [DOI] [PubMed] [Google Scholar]
- Rould M.A., Perona,J.J., Söll,D. and Steitz,T.A. (1989) Structure of E.coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science, 246, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon,A.C., Romby,P., Caillet,J., Springer,M., Rees,B., Ehresmann,C., Ehresmann,B. and Moras,D. (1999) The structure of threonyl-tRNA synthetase–tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell, 97, 371–381. [DOI] [PubMed] [Google Scholar]
- Schulman L.H. and Pelka,H. (1989) The anticodon contains a major element of the identity of arginine transfer RNAs. Science, 246, 1595–1597. [DOI] [PubMed] [Google Scholar]
- Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- Sissler M., Giegé,R. and Florentz,C. (1996) Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J., 15, 5069–5076. [PMC free article] [PubMed] [Google Scholar]
- Sissler M., Eriani,G., Martin,F., Giegé,R. and Florentz,C. (1997) Mirror image alternative interaction patterns of the same tRNA with either class I arginyl-tRNA synthetase or class II aspartyl-tRNA synthetase. Nucleic Acids Res., 25, 4899–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Himeno,H., Asahara,H., Hasegawa,T. and Shimizu,M. (1992) In vitro study of E.coli tRNAArg and tRNALys identity elements. Nucleic Acids Res., 20, 2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. (1990) WHATIF: a molecular modelling and drug design program. J. Mol. Graphics, 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Wodak S.J., Pontius,J., Vaguine,A. and Richelle,J. (1995) Validating protein structures. From consistency checking to quality assessment. In Proceedings of the CCP4 Daresbury Study Weekend: ‘Making the Most of your Model’. SERC Daresbury Laboratory, Warrington, UK, pp. 41–51. [Google Scholar]
- Woese C.R., Olsen,G.J., Ibba,M. and Soll,D. (2000) Aminoacyl-tRNA synthetases, the genetic code and the evolutionary process. Microbiol. Mol. Biol. Rev., 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]