Abstract
pH-mediated gating of Cx43 channels following an ischemic event is believed to contribute to the development of lethal cardiac arrhythmias. Studies using a soluble version of the Cx43 carboxyl-terminal domain (Cx43CT; S255–I382) have established the central role it plays in channel regulation; however, research in the authors’ laboratory suggests that this construct may not be the ideal model system. Therefore, we have developed a more ‘native-like’ construct (Cx43CT attached to the 4th transmembrane domain [TM4-Cx43CT; G178–I382]) than the soluble Cx43CT to further investigate the mechanism(s) governing this regulation. Here, we utilize circular dichroism and nuclear magnetic resonance (NMR) were used to validate the TM4-Cx43CT for studying channel gating and optimize solution conditions for structural studies. The data indicate that, unlike the soluble Cx43-CT, the TM4-Cx43CT is structurally responsive to changes in pH, suggesting the presence of the TM4 facilitates pH-induced structural alterations. Additionally, the optimal solution conditions for solving the NMR solution structure include 10% 2,2,2 trifluoroethanol and removal of the 2nd extracellular loop (G178-V196).
Keywords: Cx43, detergent micelles, NMR, TFE
INTRODUCTION
Gap junctions are specialized membrane channels that connect the cytoplasm of adjacent cells. Functionally, the channels provide an intercellular pathway for ions and small molecules, enabling the propagation and/or amplification of signaling cascades involved in cell growth, development, and whole-organ responses. Mutations in gap junction proteins are associated with numerous diseases and developmental abnormalities including neurodegenerative disorders, skin diseases, and hearing impairment (Dobrowolski and Willecke 2009). Formation of the channels occurs through the docking of connexons from adjacent cells. Each connexon is a hexameric structure composed of integral membrane proteins called connexins. Connexins are a family of proteins with 21 human isoforms that share a common topology: tetra-spanning integral membrane proteins that contain two extracellular loops (EL1, EL2), one cytoplasmic loop (CL), and cytoplasmic amino and carboxyl terminal domains (NT and CT, respectively).
Connexin43 (Cx43; named for its molecular weight in kDa) is ubiquitously expressed in humans and is the most well studied connexin isoform in terms of structure, function, and regulation. In the heart, Cx43 is the most abundant connexin and is essential for cardiac development and function (Severs et al. 2008). Regulation of Cx43 channel gating (opening and closing), such as the pH-mediated closure of Cx43 channels that occurs in response to cardiac ischemia, involves changes in the phosphorylation state and protein partner interactions of the CT (Duffy et al. 2004). However, the precise molecular mechanism by which channel regulation occurs has not been fully elucidated, primarily because of the lack of detailed structural information about the CT in its native environment (e.g., tethered to the membrane). Early structural studies of Cx43, which used cryo-electron microscopy, and the more recent x-ray crystallographic studies of Cx26 have provided a significant amount of information about channel architecture as well as connexin topology and arrangement in the channel (Unger et al. 1999b; Maeda et al. 2009). However, neither technique was able to adequately address the structure of the CT because of the dynamic and flexible nature of the domain. These same characteristics that interfere with crystallographic techniques, make nuclear magnetic resonance (NMR) an ideal tool for studying the CT.
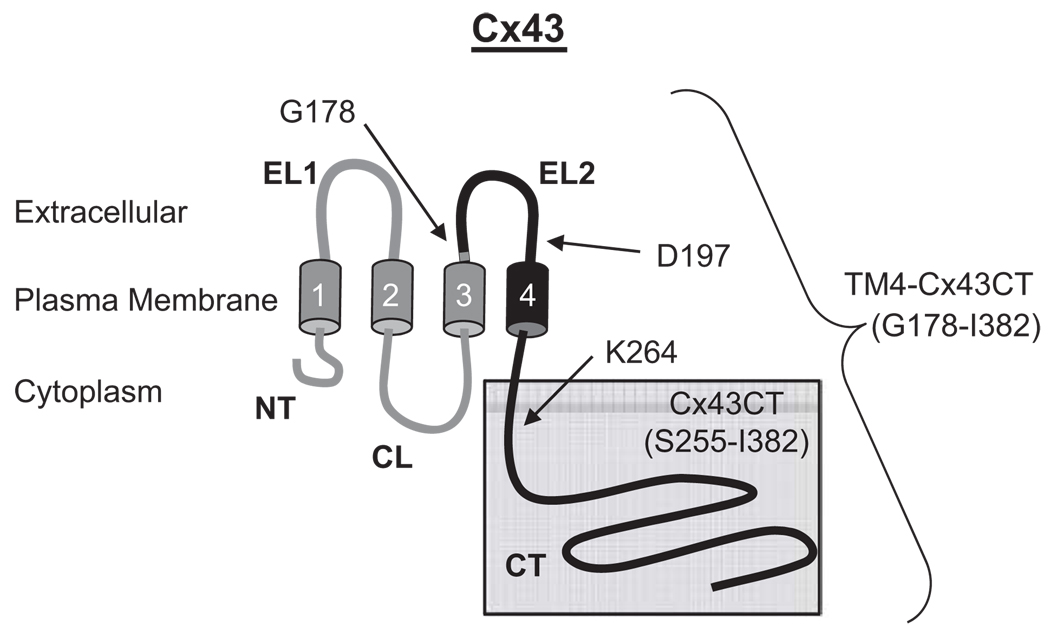
The structure of the soluble Cx43CT domain (residues S255–I382; Figure 1) was determined by NMR and has proven to be useful as a model for studying the structure-function mechanism regulating channel gating. However, several results indicate that the Cx43CT may not be the best model system (Sorgen et al. 2004a). For example, the cryo-electron microscopy studies used a CT-truncated construct of Cx43 (removed residues K264–I382). Although most of the CT domain was missing, the helical conformation of the 4th transmembrane domain was projected to extend beyond the membrane several residues into the CT domain (Unger et al. 1999a). NMR studies of the soluble Cx43CT indicate, however, that the amino-terminus of the soluble Cx43CT is highly flexible. Additionally, the NMR cross-peaks of Cx43CT residues S255–K264, which overlap with the end of the CT-truncated Cx43 construct used in the cryoelectron microscopy study, are weak, suggesting these residues are in exchange between two conformations (e.g., unstructured and α-helical). The abnormally high flexibility of the untethered amino-terminus of the soluble Cx43CT could disrupt structural stability of regions along the CT, interfere with molecular binding, or structural transitions associated with various regulatory events. Therefore, our laboratory optimized the growth, expression, and purification of a more native-like construct: the Cx43CT attached to the 4th transmembrane domain (TM4-Cx43CT, Figure 1), solubilized in detergent micelles (Kellezi et al. 2008). We demonstrated that the TM4-Cx43CT construct is folded properly and retains its ability to bind the SH3 domain of Src, an established binding partner. Additionally, circular dichroism (CD) results indicated that the TM4-Cx43CT was 46% helical; however, the TM4 portion only accounts for 10% of the total construct. This suggested that the TM4 provides structural stability to some portion of the CT, most likely the juxtamembrane region. It is also possible that the stabilizing effects may propagate further down the CT. Thus, solving the solution NMR structure of the TM4-Cx43CT will enable the extent of the TM4 structural influence on the CT to be determined. Here, we have identified the optimal solution conditions for the structural determination of the TM4-Cx43CT by NMR and provided additional evidence that the TM4-Cx43CT is a better model system than the soluble Cx43CT for studying the structure-function mechanism of pH mediated regulation of Cx43 channel gating.
Figure 1.
Schematic diagram illustrating the TM4-Cx43CT construct. The TM4-Cx43CT is colored black and the soluble Cx43CT construct is highlighted in the gray box. The abbreviations are as follows: NT, amino-terminus; CL, cytoplasmic loop; CT, carboxyl-terminus; EL1 and EL2, extracellular loops 1 and 2; 1–4, transmembrane domains 1–4.
METHODS AND MATERIALS
Plasmid Construction
Construction of the Cx43CT plasmid has been previously described (Duffy et al. 2002). TM4-Cx43CT (G178–I382) was cloned from a G2A plasmid containing the Rattus norvegicus Cx43 gene and ligated into the E. coli pET-14b expression vector (N-terminal 6× His-tag) (Novagen) using the restriction enzymes Nde I and Xho I (Kellezi et al. 2008). The TM4-Cx43CTΔEL2 (2nd extracellular loop deletion mutant; D197–I382) plasmid was constructed by introducing a second Nde1 restriction site downstream of the EL2 DNA sequence in the TM4-Cx43CT plasmid (Quick Change Lightning kit; Qiagen), resulting in the EL2 sequence (G178–V196) being flanked by the Nde I site used in the original cloning of the TM4-Cx43CT (upstream) and the new Nde I site (downstream). After Nde I digestion, the cut plasmid was gel-purified and ligated. The University of Nebraska Medical Center’s DNA Sequencing Core Facility verified all plasmid sequences.
Protein Expression and Purification
Esherichia coli strain BL21(DE3) (Novagen) was transformed with a plasmid containing the soluble Cx43CT (residues S255–I382) or the c-Src SH3 domain, and the E. coli C41(DE3) (Lucigen) strain was transformed with either the TM4-Cx43CT or TM4-Cx43CTΔEL2 plasmid. Transformed bacteria were used to inoculate Luria Bertani medium (LB) or 15N-labled enriched minimal media (Weber et al. 1992). Cultures were incubated at 37°C until they reached an optical density of 0.6 at 600 nm. Protein expression was induced by the addition of 1.0 mM isopropyl β-d-thiogalactopyranoside (final concentration). Growth was allowed to proceed for 4 h and cells were then harvested by centrifugation (1000 × g for 45 min), washed with phosphate-buffered saline (PBS), and stored at −20°C.
Purification of both the TM4-Cx43CT and TM4-Cx43CTΔEL2 was done following the procedure described in Kellezi et al. (2008). Cells were suspended in 1 × PBS buffer containing a bacterial protease inhibitor cocktail (250 µl/5 g cells; Sigma-Aldrich) and lysed by three passages through an Emulsiflex at 15,000 psi. Cell debris was removed by centrifugation (1000 × g for 30 min) and a pellet containing the inclusion bodies was collected by a high-speed centrifugation step (25,000 × g for 45 min). The pellet was resuspended in buffer A (8 M urea, 1 × PBS [pH 8.0], 1% Triton X-100, and 20 mM imidazole) and placed on a rocker at 4°C for ~2 h. The suspension was centrifuged again (25,000 × g for 45 min) and the supernatant was loaded onto a HisTrap HP affinity chromatography column using an ÄKTA FPLC (GE Healthcare). Protein elution was accomplished using a step gradient (4%, 8%, 10%, 30%, and 50% buffer B [8 M urea, 1×PBS, pH 8.0, 1% Triton X-100, and 1 M imidazole]). Prior to the elution step (30% B), the bound 6× His-tagged protein was washed with 10 column volumes of buffer A with 10% ethanol. Fractions containing purified protein were verified by Coomassie blue –stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, pooled, and dialyzed overnight at 4°C using a 10-kDa Slide-A-Lyzer dialysis cassette (Pierce) against 1 M urea, 1% Triton X-100, 1 mM dithiothreitol (DTT), and 1 mM EDTA. The precipitate was collected and centrifuged (300 × g for 5 min), washed once with water and two times with MES buffer (20 mM MES [pH 5.8], 50 mM NaCl). The precipitated protein was solubilized in MES buffer (pH 5.8) and 8% 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] (LPPG) at 42°C for 30 min.
Circular Dichroism (CD) Spectroscopy
CD experiments were performed using either an AVIV model-2055 spectrophotometer or a Jasco J-815 spectrophotometer. Both instruments are fitted with a Peltier temperature control system. CD spectra for the soluble Cx43CT (100 µM) were recorded in PBS pH 5.8 or pH 7.5. Spectra for the TM4-Cx43CT (100 µM) and TM4-Cx43CTΔEL2 (100 µM) were recorded in each of the following buffers: MES buffer (pH 5.8 and 7.5), 8% LPPG; MES buffer (pH 5.8), 8% LPPG with 5%, 10%, or 30% 2,2 trifluoroethanol (TFE). Additional CD spectra were collected for the TM4-Cx43CT (100 µM) in MES buffer (pH 5.8.) with either 8% 1-stearoyl-2-hydroxysn glycero-3-phospho-(1’ -rac-glycerol) (LSPG), 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine (LPPC), dodecylphosphocholine (DPC), or LPPG; low-salt MES buffer (20 mM MES [pH 5.8], 10 mM NaCl, 1 mM EDTA, 1 mM DTT); and phosphate buffer (50 mM potassium phosphate [pH 5.8], 1 mM EDTA, 1 mM DTT). For each sample, five scans (wavelength range: 250–190 nm; response time: 1 s; scan rate: 50 nm/min; bandwidth 1.0 nm) were collected using a 0.01-cm quartz cell, and processed using either CDtool (AVIV data) or Spectra Manager (Jasco data) (Lees et al. 2004).
Nuclear Magnetic Resonance (NMR)
NMR data were acquired at 42°C using a 600 MHz Varian INOVA NMR spectrometer fitted with a cryo-probe at the University of Nebraska Medical Center’ s NMR Shared Resource Facility. Gradient-enhanced two-dimensional 15N-HSQC experiments were used to observe backbone amide resonances of 15N-labeled TM4-Cx43CT and 15N-labeled TM4-Cx43CTΔEL2 in buffers described above. Data were acquired with 1024 complex points in the direct dimension and 128 complex points in the indirect dimension. Sweep widths were 8000 Hz in the proton dimension and 1720 Hz in the nitrogen dimension. NMR spectra were processed using NMRPipe (Delaglio et al. 1995) and analyzed using NMRView (Johnson 2004).
T1 and T2 experiments were collected with 1024 and 128 complex points in 1H and 15N respectively, with 32 scans per T1 point and a recycle delay of 1 s. All experiments used a 1H sweep width of 7800 Hz and a 15N sweep width of 1720 Hz with the 1H and 15N carriers set to 4.6 and 118 ppm, respectively. The T2 relaxation delays were 10, 30, 50, 70, 110, 150, 190, and 230 ms, with a 1-ms delay between 15N pulses in the Carr-Purcell-Meiboom-Gill sequence. The T1 longitudinal relaxation delays were 10, 20, 40, 80, 140, 200, 300, 400, 600, 800, and 1200 ms, and 2 and 4 s. Relaxation data were analyzed by the model free formalism (Lipari and Szabo) using Dasha (Orekhov et al. 1995).
RESULTS
Validation of Using the TM4-Cx43CT Construct to Study pH Gating
Our laboratory has developed a purification and reconstitution protocol to enable the biophysical characterization and structural determination by NMR methods of the TM4-Cx43CT (Kellezi et al. 2008). The rationale for using this more ‘native-like’ construct is that it will enable us to define the CT residues forming α-helical structure in the presence of a lipid environment; to define which CT residues are influenced by pH; will increase the length of the CT that we have been studying to include the second Src phosphorylation site (Y247) and the tubulin binding domain (K234–K243); and will be more native-like than the soluble Cx43CT when analyzing partner molecule interactions. The pure TM4-Cx43CT was determined to be properly folded (Kellezi et al. 2008), however, undetermined was whether this construct would show pH sensitivity, as was previously demonstrated for the soluble Cx43CT (Sorgen et al. 2004b; Duffy et al. 2002). To determine if the TM4-Cx43CT exhibits a similar behavior, CD experiments were conducted at pH 5.8 and 7.5 to investigate the effects of pH on the secondary structure of the TM4-Cx43CT.
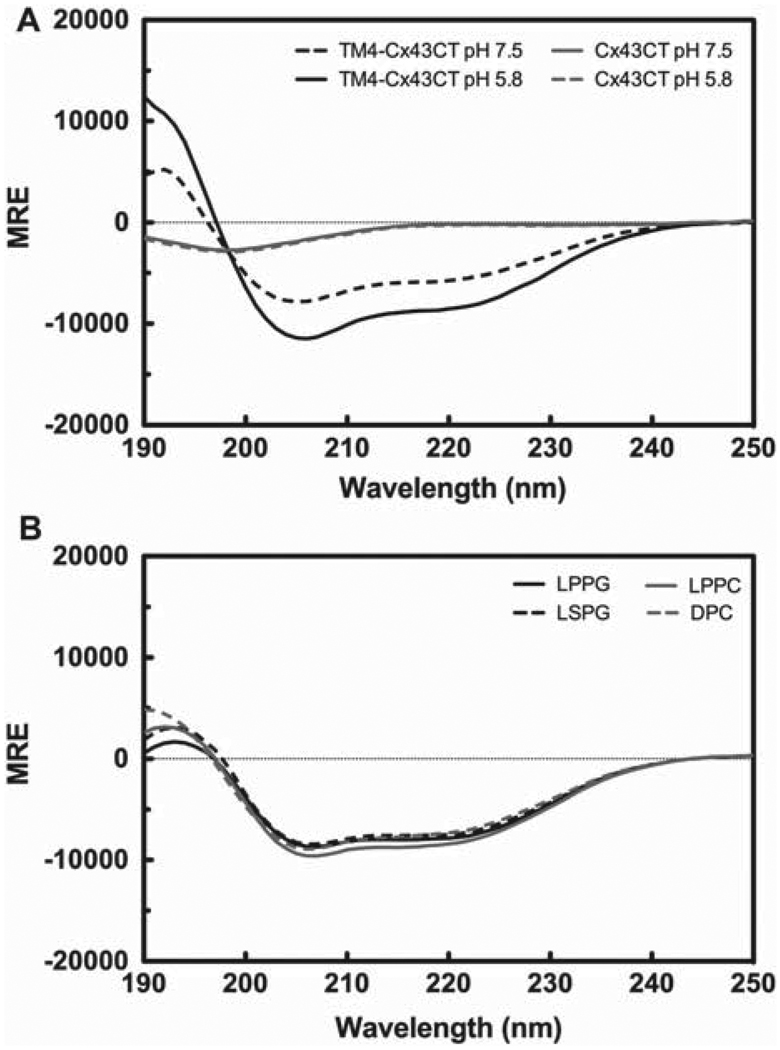
The CD spectra for the soluble Cx43CT and the TM4-Cx43CT are shown in Figure 2A. Consistent with the NMR solution structure, the CD spectral profile for the Cx43CT is indicative of a flexible, unstructured domain (characterized by a minimum at 198 nm) (Whitmore and Wallace 2008). Surprisingly, the CD spectra for the Cx43CT at pH 7.5 and 5.8 are almost identical, suggesting that the Cx43CT does not undergo a pronounced change in secondary structure in response to changes in pH. However, comparison of the CD spectrum for the TM4-Cx43CT at pH 7.5 to the spectrum at pH 5.8 indicates the TM4-Cx43CT, unlike the soluble Cx43CT, undergoes a significant conformational change in response to changes in pH. Typical of an α-helix containing protein, the CD spectra of the TM4-Cx43CT contains a double minima at 222 nm and 208 nm, and a maximum 190 nm (Whitmore and Wallace 2008). In a protein CD spectrum, changes in helical content can be quantified by evaluating changes in the magnitude of the mean residue ellipticity at 222 nm (MREλ=222). At pH 5.8, there is a 51% increase in MREλ=222 compared to the MREλ=222 at pH 7.5, signifying that changes in pH alter the helical content of the TM4-Cx43CT.
Figure 2.
Circular dichroism (CD) profile of the soluble Cx43CT and the TM4-Cx43CT. (A) CD spectra demonstrating the structural responsiveness of the soluble Cx43CT (gray) and the TM4-Cx43CT (black) to changes in pH. (B) Spectral profile of the TM4-Cx43CT in MES buffer (pH 5.8, 50 mM NaCl, 42°C) solubilized in LPPG (solid black line), LSPG (dotted black line), LPPC (solid gray line), and DPC (dotted gray line) micelles.
Optimization of the TM4-Cx43CT Solution Conditions for NMR Structural Studies
The solution conditions (20 mM MES buffer [pH 5.8], 100 mM NaCl, and 8% 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] [LPPG] at 42°C) initially used to solubilize the TM4-Cx43CT were based on a previous study that identified LPPG as a superior detergent for NMR studies (Krueger-Koplin et al. 2004). One reason for this is that the amount of TM4-Cx43CT expressed was relatively low, thus the initial trials varied only a limited number of membrane mimetics (e.g., BOG and Triton X-100) while holding other variables such as temperature, pH, and salt concentration constant. Here we expanded our optimization to include these conditions. CD experiments were performed to verify that the structural integrity of the TM4-Cx43CT was preserved under each of the conditions and NMR 15N-HSQC experiments were used to assess the effects of each condition on the spectral quality of the TM4-Cx43CT. The 15N-HSQC is a two-dimensional NMR experiment in which each residue (except proline) gives one cross peak (or chemical shift) that corresponds to the N-H group. Chemical shifts are sensitive to the chemical environment and changes in structure and/or dynamics can alter the chemical shift of an amino acid. The goal of the optimization is to identify solution conditions in which ≥ 90% of the expected cross peaks are present in a 40-min (4-scan) 15N-HSQC spectrum.
Three new detergents were selected that differed from LPPG (16-carbon chain, phosphoglycerol head group) in either chain length (LSPG; 18-carbon chain) or head group (LPPC; phosphocholine head group) or both (DPC; 12-carbon chain, phosphocholine head group). CD spectra were collected for the TM4-Cx43CT solubilized in each of the detergents (buffers described in methods section). The CD spectra (Figure 2B) for TM4-Cx43CT in LSPG, LPPC, and DPC are consistent with both the line shape and magnitude of the CD spectrum of TM4-Cx43CT in LPPG. These results indicate that TM4-Cx43CT structural integrity is maintained in each of the detergents tested.
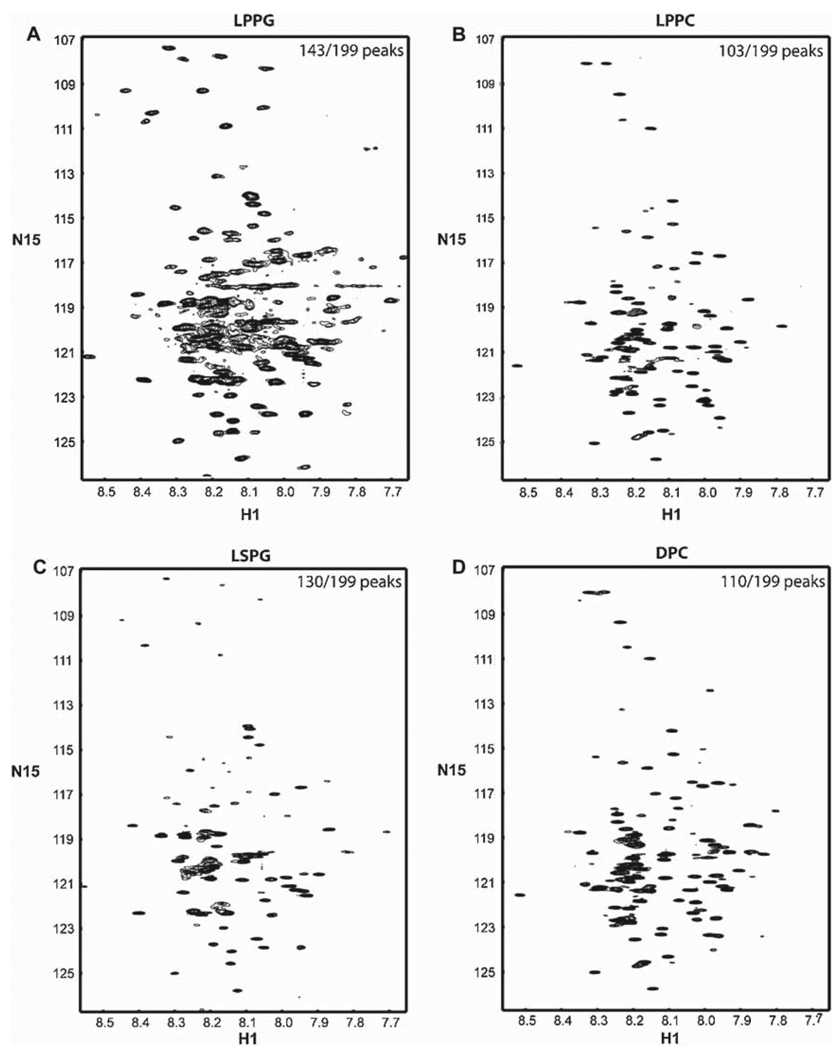
The 15N-HSQC spectra of TM4-Cx43CT in LPPG, LPPC, LSPG, and DPC are shown in Figure 3A–D. Spectral quality was determined by evaluating the number of observed versus expected cross peaks for the TM4-Cx43CT. For the detergents screened, LPPG provided the highest quality spectra, with 72% (143 out of 199) of the expected peaks being observed. The higher resolution previously reported for the 15N-HSQC spectrum of TM4-Cx43CT in LPPG was due to longer experimental time (4-h, 16-scan 15N-HSQC previously compared with 40-min, 4-scan 15N-HSQC here). LPPG was followed by LSPG with 65%, DPC with 55%, and LPPC with 52% of the expected peaks present.
Figure 3.
15N-HSQC spectra of the TM4-Cx43CT in different detergent micelles. The TM4-Cx43CT was solubilized in MES buffer (pH 5.8, 50 mM NaCl, 42°C) with (A) LPPG, (B) LPPC, (C) LSPG, and (D) DPC detergent micelles. Spectral quality was determined by the number of observed peaks versus the number of expected peaks. Peak counts are given in the top right corner of each spectrum as observed/expected.
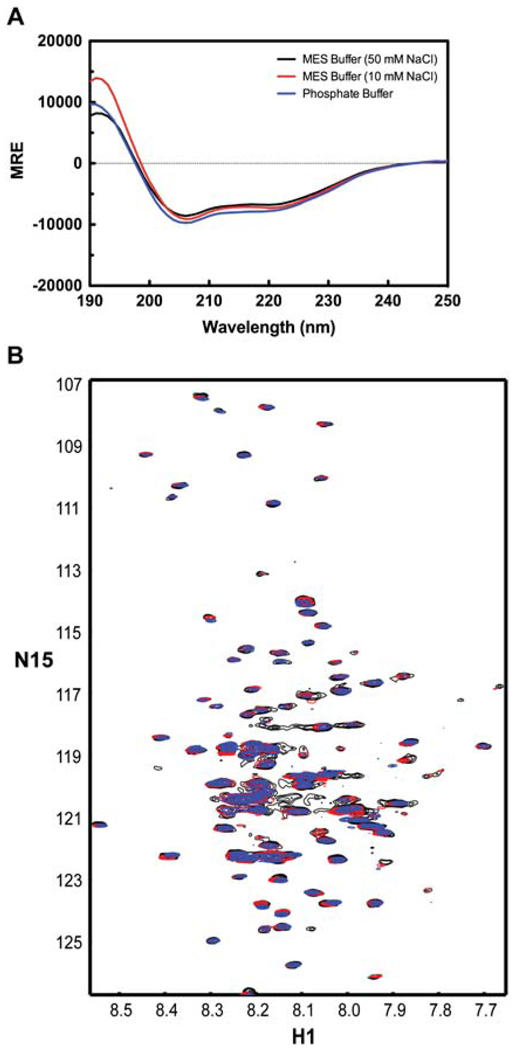
In addition to detergents, three low-conductivity buffers were also screened. CD and 15N-HSQC spectra of the TM4-Cx43CT solubilized in 8% LPPG and either MES buffer (pH 5.8) plus 50 mM NaCl, MES buffer (pH 5.8) plus 10 mM NaCl, or phosphate buffer (pH 5.8) are given in Figure 4A. Compared to the MES buffer, neither the low-salt MES buffer nor the phosphate buffer altered the CD spectral profile of the TM4-Cx43CT. Similarly, the buffers tested did not affect spectral quality compared to the MES buffer: in each buffer, 72% of the expected peaks are present (Figure 4B).
Figure 4.
Circular dichroism (CD) profile of the TM4-Cx43CT in various buffers. Spectra the TM4-Cx43CT solubilized in LPPG detergent micelles in MES buffer (pH 5.8, 50 mM NaCl, 42°C) (black), a low-salt MES buffer (pH 5.8, 10 mM NaCl, 42°C) (red), and potassium phosphate buffer (pH 5.8, 42°C) (blue). (A) CD spectra of the TM4-Cx43CT. (B) 15N-HSQC spectra of the TM4-Cx43CT. In each of the buffers, 143 peaks out of expected 199 peaks were observed.
Additional 15N-HSQC spectra of the TM4-Cx43CT in MES buffer, 8% LPPG were collected under variable temperature and pH conditions in an attempt to further improve the spectral resolution of the TM4-Cx43CT NMR spectra. For NMR spectroscopy of membrane proteins, temperatures above 30°C are typically preferred (Fernandez et al. 2003). Therefore, a temperature range of 36–48°C was tested in 5°C increments. An acidic range centered on pH 5.8 (pH 4.8–6.3 with increments of 0.5 pH units) was chosen. The particular range was selected because first, previous work has suggested protonation of His residues of Cx43 may be involved in pH sensing and/or facilitating intramolecular interactions involved in channel gating following an ischemic event: at pH 5.8, greater than 90% of the His residues are expected to be protonated; second, acidic pH minimizes proton exchange, which can improve NMR spectra; and lastly, the selected pH range does not greatly exceed the buffering capacity of the MES buffer (pKa 6.1). Neither the temperature or pH range tested greatly affected the quality of the TM4-Cx43CT 15N-HSQC spectra (data not shown). Selection of an optimum temperature and pH (42°C, pH 5.8) was based on previously conducted TM4-Cx43CT stability testing (Kellezi et al. 2008).
The optimal solution conditions for solving the NMR solution structure of the TM4-Cx43CT were determined to be MES buffer (pH 5.8), 8% LPPG at 42°C. Unfortunately, under these conditions, only 72% of the expected cross-peaks are present in the 40min (4-scan) 15N-HSQC. Therefore, NMR relaxation experiments were conducted to determine if the poor spectral resolution and missing peaks were due to the effective size of the TM4-Cx43CT-micelle complex (~106 kDa) in solution.
NMR relaxation experiments (T1 and T2) experiments were preformed to determine the correlation time (τc) of the TM4-Cx43CT. The τc is the average time it takes for a molecule to rotate, or tumble one radian from its initial position. Large molecules tumble in solution more slowly than small molecules, causing line broadening and loss of signal (cross-peaks), which is one contributing factor to the molecular size limitations of NMR. For proteins, typical τc ranges from 10−6 s (large macromolecules) to 10−9 s (small macromolecules). Rotational motion of an asymmetric molecule (e.g., rod-shaped) can be around the long axis and/or be end-over-end tumbling. The τc is affected by the type of rotational motion, but does not distinguish between them. Consequentially, asymmetric molecules generally have correlation times consistent with larger molecular weight, globular proteins. Additionally, solution conditions, including detergent concentration, and temperature, can affect the τc of a macromolecule. The τc of the TM4-Cx43CT (MES buffer, pH 5.8, 8% LPPG, 42°C) was determined to be 4.79 × 10−9 s. Although the τc is within the range of a macromolecule, the value is faster than expected for an intrinsically disordered protein. The fast τc of the TM4-Cx43CT suggests that TM4 portion, which is inserted into the LPPG micelle, is rotating freely within the micelle rather than as a large protein-micelle complex. This indicates that the poor spectral quality is not likely due to the apparent size of the TM4-Cx43CT-micelle complex. Therefore, two alternate strategies were developed to improve spectral quality: the use of a co-solvent to help stabilize α-helical structure of the CT and the removal of extraneous, unstructured residues from the TM4-Cx43CT.
The CD results for the TM4-Cx43CT indicate an increase in α-helical content in response to a decrease in pH. The low spectral dispersion of the 15N-HSQC and missing cross-peaks suggest that the α-helical region(s) along the CT of the TM4-Cx43CT are highly dynamic and likely in exchange between structured and unstructured conformations. Therefore, stabilization of the helical region(s) along the CT in a more rigid conformation should increase the spectral dispersion, resulting in improved spectral resolution and signal strength.
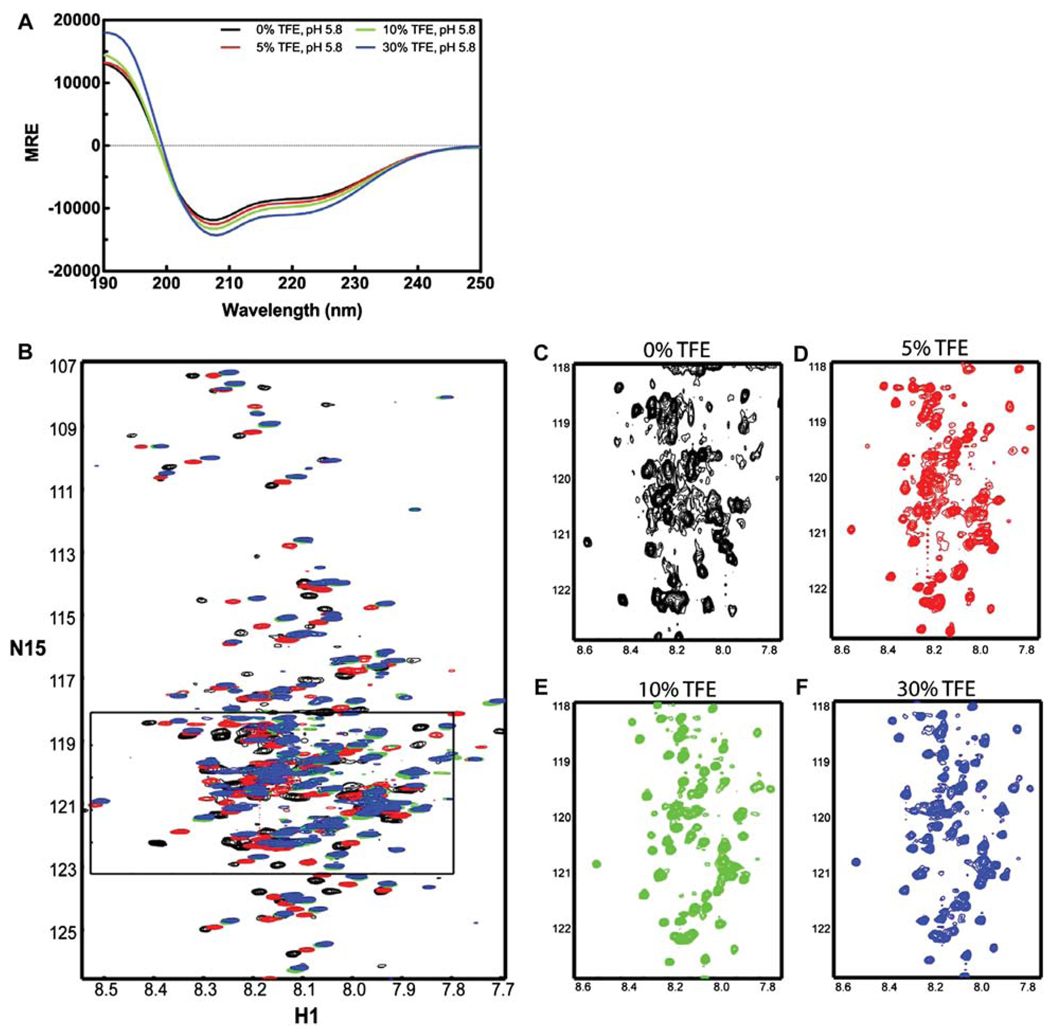
The co-solvent 2, 2, 2-trifluoroethanol (TFE) is a helix-stabilizing compound that is readily used in protein folding and structural studies, as well as a tool for probing the helical propensity of proteins and peptides (Fort and Spray 2009; Libich and Harauz 2008). At low levels (<50%), TFE will stabilize regions with intrinsic α-helical character and, at high levels (>50%), can induce an α-helical conformation (Reiersen and Rees 2000). The increase in helical content in the TM4-Cx43CT in response to a decrease in pH (Figure 2A) indicates region(s) along the CT have an inherent propensity to form an α-helix. TFE stabilizes helical conformations in regions with the highest helical propensity first. Therefore, the TM4-Cx-43CT (pH 5.8) was titrated with 5%, 10%, and 30% TFE (Figure 5A) in order to find the lowest percent of TFE that improves NMR spectral resolution without altering the CD spectra line shape and magnitude. As expected, the α-helical content of the TM4-Cx43CT increases with increasing TFE amounts. Relative to 0% TFE, there is a 7% and 14% increase in MREλ=222 at 5% and 10% TFE, respectively, whereas there is a 29% increase in MREλ=222 at 30% TFE.
Figure 5.
Spectra of the TM4-Cx43CT in TFE. TM4-Cx43CT solubilized in LPPG micelles (MES buffer, pH 5.8, 50 mM NaCl, 42°C) titrated with 0% (black) 5% (red), 10% (green), and 30% (blue) TFE. (A) CD spectra collected for the TM4-Cx43CT TFE titration. (B) Overlay of the NMR spectra collected for the TM4-Cx43CT titrated with TFE. (C–F) Close-up view of each spectrum (black rectangle from A).
Figure 5B is an overlay of 15N-HSQC spectra of the TM4-Cx43CT in 0%, 5%, 10%, and 30% TFE. A magnified view of each of the individual spectra is provided in Figure 5C–F. Although the magnitude of CD spectrum for the TM4-Cx43CT at pH 5.8 with 5% TFE was most similar to the magnitude of the CD spectrum at pH 5.8 (0% TFE), 5% TFE did not improve the NMR spectrum. However, in the presence of 10% TFE, the 15N-HSQC spectrum of the TM4-Cx43CT was greatly improved (91% of the expected cross-peaks were present). Use of 30% TFE did not improve the spectral quality.
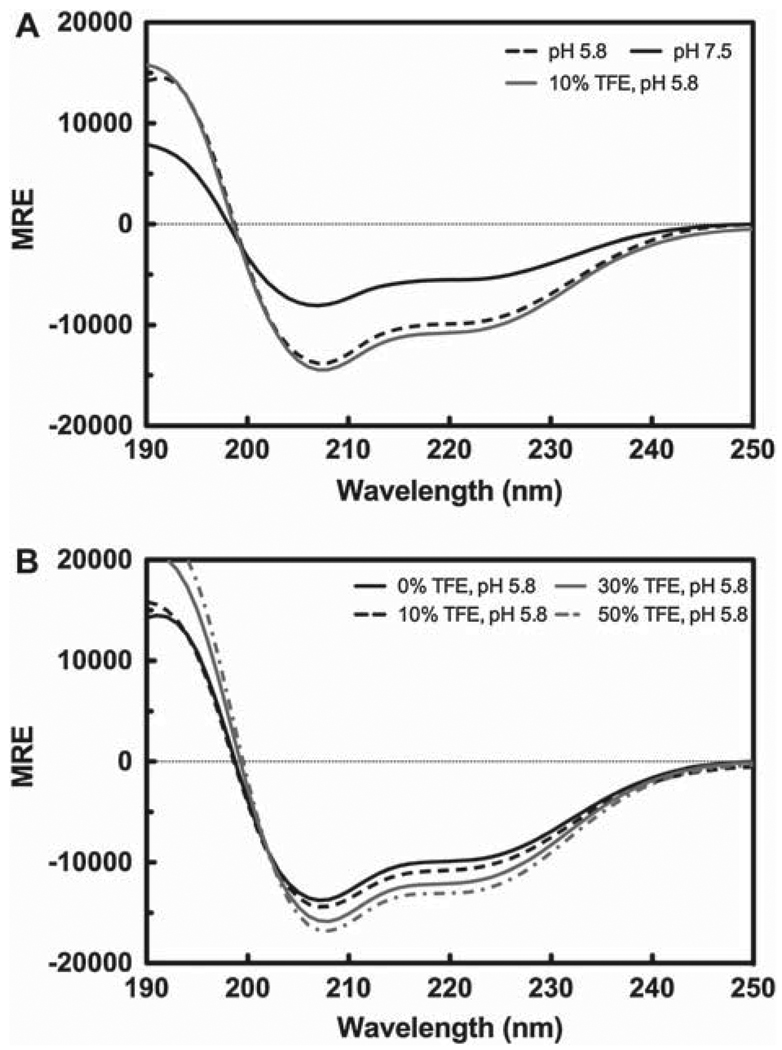
Another strategy to decrease the spectral complexity of the TM4-Cx43CT was to remove the EL2 portion (residues G178–V196) of the TM4-Cx43CT. The crystal structure of Cx26 indicates that the EL2 is primarily unstructured (Maeda et al. 2009). It is anticipated that the disordered EL2 residues of the TM4-Cx43CT construct is contributing to NMR spectral overlap. After removal of EL2 residues, CD was used to validate that the TM4-Cx43CTΔEL2 construct retains pH sensitivity. Figure 6A is the CD spectra of the TM4-Cx43CTΔEL2 at pH 7.5 and 5.8. At pH 5.8 there is a 78% increase in MREλ=222 as compared to the MREλ=222 at pH 7.5. Titration of the TM4-Cx43CTΔEL2 with TFE also yielded results similar to those obtained for the TM4-Cx43CT. The increase in MREλ=222 for the TM4-Cx43CTΔEL2 (pH 5.8) in 10%, 30%, and 50% TFE was 8%, 22%, and 32%, respectively (Figure 6B). The CD spectrum of the TM4-Cx43CTΔEL2 at pH 5.8 and 10% TFE is also given in Figure 6A to show that the CD spectra at pH 5.8 with 0% and 10% TFE are nearly identical, suggesting that TFE is stabilizing the helical region involved in the pH mediated response.
Figure 6.
Circular dichroism (CD) profile of the TM4-Cx43CTΔEL2. CD spectra of the TM4-Cx43CTΔEL2 solubilized in LPPG micelles (MES buffer pH 5.8, 50 mM NaCl, 42°C). (A) CD spectra comparing the secondary structure of the TM4-Cx43CTΔEL2 at pH 7.5 (solid black line) to pH 5.8 (dotted black line) and to pH 5.8 and 10% TFE (solid gray line). (B) CD spectra for the TM4-Cx43CTΔEL titrated with 0% (solid black line), 10% (dotted black line), 30% (solid gray line), and 50% (dotted gray line).
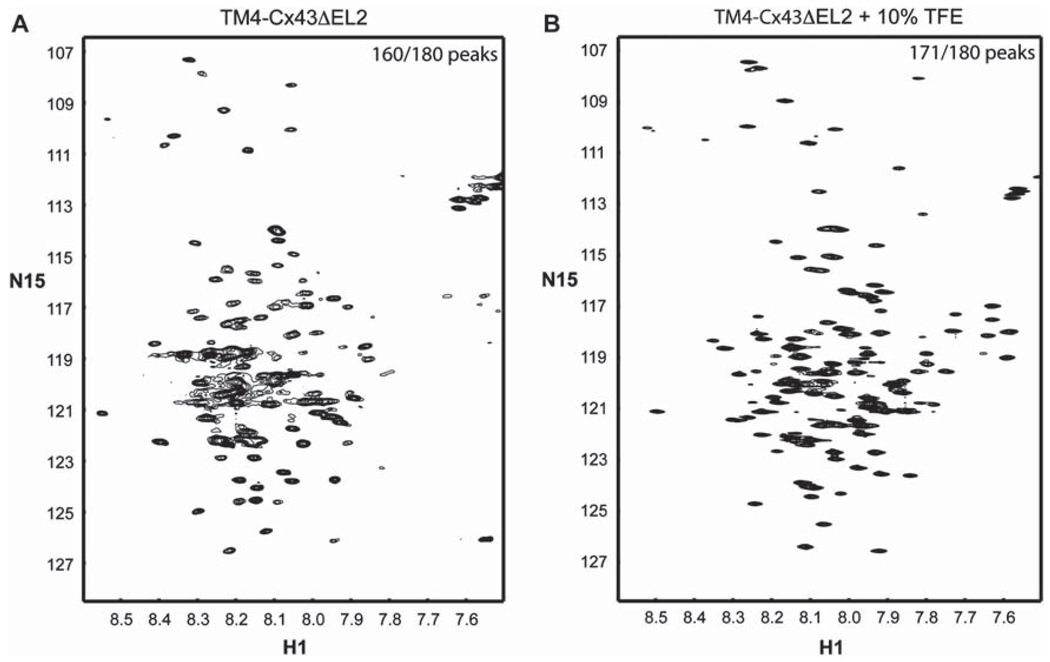
Figure 7A is the 15N-HSQC of the TM4-Cx43CTΔEL2 in MES buffer (pH 5.8), 8% LPPG at 42°C. Removal of the EL2 improved the spectra such that 88% of the expected cross-peaks are present (compared to 72% for the TM4-Cx43CT). Because the addition of TFE improved the quality of the NMR spectrum of TM4-Cx43CT, a 15N-HSQC spectrum of the TM4-Cx43CTΔEL2 in 10% TFE was collected (Figure 7B). Utilization of the TM4-Cx43CTΔEL2 in the presence of 10% TFE resulted in 95% of the expected cross-peaks in the 15N-HSQC spectrum, indicating that the optimum protein construct and solution conditions for solving the structure by NMR is the TM4-Cx43CTΔEL2 in MES buffer (pH 5.8), 8% LPPG, 10% TFE at 42°C.
Figure 7.
15N-HSQC spectra of the TM4-Cx43CTΔEL2 solubilized in LPPG micelles. (A) TM4-Cx43CTΔEL2 spectrum collected under conditions optimized for the TM4-Cx43CT (MES buffer [pH 5.8], 8% LPPG, 42°C). (B) Spectrum of the TM4-Cx43CTΔEL2 in the buffer from A plus 10% TFE. Observed versus expected peaks are given in the top right corner of each of the spectra.
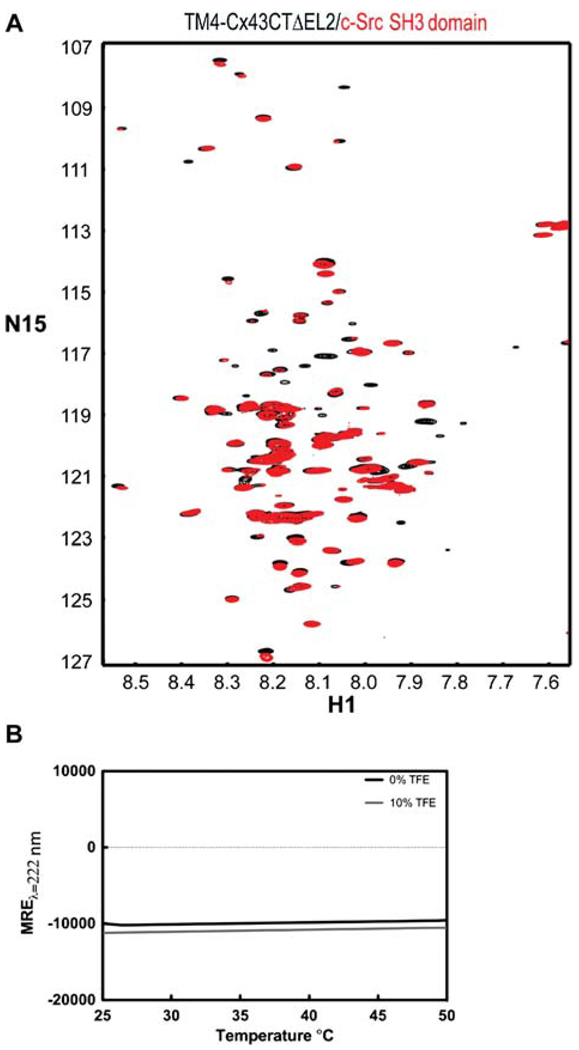
To validate the biological relevance of determining the solution structure of the TM4-Cx43CTΔEL2 under these solution conditions (particularly temperature and detergent concentration), we used NMR to assess the ability of the construct to interact with a known binding partner and CD to determine the effect of temperature on the structure of the construct. Figure 8A is an overlay of 15N-HSQC spectra of the TM4-Cx43ΔEL2 in the absence and presence of the c-Src SH3 domain. Changes in chemical shifts (red no longer over black) and peak intensities along with missing peaks in the 15N-TM4-Cx43CTΔEL2 spectrum in the presence of the SH3 domain are all indicative of a protein-protein interaction. It should be noted that the SH3 precipitates in the presence of TFE (even at 5%), therefore it was not possible to use TFE in the binding assay. Figure 8B is the CD spectra (MREλ=222) of the TM4-Cx43CTΔEL2 (pH 5.8) with 0% and 10% TFE as a function of temperature. The difference in MREλ=222 at 37°C compared to 42°C is 1.2% and 1.4% for the TM4-Cx43CTΔEL2 with 0% TFE and 10% TFE, respectively. This result indicates that the structure of the TM4-Cx43CTΔEL2 is almost the same at 37°C (biologically relevant temperature) and 42°C (optimal NMR temperature).
Figure 8.
Effects of temperature on native binding interactions and secondary structure of the TM4-Cx43CTΔEL2. (A) Overlay of 15N-HSQC spectra of TM4Cx43CTΔEL2 solubilized in LPPG micelles (MES buffer, pH 5.8, 50 mM NaCl, 42°C) alone (black) and in the presence of c-Src SH3 domain (red), a known binding partner of Cx43. (B) CD spectra of the TM4Cx43CTΔEL2 solubilized in LPPG micelles (MES buffer, pH 5.8, 50 mM NaCl) (black line) and with the addition of 10% TFE (gray line) collected at 222 nm to monitor the change in helical content as a function of temperature.
DISCUSSION
In this study we have optimized the solution conditions to solve the structure of the TM4-Cx43CT by NMR. The initial optimization strategy, which focused primarily on detergents and buffers, yielded solution conditions that were similar to those identified previously (Kellezi et al. 2008). However, further optimization strategies, which included the addition of 10% TFE and removal of the EL2 residues from the original TM4-Cx43CT construct improved the spectral resolution and dispersion such that 95% of the expected TM4-Cx43CTΔEL2 peaks are present in the 15N-HSQC spectrum. Additionally, our studies validated that the TM4-Cx43CTΔEL2 is a more ideal construct than the previously utilized soluble Cx43CT domain to characterize pH-mediated channel gating: the more native like construct was more structurally responsive (i.e., increased α-helical content) to changes in pH.
Validation for Using the TM4-Cx43CT
The role of the Cx43CT in pH-mediated channel gating has been well established (Delmar et al. 2004). pH-mediated channel gating is believed to occur through a ‘particle-receptor’ type mechanism whereby, under acidic intracellular conditions, the ‘particle’ (the CT domain) interacts with the ‘receptor’ (the cytoplasmic loop domain), resulting in channel closure (Delmar et al. 2004). Our previous studies have identified a pH-dependent interaction between the soluble Cx43CT and a peptide that corresponds to the second half of the Cx43CL (D119–K144; Cx43L2) (Hirst-Jensen et al. 2007; Duffy et al. 2002). Furthermore, under acidic conditions, the Cx43L2 adopts an α-helical conformation (residues N122–Q129 and K136–G143) and this structural rearrangement is believed to be necessary for this interaction (Duffy et al. 2002). The NMR and CD spectra of a soluble and more ‘native-like’ Cx43CL construct (M100-Y155) indicated the structure was completely random coiled (unpublished data). However, titration of the CL with TFE indicates the Cx43CL has a strong α-helical propensity (unpublished data). The data suggest the increased number of disordered residues in the first half of the Cx43CL is destabilizing the α-helices in the second half of the Cx43CL. We hypothesize that these α-helices in the second half of the Cx43CL will be stabilized when attached to the 2ndand 3rdtransmembrane domains. Kellezi et al. (2008) showed that the helical content of the TM4-Cx43CT could not be attributed solely to the addition of the transmembrane domain to the soluble CT domain alone. It is likely that the larger than expected helical content is the result of the trans-membrane domain stabilizing the juxtamembrane region of the Cx43CT domain. Our data support the hypothesis that tethering of a soluble domain to the membrane can stabilize secondary structure for soluble Cx43 domains. Tethering of the soluble domain also affected the structural sensitivity of Cx43CT domain to changes in pH. The increase in α-helical content of the more native-like TM4-Cx43CT in response to acidification is expected to be necessary for an increased binding affinity for the Cx43CL. Additionally, our laboratory has identified that under acidic conditions, the soluble Cx43CT domain forms a homodimer (Sorgen et al. 2004b; Hirst-Jensen et al. 2007). Although the functional consequence(s) for Cx43CT dimerization in vivo is currently unknown, we would hypothesize that the tethering of the Cx43CT to the membrane would also stabilize the interaction between two Cx43CT molecules.
Optimization of Solution Conditions for the TM4-Cx43CT
One of the biggest challenges for the structural determination of an integral membrane protein by solution NMR is finding a suitable membrane mimetic that does not exceed the size limitations of solution NMR, which is currently approaching 100 kDa (Fernandez and Wuthrich 2003). As the complex size increases, signal is lost due to line broadening and poor spectral resolution. Therefore, due to their relatively smaller size, detergent micelles are generally a more suitable membrane mimetic for NMR studies than the larger bicelles or liposomes (Sanders and Sonnichsen 2006). Although no single detergent has been shown to be overwhelmingly helpful, one detergent, LPPG, has proven to be effective as a general membrane mimetic for the study of integral membrane proteins by NMR (Krueger-Koplin et al. 2004). Previous work suggests that, unlike other detergent micelles, LPPG micelles may allow the membrane spanning regions of proteins to rotate freely within the micelle rather than as a protein-micelle complex. This could be due to the large hydrophobic core volume associated with LPPG micelles (72 nm 3compared to 27 nm 3for DPC micelles) (Columbus et al. 2009). Solubilization of the TM4-Cx43CT in LPPG micelles provided the highest quality NMR spectra relative to the other detergents tested (e.g., LPPC, LSPG and DPC). The τc obtained suggests that the TM4-Cx43CT is not rigidly anchored in the micelle: the TM4 portion inserted in the micelle is likely rotating freely within the micelle. However, the spectral quality, as determined by the percent of expected peaks observed in the spectrum, was low even in LPPG. Further optimization of buffering system, temperature, and pH provided little improvements in spectral quality beyond what was previously determined (Kellezi et al. 2008). In a more aggressive optimization strategy, two factors, which had the most profound impact on spectral quality, were identified: the use of TFE and the removal of the EL2.
TFE is a co-solvent that is thought to stabilize secondary structure either directly by associating with the protein/peptide or indirectly by destabilizing the disordered form of the protein/peptide (Reiersen and Rees 2000). Regardless of its mode of action, addition of low amounts of TFE can stabilize protein secondary structure. TFE has been used in a multitude of studies ranging from investigating protein folding pathways, to evaluating disease-related effects of amino acid mutations on structural propensities (Fort and Spray 2009; Libich and Harauz 2008; Main and Jackson 1999). The underlying assumption with each of these studies is that, at low levels (<50%), TFE stabilizes structure in regions that contain an innate proclivity for secondary structure. Here, working under the same hypothesis, we utilized TFE to stabilize the flexible, dynamic helical region of the TM4-Cx43CT. The inclusion of 10% TFE yielded 15N-HSQC spectra with greatly improved spectral resolution and chemical shift dispersion that is more consistent with the structural characteristics of the TM4-Cx43CT elucidated by CD. Furthermore, the CD spectra collected for the TFE titrations at pH 5.8 indicate that, at 10% TFE, the α-helical content is similar to that seen at pH 5.8 (0% TFE), supporting the notion that TFE is stabilizing the dynamic helical region(s) along the CT in the TM4-Cx43CT. In the presence of 10% TFE at pH 5.8, 91% of the expected TM4-Cx43CT cross-peaks were observed in 15N-HSQC spectra collected over 40 min. Typically, observation of ≥ 95% of the expected cross-peaks in a short 2D 15N-HSQC experiment translates to observation of all expected peaks in the 3D NMR experiments used for residue assignments and structural determination. Inclusion of TFE produced the most substantial improvements in spectral quality.
Further refinement of the TM4-Cx43CT protein construct by the removal of extraneous and unstructured residues also improved spectral quality by decreasing spectral overlap and low signal intensities inherent with disordered residues (88% of expected cross-peaks were observed in the TM4-Cx43CTΔEL2). Removal of the EL2 residues did not affect the pH sensitivity of the protein or its ability to interact with the c-Src SH3 domain. Additionally, the secondary structure of the TM4-Cx43CTΔEL2 is stable at 42°C compared to 37°C. Similar to the full-length TM4-Cx43CT construct, the addition of 10% TFE to the TM4-Cx43CTΔEL2 resulted in structural stabilization of the protein within the range of what is exhibited by changes in pH. The increased structural stability of the TM4-Cx43CTΔEL2 by the addition of 10% TFE translated to higher resolution and chemical shift dispersion in 15N-HSQC spectra, resulting in 95% of the expected peaks being observed. Therefore, the optimal conditions for determining resonance assignments and solving the NMR solution structure of the TM4-Cx43CTΔEL2, as well as studying pH-mediated interactions with the Cx43CL domain is MES buffer (pH 5.8), 8% LPPG, 10% TFE at 42°C in the presence of 10% TFE.
Acknowledgments
This work was supported by United States Public Health Service grant GM072631 and the Nebraska Research Initiative for the Nebraska Center for Structural Biology.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- Columbus L, Lipfert J, Jambunathan K, Fox DA, Sim AY, Doniach S, Lesley SA. Mixing and matching detergents for membrane protein NMR structure determination. J Am Chem Soc. 2009;131:7320–7326. doi: 10.1021/ja808776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Delmar M, Coombs W, Sorgen P, Duffy HS, Taffet SM. Structural bases for the chemical regulation of Connexin43 channels. Cardiovasc Res. 2004;62:268–275. doi: 10.1016/j.cardiores.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Dobrowolski R, Willecke K. Connexin-caused genetic diseases and corresponding mouse models. Antioxid Redox Signal. 2009;11:283–295. doi: 10.1089/ars.2008.2128. [DOI] [PubMed] [Google Scholar]
- Duffy HS, Ashton AW, O’Donnell P, Coombs W, Taffet SM, Delmar M, Spray DC. Regulation of connexin43 protein complexes by intracellular acidification. Circ Res. 2004;94:215–222. doi: 10.1161/01.RES.0000113924.06926.11. [DOI] [PubMed] [Google Scholar]
- Duffy HS, Sorgen PL, Girvin ME, O’Donnell P, Coombs W, Taffet SM, Delmar M, Spray DC. pH-dependent intramolecular binding and structure involving Cx43 cytoplasmic domains. J Biol Chem. 2002;277:36706–36714. doi: 10.1074/jbc.M207016200. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Wuthrich K. NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 2003;555:144–150. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- Fort AG, Spray DC. Trifluoroethanol reveals helical propensity at analogous positions in cytoplasmic domains of three connexins. Biopolymers. 2009;92:173–182. doi: 10.1002/bip.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst-Jensen BJ, Sahoo P, Kieken F, Delmar M, Sorgen PL. Characterization of the pH-dependent interaction between the gap junction protein connexin43 carboxyl terminus and cytoplasmic loop domains. J Biol Chem. 2007;282:5801–5813. doi: 10.1074/jbc.M605233200. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kellezi A, Grosely R, Kieken F, Borgstahl GE, Sorgen PL. Purification and reconstitution of the connexin43 carboxyl terminus attached to the 4th transmembrane domain in detergent micelles. Protein Expr Purif. 2008;59:215–222. doi: 10.1016/j.pep.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- Lees JG, Smith BR, Wien F, Miles AJ, Wallace BA. CDtool—An integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal Biochem. 2004;332:285–289. doi: 10.1016/j.ab.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Libich DS, Harauz G. Solution NMR and CD spectroscopy of an intrinsically disordered, peripheral membrane protein: Evaluation of aqueous and membrane-mimetic solvent conditions for studying the conformational adaptability of the 18.5 kDa isoform of myelin basic protein (MBP) Eur Biophys J. 2008;37:1015–1029. doi: 10.1007/s00249-008-0334-8. [DOI] [PubMed] [Google Scholar]
- Lipari G, Szabo A. Nuclear magnetic resonance relaxation in nucleic acid fragments: Models for internal motion. Biochemistry. 1981;20:6250–6256. doi: 10.1021/bi00524a053. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Main ER, Jackson SE. Does trifluoroethanol affect folding pathways and can it be used as a probe of structure in transition states? Nat Struct Biol. 1999;6:831–835. doi: 10.1038/12287. [DOI] [PubMed] [Google Scholar]
- Orekhov VYu, Nolde DE, Golovanov AP, Korzhnev DM, Arseniev AS. Processing of heteronuclear NMR relaxation data with the new software DASHA. Appl Magn Reson. 1995;9:581–588. [Google Scholar]
- Reiersen H, Rees AR. Trifluoroethanol may form a solvent matrix for assisted hydrophobic interactions between peptide side chains. Protein Eng. 2000;13:739–743. doi: 10.1093/protein/13.11.739. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: Practice and challenges. Magn Reson Chem. 2006;44(Spec No.):S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Sahoo P, Coombs W, Delmar M, Spray DC. Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1. J Biol Chem. 2004a;279:54695–54701. doi: 10.1074/jbc.M409552200. [DOI] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Spray DC, Delmar M. pH-dependent dimerization of the carboxyl terminal domain of Cx43. Biophys J. 2004b;87:574–581. doi: 10.1529/biophysj.103.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Expression, two-dimensional crystallization, and electron cryo-crystallography of recombinant gap junction membrane channels. J Struct Biol. 1999a;128:98–105. doi: 10.1006/jsbi.1999.4184. [DOI] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999b;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Weber DJ, Gittis AG, Mullen GP, Abeygunawardana C, Lattman EE, Mildvan AS. NMR docking of a substrate into the X-ray structure of staphylococcal nuclease. Proteins. 1992;13:275–287. doi: 10.1002/prot.340130402. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]