Abstract
ADP-ribosylation factors, a family of small GTPases, are believed to be key regulators of intracellular membrane traffic. However, many biochemical in vitro experiments have led to different models for their involvement in various steps of vesicular transport, and their precise role in living cells is still unclear. We have taken advantage of the powerful yeast genetic system and screened for temperature-sensitive (ts) mutants of the ARF1 gene from Saccharomyces cerevisiae. By random mutagenesis of the whole open reading frame of ARF1 by error-prone PCR, we isolated eight mutants and examined their phenotypes. arf1 ts mutants showed a variety of transport defects and morphological alterations in an allele-specific manner. Furthermore, intragenic complementation was observed between certain pairs of mutant alleles, both for cell growth and intracellular transport. These results demonstrate that the single Arf1 protein is indeed involved in many different steps of intracellular transport in vivo and that its multiple roles may be dissected by the mutant alleles we constructed.
INTRODUCTION
ADP-ribosylation factors (Arfs) constitute a ubiquitous subfamily of the small GTPases in eukaryotes, which were originally identified in animal cells as cofactors of the cholera toxin-catalyzed ADP ribosylation of Gsα in vitro (Schleifer et al., 1982; Kahn and Gilman, 1984). They are now recognized as essential components in vesicular trafficking pathways. A number of studies have led to the model that Arf regulates the cycle of budding driven by coatomer (COPI coat protein complex). Orci et al. (1986) proposed a role of coatomer in the anterograde traffic within the Golgi complex, and Arf itself was purified as an inhibitory factor of intra-Golgi transport in the presence of a nonhydrolyzable analogue of GTP (Serafini et al., 1991; Taylor et al., 1992). However, many recent lines of evidence indicate that Arf proteins can regulate the formation of vesicles involved in a wide variety of transport steps. Letourneur et al. (1994) showed that the COPI complex, including Arf, plays an essential role in the Golgi-to-endoplasmic reticulum (ER) retrieval pathway by the following two lines of evidence. First, COPI physically interacts with the dilysine ER retrieval motif in vitro (Cosson and Letourneur, 1994). Second, yeast cells with a mutation in subunits of COPI showed a defect in the retrieval of dilysine-tagged proteins back to the ER (Letourneur et al., 1994; Cosson et al., 1996). On the other hand, evidence also exists supporting a role for Arf in the ER-to-Golgi anterograde transport. For example, mutants of mammalian Arf1 restricted to the GDP- or GTP-bound form inhibit ER-to-Golgi transport in vitro (Rowe et al., 1996). Bednarek et al. (1995) showed that purified COPI and Arf can promote vesicle budding from the ER in vitro. Furthermore, some studies have shown that Arf is also involved in the transport reactions along the endocytic pathway (Lenhard et al., 1992; D'Souza-Schorey and Stahl, 1995; Gaynor et al., 1998). There is also evidence that Arf regulates four adaptin complexes, AP-1 (Stamnes and Rothman, 1993; Traub et al., 1993), AP-2 (West et al., 1997), AP-3 (Ooi et al., 1998), and AP-4 (Hirst et al., 1999).
Despite many experiments performed, the understanding about the role of Arf is still confusing and has often been a subject of controversy. Does Arf indeed play various roles in many different steps of intracellular transport in living cells? Does Arf itself manage all of these reactions? We have to point out that in vivo information about Arf is very limited to answer these questions. This is in striking contrast to Sar1 GTPase (Nakano and Muramatsu, 1989), a relative of Arf proteins, which plays an essential role exclusively in the ER-to-Golgi anterograde traffic by virtue of the assembly of another coat protein complex, COPII, on the ER membrane (Barlowe et al. 1994; Oka and Nakano, 1994). Temperature-sensitive (ts) mutants of SAR1 have been quite useful for the analysis of its in vivo roles (Nakano et al., 1994; Yamanushi et al., 1996; Saito et al., 1999). In the case of Arf, however, only one ts mutant allele of ARF1, arf1-3, has been well characterized thus far (Zhang et al., 1998). If Arf1p itself in fact regulates multiple transport steps, various mutants that are defective in particular vesicular transport reactions may be isolated.
In the present study, we have made use of the powerful yeast genetic system and isolated additional eight ts alleles of ARF1. In vivo analysis of these mutants has provided important hints for understanding the multiple functions of Arf in the cell.
MATERIALS AND METHODS
Strains, Plasmids, and Media
Yeast cells were grown in YPD (1% [wt/vol] Bacto yeast extract [Difco Laboratories, Detroit, MI], 2% [wt/vol] polypeptone [Nihon Seiyaku, Tokyo, Japan], and 2% [wt/vol] glucose) or in MVD (0.67% yeast nitrogen base without amino acids [Difco Laboratories] and 2% glucose) medium supplemented appropriately. MCD medium is MVD containing 0.5% casamino acids (Difco Laboratories). Yeast single-copy plasmids, pRS314 and pRS316, and replication plasmids, pJJ215 and pASZ10, have been described elsewhere (Sikorski and Hieter, 1989; Jones and Prakash, 1990; Stotz and Linder, 1990). pNY16-A1 was generated by cloning the 1.8-kb EcoRI-PstI fragment of the wild-type ARF1 into pRS316. pNY14-A1S was constructed by inserting the 1.7-kb NheI-PstI fragment of the wild-type ARF1 into the multicloning sites of pRS314, and one base substitution adjacent to the start codon of ARF1 was introduced to produce an EcoRI site by site-directed mutagenesis. This construct was confirmed for its ability to complement arf1 null strains. pSKY5RER1-0 is a single-copy plasmid expressing a green fluorescent protein (GFP)-Rer1p protein, in which EGFP1 (CLONTECH, Palo Alto, CA) is fused to the N terminus of Rer1p (Sato et al., 1995), under control of the TDH3 promoter. Saccharomyces cerevisiae strains used in this study are listed in Table 1. To examine intragenic complementation, arf1 ts mutants were crossed as follows. MATa and MATα cells of each mutant were transformed with pRS314 and pRS316, respectively, crossed with each other, plated on YPD for 12 h, and then streaked on MCD (−Ura, −Trp) plates to select diploids.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YPH499 | MATa ura3 lys2 ade2 trp1 his3 leu2 | Sikorski et al., 1989 |
| YPH501 | MATa/MATα ura3/ura3 lys2/lys2 ade2/ade2 trp1/trp1 his3/his3 leu2/leu2 | Sikorski et al., 1989 |
| NYY539-1 | MATa arf1∷HIS3 arf2∷HIS3 ura3 lys2 ade2 trp1 his3 leu2 [pNY16-A1] | This study |
| NYY539-2 | MATα arf1∷HIS3 arf2∷HIS3 ura3 lys2 ade2 trp1 his3 leu2 [pNY16-A1] | This study |
| NYY0-1 | MATa ade2∷ARF1∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY0-2 | MATα ade2∷ARF1∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY11-1 | MATa ade2∷arf1-11∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY11-2 | MATα ade2∷arf1-11∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY12-1 | MATa ade2∷arf1-12∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY12-2 | MATα ade2∷arf1-12∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY13-1 | MATa ade2∷arf1-13∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY13-2 | MATα ade2∷arf1-13∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY14-1 | MATa ade2∷arf1-14∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY14-2 | MATα ade2∷arf1-14∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY15-1 | MATa ade2∷arf1-15∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY15-2 | MATα ade2∷arf1-15∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY16-1 | MATa ade2∷arf1-16∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY16-2 | MATα ade2∷arf1-16∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY17-1 | MATa ade2∷arf1-17∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY17-2 | MATα ade2∷arf1-17∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY18-1 | MATa ade2∷arf1-18∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| NYY18-2 | MATα ade2∷arf1-18∷ADE2 arf1∷HIS3 arf2∷HIS3 ura3 lys2 trp1 his3 leu2 | This study |
| SEY5016 | MATα sec1-1 ura3 leu2 | Scott Emr |
| MBY3-15A | MATα sec13-1 ura3 leu2 his3 | Nakano et al., 1989 |
| YW10-2B | MATa slp1∷LEU2 ade2 | Wada et al., 1990 |
| KUY166 | MATa vps1∷HIS3 ura3 lys2 ade2 trp1 his3 leu2 | Umebayashi and Nakano, unpublished |
| SNH23-7D | MATα rer2-2 mfα1∷ADE2 mfα2∷TRP1 bar1∷HIS3 ade2 trp1 his3 leu2 ura3 lys2 | Sato et al., 1999 |
PCR Mutagenesis
To screen for ts alleles, the whole open reading frame (ORF) of the ARF1 gene was randomly mutagenized by the error-prone PCR method. The fidelity of PCR was reduced by increasing the concentration of MnCl2 in the reaction mixture as previously described (Cadwell et al., 1992). The region containing the wild-type ARF1 gene in pNY14-A1S was replaced with the mutated fragment by EcoRI and AflII digestion. To observe the phenotypes of mutant ARF1 on the plasmid, it was necessary to disrupt both ARF1 and ARF2 in chromosomes because these genes are functionally redundant. We constructed deletion-insertion mutations in ARF1 and ARF2, by inserting the 1764-bp BamHI fragment of pJJ215 containing HIS3 into the XbaI-NcoI sites of ARF1 and the XbaI-XbaI sites of ARF2, respectively. The obtained Δarf1 Δarf2 double-null mutant, whose growth depends on the wild-type ARF1 on a URA3-based plasmid, was named NYY539. NYY539 strain was transformed with the obtained library of mutagenized ARF1. The transformants were plated on the medium containing 5-fluoroorotic acid to get rid of the wild-type ARF1, checked for temperature sensitivity, and selected.
To stably observe the phenotypes of arf1 ts mutants, each mutant copy of arf1 was integrated into the chromosomal ADE2 site. The NheI-SalI fragments of ARF1 and mutant arf1 genes were subcloned into the EcoRI site of pASZ10. The resulting plasmids were digested with HpaI and introduced into NYY539. Ade+ transformants were picked up, plated on the medium containing 5-fluoroorotic acid, and selected. These stains were confirmed for the integration by genomic Southern blotting with the ECL gene detection systems (Amersham, Tokyo, Japan).
Antibodies and Immunoblotting
The antibody recognizing Arf1p was raised in a rabbit by using a C-terminal peptide CATSGEGLYEGLEWL as an antigen as previously described (Stearns et al., 1990). Rabbit anti-carboxypeptidase Y (CPY) polyclonal antibody was prepared as described previously (Stevens et al., 1982). Invertase antisera were kind gifts from Nobuhiro Nakamura and Katsuyoshi Mihara of Kyushu University (Fukuoka, Japan) and from Erin Gaynor and Scott Emr of the University of California, San Diego. Vacuolar alkaline phosphatase (ALP) and immunoglobulin heavy-chain–binding protein (BiP) antibodies were gifts from Yoh Wada of Osaka University (Osaka, Japan) and Masao Tokunaga of Kagoshima University (Kagoshima, Japan), respectively. Colony immunoblotting to examine the secretion of CPY and BiP was performed as described previously (Roberts et al., 1991).
Cell Labeling and Immunoprecipitation
Metabolic labeling of yeast cells, preparation of cell extracts, and immunoprecipitation were performed essentially as previously described (Rothblatt and Schekman, 1989; Nishikawa and Nakano, 1991; Gaynor and Emr, 1997; Peyroche et al., 1999). Briefly, log-phase yeast cells were labeled with 25 μCi of Tran35S-label (ICN Biochemicals, Costa Mesa, CA) per 1 × 107 cells and chased for appropriate times. Objective proteins were recovered from cell lysates by immunoprecipitation and resolved on SDS-polyacrylamide gels. The gels were treated with Amplify (Amersham), dried, and fluorographed. Radioimages were observed with an image analyzer BAS-2500 (Fuji Film, Tokyo, Japan). To assay internal and external invertase, cells were converted to spheroplasts with Zymolyase (Seikagaku Kogyo, Tokyo, Japan; 25 U/ml cell suspension) and incubated at 30°C for 30 min. After pulse-chase, cell suspensions were centrifuged and separated into spheroplasts and media as intracellular and extracellular fractions, respectively. To assay total proteins secreted to the medium, cells were labeled in media containing 100 μg/ml α2-macroglobulin and 300 μg/ml bovine serum albumin. Media proteins were precipitated by the addition of trichloroacetic acid (TCA) to the final concentration of 10% (wt/vol), kept on ice for 20 min, and centrifuged at 16,000 rpm for 5 min. After the wash, once with cold 5% (wt/vol) TCA and twice with cold acetone, pellets were solubilized in SDS-PAGE sampling buffer containing 5% β-mercaptoethanol, boiled, and cleared by centrifugation at 16,000 × g for 10 min. Proteins equivalent to 2.5 × 106 cells were analyzed by SDS-PAGE with a 7.5% polyacrylamide gel.
Electron Microscopy
Preparation of thin sections of yeast cells was carried out by the freeze-substitution fixation method as described by Sun et al. (1992). After brief centrifugation of cultures, pellets of cells were mounted on copper meshes to form a thin layer and plunged into liquid propane. Frozen cells were transferred to 4% OsO4 in anhydrous acetone that had been precooled in a dry ice/acetone bath and kept at −80°C for 48 h. Samples were held at −20°C for 2 h, at 4°C for 2 h, and then at room temperature for 2 h. After a wash with anhydrous acetone, samples were embedded in Spurr's resin (Nisshin EM, Tokyo, Japan). Thin sections were stained with uranyl acetate and lead citrate and observed under a JEM-2000FXII electron microscope ( JEOL, Tokyo, Japan).
Fluorescence Microscopy and Image Processing
Cell labeling with FM4-64 (Molecular Probes, Eugene, OR) was performed as described previously (Vida and Emr, 1995; Gaynor et al., 1998). Cells were grown to a logarithmic phase in YPD medium at 23°C, and then half of the culture was shifted to 37°C and incubated for an additional 1 h. Cells were harvested and resuspended at ∼2 × 108/ml in YPD medium prewarmed to 23 or 37°C and incubated for 10 min. FM4-64 was added to the final concentration of 50 μM from a stock solution of 2 mM in dimethyl sulfoxide, and after incubation for another 10 min, the cells were harvested and resuspended in fresh prewarmed medium and the incubation continued. Images of the cells were acquired with a fluorescence microscope, BX60 (Olympus, Tokyo, Japan), at ∼15–20 min and 1 h after the addition of fresh medium. GFP-RER1-expressing cells were grown to a log phase, preincubated at 23 or 37°C for 30 min, and subjected to microscopic observation. When images were to be obtained at 37°C, the temperature on the stage of the microscope was carefully controlled with a stage incubator (Tokai Hit, Shizuoka, Japan). Confocal digital images were acquired using a CSU10 confocal system (Yokogawa Electric, Tokyo, Japan) controlled and analyzed by IPLab Spectrum software (Scanalytics, Fairfax, VA).
RESULTS
Isolation of arf1 ts Mutants
S. cerevisiae has three isotypes of Arf proteins (Arf1p, Arf2p, and Arf3p). Arf1p and Arf2p are 96% identical in amino acid sequence and are thought to be redundant in function by the following two lines of evidence. First, cells disrupted for either ARF1 or ARF2 are viable, but the deletion of the both is lethal, indicating that this pair of genes provides essential function(s) that cannot be carried out by ARF3. Second, the increase of the ARF2 level was sufficient to complement the phenotypes of Δarf1 (slow growth, cold sensitivity, and fluoride supersensitivity). The level of protein produced from ARF1 is ∼10-fold higher than that from ARF2, which explains why Δarf2 displays no growth phenotype (Stearns et al., 1990). The third Arf protein, Arf3p, is structurally distant from Arf1p/Arf2p; its deletion by itself or in combination with Δarf1 or Δarf2 is viable, and Δarf3 cells do not show a severe defect of protein transport (Lee et al., 1994). Thus, we decided to focus on Arf1p and examine the effect of mutations in ARF1 with the deletion of ARF2.
To screen for ts alleles, we used the error-prone PCR method and randomly mutagenized the whole ORF of the ARF1 gene. The mutated gene was introduced to yeast cells under the Δarf1 Δarf2 background. Among 2000 colonies we screened, 11 gave a ts phenotype reproducibly. Plasmids were recovered and subjected to determination of the mutation points in DNA and amino acid sequences. Most of the ts plasmids had more than one mutation in the ARF1 ORF. To examine which mutation gave the ts phenotype, we separated the mutations by restriction enzyme digestions and tested the ts growth again.
As shown in Table 2, six single mutations in ARF1 (arf1-12, arf1-13, arf1-14, arf1-15, arf1-17, and arf1-18) conferred ts growth to cells under the Δarf1 Δarf2 background. In the cases of arf1-11 and arf1-16, the original double and triple mutations gave greater temperature sensitivity than the single mutations. We selected these alleles as well and analyzed the total eight ts mutants further. In all experiments hereafter, we used strains in which ARF1 and ARF2 loci were disrupted, and one of the eight ts mutant arf1 alleles was integrated at the ADE2 locus in the chromosome. The wild-type ARF1 was also integrated at the ADE2 locus in the same background and was always used as a control to exclude the possibility that the disruption of ARF2 had any effect. The growth of these mutant cells on plates at various temperatures is summarized in Table 2. Although the degree of temperature sensitivity differs, they all ceased growth almost completely at 37°C.
Table 2.
The newly obtained eight arf1 ts mutant alleles: mutation points in the amino acid sequence and growth phenotypes
| Mutation points | Growth
|
||||||
|---|---|---|---|---|---|---|---|
| 15°C | 23°C | 26.5°C | 30°C | 35°C | 37°C | ||
| Wild type | +++ | +++ | +++ | +++ | +++ | +++ | |
| Δarf2 | +++ | +++ | +++ | +++ | +++ | +++ | |
| arf1-11 | K38T E132D L173S | +++ | +++ | +++ | +/− | − | − |
| arf1-12 | K38T | +++ | +++ | +++ | +++ | +++ | +/− |
| arf1-13 | L173S | +++ | +++ | +++ | +++ | +++ | +/− |
| arf1-14 | F51I | +++ | +++ | +++ | +++ | + | − |
| arf1-15 | L25P | +++ | +++ | +++ | +++ | +++ | +/− |
| arf1-16 | E41V D129E | ++ | ++ | ++ | +/− | − | − |
| arf1-17 | E41V | +++ | +++ | +++ | +++ | +++ | − |
| arf1-18 | H80P | +++ | +++ | +++ | +++ | +++ | +/− |
Growth was examined on YPD plates. +++, wild-type growth; ++, growth slower than wild type; +, very slow growth (minimal single-colony formation); +/−, almost no growth (no single-colony formation); −, completely no growth.
The mutation points of the eight ts mutants are shown in Table 2. It should be noted that arf1-11 contains the two point mutations that are present in arf1-12 (K38T) and arf1-13 (L173S) plus E132D. The E132D mutation is not silent because the triple mutant showed much clearer ts growth than the K38T L173S double mutant. The D129E mutation in arf1-16 (E41V D129E) is not silent either because arf1-16 is much tighter than arf1-17 (E41V alone). Immunoblotting analysis with the anti-Arf1p antibody indicated that the products of the mutant arf1 genes were stable up to at least 3 h after a temperature shift to 37°C (Yahara and Nakano, unpublished data). Cell viabilities of two tight alleles, arf1-11 and arf1-16, decreased quickly at 37°C (∼50% at 4 h). Other alleles remained 40–60% viable at 37°C for 24 h. All of these arf1 ts alleles were recessive to the wild type, because the heteroallelic diploids constructed between any arf1 mutant and wild-type cells grew at 37°C, and a single-copy plasmid of wild-type ARF1 rescued the ts growth of all arf1 mutants (Yahara and Nakano, unpublished data).
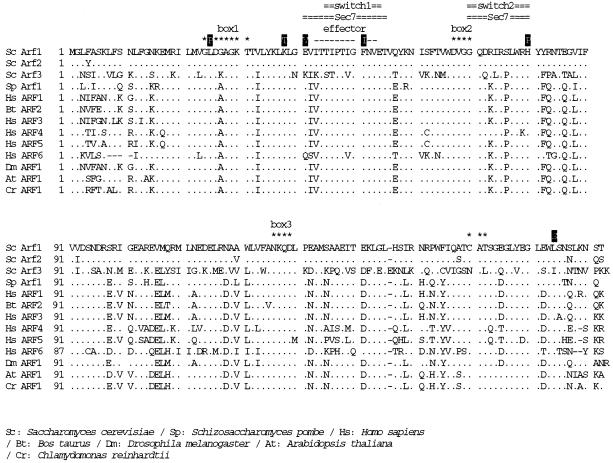
The mutation points in the amino acid sequence that cause ts phenotype by a single mutation are mapped in Figure 1 with a comparison of Arf sequences from various organisms. All of them were on well-conserved residues.
Figure 1.
The mutation points of yeast Arf1p, which cause ts phenotype as a single mutation, and the sequence comparison of Arf1p from various organisms. Dots indicate identity with S. cerevisiae Arf1p. Consensus GTP-binding (DVGG, NKQD, and CAT) and hydrolysis (GX4GK) sequences are marked with asterisks. The putative effector domain is shown by dashes. The site of Sec7 domain interaction on human ARF1 and the switch 1 and switch 2 GTPase regions (Mossessova et al., 1998) are marked with double lines. Sc, S. cerevisiae; Sp, Schizoaccharomyces pombe; Hs, Homo sapiens; Bt, Bos taurus; Dm, Drosophila melanogaster; At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii. The database accession numbers from GenBank are as follows: Sc Arf1, P11076; Sc Arf2, P19146; Sc Arf3, P40994; Sp Arf1, AL031534.1; Hs ARF1, NM 001658.1; Bt ARF2, P16500; Hs ARF3, NM 001659.1; Hs ARF4, NM 001660.1; Hs ARF5, NM 001662.1; Hs ARF6, NM 001663.1; Dm ARF1, P35676; At ARF1, P36397; Cr ARF1, U27120.1.
Analysis of Protein Transport in arf1 ts Mutants
The first question we wanted to address concerning these arf1 mutants was whether any of them show a particular transport defect when the cells were shifted to the restricted temperature. By looking at various types of marker molecules, we examined protein transport in vacuolar, secretory, and endocytic pathways as follows.
Intracellular Transport of CPY and ALP (ER→Golgi→Vacuole).
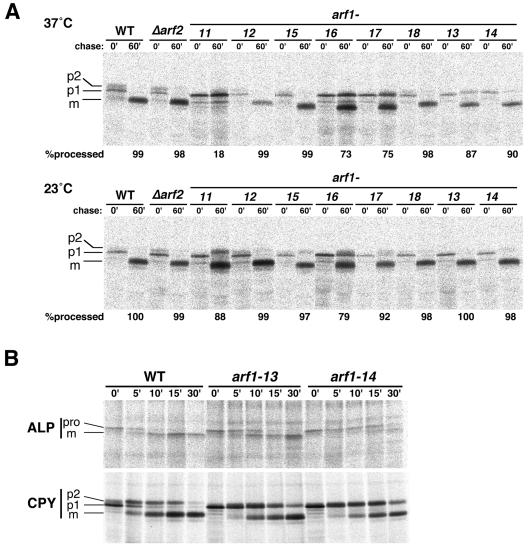
First, we performed pulse-chase experiments on a vacuolar soluble protein, CPY, to analyze the anterograde transport from the ER via the Golgi apparatus to the vacuole. The fate of CPY in the biosynthetic pathway has been very well characterized; the ER form (p1) with core oligosaccharides is modified to the Golgi form (p2) by elongation of sugar chains and finally converted to the mature form in the vacuole by proteolytic processing (Stevens et al., 1982). Wild-type and mutant cells were preincubated at the permissive (23°C) or restricted (37°C) temperature for 30 min and subjected to 35S-pulse–labeling and chase at the same temperature. CPY was recovered by immunoprecipitation and analyzed by SDS-PAGE and fluorography. As shown in Figure 2A, the conversion of CPY from p1 through p2 to the mature form (m) was completed much earlier than 60 min in controls (the wild-type cells [WT] and the integrant of wild-type ARF1 in the Δarf1 Δarf2 background [Δarf2]). In arf1-13, arf1-14, arf1-16, and arf1-17 alleles, the transport of CPY was partially delayed at the restrictive temperature (37°C), as seen by the residual p1 and p2 forms at 60 min. The mature form appeared to be underglycosylated in these alleles, especially in arf1-16 and arf1-17. Interestingly, arf1-11 cells showed an almost complete block in the p1 stage at 37°C. This accumulating p1 CPY was not immunoprecipitable with the anti-α1,6-mannose antibody (Yahara and Nakano, unpublished data), strongly suggesting that the transport of CPY was arrested in the ER in the arf1-11 mutant. In arf1-12, arf1-15, and arf1-18, CPY was transported to the vacuole with kinetics similar to the wild-type cells. At the permissive temperature (23°C), the transport of CPY was normal in most of the mutants except for arf1-11 and arf1-16, which showed some retardation.
Figure 2.
Pulse-chase experiments to follow the intracellular transport of CPY and ALP. (A) Wild-type cells (YPH499, WT), integrants of wild-type ARF1 (NYY0, Δarf2), and arf1 ts mutant alleles (arf1-11–arf1-18) were grown to an early log phase at 23°C, preincubated at 37°C for 30 min, labeled with Tran35S-label for 4 min, and chased. Aliquots were withdrawn at the indicated times, subjected to sampling for immunoprecipitation with the anti-CPY antiserum, and visualized by SDS-PAGE and fluorography. p1, ER-precursor form; p2, Golgi-precursor form; m, mature form. Quantitation of CPY processing at the 60-min chase point is indicated as the percentage of the total. (B) Wild-type, arf1-13 and arf1-14 cells were subjected to a similar but more precise pulse-chase and immunoprecipitation experiment with anti-ALP and anti-CPY antisera. The migration positions of the precursor (pro) and mature (m) forms of ALP are also indicated.
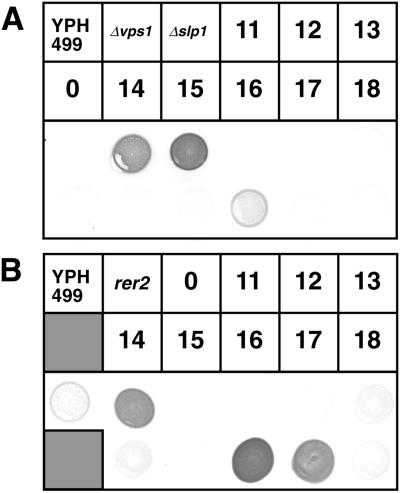
Certain aberrations in Golgi functions are known to cause missecretion of CPY to the medium, which is seen in a large number of vps (vacuolar sorting defective) mutants. To test whether such a Vps− phenotype is observed in arf1 ts mutants, we performed a colony-blotting assay. As shown in Figure 3A, obvious secretion of immunoreactive CPY was not detected in most of the arf1 ts mutants except arf1-16. arf1-16 did secrete CPY to the medium, although the degree of missecretion was not as remarkable as in vps controls, Δvps1 and Δslp1 (Δvps33) (Wada et al., 1990; Vater et al., 1992). We further asked whether an ER-resident protein, BiP, is also missecreted in arf1 ts mutants. Interestingly, arf1-16 as well as arf1-17 secreted a large amount of BiP, which is comparable to the BiP secretion defect of an ER localization mutant, rer2 (Sato et al., 1999) (Figure 3B).
Figure 3.
Colony-blotting analysis of CPY and BiP missecretion. Integrants of wild-type ARF1 (NYY0:0) and arf1 ts mutant alleles (arf1-11, -12, -13, -14, -15, -16, -17, and -18) were grown on YPD for 1.5 d. Prewetted nitrocellulose filters were overlaid onto the streaks of the cells, incubated at a semipermissive temperature (35°C) for 20 h, and let react with anti-CPY or anti-BiP antisera to reveal secreted CPY (A) and BiP (B). Δvps1 (KUY165), Δslp1 (YW10–2B), and rer2 (SNH23-7D) were used as positive controls.
The transport kinetics of another vacuolar protein, ALP, was also examined. ALP, the product of the PHO8 gene, is a type II membrane protein that undergoes a PEP4-dependent cleavage when reaching the vacuole (Klionsky and Emr, 1989). Previous studies have demonstrated that CPY and ALP traffic to the vacuole by way of two different routes, namely, the CPY pathway via the prevacuolar late endosome and the ALP pathway, which bypasses the endosome (Cowles et al., 1997; Conibear and Stevens, 1998). The arf1 mutant cells showed a variety of defects in the maturation of ALP, in a similar manner to that of CPY, i.e., the processing was almost completely blocked in arf1-11, was partially blocked in arf1-13, arf1-14, and arf1-16, and went on normally in arf1-12, arf1-15, and arf1-18. The data of arf1-13 and arf1-14 are shown in Figure 2B. Here we noticed that the delay of ALP transport was much more severe in arf1-14 than in arf1-13, whereas such a difference was not obvious in the kinetics of CPY processing.
Intracellular Transport of Invertase (ER→Golgi→Periplasm).
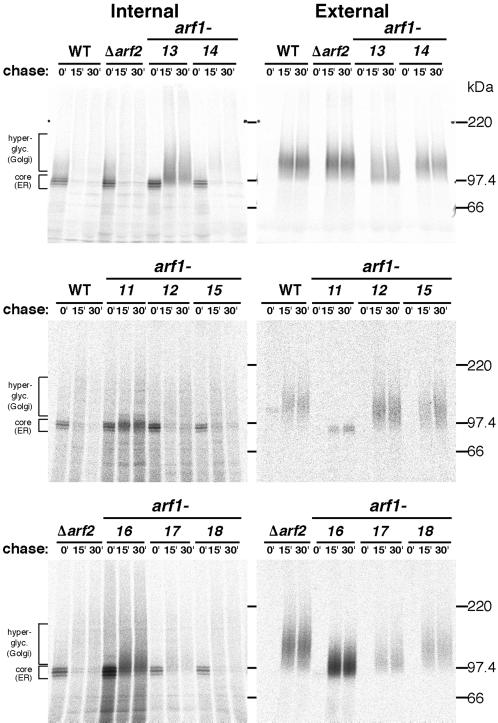
Next, the secretion of invertase was examined. Because invertase acquires 9–10 N-linked oligosaccharide chains, which are heterogeneously modified in the Golgi apparatus, it becomes highly glycosylated when secreted and thus gives a high molecular mass smear in SDS gels. The cells were converted to spheroplasts, incubated at 37°C for 30 min, pulse-labeled and chased at 37°C, and separated into intracellular and extracellular (periplasmic) fractions from which invertase was immunoprecipitated. The results are shown in Figure 4. In the controls (WT and Δarf2), most of invertase was secreted during a 15-min chase. In arf1-12, arf1-14, arf1-15, and arf1-18, invertase was secreted with kinetics similar to the wild-type cells. In arf1-13, ∼50% of invertase remained intracellular even after 30 min. In addition, the secreted form of invertase seemed to be underglycosylated. In arf1-11 cells, invertase was mostly retained intracellularly with the ER form (core), and a small amount of the ER-form invertase appeared to be secreted. arf1-16 and arf1-17 cells showed marked defects in glycosylation and secreted underglycosylated invertase.
Figure 4.
Pulse-chase experiments of invertase secretion. Wild-type cells (YPH499, WT), integrants of wild-type ARF1 (NYY0, Δarf2), and arf1 ts mutant alleles (arf1-11–arf1-18) were grown to an early log phase at 23°C, converted to spheroplasts, induced to produce invertase, preincubated at 37°C for 30 min, and labeled with Tran35S-label for 4 min. Aliquots were withdrawn at the indicated times and then separated by centrifugation into internal and external fractions. Invertase was recovered by immunoprecipitation and visualized by SDS-PAGE and fluorography.
General Secretion (ER→Golgi→Medium).
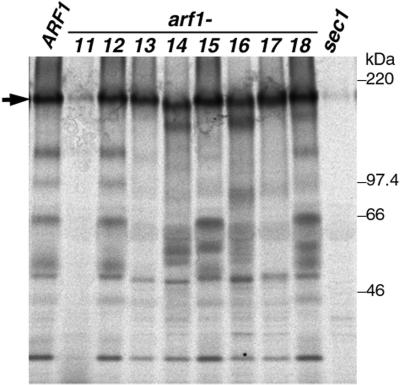
We also assayed the general secretion competence of the arf1 mutants by examining the bulk secretion of proteins into the medium. Whole cells were incubated at 37°C for 30 min, pulse-labeled, and chased for 30 min. Cells and media were separated by centrifugation, and proteins in the media were precipitated with TCA and resolved by SDS-PAGE. As shown in Figure 5, several bands were apparent in the medium in the control. Gaynor and Emr (1997) showed a similar result and revealed that the particularly abundant protein migrating at ∼150 kDa (arrow) is HSP150. Virtually no proteins were secreted to the medium by either sec23-1 or sec1-1 mutant cells, which have a defect in the canonical secretory pathway (Gaynor and Emr, 1997; Figure 5, sec1).
Figure 5.
General secretion by arf1 ts mutants. Wild-type control (NYY0, ARF1) and arf1 ts mutant cells (arf1-11–arf1-18) were grown to an early log phase at 23°C, preincubated at 37°C for 30 min, labeled with Tran35S-label for 10 min, and then chased for 30 min. Cells and media were separated by centrifugation, and proteins secreted into the media were precipitated with TCA and analyzed by SDS-PAGE and fluorography. The secreted HSP150 is indicated by an arrow. SEY5016 (sec1) was used as a control in this experiment.
In arf1-11 cells, the secretion of these proteins was also almost completely blocked, again implying defects of this mutant in the anterograde transport in the secretory pathway. arf1-13, arf1-14, arf1-15, arf1-16, and arf1-17 cells showed various band patterns of secreted proteins, suggesting that in these arf1 mutants some proteins were efficiently secreted to the medium, whereas others were not. The secreted HSP150 was underglycosylated in arf1-14 and arf1-16 cells. arf1-12 and arf1-18 cells looked normal in the secretion of these proteins.
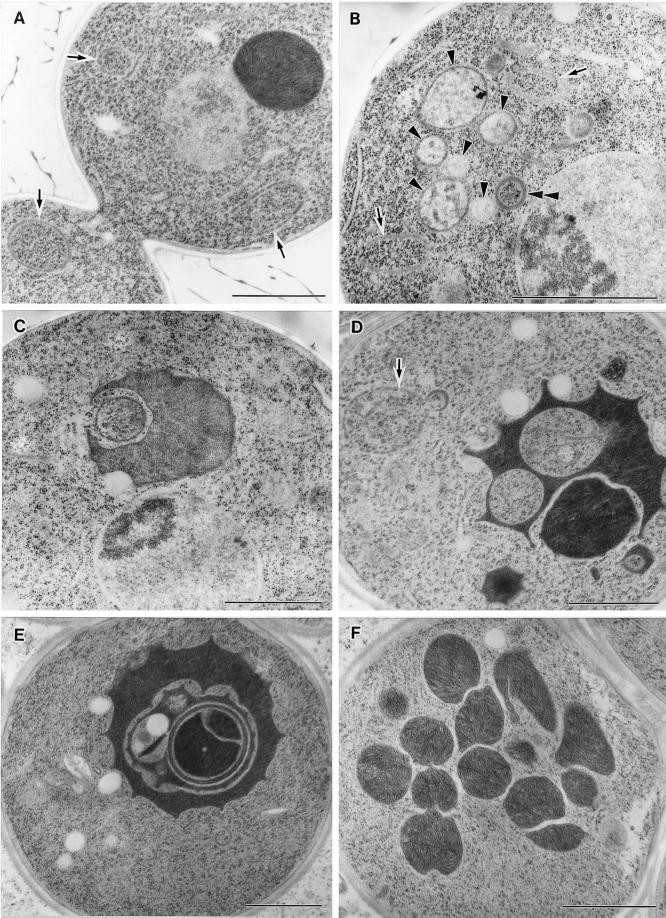
The observations that arf1-11 cells accumulated ER precursor forms of CPY and invertase and were defective in general secretion at 37°C led us to postulate that this mutant has a lesion in the ER-to-Golgi transport. To confirm this, we examined the morphological alterations of the mutant cells by electron microscopy. The wild-type and arf1-11 cells were incubated at 37°C for 1 h and then processed for freeze-substitution electron microscopy. As shown in Figure 6, arf1-11 mutant cells accumulated ER membranes (Figure 6B) just like the ER-to-Golgi sec mutants and sar1 ts mutants (Novick et al., 1980; Yamanushi et al., 1996). Incubation at 37°C for 4 h led to further exaggeration of the ER (Figure 6C).
Figure 6.
Electron microscopic observation of arf1-11 ts mutant cells. Wild-type (YPH499, WT) (A) and arf1-11 ts mutant cells (B and C) were grown to an early log phase at 23°C, incubated at 37°C for 1 h (B) and 4 h (A and C), prepared for electron microscopy by the freeze-substitution fixation method, and observed under an electron microscope. Bars, 1.0 μm.
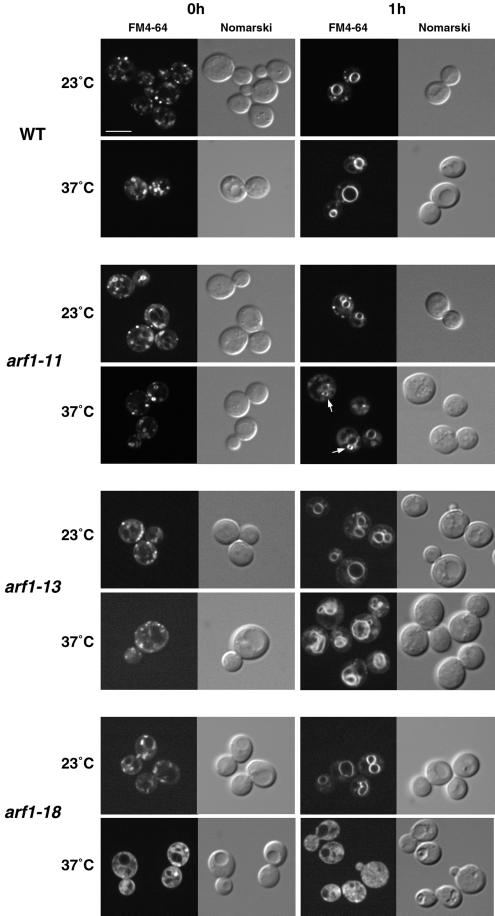
FM4-64 Staining (Plasma Membrane→Endosome→Vacuole).
Arf has also been implicated in endosomal dynamics. We examined the behaviors of endocytic organelles by vital staining with a lipophilic styryl dye, FM4-64 [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)-hexa-trienyl)-pyridinium dibromide]. FM4-64 has been used as an endocytic tracer that travels from the plasma membrane to the vacuolar membrane via endosomes in yeast (Vida and Emr, 1995; Gaynor et al., 1998). We analyzed uptake of FM4-64 by the arf1 mutant cells. The cells were preincubated at either 23 or 37°C for 1 h, exposed to 50 μM FM4-64 for 10 min, and resuspended in fresh medium prewarmed to the same temperature. Aliquots were taken at 0 and 1 h and observed under a confocal laser scanning microscope. As shown in Figure 7 (top), small discrete fluorescent dots, which probably represent early endosomes, appeared in the cytoplasm of wild-type cells within ∼15 min after labeling with the dye (0 h). After 1 h, FM4-64 stained the vacuolar membrane. In arf1-12, arf1-14, arf1-15, arf1-16, and arf1-17 cells, the change of the FM4-64 staining pattern during 1 h was indistinguishable from that of the wild type at either 23 or 37°C (Yahara and Nakano, unpublished data).
Figure 7.
FM4-64 staining of endocytic membranes in arf1 ts mutants. Wild-type (YPH499, WT) and arf1 ts mutant cells (arf1-11, arf1-13, and arf1-18) were grown to a logarithmic phase in YPD medium at 23°C, and then half of the culture was shifted to 37°C and incubated for 1 h. Cells were harvested and resuspended at ∼2 × 108/ml in YPD medium prewarmed to 23 or 37°C and incubated for 10 min. FM4-64 was added to the final concentration of 50 μM. After incubation for another 10 min, the cells were harvested and resuspended in fresh prewarmed medium and the incubation continued. Images of the cells were acquired at ∼15–20 min (0 h) and 1 h after the addition of fresh medium and visualized by fluorescence confocal (left) and Nomarski (right) microscopy. Bar, 5 μm.
In arf1-11 cells, ring structures were observed at 37°C (arrows), which were distinct from the larger, more faintly stained vacuoles. The frequency of the appearance of these ring structures at 1 h was much higher in arf1-11 cells (∼40% of the cells) than in the wild type (<1%).
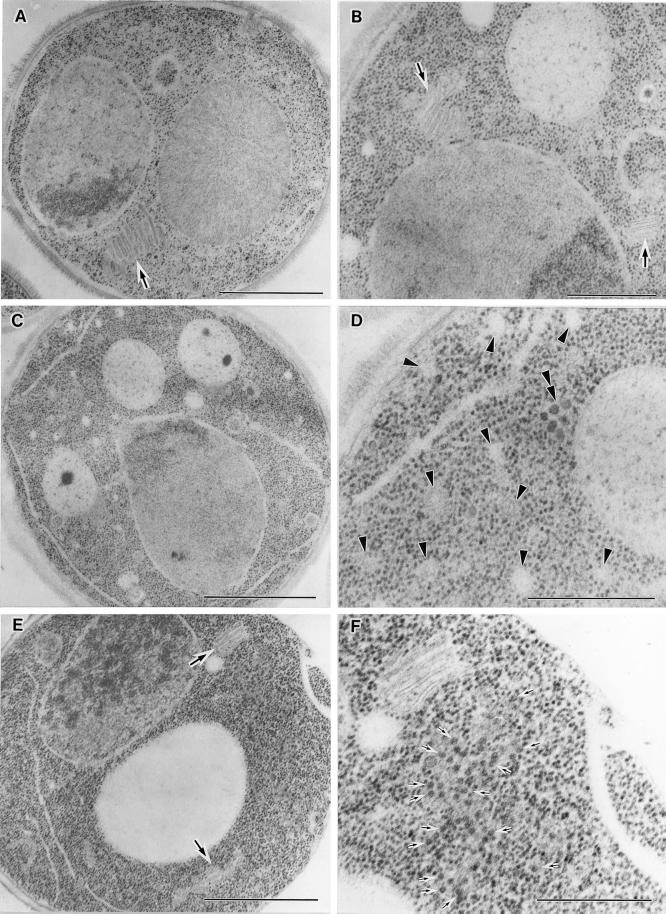
arf1-13 cells showed a different type of anomaly in the staining of FM4-64 at 37°C. The dye appeared to have reached vacuoles by 1 h, but the morphology of vacuolar membranes was quite irregular. They were often distorted and sometimes fragmented. To examine this disorder in more detail, we made electron microscopic observations of the arf1-13 mutant (Figure 8). Even at 23°C, arf1-13 cells often showed discontinuous ring-like structures indicated by arrows (Figure 8A). Similar structures were also seen when the cells were incubated at 37°C (Figure 8, B and D). When the cells were incubated at 37°C for 1 h, however, a even more prominent phenotype was accumulation of membranes, which appeared to contain internal vesicle-like structures (Figure 8B). They may correspond to late endosomes or multivesicular bodies (MVBs) as reported by Odorizzi et al. (1998). A small portion of cells also revealed morphological alteration of vacuoles (Figure 8C). Further incubation of arf1-13 cells at 37°C (4 h) led to clear appearance of electron-dense vacuoles which exhibited distorted shapes. The vacuole membranes showed intricate patterns, which often look like multilamellar structures (Figure 8E) and were sometimes fragmented (Figure 8F). It should be noted that the cells showing deformed vacuoles in the FM4-64 experiment (Figure 7) were incubated at 37°C for 2 h including preincubation. In fact, longer incubation (4 h) at 37°C resulted in more marked aberration of vacuoles as seen by FM4-64 staining (Yahara and Nakano, unpublished data). Thus, the arf1-13 cells seem to accumulate MVB-like structures first and then gradually proceed to vacuole deformation.
Figure 8.
Electron microscopic observation of the arf1-13 ts mutant. arf1-13 ts mutant cells were grown to an early log phase at 23°C, further incubated at 23°C (A) or at 37°C for 1 h (B and C) and 4 h (D, E, and F), and then subjected to freeze-fixation electron microscopy. In A, B and D, ring structures are indicated by arrows. Arrowheads in B indicate MVB-like structures. The vesicle marked with a double arrowhead harbors a double membrane. Bars, 1.0 μm.
arf1-18 showed a most striking phenotype in the staining with FM4-64 (Figure 7). When the cells were incubated at 37°C, no obvious staining of membrane structures was observed and the dye appeared to be diffused in the cytoplasm. Because it is very unlikely that FM4-64, which is very lipophilic, can stay soluble in the cytosol, this pattern may indicate that very small vesicular structures which are enriched with the dye are scattering in the cytoplasm (see below).
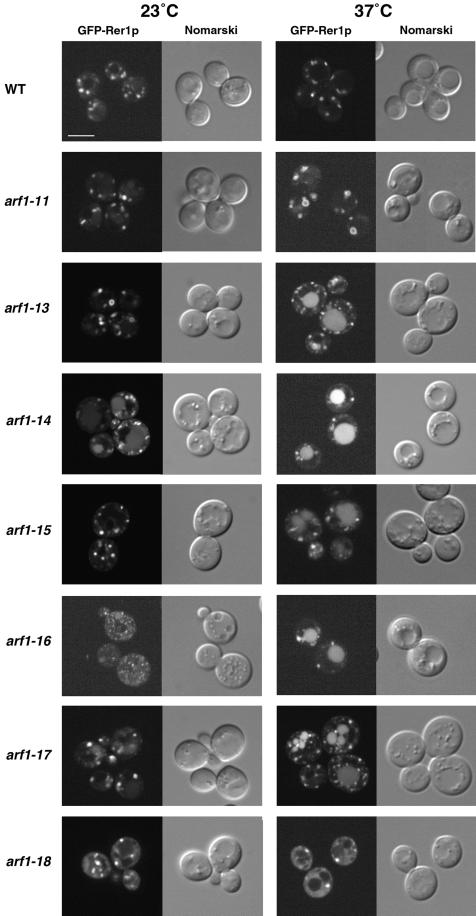
Localization of GFP-Rer1p
As mentioned in the introduction, the COPI coat complex whose assembly is regulated by Arf has been shown to play an important role in the Golgi-to-ER retrograde transport. To directly monitor this transport reaction in the arf1 ts mutant cells, we decided to adopt a morphological method as follows. Ken Sato, one of the authors of this paper, has been working on Rer1p, a membrane protein in the cis-Golgi. Rer1p is required for the correct localization of a set of ER membrane proteins such as Sec12p, Sec63p, and Sec71p by a retrieval mechanism utilizing COPI vesicles (Nishikawa and Nakano, 1993; Sato et al., 1995, 1997). To observe the dynamic behavior of Rer1p in living yeast cells, Sato constructed a fusion protein between Rer1p and the green fluorescence protein (GFP-Rer1p) and found that its localization is determined by the equilibrium of very active membrane recycling (Sato, Sato, and Nakano, unpublished data). In wild-type cells, GFP-Rer1p is normally localized to the Golgi apparatus. If the ER-to-Golgi anterograde transport is blocked by mutations in COPII, recycling of GFP-Rer1p to the ER continues and eventually the protein is completely relocated to the ER. If the Golgi-to-ER retrograde transport is inhibited by mutations in COPI, on the other hand, GFP-Rer1p is mostly mistransported to vacuoles.
These findings are immediately applicable to arf1 ts mutants. For example, if a particular ts allele of arf1 is defective in the anterograde ER-to-Golgi transport but not in the retrograde traffic like COPII mutants, GFP-Rer1p should relocate to the ER. The GFP-Rer1p plasmid was introduced into the wild-type and arf1 ts mutant cells, and the transformants were observed under a confocal laser scanning microscope after 30 min of preincubation at 23 or 37°C. As shown in Figure 9, wild-type cells displayed large punctate structures, a typical pattern of the yeast Golgi apparatus. Quite interestingly, the arf1-11 mutant, which we concluded was defective in the ER-to-Golgi anterograde transport, did not change the localization of GFP-Rer1p to the ER pattern at the restrictive temperature. The ts phenotype of arf1-11 was not suppressed by the GFP-Rer1p plasmid (Yahara, Sato, and Nakano, unpublished data). This observation strongly argues that arf1-11 has a defect in the Golgi-to-ER retrograde transport as well. Furthermore, ring-like structures (Figure 9, arrows), which were never seen in wild-type cells, were observed very frequently (∼50% of the cells). Weak staining of vacuoles was also observed occasionally.
Figure 9.
Localization of GFP-Rer1p in arf1 ts mutants. Wild-type (YPH499, WT) and the indicated arf1 ts mutant cells were grown to an early log phase at 23°C, and then half of the culture was shifted to 37°C and incubated for 30 min. Images were visualized by fluorescence confocal (left) and Nomarski (right) microscopy. Bar, 5 μm.
In arf1-13, arf1-14, arf1-15, arf1-16, and arf1-17 cells, mislocalization of GFP-Rer1p to vacuoles was obvious. Such behavior of GFP-Rer1p is similar to the case of a COPI mutant, ret1-1 (Sato, Sato, and Nakano, unpublished data), suggesting that these alleles are impaired in the Golgi-to-ER traffic. arf1-14, arf1-16, and arf1-17 cells showed weak staining of vacuoles even at the permissive temperature. arf1-13 cells exhibited ring-like structures of GFP-Rer1p at 23°C. Electron microscopic observation of arf1-13 also revealed the appearance of large ring structures frequently at 23°C and occasionally at 37°C (Figure 8, A, B, and D, arrows).
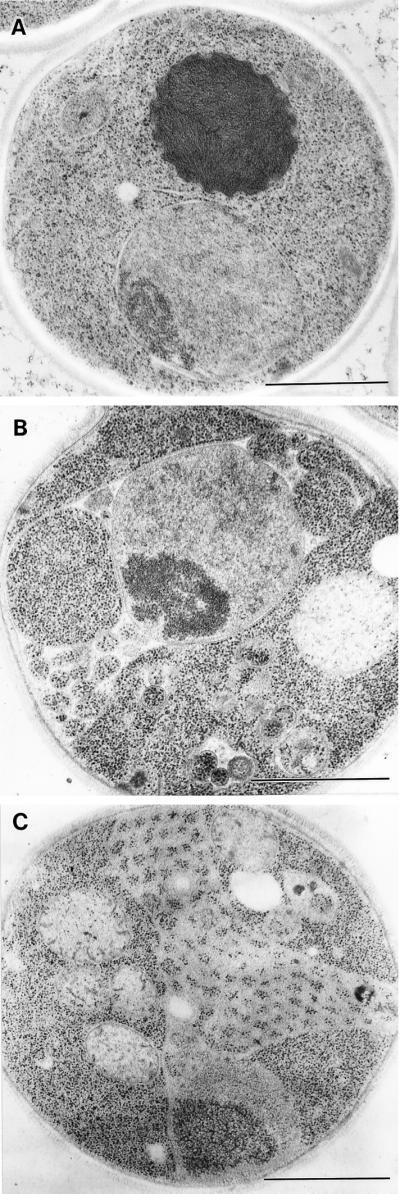
arf1-18 cells showed a dispersed pattern of GFP-Rer1p. Even at the permissive temperature, diffuse staining of cytoplasm was observed together with a few bright spots. At the restrictive temperature, the cytoplasmic staining became clearer and the number of the bright spots decreased. This observation reminds us of the FM4-64 staining of this mutant, which also showed a dispersed pattern at 37°C. To examine whether such seemingly cytosolic staining was due to the degradation of GFP-Rer1p, which would release the soluble GFP moiety into the cytosol, we examined the wild-type and arf1-18 cells by immunoblotting with an anti-GFP antibody. However, no degradation was detected (Yahara, Sato, and Nakano, unpublished data). We further asked whether this mutant accumulates any abnormal membrane structures in the cells by electron microscopy. The result is shown in Figure 10. We observed exaggeration of the ER and Golgi at both 23 and 37°C (Figure 10, A, B, and E). The Golgi apparatus sometimes formed large stacks (Figure 10, A, B, and E, large arrows). At 37°C, a closer look manifests accumulation of various kinds of vesicles, such as structures indicated by arrowheads and the double arrowhead in Figure 10D and small arrows in Figure 10F. Membranes are not always visible for the structures marked with arrowheads, but the exclusion of ribosomes is obvious. The diameter of vesicles indicated by small arrows is as small as 20–25 nm (Figure 10F). The origin of these vesicles remains to be studied.
Figure 10.
Electron microscopic observation of the arf1-18 ts mutant. arf1-18 ts mutant cells were grown to an early log phase at 23°C, further incubated at 23°C (A) or at 37°C for 1 h (B, C, and D) and 2 h (E and F), and then subjected to electron microscopy. In A, B, and E, large arrows mark Golgi structures. In D and F, various kind of vesicles are indicated by arrowheads, a double arrowhead, and small arrows. Bars: A, C, and E, 1.0 μm; B, D, and F, 500 nm.
Intragenic Complementation between arf1 Alleles
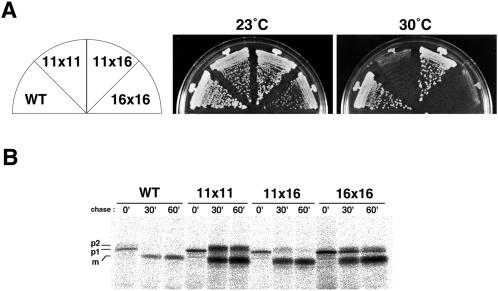
As hitherto described, arf1 ts mutants exhibit a variety of transport phenotypes in an allele-specific manner. If Arf1 indeed executes multiple functions and different ts mutants have different lesions, intragenic complementation between the alleles could be observed. Such a genetic test has proved useful for the analysis of multifunctional genes (e.g., HIS4 and CMD1; Bigelis et al., 1981; Ohya and Botstein, 1994), because a mutation affecting only one step may well complement another mutation affecting a different step. We prepared MATa and MATα strains from each arf1 ts allele and crossed all possible combinations except for leaky alleles, arf1-12, and arf1-15. Heteroallelic diploids were selected together with homoallelic diploids as controls, and we examined them for their growth at restrictive temperatures. The results are summarized in Table 3, and one example of plate growth is shown in Figure 11A. From Table 3, it is clear that the six arf1 alleles can be divided into three groups: 1) arf1-11, arf1-13, and arf1-14; 2) arf1-16 and arf1-17; and 3) arf1-18. Basically, intragenic complementation indicated by '+' was observed between alleles of different groups but not of the same group. Only one exception was arf1-13 × arf1-16, which failed to complement each other. In the case of arf1-11 × arf1-16, the heterozygous diploid grew well at 30°C but not at 37°C. This pair is interesting because the temperature sensitivity of these two alleles is very tight and both show a defect in intracellular transport of CPY. We examined whether the arf1-11 × arf1-16 heterozygous diploid had complemented the transport defect of each allele at 30°C by a pulse-chase experiment of CPY (Figure 11B). The homozygous diploids of arf1-11 (11 × 11) and arf1-16 (16 × 16) both showed retardation of conversion from p1 to p2 and from p2 to m. The heterozygote (11 × 16), however, recovered almost normal conversion of these CPY forms, indicating that arf1-11 and arf1-16 in fact complemented each other in terms of the intracellular transport of CPY.
Table 3.
Intragenic complementation between ts arf1 mutants
| 0α | 11α | 13α | 14α | 16α | 17α | 18α | |
|---|---|---|---|---|---|---|---|
| 0a | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 11a | +++ | − | − | − | +++a | + | ++ |
| 13a | +++ | − | − | − | − | + | +++ |
| 14a | +++ | − | − | − | + | +++ | +++ |
| 16a | +++ | +++a | − | + | − | − | +++ |
| 17a | +++ | + | + | +++ | − | − | +++ |
| 18a | +++ | ++ | +++ | +++ | +++ | +++ | − |
MATa and MATα cells of wild-type ARF1 (0) and arf1 ts mutants (arf1-11, arf1-13, arf1-14, arf1-16, arf1-17, and arf1-18) were crossed in all combinations. Diploids were selected and grown on YPD plates at 37°C for 4 d except for arf1-11 × arf1-16 (
, 30°C). +++, wild-type growth; ++, growth slower than wild type; +, very slow growth (minimal single-colony formation); −, completely no growth.
Figure 11.
Intragenic complementation between ts arf1 mutants. (A) Wild-type (WT), arf1-11 (11), and arf1-16 (16) cells were crossed with each other. Diploids were selected by complementary auxotrophic markers in the strains and grown on YPD plates at the indicated temperatures for 4 d. (B) Intragenic complementation as for intracellular transport of CPY between arf1-11 and arf1-16. The diploid cells were pulse-chased at 30°C as described in the legend of Figure 2.
DISCUSSION
The aim of this study was to examine in vivo whether a single Arf1 protein can execute multiple functions in a yeast cell. The answer was clearly yes. The ts mutant alleles of arf1 that we constructed showed a variety of phenotypes in terms of intracellular protein transport and membrane organizations in an allele-specific manner. Even more compelling evidence was the intragenic complementation between arf1 alleles. A particular type of arf1 mutation could complement another allelic mutation in diploid cells, indicating that these two mutations caused lesions in different functions of Arf1p.
Classification of arf1 ts Mutants
Among the eight ts alleles of arf1 that we obtained, six have been divided into three intragenic complementation groups: 1) arf1-11, arf1-13, and arf1-14; 2) arf1-16 and arf1-17; and 3) arf1-18 (Table 3). In general, two mutant alleles from different complementation groups can remedy each other in diploid cells. Such complementation is not only for ts growth. The ts transport defect of CPY, which is clear in arf1-11 and arf1-16, is almost completely cured in the arf1-11 × arf1-16 diploid at 30°C (Figure 11B). These results strongly suggest that the seemingly similar phenotypes are caused by different types of lesions.
Arf1 of different mutant alleles might complement each other by forming a dimer complex. In fact, mammalian Arf1 crystallizes as a dimer (Amor et al., 1994) and a photo-cross–linking experiment suggests the presence of Arf homodimers in solution as well (Zhao et al., 1999). Even though dimerization occurs, however, the distinct phenotypes of different alleles cannot be explained by the single action of a single dimer. Pleiotropic phenotypes would be only understood as the consequence of differentiated interaction with other components.
Phenotypes of arf1 ts Mutants
It is not easy to briefly describe the phenotypes of all mutant alleles of arf1. Many of them show partial defects in some transport processes but are not completely blocked. It should be pointed out, however, that the degree of a transport defect may not necessarily indicate the degree of deficiency in that particular step. For example, missorting of a subset of cargo receptors may result in secondary defects of transport, as has been discussed by Gaynor and Emr (1997) for the pleiotropic defect of the general secretion in sec21, the mutant of COPI γ subunit. Because COPI vesicles can be used in the Golgi-to-ER retrograde transport and in the trans-to-cis Golgi intercisternal retrograde transport, imbalance of either the ER-Golgi recycling or the cis-trans recycling in the Golgi could result in mistargeting of sorting receptors in the ER or in the Golgi. Rer1p, the retrieval receptor for Sec12p and many other ER membrane proteins, appears to be correctly localized to the cis-Golgi only when these two recycling reactions balance (Sato, Sato, and Nakano, unpublished data). GFP-Rer1p is mistargeted to the vacuole in several arf1 ts mutants, suggesting that early- or late-recycling processes may be impaired in those mutants.
Paying attention to these precautions, we will summarize and discuss the properties of the three class of mutations as follows.
Group 1 (arf1-11, arf1-13, and arf1-14).
arf1-11 is the only allele among eight that shows a very clear defect in the ER-to-Golgi transport. It accumulates ER precursors of CPY and invertase and exaggerates ER membranes at the restrictive temperature (Figures 2, 4, and 6). However, it is distinct from the typical sec mutants that are defective in the ER-to-Golgi anterograde traffic. GFP-Rer1p, a visible marker of the yeast cis-Golgi, is in a dynamic equilibrium of ER-Golgi vesicle recycling and relocates to the ER in early sec mutants such as sec12, sec13, and sec23 (Sato, Sato, and Nakano, unpublished data). In the arf1-11 mutant, however, the fluorescence of GFP-Rer1p never changes to the ER pattern (Figure 9). This suggests that arf1-11 has a defect in the Golgi-to-ER retrograde traffic as well, like the case of ret1 and sec21 mutants of yeast COPI. Some mutants of COPI are known to cause a tight block of the ER-to-Golgi anterograde traffic, which is largely explained as the indirect consequence of the impaired recycling of ER-resident proteins that are required for vesicle budding (Gaynor and Emr, 1997). This reasoning may well apply to arf1-11.
It is also noteworthy that, in the arf1-11 cells, GFP-Rer1p is not mistargeted to the vacuole either but accumulates in ring-like structures (Figure 9). arf1-11 cells also show anomaly in endocytosis of FM4-64. The uptake of the dye is not completely blocked but tends to retard in ring-like structures (Figure 7). These structures resemble the “class E compartment,” which is exaggerated in a certain class of vacuolar protein-sorting mutants, such as vps4 and vps27, and is believed to correspond to the late endosomes/prevacuolar compartment of yeast (Raymond et al., 1992; Vida et al., 1995; Lewis et al., 2000). These observations suggest that arf1-11 fails to function correctly at least in two steps of membrane traffic, ER-Golgi recycling and the exit from the prevacuolar compartment. arf1-11 cells do not show the typical Vps− phenotype, missecretion of CPY. This could be because CPY is largely arrested in the ER. However, CPY is not secreted even at semirestrictive temperatures, 30 and 35°C, at which the transport of CPY was not completely blocked. The lesion of arf1-13 in the prevacuolar compartment is apparently distinct from that of class E vps mutants.
arf1-13 and arf1-14 cells commonly show partial defects in intracellular transport of CPY, general secretion, and localization of GFP-Rer1p, but they do differ in some behavior. A remarkable phenotype of arf1-13 is the abnormal shape of vacuoles. Not only vital staining with FM4-64 but also rapid freeze-substitution electron microscopy reveal amazing deformation of vacuoles in this mutant (Figures 7 and 8). The anomalous vacuoles sometimes look like intermediate structures of autophagy. This phenotype, however, becomes obvious only after a long incubation at 37°C. At earlier points, accumulation of MVB-like structures is more prominent. The MVB-like structures might be taken up by vacuoles in the long run at the restrictive temperature. arf1-14 cells are normal in vacuolar morphology. It is also interesting that arf1-13 but not arf1-14 shows a defect in invertase secretion, whereas ALP transport is more defective in arf1-14 than in arf1-13 (Figures 2B and 4). The involvement of Arf1p in the post-Golgi transport routes, secretory, and AP-1 and AP-3 pathways (Cowles et al., 1997; Stepp et al., 1997), may be differentiated and these two mutant alleles would provide very useful tools to dissect the molecular mechanisms of route selection.
Group 2 (arf1-16 and arf1-17).
In both arf1-16 and arf1-17 alleles, glutamic acid at position 41 is mutated to valine. Recently, Al-Awar et al. (2000) showed that Q37 of mammalian ARF6, corresponding to E41 of yeast Arf1p, is critical for its effector function, formation of protrusion, suggesting that this position represents a site of interaction with effector molecules. arf1-16 has one additional mutation, D129E, which is in the consensus GTP-binding domain, NKXD. The common phenotypes of arf1-16 and arf1-17 are retardation of ER-to-Golgi transport of CPY, secretion of underglycosylated invertase, and mislocalization of GFP-Rer1p to the vacuole.
Another interesting feature of this group is the missecretion of BiP, a soluble ER protein that is retained in the ER and sent back from the Golgi by the HDEL recognition system (Figure 3). This may suggest that the group 2 mutants have lesions in ER retrieval, not only by COPI- and Rer1p-dependent systems but also in the HDEL-receptor–mediated mechanism. arf1-16 shows a weak Vps− phenotype as well.
Group 3 (arf1-18).
The arf1-18 mutant looks normal in the transport of CPY and invertase and in general secretion. However, the fluorescence of FM4-64 and GFP-Rer1p displays a quite strange diffuse pattern in the cytoplasm (Figures 7 and 9). This cannot be due to the cytosolic distribution of these molecules. FM4-64 is a very hydrophobic dye. Rer1p is an integral membrane protein with four transmembrane domains. Release of the soluble GFP by degradation of GFP-Rer1p is not taking place in the mutant cells. Electron microscopic observation of arf1-18 cells reveals various kinds of vesicles, such as numerous small vesicles of 20- to 25-nm scattering in the cytoplasm (Figure 10). Presumably, these vesicles give rise to the diffuse signals of FM4-64 and GFP-Rer1p. The origin of these vesicles has yet to be elucidated. Similar diffuse staining with FM4-64 has been observed in sec1 and sec15 cells, which accumulate secretory vesicles (Vida et al., 1995), and in the mutant cells lacking Tlg1p, Tlg2p, Pep12p, and Vam3p, members of the t-SNARE family (Holthuis et al., 1998). arf1-18 might be somehow affected in the vesicle fusion reaction, although the traffic remains normal in the vacuolar and secretory pathways.
The arf1-18 cells sometimes build up a large stack of the Golgi apparatus unlike normal yeast cells (Figure 10). This phenotype is reminiscent of that of sec7 (Novick et al., 1981; Rambourg et al., 1993), a mutant of a guanine nucleotide exchange factor (GEF) of Arf (see below), but its meaning is unknown.
Regulation of Multiple Functions of Arf1p
As described above, arf1 ts mutants show a variety of different phenotypes, which cannot be explained only by the degrees of a defect in a single function. This convinces us to conclude that Arf1p in fact regulates multiple steps of intracellular protein transport.
Now, a big question remains as to how a single Arf can fulfill such divergent functions. The three-dimensional structure of human Arf1 has been solved by x-ray crystallographic analysis (Amor et al., 1994). We have tried to map into this three-dimensional structure the mutated residues of yeast Arf1p, which are all conserved between yeast and human. The positions of L25, E41, F51, H80, and L173 are well separated from each other. It is plausible that these residues may be involved in the interaction with different regulators of Arf1p.
Many regulators of Arf functions have been determined to date. As GEFs and GTPase-activating proteins (GAPs), many proteins have been identified from mammals and yeast (Jackson and Casanova, 2000; Donaldson, 2000). In S. cerevisiae, at least four Arf GEFs, Gea1p, Gea2p, Sec7p, and Syt1p (Peyroche et al., 1996; Sata et al., 1998; Jones et al., 1999), and six putative Arf GAPs, Gcs1p, Glo3p, Sat1p, Sat2p, Sps18p, and Gts1p (Ireland et al., 1994; Poon et al., 1996, 1999; Zhang et al., 1998; Dogic et al., 1999) exist. Mutations of these genes give different phenotypes, suggesting that these GEFs and GAPs act on Arf1p at different times and at different places. After we submitted the original manuscript of this paper, Eugster et al. (2000) showed that COPI subunits (Sec21p and Sec27p) interact with the Arf-GAP Glo3p but not Gcs1p, whereas both Glo3p and Gcs1p have the GAP activity on Arf1p in vitro and interact with the GTP-stabilized mutant Q71L-Arfp in vivo. Dogic et al. (1999) showed in the analysis of deletions of the six Arf-GAP genes that Δglo3 alone results in a defect in the retrieval of KKXX-proteins from the Golgi. These reports support the view that different Arf-GAPs (and Arf-GEFs) act on Arf differently. Differential actions of GEFs and GAPs will explain the different phenotypes of the arf1 ts mutants, and detailed analysis of genetic interactions between these genes and our mutants are now underway.
Interaction with so-called effectors may also feed back to the function of Arf proteins. Attempts to identify Arf effectors have been extensively made in mammalian systems. For example, Arfaptin1 and Arfaptin2 have been isolated as putative effectors by a yeast two-hybrid screening of a human HL60 cDNA library, using dominant active human Arf3 (Q71L) as bait (Kanoh et al., 1997). Phospholipase D (Cockcroft et al., 1994; Roth et al., 1999), phosphatidylinositol 4.5-bisphosphate (Terui et al., 1994), and phosphatidylinositol 4-phosphate 5-kinase (Honda et al., 1999) are also considered as targets of Arf in mammalian cells. Very recently, GGAs, Golgi-localizing, γ-adaptin ear homology domain, ARF-binding proteins, have also been identified as novel effectors of Arf in mammals and yeast (Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000; Takatsu et al., 2000). The arf1 ts mutants that we constructed may have different abilities to interact with such a variety of targets and thus express different downstream activities.
Genetic Approaches to Further Understand the Multiple Roles of Arf1p
Genetics is again quite powerful in identifying regulators of Arf1p in its various functions. Chen and Graham (1998) discovered that Δarf1 exhibits synthetic lethality with deletion mutations of DRS2, a gene encoding a P-type ATPase, and CHC1, the gene for clathrin heavy chain, respectively (Chen and Graham, 1998; Chen et al., 1999). The mutants of DRS2 and CHC1 accumulate ring-like structures, which are similar to those observed in arf1-13. Δdrs2 shows no genetic interaction with COPI mutants (sec21-1, sec27–1, and ret1-1), which are synthetic lethal with Δarf1 (Gaynor et al., 1998; Chen et al., 1999), suggesting that Arf1p functions with Drs2p and COPI independently. Interestingly, some of our arf1 ts mutants appear to show synthetic lethality with Δdrs2 as well (Yahara and Nakano, unpublished observations).
It is obvious that our collection of new arf1 ts alleles will enable extensive genetic screening of interacting molecules in an allele-dependent manner. Now we have started screening of multicopy suppressers for each arf1 ts mutant and have already obtained candidate clones that may suppress the temperature sensitivity of arf1 ts mutants in an allele-specific manner. Our mutants will be thus extremely useful to understand how the multiple roles of Arf are executed at different places in a cell.
ACKNOWLEDGMENTS
We are thankful to Scott Emr and Erin Gaynor of the University of California, San Diego, Katsuyoshi Mihara and Nobuhiro Nakamura of Kyushu University, Yoh Wada of Osaka University, and Masao Tokunaga of Kagoshima University for excellent antibodies. We are also grateful to members of the Nakano laboratory and Akio Toh-e of the University of Tokyo for many helpful discussions during the course of this work. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, by a research grant from the Human Frontier Science Program Organization, and by grants from the Biodesign and Bioarchitect Projects of RIKEN. N.Y. is a recipient of the Junior Research Associate fellowship of RIKEN.
Abbreviations used:
- Arf
ADP-ribosylation factor
- ALP
vacuolar alkaline phosphatase
- BiP
immunoglobulin heavy-chain–binding protein
- CPY
carboxypeptidase Y
- ER
endoplasmic reticulum
- GAP
GTPase-activating protein
- GEF
guanine-nucleotide exchange factor
- GFP
green fluorescent protein
- MVB
multivesicular body
- ORF
open reading frame
- TCA
trichloroacetic acid
- ts
temperature sensitive
REFERENCES
- Al-Awar O, Radhakrishna H, Powell NN, Donaldson JG. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol Cell Biol. 2000;20:5998–6007. doi: 10.1128/mcb.20.16.5998-6007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor JC, Harrison DH, Kahn RA, Ringe D. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature. 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Bigelis R, Keesey JK, Fink GR. The yeast his4 multifunctional protein. J Biol Chem. 1981;256:5144–5152. [PubMed] [Google Scholar]
- Boman AL, Zhang C-j, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Chen CY, Graham TR. An arf1Δ synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics. 1998;150:577–589. doi: 10.1093/genetics/150.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S, Thomas GM, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty NF, Truong O, Hsuan JJ. Phospholipase D: a downstream effector of ARF in granulocytes. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Cosson P, Demolliere C, Hennecke S, Duden R, Letourneur F. δ-, and ζ-COP,. two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogic D, de Chassey B, Pick E, Cassel D, Lefkir Y, Hennecke S, Cosson P, Letourneur F. The ADP-ribosylation factor GTPase-activating protein Glo3p is involved in ER retrieval. Eur J Cell Biol. 1999;78:305–310. doi: 10.1016/s0171-9335(99)80064-8. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Filling in the GAPs in the ADP-ribosylation factor story. Proc Natl Acad Sci USA. 2000;97:3792–3794. doi: 10.1073/pnas.97.8.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Stahl PD. Myristoylation required for the intracellular localization and endocytic function of ARF6. Exp Cell Res. 1995;221:153–159. doi: 10.1006/excr.1995.1362. [DOI] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Chen CY, Emr SD, Graham TR. ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol Biol Cell. 1998;9:653–670. doi: 10.1091/mbc.9.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Bright NA, Rous B, Robinson MS. Characterization of a fourth adaptor-related protein complex. Mol Biol Cell. 1999;10:2787–2802. doi: 10.1091/mbc.10.8.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JCM, Nichols BJ, Pelham HRB. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol Biol Cell. 1998;12:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- Ireland LS, Johnston GC, Drebot MA, Dhillon N, DeMaggio AJ, Hoekstra MF, Singer RA. A member of a novel family of yeast 'Zn-finger' proteins mediates the transition from stationary phase to cell proliferation. EMBO J. 1994;15:3812–3821. doi: 10.1002/j.1460-2075.1994.tb06692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics. 1999;152:1543–1556. doi: 10.1093/genetics/152.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG. Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J Biol Chem. 1984;259:6228–6234. [PubMed] [Google Scholar]
- Kanoh H, Williger BT, Exton JH. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J Biol Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJ, Stevens LA, Kao YL, Moss J, Vaughan M. Characterization of a glucose-repressible ADP-ribosylation factor 3 (ARF3) from Saccharomyces cerevisiae. J Biol Chem. 1994;269:20931–20937. [PubMed] [Google Scholar]
- Lenhard JM, Kahn RA, Stahl PD. Evidence for ADP-ribosylation factor (ARF) as a regulator of in vitro endosome-endosome fusion. J Biol Chem. 1992;267:13047–13052. [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HRB. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Gulbis JM, Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human Arno and analysis of the interaction with ARF GTPase. Cell. 1998;92:415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- Nakano A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Ohtsuka H, Yamagishi M, Yamamoto E, Kimura K, Nishikawa S, Oka T. Mutational analysis of the Sar1 protein, a small GTPase which is essential for vesicular transport from the endoplasmic reticulum. J Biochem. 1994;116:243–247. doi: 10.1093/oxfordjournals.jbchem.a124513. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Nakano A. The GTP-binding Sar1 protein is localized to the early compartment of the yeast secretory pathway. Biochim Biophys Acta. 1991;1093:135–143. doi: 10.1016/0167-4889(91)90114-d. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Nakano A. Identification of a gene required for membrane protein retention in the early secretory pathway. Proc Natl Acad Sci USA. 1993;90:8179–8183. doi: 10.1073/pnas.90.17.8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Fero S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translation events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Ohya Y, Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994;263:963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- Oka T, Nakano A. Inhibition of GTP hydrolysis by Sar1p causes accumulation of vesicles that are a functional intermediate of the ER-to-Golgi transport in yeast. J Cell Biol. 1994;124:425–434. doi: 10.1083/jcb.124.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Glick BS, Rothman JE. A new type of coated vesicular carrier that appears not to contain clathrin: its possible role in protein transport within the Golgi stack. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:481–484. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer RA, Johnston GC. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Képès F. Modulation of the Golgi apparatus in Saccharomyces cerevisiae sec7 mutants as seen by three-dimensional electron microscopy. Anat Rec. 1993;237:441–452. doi: 10.1002/ar.1092370402. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;12:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Roth MG, Bi K, Ktistakis NT, Yu S. Phospholipase D as an effector for ADP-ribosylation factor in the regulation of vesicular traffic. Chem Phys Lipids. 1999;98:141–152. doi: 10.1016/s0009-3084(99)00026-2. [DOI] [PubMed] [Google Scholar]
- Rothblatt J, Schekman R. A hitchhiker's guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. doi: 10.1016/s0091-679x(08)61165-6. [DOI] [PubMed] [Google Scholar]
- Rowe T, Aridor M, McCaffery JM, Plutner H, Nuoffer C, Balch WE. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J Cell Biol. 1996;135:895–911. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yamanushi T, Oka T, Nakano A. Identification of SEC12, SED4, truncated SEC16 and EKS1/HRD3 as multicopy suppressors of ts mutants of Sar1 GTPase. J Biochem. 1999;125:130–137. doi: 10.1093/oxfordjournals.jbchem.a022249. [DOI] [PubMed] [Google Scholar]
- Sata M, Donaldson JG, Moss J, Vaughan M. Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc Natl Acad Sci USA. 1998;95:4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nishikawa S, Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol Biol Cell. 1995;6:1459–1477. doi: 10.1091/mbc.6.11.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc Natl Acad Sci USA. 1997;94:9693–9698. doi: 10.1073/pnas.94.18.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K, Nakano A. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol Cell Biol. 1999;19:471–483. doi: 10.1128/mcb.19.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer LS, Kahn RA, Hanski E, Northup JK, Sternweis PC, Gilman A. Requirements for cholera toxin-dependent ADP-ribosylation of the purified regulatory component of adenylate cyclase. J Biol Chem. 1982;257:20–23. [PubMed] [Google Scholar]
- Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- Sun GH, Hirata A, Ohya Y, Anraku Y. Mutations in yeast calmodulin cause defects in spindle pole body functions and nuclear integrity. J Cell Biol. 1992;119:1625–1639. doi: 10.1083/jcb.119.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Yoshino K, Nakayama K. Adaptor gamma ear homology domain conserved in gamma-adaptin and GGA proteins that interact with gamma-synergin. Biochem Biophys Res Commun. 2000;271:719–725. doi: 10.1006/bbrc.2000.2700. [DOI] [PubMed] [Google Scholar]
- Taylor TC, Kahn RA, Melançon P. Two distinct members of the ADP-ribosylation factor family of GTP-binding proteins regulate cell-free intra-Golgi transport. Cell. 1992;70:69–79. doi: 10.1016/0092-8674(92)90534-j. [DOI] [PubMed] [Google Scholar]
- Terui T, Kahn RA, Randazzo PA. Effects of acid phospholipids on nucleotide exchange properties of ADP-ribosylation factor 1: evidence for specific interaction with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1994;269:28130–28135. [PubMed] [Google Scholar]
- Traub LM, Ostrom JA, Kornfeld S. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Kitamoto K, Kanbe T, Tanaka K, Anraku Y. The SLP1 gene of Saccharomyces cerevisiae is essential for vacuolar morphogenesis and function. Mol Cell Biol. 1990;10:2214–2223. doi: 10.1128/mcb.10.5.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Bright NA, Robinson MS. The role of ADP-ribosylation factor and phospholipase D in adaptor recruitment. J Cell Biol. 1997;138:1239–1254. doi: 10.1083/jcb.138.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanushi T, Hirata A, Oka T, Nakano A. Characterization of yeast sar1 temperature-sensitive mutants, which are defective in protein transport from the endoplasmic reticulum. J Biochem. 1996;120:452–458. doi: 10.1093/oxfordjournals.jbchem.a021432. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cavenagh MM, Kahn RA. A family of Arf effectors defined as suppressors of loss of Arf function in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:19792–19796. doi: 10.1074/jbc.273.31.19792. [DOI] [PubMed] [Google Scholar]
- Zhao L, Helms JB, Brunner J, Wieland FT. GTP-dependent binding of ADP-ribosylation factor to coatomer in close proximity to the binding site for dilysine retrieval motifs and p23. J Biol Chem. 1999;274:14198–14203. doi: 10.1074/jbc.274.20.14198. [DOI] [PubMed] [Google Scholar]