Abstract
Bats are known to harbor a number of emerging and re-emerging zoonotic viruses, many of which are highly pathogenic in other mammals but result in no clinical symptoms in bats. The ability of bats to coexist with viruses may be the result of rapid control of viral replication early in the immune response. IFNs provide the first line of defense against viral infection in vertebrates. Type III IFNs (IFN-λs) are a recently identified IFN family that share similar antiviral activities with type I IFNs. To our knowledge, we demonstrate the first functional analysis of type III IFNs from any species of bat, with the investigation of two IFN-λ genes from the pteropid bat, Pteropus alecto. Our results demonstrate that bat type III IFN has similar antiviral activity to type I and III IFNs from other mammals. In addition, the two bat type III IFNs are differentially induced relative to each other and to type I IFNs after treatment or transfection with synthetic dsRNA. Infection with the bat paramyxovirus, Tioman virus, resulted in no upregulation of type I IFN production in bat splenocytes but was capable of inducing a type III IFN response in three of the four bats tested. To our knowledge, this is the first report to describe the simultaneous suppression of type I IFN and induction of type III IFN after virus infection. These results may have important implications for the role of type III IFNs in the ability of bats to coexist with viruses.
As the only flying mammal, bats are the putative natural host reservoir for a number of zoonotic viruses, many of which cause significant morbidity and mortality in humans and other mammals (1). Most of the highly lethal viruses of bat origin that are pathogenic in humans and other mammals appear to cause no clinical signs of disease in bats under natural or experimental infection (1–3). Bats have been implicated in the spillover of some of the most deadly viruses known, including rabies and rabies-like lyssaviruses, Hendra virus, Nipah virus, severe acute respiratory syndrome coronavirus, Ebola virus, Melaka virus, and Marburg virus (4–13). Thus, bats appear to be able to coexist with viruses and may have evolved mechanisms to rapidly control viral replication while still allowing transmission and occasional spillover to susceptible species. Despite the central role of bats as reservoirs for viral diseases, there is a paucity of information on any aspect of bat immunology. Research on bat immunology, especially on antiviral immunology, is needed to address the question of how bats can coexist asymptomatically with viruses and also to seek potential therapeutic strategies targeting these viruses.
One of the earliest immune responses to be initiated after viral infection is the production of IFN. IFNs provide the first line of defense against viral infection and play a role in shaping the adaptive immune response (14). Three classes of IFN have been identified, designated types I, II, and III, which differ in their amino acid sequences and the receptor complex they signal through. Type I (including α and β) and III (λ) IFNs are induced directly in response to viral infection, and thus play an important role in innate immunity. Type I IFNs engage the ubiquitously expressed IFN-α receptor (IFNAR) complex that is composed of IFNAR1 and IFNAR2 (15). Type III IFNs signal through receptors containing IFN-λR1 (also known as IL-28Ra) and IL-10R2 (also known as IL-10Rb). The type II class of IFN comprises the single IFN-γ gene product that binds the IFN-γR complex (16). Unlike the type I and III IFNs, IFN-γ is predominantly involved in the Th1 immune response, and in coordinating and establishing an antiviral state for longer term viral control (14).
The type III IFN system was discovered in 2003 by two independent groups who identified three human IFN-λ genes, IFN-λ1, -λ2, and -λ3, also known as IL29, IL28a, and IL28b in the human genome sequence based on limited sequence similarity to type I IFNs and IL-10 family members (17, 18). Unlike type I IFNs, which are single-exon genes, type III IFNs are encoded by five to six exons. Since their discovery in humans, type III IFNs have been characterized in mice, zebrafish, chicken, and Xenopus (19–22). Mice have two functional IFN-λ genes and one pseudogene, whereas zebrafish and chickens express only a single IFN-λ gene product. Five IFN-λ genes are present in the Xenopus genome, but only four are functional (22). Numerous type III IFNs have also been identified in the publicly available whole-genome sequence data from a variety of other species (23), but for the majority of species, no functional analysis has been performed.
Like type I IFNs, type III IFNs have antiviral activities in all species in which they have been characterized (15, 18, 20, 24). Both type I and III IFNs share similar production and signaling pathways, and result in the production of hundreds of IFN-stimulated genes (ISGs) that, in turn, are responsible for much of the antiviral action of IFNs (25–27). Although it is unclear why two IFN systems with similar antiviral activities have evolved, differences in their receptor distribution suggest that the type I and III IFNs do not merely duplicate each other. The type III IFNR is expressed predominantly by epithelial cells consistent with a more specialized role in the immediate immune response in tissues that represent the sites of virus entry (17, 18, 25, 28). In most circumstances, type I and III IFNs are simultaneously expressed (26, 27). However, recent evidence suggests that there may be differences in the mechanisms involved in the regulation of these cytokines resulting in differential expression in some circumstances (29, 30). However, because of their recent discovery, the type III IFNs are poorly characterized, and more work remains to determine whether type III IFNs have functions not shared with type I IFN, and whether these two types of IFNs exert antiviral activity with different kinetics (5, 25).
The black flying fox, Pteropus alecto, has become a model bat species for studies on host–virus interactions (31, 32). To gain insight into the mechanisms responsible for the control of viral replication in bats, we have begun an investigation of innate and adaptive immunity in this species, including the recent characterization of IgH diversity (33) and the repertoire of TLRs (34). In this article, we describe the characterization of type III IFNs in P. alecto. Type I IFNs were recently described in two species of fruit bats, the Egyptian Rousette, Rousettus aegyptiacus, and the Malaysian flying fox, Pteropus vampyrus (35, 36). Evidence for the presence of type III IFNs in bats has been reported in an earlier investigation describing the in silico identification of IFN-λ genes in the publicly available microbat genome (23). However, before this study, no reports have described the characterization of bat IFN-λ genes. Our results demonstrate that P. alecto has two expressed IFN-λ genes that are conserved with other mammalian IFN-λ sequences. Similar to other mammalian IFNs, P. alecto IFN-λ displays antiviral activity in vitro and induces the production of ISGs. Furthermore, these results provide evidence for differential induction of type III IFNs relative to each other and to type I IFNs after dsRNA stimulation and viral infection. These results are consistent with type III IFNs playing an important role in the early innate immune response to viral infections in bats.
Materials and Methods
Cell lines
The establishment and culture conditions for the P. alecto cell lines have been described previously (31). The cell lines used in this study included two immortalized and cloned cell lines, lung PaLuT02 and fetus PaFeT05, and seven nonimmortalized primary cell lines of lung, liver, heart, kidney, small intestine, brain, and salivary gland origin, respectively. All of the bat cell lines consist of adherent tissue cell types. Bat cell lines were cultured in DMEM/F12-Hams (Sigma), each supplemented with 15% FCS (Hyclone), 100 U/ml penicillin, 100 mg/ml streptomycin, and 50 mg/ml gentamicin (Sigma). Vero cells were cultured in DMEM supplemented with 10% FCS. All cells were maintained in a humidified atmosphere of 5% CO2 in air at 37°C.
Isolation of bat splenocytes
Wild caught P. alecto bats were trapped in Southern Queensland, Australia, and transported alive by air to the Australian Animal Health Laboratory in Victoria, where they were euthanized for dissection using methods approved by the Australian Animal Health Laboratory Animal Ethics Committee. Spleen cell suspensions were prepared by pressing spleen tissue through a cell strainer using a syringe plunger. Mononuclear splenocytes were isolated by density centrifugation over Lymphoprep (Axis-Shield). Culture media for mononuclear splenocytes consisted of DMEM supplemented with 10% FCS, 15 mM HEPES, 15 mM l-gluta-mine, 100 mg/ml penicillin, and 100 mg/ml streptomycin.
Genome analyses
IFN-λ, ISG56, and retinoic acid-inducible gene I (RIG-I) were identified in the whole genome sequence of the Malaysian flying fox, P. vampyrus, available in the Ensembl database (assembly pteVam1, 2.63× coverage, July 2008) using a combination of key word searches and the BLAT, BLASTN, and BLASTX algorithms. GenBank TRACE archives were also used to search for the P. vampyrus IFN-λ genes. For comparative purposes, the current genome assemblies from human (NCBI 36), macaque (MMUL_1), gorilla (gorGor3), mouse (NCBI m37), chicken (WASHUC2), rabbit (RABBIT), dog (CanFam2.0), cat (CAT), horse (preEnsembl Equ-Cab2), hedgehog (eriEur1), elephant (BROAD E1), armadillo (ARMA), platypus (OANA5), opossum (BROADO5), Xenopus tropicalis (JGI4.1), and zebrafish (Zv8) were searched for IFN-λ genes using the BLAST algorithms.
Virus culture
Tioman virus (TioPV), a bat paramyxovirus (37), and Pulau virus (PulV), a bat orthoreovirus (38), were grown in Vero cells for 48 h in a humidified atmosphere of 5% CO2 in air at 37°C. The virus-containing supernatant was collected, and the 50% end point of tissue culture infective dose (TCID50) was determined. TioPV grew to a titer of 2.5 × 106 TCID50/ml, whereas PulV had a titer of 2.9 × 105 TCID50/ml.
Culture of cell lines and cells with polyinosinic-polycytidylic acid
The P. alecto lung cell line, PaLuT02, was seeded at 1 × 106 cells/well in six-well tissue culture plates (Nunc). The cells were stimulated by transfection with 33 μg/ml polyinosinic-polycytidylic acid (polyI:C; Invivogen) using 10 μl Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions and incubated in a humidified atmosphere of 5% CO2 in air at 37°C. Cells were harvested in RLT buffer (Qiagen) at 0, 0.5, 1.5, 3, 6, 9, 15, and 24 h after transfection and stored at –80°C until the RNA was extracted. The seven bat primary cell lines from lung, liver, heart, kidney, small intestine, brain, and salivary gland were cultured at a concentration of 1 × 106 cells/well in six-well tissue culture plates and stimulated with 33 μg/ml polyI:C with (transfected) or without transfection reagent (treated) for 3 h. After stimulation, the cells were collected into buffer RLT (Qiagen) for RNA extraction.
The primary splenocytes were cultured at a concentration of 5 × 106 cells/well in six-well tissue culture plates. The cells were stimulated by treatment or transfection with polyI:C at a concentration of 10 μg/ml and incubated in a humidified atmosphere of 5% CO2 in air at 37°C. Cells were harvested from duplicate wells at 3, 6, and 24 h after treatment and lysed in RLT buffer. A concentration of 10 μg/ml polyI:C was chosen for these experiments after optimizing the dose of polyI:C in these cells.
TioPV infection of bat splenocytes
For virus infection in splenocytes, 5 × 106 cells/well were seeded into 24-well tissue culture plates. Cells were infected with TioPV at a multiplicity of infection of 0.1 and incubated in a humidified atmosphere of 5% CO2 in air at 37°C for 3 h. Cells were harvested and lysed in RLT buffer (Qiagen) for RNA extraction.
PCR amplification of IFN-λ genes
As IFNs are induced after stimulation with dsRNA, total RNA extracted from the P. alecto lung cell line, PaLuT02, 3 h after transfection with polyI:C was used as a template for the amplification of IFN-λ genes. Samples were homogenized using Qiashredders (Qiagen). Total RNA was extracted using the RNeasy mini kit (Qiagen) with on-column DNase I treatment (Qiagen) to remove traces of genomic DNA. Full-length coding sequences for IFN-λ were obtained using 5′ and 3′ RACE PCRs using the GeneRacer Kit (Invitrogen, Carlsbad, CA) with Long-amp DNA polymerase (New England Biosystems) and the manufacturer's recommended PCR conditions on total RNA. Primers were designed based on sequences obtained from the P. vampyrus whole-genome sequence deposited in the Ensembl Genome Browser and are listed in Table I. RT-PCR using primers based on the RACE sequences to amplify full-length cDNAs was also performed on RNA extracted from freshly isolated bat splenocytes and polyI:C transfected PaLuT02 cells. To confirm the length of intron 2 of IFN-λ1, we performed PCR on genomic DNA extracted from P. alecto spleen and liver.
Table I.
Primers used in this study
| Gene | Primera | Sequence 5′—3′ | Application |
|---|---|---|---|
| IFN-λ1 | IFN-λ1-1F | CTTGGCCGGAGCAAGCCCTGTTC | RACE |
| IFN-λ1-1R | CTTGTAAGGAGGCGGAAGAGGTTGA | RACE | |
| IFN-λ1-2F | ATCTCTGCTACCAACGGAGTGGAA | RACE | |
| IFN-λ1-2R | GGGACTTGCTGTGGGCTGAGGTAA | RACE | |
| IFN-λ1-3F | GCCCCAAAGAAAGTGTCTCAAG | qRT-PCR | |
| IFN-λ1-3R | GGCGGAAGAGGTTGAATGTG | qRT-PCR | |
| IFN-λ1-4F | AGTTGCGATTTGGCGATGGCTGTA | Full-length amplification | |
| IFN-λ1-4R | AGTCAGGGCTGCAGCTCCATAAA | Full-length amplification | |
| IFN-λ2 | IFN-λ2 -1F | CGGCTTGCACGCTGCTTCTGATG | RACE |
| IFN-λ2 -1R | GATGAGGAGGCGGAAGAGGTTGAA | RACE | |
| IFN-λ2 -2F | GGCTACAAGGGGCTGCCACATGT | RACE | |
| IFN-λ2 -2R | GCCTGGAGTCTGGCGTGGATGTG | RACE | |
| IFN-λ2 -3F | CATGTCCCAGTTCAGGTCTCTGT | qRT-PCR | |
| IFN-λ2 -3R | CATCCTTGGCCCTCTTGAAA | qRT-PCR | |
| IFN-λ2 -4F | GTAAGCGGCCGCACCATGGAGCTGGGCGCGGCC | Protein expression | |
| IFN-λ2 -4R | AGAGGGGCCCTCAGACACACAGGTTTCTGCTGGCG | Protein expression | |
| IFN-λ2 -5F | GCCGAGGAAGACTGAGCGAGA | Full-length amplification | |
| IFN-λ2 -5R | GTACATAAATATATAAATAGG | Full-length amplification | |
| GAPDH | GAPDH-1F | TCCTCCCGTTCTACAGACAG | qRT-PCR |
| GAPDH-1R | ACAAGGTAGGGCTCCCTAAG | qRT-PCR | |
| RIG-I | RIG-I-1F | CTCAGGTCGTTGGGCTGACT | qRT-PCR |
| RIG-I-1R | CCAAGGCTTCACCTGTGCTT | qRT-PCR | |
| ISG56 | ISG56-1F | CTTGAGCATCCTCGGGTTCATC | qRT-PCR |
| ISG56-1R | AAGTCAGCAGCCAGGTTTAGGG | qRT-PCR | |
| IFN-α | IFN-α-1F | GAGACTCCCCTGCTGGATGA | qRT-PCR |
| IFN-α-1R | ATAGAGGGTGATTCTCTGGAAGTATTTC | qRT-PCR | |
| IFN-β | IFN-β-1F | CTCTAGCACTGGCTGGAATGAA | qRT-PCR |
| IFN-β-1R | TGCCCACCGAGTGTCTCA | qRT-PCR |
F, forward; R, reverse.
Sequencing and sequence analysis
PCR and RACE-PCR products were cloned into the pCR4-TOPO vector using the TOPO TA Cloning Kit for sequencing (Invitrogen). M13 primers were used for sequencing using BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems, Foster City, CA) with nonisotopic dye terminators in 20-μl reactions, according to the manufacturer's instructions, and analyzed on an Applied Biosystems 3130 XL Genetic Analyzer. Sequences were assembled manually using Seqman PRO (Lasergene) and Clone Manager 9.0 (Sci-Ed Software), and compared with sequences in the GenBank database using the BLAST algorithm (20). Sequence alignments were performed using the Clustal X program (39) and visualized using GeneDoc (http://www.nrbsc.org/gfx/genedoc/index.html). Signal peptides were identified by SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP). To determine the exon-intron boundaries of the P. alecto IFN-λ genes, we aligned the full-length coding sequences with the corresponding genomic sequences in the P. vampyrus whole-genome sequence. Intron-exon maps were drawn using Fancy Gene v1.4 (http://host13.bioinfo3.ifom-ieo-campus.it/fancygene/).
Phylogenetic and pairwise analysis
Based on the amino acid alignments, phylogenetic trees were constructed by the neighbor joining method of Saitou and Nei (40), maximum parsimony, and minimum evolution using the MEGA 4.1 program with 1000 bootstrap replicates (41). The GenBank accession numbers for sequences used in the phylogenetic analysis are as follows: IFN-λ1—AAI26184.1, Homo sapiens (human); NP_001108325.1, Canis lupus familiaris (dog); XP_001501239.1, Equus caballus (horse); NP_001136309.1, Sus scrofa (pig); IFN-λ2/3—AAN28263.1 and AAN28264.1 (human); XP_855366.1 (dog); NP_001159962.1 (pig); AAI50797.1 and AAX58715.1, Mus musculus (mouse); XP_001368442.1, Monodelphis domestica (opossum); XP_001517931.1, Ornithorhynchus anatinus (platypus); NP_001121968.1, Gallus gallus (chicken); NP_001165235.1, Xenopus (Silurana) tropicalis (frog); IFN-λ (unclassified)—XP_002711513.1, Oryctolagus cuniculus (domestic rabbit); AAM95448.1, Danio rerio (zebrafish).
Generation of an IFN-λ–producing stable cell line (λ2-293T)
The bat IFN-λ2 gene was cloned into the pQCXIH retroviral expression vector (Clontech) using the primers listed in Table I. Correct cloning was confirmed by sequencing. The IFN-λ2–containing plasmid was then transfected into 293T cells by calcium phosphate transfection (Invitrogen). Forty-eight hours after transfection, the transformed cells were selected by the addition of 100 μg/ml hygromycin. Antibiotic-resistant cells were selected using 200 μg/ml hygromycin. Quantitative RT-PCR (qRT-PCR) targeting IFN-λ2 gene was used to measure the mRNA level of IFN-λ2 in the cell line.
Antiviral activity and ISG production in response to recombinant bat IFN-λ
The activity of the recombinant IFN-λ protein was determined by its ability to induce the production of ISGs and inhibit virus-mediated cytolysis. The ISG response was determined using ISG56 and RIG-I, both of which are known to be induced by type I IFNs in mammals (42, 43). Supernatant collected from λ2-293T cells was serial diluted and used to treat the P. alecto fetus cell line, PaFeT05. The PeFeT05 cell line was chosen for these experiments after it was identified as being capable of the best ISG response after IFN-λ stimulation. Supernatant from normal cultured 293T cells was used as mock control. Cells were incubated at 37°C for 6 h and collected into buffer RLT (Qiagen) for extraction of total RNA. ISG56 and RIG-I expression were determined by qRT-PCR.
PaFeT05 cells were seeded into 96-well tissue culture plates at 1 × 105 cells/well and incubated with serial dilutions of supernatant from λ2-293T or 293T cells at 37°C for 24 h to test the antiviral activity of the IFN-λ2 protein. Medium was then replaced with PulV-containing supernatant at a multiplicity of infection of 1.0 and incubated for 1 h at 37°C. Virus-containing medium was then replaced with culture medium and incubated for 24 h in a humidified atmosphere of 5% CO2 in air at 37°C. Culture supernatant was collected for TCID50 testing in Vero cells. The PulV-infected cells were immersed in absolute methanol for 10 min at –20°C to fix and inactivate the virus. PulV was chosen for these experiments because infection with this virus produced a very high cytopathic effect (CPE) compared with other viruses tested, including TioPV.
Quantitative reverse transcription PCR
qRT-PCR was performed on total RNA extracted from cells and cell lines as described previously (34). In brief, total RNA was extracted as described for RACE PCR and used for the synthesis of cDNA using the Quantitect reverse transcription kit for real-time PCR (Qiagen). qRT-PCR primers were designed using Primer Express 3.0 (Applied Biosystems) with default parameter settings and are listed in Table I. The 18s rRNA primers have been described previously (34). Reactions were carried out using EXPRESS SYBR GreenER qPCR Supermix Universal (Invitrogen) and an Applied Biosystems 7500 Fast Real-Time qPCR instrument. For each reaction, 2 μl of 1:5 diluted cDNA and a final concentration of 200 nmol of each primer was used. The cycling profile consisted of an initial denaturation at 90°C for 1 min followed by 40 cycles of 90°C for 15 s, 60°C for 1 min, followed by melt curve analysis. Expression level of the target genes were calculated using either standard curves method (IFNs, GAPDH, 18s rRNA) or fold induction compared with the mock (ISG56 and RIG-I). All data were normalized relative to the housekeeping genes, 18s rRNA or GAPDH.
Results
Characterization of P. alecto type III IFNs
Although no whole genome is available for P. alecto, a low coverage genome sequence from the closely related pteropid bat, P. vampyrus, is publicly available in the Ensembl database. Analysis of the P. vampyrus genome using a combination of keyword and BLAST searches resulted in the identification of three IFN-λ genes with similarity to human IFN-λ1, -λ2, and -λ3 (IL-28A, IL-28B, and IL-29). Of the three P. vampyrus genes, IFN-λ1 contained an 80-nucleotide gap within its coding sequence. This gap is an artifact of the low coverage nature of the P. vampyrus genome assembly and not a reflection of the actual gene and, therefore, could not be included in further phylogenetic or sequence analysis. IFN-λ2 and -λ3 shared very high sequence similarity, differing by only five nucleotides within their coding regions. The P. vampyrus sequences were used to design oligo-nucleotide primers to amplify full-length cDNAs from P. alecto using RACE PCR (Table I). The cloned and immortalized lung cell line PaLuT02 has previously been demonstrated to produce IFN-α and IFN-β in response to transfection with the dsRNA mimic, polyI:C (31). To determine the complete gene structure of the P. alecto IFN-λ genes, 5′ and 3′ RACE PCR was performed using RNA extracted from polyI:C transfected PaLuT02 cells, resulting in the amplification of two IFN-λ genes. RT-PCR using primers based on the 5′ and 3′ RACE sequences to amplify full-length cDNAs was also performed using RNA from both freshly isolated bat splenocytes and polyI:C transfected PaLuT02 cells. These sequences further confirmed the transcription of only two IFN-λ loci in P. alecto despite the identification of three loci in the P. vampyrus genome sequence. The P. alecto IFN-λ sequences were designated IFN-λ1 and IFN-λ2 based on their sequence similarity to previously described human and mouse IFN-λ genes (Table II). These sequences have been deposited in GenBank under accession numbers HQ201955 (-λ2) and HQ201956 (-λ1).
Table II.
Percentage of similarity between bat, human, and mouse type III IFNs
| Bat IFN-λ1 | Bat IFN-λ2 | Human IFN-λ1 | Human IFN-λ2 | Human IFN-λ3 | Mouse IFN-λ2 | Mouse IFN-λ3 | |
|---|---|---|---|---|---|---|---|
| Bat IFNλ1 | 63 | 64 | 60 | 61 | 56 | 53 | |
| Bat IFNλ2 | 71 | 61 | 67 | 68 | 56 | 53 | |
| Human IFN-λ1 | 72 | 68 | 66 | 67 | 53 | 49 | |
| Human IFN-λ2 | 70 | 78 | 73 | 96 | 61 | 58 | |
| Human IFN-λ3 | 70 | 78 | 73 | 98 | 62 | 59 | |
| Mouse IFN-λ2 | 67 | 69 | 65 | 72 | 72 | 93 | |
| Mouse IFN-λ3 | 65 | 67 | 63 | 70 | 70 | 97 |
Bold numbers indicate amino acid identity; non-bold numbers represent nucleotide identity.
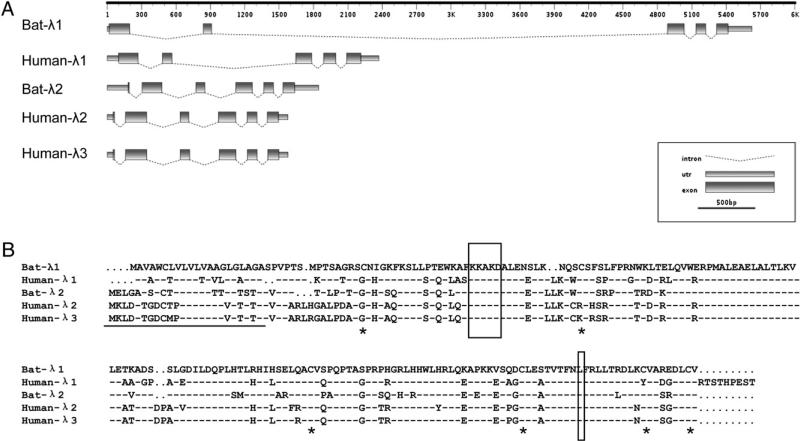
Using the P. alecto cDNA sequences, we were able to determine the complete exon and intron organization of each locus by aligning the complete cDNA sequences with the corresponding genomic sequences identified in the whole genome of P. vampyrus (Fig. 1A). Bat IFN-λ2 showed a similar genomic structure to human IFN-λ2 and -λ3, with six exons and five introns, including an additional 5′ exon of only 10 nucleotides that contained the initial start methionine (18, 23). IFN-λ1 had five exons and four introns, which is similar to human IFN-λ1. An unusual feature of the bat IFN-λ1 gene is the presence of a 4-kb intron separating exons 2 and 3 compared with the typical size of 1.1 kb for this intron in other species (Fig. 1A). Identification of IFN-λ genes in the whole-genome sequences of a variety of other species available in the Ensembl database confirmed the presence of an ~1.1-kb intron in all other species available in Ensembl. This 4-kb intron was further confirmed by PCR using P. alecto genomic DNA.
FIGURE 1.
Comparison of bat type III IFNs with human type III IFNs. A, The gene organization of bat IFN-λ genes compared with that of corresponding human IFN-λ genes. The intron-exon structures of IFN-λs were predicted by nucleotide alignment of IFN-λ mRNA with the corresponding sequences in either the P. vampyrus or human genome sequence. Putative untranslated regions (UTRs) and exons are drawn as rectangles; introns are shown as dotted lines. B, Alignment of the deduced amino acid sequence of bat IFN-λ genes with human IFN-λ genes. The predicted signal peptides are underlined. Two of the conserved regions that are responsible for binding to IFN-λR1 are boxed. Six conserved cysteine residues are indicated by asterisks. Dashes indicate similarity; dots indicate gaps.
The deduced protein encoded by IFN-λ1 and IFN-λ2 shown in Fig. 1B contained many features that are conserved in IFN-λ genes from other species, including conserved signal peptide cleavage sites and several residues that are responsible for binding to the type III IFNR (44) (Fig. 1B). Six conserved cysteine residues important for disulphide bond formation are also conserved in the bat IFN-λ sequences (Fig. 1B).
Bat IFN-λ1 and IFN-λ2 genes are closely related to mammalian IFN-λ genes
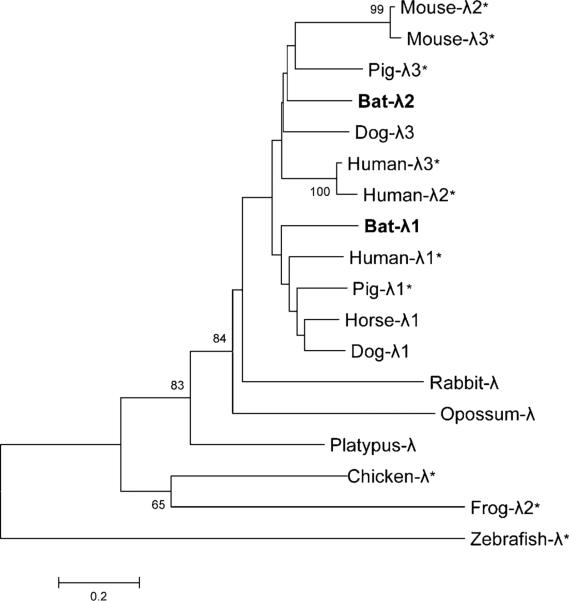
Phylogenetic analysis was performed on bat IFN-λs with a group of IFN-λ sequences from a variety of other vertebrates (Fig. 2). Bat IFN-λ1 and -λ2 clustered with mammalian -λ1 and -λ2/3, respectively. The tree shown in Fig. 2 was reconstructed using the neighbor joining method; however, identical results were found when maximum parsimony and minimum evolution were used (data not shown).
FIGURE 2.
Phylogenetic analysis based on amino acid alignments of bat type III IFNs with representative vertebrate species. Branch support is indicated as the percentage of 1000 bootstrap replicates and is shown where support is >60%. The bat IFN-λs are highlighted in bold. The accession numbers of IFN-λ proteins used in this tree can be found in Materials and Methods. Asterisks indicate the proteins that have been characterized; all other sequences are predicted based on whole-genome sequences.
Bat IFN-λ1 and IFN-λ2 mRNAs are produced early in the response to polyI:C transfection
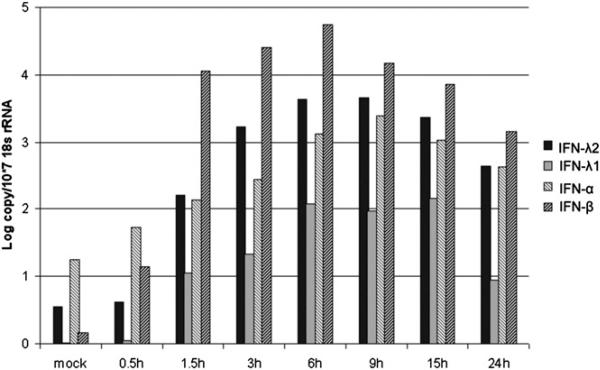
To determine the production kinetics of bat IFN-λs, the cloned and immortalized P. alecto lung cell line PaLuT02 was transfected with polyI:C, and total cellular RNA was isolated at different time points after transfection. The production kinetics of the two type III IFNs was compared with that of type I IFNs (Fig. 3). The type I IFNs were induced as early as 0.5 h, closely followed by the induction of type III IFNs at 1.5 h after transfection. Both type I and III IFNs peaked at 6 h, decreasing by 24 h. IFN-λ2 induction by polyI:C was ~100-fold greater than IFN-λ1 at each time point tested (Fig. 3).
FIGURE 3.
Production time course of bat type I and III IFNs on polyI:C transfection in the bat lung PaLuT02 cell line. Cells were stimulated with polyI:C and collected at the indicated time points. IFN-λ mRNA was measured by qRT-PCR. The data were normalized against the housekeeping gene 18s rRNA.
Primary bat cell lines display differential type I and III IFN expression patterns after stimulation with polyI:C
To examine the tissue distribution of IFN-λ induction, we examined the expression of bat IFN-λ1 and IFN-λ2 in P. alecto primary cell lines from a variety of tissues after polyI:C stimulation and compared them with the induction of IFN-β. Seven nonimmortalized primary cell lines from a range of tissues including lung, liver, heart, kidney, small intestine, brain, and salivary gland were tested. Each of the primary cell lines was a mixture of different types of adherent cells; therefore, they were more closely representative of the original tissue from which they were derived compared with the cloned and immortalized cell types used for the characterization of bat IFN-λ genes (31). Based on the kinetics of IFN-λ induction in PaLuT02 cells, all seven primary bat cell lines were stimulated (both treated and transfected) with polyI:C for 3 h. The use of these two different stimulations was used to deliver polyI:C either to extracellular/endosomal compartments (treated) or intracellularly (transfected).
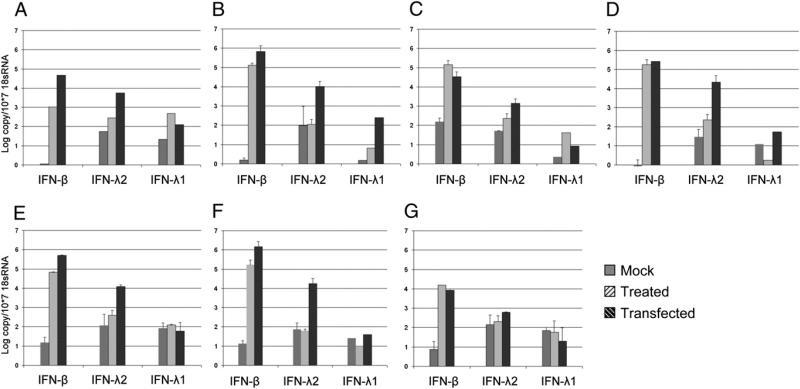
In both polyI:C treated and transfected cells lines, the induction of IFN-β was higher than IFN-λ1 or IFN-λ2. The kinetics of the IFN-β response was similar between polyI:C treated and transfected cells with the exception of the lung cell line, which displayed around 50 times higher induction of IFN-β after polyI:C transfection (Fig. 4). The mRNA expression of the two bat IFN-λ genes demonstrated a differential expression pattern between treated and transfected cells and between the different cell lines. IFN-λ2 was induced in polyI:C transfected cells to a 1–2 log copy change compared with the baseline. However, in polyI:C treated cells, only minor or no change in IFN-λ2 was observed. IFN-λ1 was induced only in the lung, heart, and liver cell lines after either transfection or treatment with polyI:C, and IFN-λ1 expression was the lowest among the three IFNs in all the primary cell lines tested. In the cell lines in which IFN-λ1 could be induced, the level of IFN-λ1 was about 100 times lower than IFN-λ2. Thus, in bat primary cell lines, a differential expression pattern was observed between type I and III IFNs, and between the two IFN-λ genes after stimulation with polyI:C.
FIGURE 4.
Expression of type I and type III IFNs on polyI:C stimulation in bat primary cell lines. Primary cell lines from lung (A), liver (B), heart (C), kidney (D), small intestine (E), brain (F), and salivary gland (G) were stimulated with 33 μg/ml polyI:C for 3 h. IFN mRNA was measured by qRT-PCR and normalized against 18s RNA. A–G, Data are mean values of two separate experiments, and the error bars represent SEs.
IFN-λ2 is highly upregulated in polyI:C treated bat splenocytes
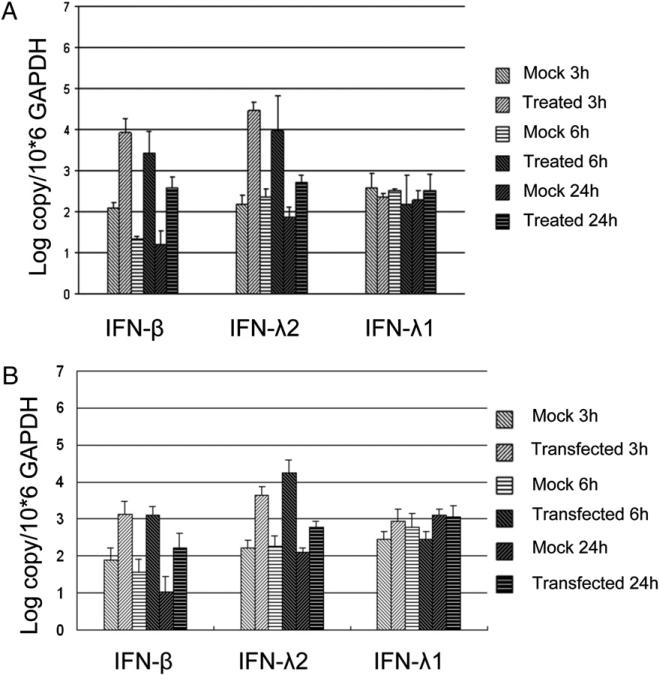
The spleen is one of the most important immune tissues and contains large numbers of immune-related cells. To determine whether there were differences in the induction of IFNs between our bat cell lines and freshly isolated cells, we next analyzed the IFN response of bat splenocytes after polyI:C stimulation. The freshly isolated bat splenocytes were stimulated by treatment and transfection with polyI:C for 3, 6, and 24 h. In contrast with the response of the primary cell lines, both polyI:C treatment and transfection resulted in a higher induction of IFN-λ2 compared with IFN-β in freshly isolated cells at 3 and 6 h (Fig. 5). Similar to the primary cell lines, IFN-λ1 was not induced in bat splenocytes on polyI:C treatment or transfection.
FIGURE 5.
Expression of type I and type III IFNs on polyI:C stimulation in fresh bat splenocytes. The fresh splenocytes were treated (A) or transfected (B) with 10 μg/ml polyI:C and collected at the indicated time points. IFN mRNA was measured in qRT-PCR and normalized against the housekeeping gene GAPDH. The data shown represent the mean of two and three individual bats, respectively, for the treated and transfected cells, and the error bars indicate SDs.
IFN-λs are upregulated in bat splenocytes after TioPV infection
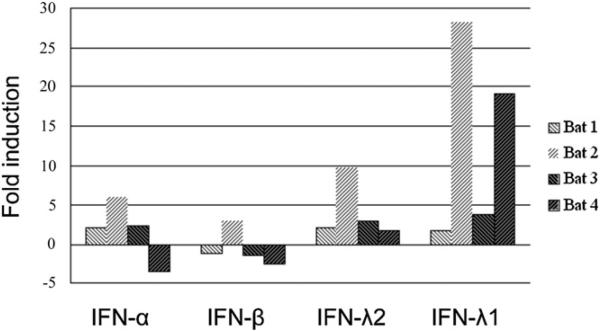
To examine the production patterns of bat IFNs in response to viral challenge, we used TioPV to infect freshly isolated bat splenocytes. Fruit bats are the natural host reservoir of TioPV, which is a paramyxovirus with a typical nonsegmented negative sense RNA genome (37). Four individual bats were sacrificed, and fresh splenocytes were challenged with TioPV for 3 h. Successful infection of TioPV was confirmed by RT-PCR (data not shown). As shown in Fig. 6, TioPV infection of primary bat splenocytes resulted in no upregulation of either IFN-α or IFN-β. Of the four bats tested, one bat displayed an IFN-α and IFN-β response that was lower than the baseline level (bat 4), and a further two bats (bats 1 and 3) displayed an IFN-β response lower than baseline. In contrast, IFN-λ, particularly IFN-λ1, was upregulated to a higher level compared with type I IFN in two of the four bats (bats 2 and 4; Fig. 6). Although no upregulation in IFN-λ occurred in bat 1 at 3 h post-infection, by 6 h this bat displayed an increase in type III IFN production above that of the type I IFNs, and this response was highest for IFN-λ1 (Supplementary Fig. 1). Only bat 3 showed no upregulation in type III IFNs after TioPV infection. Overall, these results are in contrast with those obtained after stimulation with polyI:C, which caused little or no induction of IFN-λ1 in the cell lines or splenocytes (Fig. 5).
FIGURE 6.
Expression of type I and III IFNs in fresh bat splenocytes in response to TioPV. Fresh splenocytes from four individual bats (bats 1–4) were challenged with TioPV for 3 h at a multiplicity of infection 0.1. The cellular RNA was extracted and IFN mRNA level was measured by qRT-PCR. Data are shown as fold change of expression obtained from comparison of the virus challenged and mock challenge group. The gene expression level had been normalized against GAPDH.
Bat IFN-λ2 was inhibitory to PulV replication in cloned bat cells
To assess the functional activity of bat IFN-λ, we constructed a stable cell line expressing IFN-λ2 in 293T cells (λ2-293T). The IFN-λ2 gene was chosen for these experiments because of its high induction in bat cells and cell lines after polyI:C stimulation. Successful construction was confirmed by detection of IFN-λ2 in λ2-293T cell lines by qRT-PCR (data not shown).
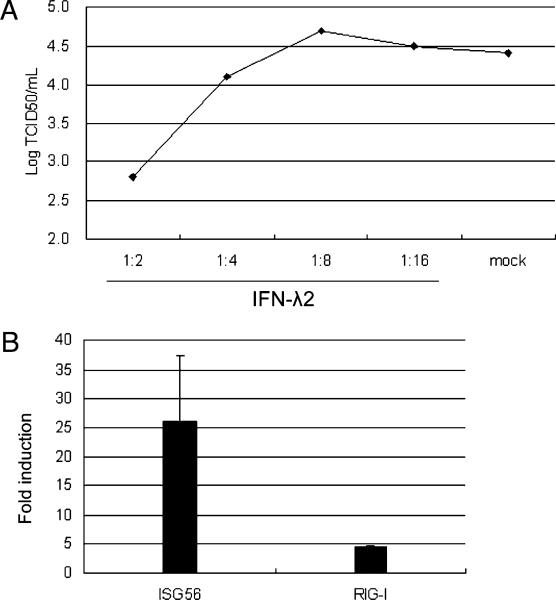
IFN-λs exerts antiviral activities in other species against a variety of different viruses (17, 18, 21, 24). To assess whether bat IFN-λ could also protect cells from virus infection, the antiviral activity of bat IFN-λ on PulV-infected cells was examined. PulV is an orthoreovirus identified in fruit bats (38). PulV was chosen for these experiments because of its ability to infect and cause obvious CPE in the PaFeT05 cells. This combination of cells and virus was therefore considered to be most informative in terms of the ability of bat IFN-λ to inhibit viral replication. Supernatant collected from λ2-293T cells was serial diluted and used to stimulate PaFeT05 cells. Supernatant from normal cultured 293T cells was used as mock in this experiment. As shown in Fig. 7A, TCID50 demonstrated that PulV growth was clearly inhibited in the IFN-λ2–treated cells, and this inhibition was dose dependent. Antiviral activity was significantly higher in cells treated with a 1:2 dilution of IFN-λ2 compared with mock-treated cells (p = 0.004), and this effect had disappeared by a dilution of 1:8 of IFN-λ2.
FIGURE 7.
Bat IFN-λ2 displays IFN-λike activities. A, Protection of bat PaFeT05 cells from PulV infection after pretreatment with bat IFN-λ2. Cells were treated with 1:2, 1:4, 1:8, and 1:16 diluted IFN-λ2 containing (or 1:2 diluted mock containing) 293T cell medium for 24 h and then infected with PulV for another 24 h. PulV replication levels were determined by measurement of PulV in the supernatant by TCID50. B, Induction of ISGs by bat IFN-λ2 in cloned PaFeT05 cells. The cells were stimulated with 1:2 diluted IFN-λ2 containing (or mock containing) 293T cell medium for 6 h. The cellular RNA was extracted and mRNA level of ISG56 and RIG-I was measured by qRT-PCR. Data are shown as fold change of expression obtained from comparison of the IFN-λ2 stimulated and normal 293T culture medium. The gene expression level had been normalized against GAPDH.
Bat IFN-λ2 induces the production of ISGs
The IFN-λ2 concentration showing the greatest antiviral effect was tested for its ability to induce ISGs. As shown in Fig. 7B, bat IFN-λ resulted in up to 25-fold induction of ISG56 and up to 4-fold induction of RIG-I.
Discussion
Bats are rich reservoirs for emerging viruses, including many that are highly pathogenic to humans and other mammals (1). One hypothesis for the asymptomatic nature of viral infection in bats is that viral replication is controlled rapidly after infection by the innate immune response. Toward understanding the innate immune response of bats, we report, to our knowledge, the first characterization of type III IFNs from any bat species, providing evidence for induction and antiviral activity of type III IFNs in pteropid bats. These results represent an important step in understanding the mechanisms that contribute to the ability of bats to coexist with viruses.
The Malaysian flying fox, P. vampyrus, is the first pteropid bat to have its genome sequenced and is closely related to P. alecto. Three IFN-λ genes with similarity to human IFN-λ1, IFN-λ2, and IFN-λ3 were identified in the genome sequence of P. vampyrus, but only two appear to be transcribed in P. alecto. Phylogenetic analysis revealed that the P. alecto IFN-λ1 and IFN-λ2 genes were orthologous to other mammalian IFN-λ1 and IFN-λ2/3 genes, respectively. Although both IFN-λ2 and IFN-λ3 were annotated in the P. vampyrus whole-genome sequence, this genome has low sequence coverage (2.63×), which can cause some genes to appear as multiple loci when they actually belong to a single locus (23). The pteropid bat IFN-λ2 and IFN-λ3 genes therefore either represent the same locus or highly similar loci, of which IFN-λ2 is likely a pseudogene in pteropid bats. The latter situation would be similar to other mammals and nonmammals including mice that have two transcribed IFN-λ genes and one pseudogene and the amphibian, Xenopus tropicalis, which has four transcribed IFN-λ genes and one pseudogene (21, 22).
Type I and III IFNs are believed to have a common evolutionary origin, with IFN-λ considered to be the ancestral IFN system of vertebrates (45). The genomic organization of type III IFNs appear to be highly conserved among mammals and nonmammals (22). However, an unusual feature of the bat IFN-λ1 gene structure not shared by other vertebrates is the presence of an unusually long intron separating exons 2 and 3, spanning 4 kb. Comparative analysis of the current genome assemblies from a variety of other vertebrates present in Ensembl revealed the presence of an intron spanning only 1.1 kb in all species examined. The presence of an unusually long intron in the bat IFN-λ1 locus is striking considering the genomes of bats are constrained in size in comparison with other mammals (46). Although the kinetics of IFN induction in response to polyI:C appeared to be similar between the two bat IFN-λs, it is possible that the evolution of such an intron may have functional significance in the regulation of the IFN response in vivo.
The innate immune system has evolved two pathways of viral RNA recognition, one of which is present in the endosome and one in the cytosol of cells. TLR3 is expressed in endosomes and detects dsRNA after endocytosis, whereas the RIG-I–like helicases are expressed in the cytoplasm and detect viral RNA generated during replication (14). In the primary bat cell lines, IFN-λ2 was up-regulated only in polyI:C transfected cells, whereas both treatment and transfection caused a significant upregulation of IFN-β. The high induction of type I IFNs by both treatment and transfection with polyI:C is consistent with the activation of both endosomal and cytosolic pattern recognition receptors (PRRs). In contrast, IFN-λ2 was induced only after transfection of polyI:C, which would result in the activation of IFN predominantly as a consequence of recognition by cytosolic PRRs such as RIG-I. This result contrasts with that of mice that appear to use both cytosolic and endosomic PRRs for the induction of type III IFNs (26, 47). The differential expression pattern of the two type III IFNs also differs from other species in which both IFN-λ1 and IFN-λ2/3 are induced to a similar level (5, 26, 48). These results support the hypothesis that rather than being redundant to each other, the type I and III IFN systems perform complementary functions (49). However, further study of the production pathways of bat IFN-λ are required to elucidate the mechanisms responsible for the induction of this IFN.
Viruses have evolved a variety of mechanisms to evade the host's IFN response, resulting in the inhibition of IFN production (14). Because many of the viruses identified in bats are ssRNA viruses, we examined the response of bat cells to the ssRNA virus, TioPV. TioPV is a member of the paramyxovirus family that includes the highly pathogenic henipaviruses, Hendra and Nipah, which are carried by P. alecto and P. vampyrus, respectively (33). Although TioPV has not been identified in P. alecto, its natural host reservoir, Pteropus hypomelanus is closely related to P. alecto. Infection of P. alecto splenocytes with TioPV caused no upregulation of either of the type I IFNs, resulting in lower than baseline levels of IFN-β in three of the four bats tested. For the type III IFNs, IFN-λ1 was upregulated in two of the four bats at 3 h postinfection, and by 6 h a third bat had responded with an increase in IFN-λ1. As these bats were wild caught individuals of unknown age and history of viral exposure, some variation between bats is not surprising. Although additional experiments are necessary to confirm these results in a larger number of bats and in response to different viruses, to our knowledge, this is the first report describing the simultaneous suppression of type I IFN and induction of type III IFN after virus infection. Furthermore, IFN-λ1, which was not induced on polyI:C stimulation of bat splenocytes, was the type III IFN that underwent the highest upregulation in three of the four bats after viral challenge. One explanation for the upregulation of IFN-λ1 by TioPV and the absence of a response observed after polyI:C stimulation is the activation of different PRRs resulting in differential activation of the IFN system. In contrast with double-stranded polyI:C, TioPV is an ssRNA virus and would be recognized by endosomal PRRs, TLR7 and TLR8. Thus, at least in the case of TioPV infection, our results are consistent with IFN-λ1 induction through TLR7/8 activation, thus providing evidence that the two bat IFN-λ genes are activated by different mechanisms. Ank et al. (50) speculated that IFN-λ may be important during infection with viruses that rapidly close down production of type I IFNs by host cells and may act as a slow-acting defense system designed to prolong IFN-mediated antiviral activity. Whether a similar pattern will be observed after infection with other bat viruses, including Hendra virus, remains to be determined, but these results may represent an important antiviral strategy used by bats that may be less susceptible to suppression by viral immune evasion mechanisms.
To determine whether bat IFN-λ has similar antiviral activity to type I and III IFNs from other species, we assessed the ability of bat IFN-λ to protect cells from viral infection. PulV was chosen for these experiments because of the obvious CPE it is capable of producing in bat cell lines. PulV is a dsRNA orthoreovirus, first identified in fruit bats P. hypomelanus on Tioman Island off the coast of Malaysia (38). Bat IFN-λ2 protected cells from viral infection, demonstrating antiviral activity against PulV infection. To our knowledge, this is the first IFN protection assay ever reported using a bat virus. In addition, the ISGs, ISG56 and RIG-I, were induced by rIFN-λ2 in the P. alecto cell line, demonstrating that bat IFN-λ2 likely induces its antiviral activity through a similar mechanism to type I and III IFNs from other species. Together with our results demonstrating the induction of type III IFN after TioPV infection, these results demonstrate that bat IFN-λ may play an important role in inhibiting viral replication in bats.
As natural reservoirs for a variety of viral diseases, bats provide an important model for examining the evolution of antiviral immunity. Studies on antiviral immunity in bats have the potential to provide insights into new antiviral strategies and result in the identification of new therapeutic targets. Our results provide the first evidence, to our knowledge, for a type III IFN response in any species of bat, demonstrating that bat type III IFNs have antiviral activities similar to type I and III IFNs from other species. The induction of type III IFNs in bat cells after viral infection may provide an alternate antiviral strategy after the suppression of the type I IFN response and may have implications for the ability of bats to coexist with viruses. However, further work is required to fully elucidate the IFN response and induction pathways in bats to a wide variety of bat viruses to understand the role of IFNs in the asymptomatic nature of viral infections in bats.
Supplementary Material
Acknowledgments
We thank Craig Smith, Hume Field, and Susanne Wilson for provision of bat spleen material used for this study.
This work was supported by a Commonwealth Scientific and Industrial Research Organization Chief Executive Officer Science Leaders award (to L.-F.W.), National Institutes of Health Grant P20RR018754 from the Institutional Development Award Programme of the National Center for Research Resources (to M.L.B.), and a scholarship from the Chinese Academy of Sciences (to P.Z.).
Abbreviations used in this article
- CPE
cytopathic effect
- IFNAR
IFN-α receptor
- ISG
IFN-stimulated gene
- polyI:C
polyinosinic-polycytidylic acid
- PRR
pattern recognition receptor
- PulV
Pulau virus
- qRT-PCR
quantitative RT-PCR
- RIG-I
retinoic acid-inducible gene I
- TCID50
50% end point of tissue culture infective dose
- TioPV
Tioman virus
Footnotes
The sequences presented in this article have been submitted to GenBank under accession numbers HQ201955 (-λ2) and HQ201956 (-λ1).
The online version of this article contains supplemental material.
Disclosures: The authors have no financial conflicts of interest.
References
- 1.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Middleton DJ, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, Westbury HA, Halpin K, Daniels PW. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J. Comp. Pathol. 2007;136:266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Williamson MM, Hooper PT, Selleck PW, Gleeson LJ, Daniels PW, Westbury HA, Murray PK. Transmission studies of Hendra virus (equine morbillivirus) in fruit bats, horses and cats. Aust. Vet. J. 1998;76:813–818. doi: 10.1111/j.1751-0813.1998.tb12335.x. [DOI] [PubMed] [Google Scholar]
- 4.van der Poel WH, Lina PH, Kramps JA. Public health awareness of emerging zoonotic viruses of bats: a European perspective. Vector Borne Zoonotic Dis. 2006;6:315–324. doi: 10.1089/vbz.2006.6.315. [DOI] [PubMed] [Google Scholar]
- 5.Witte K, Witte E, Sabat R, Wolk K. IL-28A, IL-28B, and IL-29: promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 2010;21:237–251. doi: 10.1016/j.cytogfr.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- 7.Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, Rodwell B, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 8.Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 9.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, et al. Bats are natural reservoirs of SARS-like corona-viruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 11.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 12.Chua KB, Crameri G, Hyatt A, Yu M, Tompang MR, Rosli J, McEachern J, Crameri S, Kumarasamy V, Eaton BT, Wang LF. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. USA. 2007;104:11424–11429. doi: 10.1073/pnas.0701372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 15.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 16.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 17.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 19.Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, Reuhl K, Dickensheets H, Sheikh F, Donnelly RP, et al. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Karpala AJ, Morris KR, Broadway MM, McWaters PG, O'Neil TE, Goossens KE, Lowenthal JW, Bean AG. Molecular cloning, expression, and characterization of chicken IFN -lambda. J. Interferon Cytokine Res. 2008;28:341–350. doi: 10.1089/jir.2007.0117. [DOI] [PubMed] [Google Scholar]
- 22.Qi Z, Nie P, Secombes CJ, Zou J. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J. Immunol. 2010;184:5038–5046. doi: 10.4049/jimmunol.0903374. [DOI] [PubMed] [Google Scholar]
- 23.Fox BA, Sheppard PO, O'Hara PJ. The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-lambda family. PLoS ONE. 2009;4:e4933. doi: 10.1371/journal.pone.0004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartlett NW, Buttigieg K, Kotenko SV, Smith GL. Murine interferon lambdas (type III interferons) exhibit potent antiviral activity in vivo in a poxvirus infection model. J. Gen. Virol. 2005;86:1589–1596. doi: 10.1099/vir.0.80904-0. [DOI] [PubMed] [Google Scholar]
- 25.Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors. 2009;35:82–87. doi: 10.1002/biof.19. [DOI] [PubMed] [Google Scholar]
- 26.Ank N, West H, Paludan SR. IFN-lambda: novel antiviral cytokines. J. Interferon Cytokine Res. 2006;26:373–379. doi: 10.1089/jir.2006.26.373. [DOI] [PubMed] [Google Scholar]
- 27.Uzé G, Monneron D. IL-28 and IL-29: newcomers to the interferon family. Biochimie. 2007;89:729–734. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-λ (IFN-λ)is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iversen MB, Ank N, Melchjorsen J, Paludan SR. Expression of type III interferon (IFN) in the vaginal mucosa is mediated primarily by dendritic cells and displays stronger dependence on NF-kappaB than type I IFNs. J. Virol. 2010;84:4579–4586. doi: 10.1128/JVI.02591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prescott J, Hall P, Acuna-Retamar M, Ye C, Wathelet MG, Ebihara H, Feldmann H, Hjelle B. New World hantaviruses activate IFNlambda production in type I IFN-deficient vero E6 cells. PLoS ONE. 2010;5:e11159. doi: 10.1371/journal.pone.0011159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crameri G, Todd S, Grimley S, McEachern JA, Marsh GA, Smith C, Tachedjian M, De Jong C, Virtue ER, Yu M, et al. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS ONE. 2009;4:e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Hurk AF, Smith CS, Field HE, Smith IL, Northill JA, Taylor CT, Jansen CC, Smith GA, Mackenzie JS. Transmission of Japanese Encephalitis virus from the black flying fox, Pteropus alecto, to Culex annulirostris mosquitoes, despite the absence of detectable viremia. Am. J. Trop. Med. Hyg. 2009;81:457–462. [PubMed] [Google Scholar]
- 33.Baker ML, Tachedjian M, Wang LF. Immunoglobulin heavy chain diversity in Pteropid bats: evidence for a diverse and highly specific antigen binding repertoire. Immunogenetics. 2010;62:173–184. doi: 10.1007/s00251-010-0425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowled C, Baker M, Tachedjian M, Zhou P, Bulach D, Wang LF. Molecular characterisation of Toll-like receptors in the black flying fox Pteropus alecto. Dev. Comp. Immunol. 2011;35:7–18. doi: 10.1016/j.dci.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kepler TB, Sample C, Hudak K, Roach J, Haines A, Walsh A, Ramsburg EA. Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics. 2010;11:444. doi: 10.1186/1471-2164-11-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omatsu T, Bak EJ, Ishii Y, Kyuwa S, Tohya Y, Akashi H, Yoshikawa Y. Induction and sequencing of Rousette bat interferon alpha and beta genes. Vet. Immunol. Immunopathol. 2008;124:169–176. doi: 10.1016/j.vetimm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chua KB, Wang LF, Lam SK, Crameri G, Yu M, Wise T, Boyle D, Hyatt AD, Eaton BT. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology. 2001;283:215–229. doi: 10.1006/viro.2000.0882. [DOI] [PubMed] [Google Scholar]
- 38.Pritchard LI, Chua KB, Cummins D, Hyatt A, Crameri G, Eaton BT, Wang LF. Pulau virus; a new member of the Nelson Bay orthoreovirus species isolated from fruit bats in Malaysia. Arch. Virol. 2006;151:229–239. doi: 10.1007/s00705-005-0644-4. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 42.Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 43.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 44.Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J. Biol. Chem. 2009;284:20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levraud JP, Boudinot P, Colin I, Benmansour A, Peyrieras N, Herbomel P, Lutfalla G. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J. Immunol. 2007;178:4385–4394. doi: 10.4049/jimmunol.178.7.4385. [DOI] [PubMed] [Google Scholar]
- 46.Smith JD, Gregory TR. The genome sizes of megabats (Chiroptera: Pteropodidae) are remarkably constrained. Biol. Lett. 2009;5:347–351. doi: 10.1098/rsbl.2009.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 48.Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uzé G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.