Abstract
To provide accurate biological hypotheses and inform upon global properties of cellular networks, systematic identification of protein–protein interactions has to meet high-quality standards. We present an expanded Caenorhabditis elegans protein-protein interaction network, or “interactome” map derived from testing a matrix of ~ 10,000 × ~ 10,000 proteins using a highly specific high-throughput yeast two-hybrid system. Through a new quality control empirical framework, We show that the resulting dataset (Worm Interactome 2007 or WI-2007) is similar in quality to low-throughput data curated from the literature. Previous interaction datasets have been filtered and integrated with WI-2007 to generate a high confidence consolidated map (Worm Interactome version 8 or WI8). This work allows us to estimate the size of the worm interactome at ~ 116,000 interactions. Comparison with other types of functional genomic data shows the complementarity of distinct experimental approaches in predicting different functional relationship features between genes or proteins.
The interactome of an organism is the network formed by the complete set of binary physical interactions that can occur between all proteins. Low-throughput protein-protein interaction experiments are of immense value to understand cellular processes at the molecular level. However, the development of high-throughput approaches can substantially increase the pace and scale of discovery, while permitting the implementation of standardized and systematic quality control. Initial steps towards binary interactome mapping in metazoans have been undertaken1–5, and the resulting partial interactome maps: i) provide insights into the organization of biological networks, ii) assist in determining functions of many proteins and complexes, and iii) identify hundreds of novel connections to proteins associated with human diseases.
High-throughput interactome mapping is particularly needed for C. elegans, a major model organism for which the set of protein-protein interactions derived from small-scale experiments and accessible in public databases is limited to less than 500. The first proteome-scale version of the Worm Interactome (WI5)3 combines several sources of protein-protein interactions: literature-curated interactions, yeast two-hybrid (Y2H) “module” maps each devoted to a specific biological process1,6–11, “interolog” interactions, i.e., predicted pairs of interactors whose respective orthologs interact in another organism, and lastly, Y2H interactions derived from a high-throughput screen performed with ~ 2,000 “metazoan” proteins as baits3 (WI-2004). WI5 represents a key resource to elaborate biological hypotheses and investigate the properties of the C. elegans interaction network. However, WI5 includes non-binary interactions derived from the literature, non-experimentally confirmed interologs, and some lower-confidence Y2H interactions.
Our updated Worm Interactome map (WI8) combines the implementation of several techniques and strategies that are critical for generating high-quality protein-protein interaction data on a proteome scale. First, we expanded the worm interactome map by screening a matrix of ~ 10,000 × ~ 10,000 proteins. Second, we developed new standards to deliver a dataset of unprecedented quality. These standards involve a highly stringent high-throughput yeast two-hybrid (HT-Y2H) assay, strict methods for filtering and updating existing datasets, independent measurement of technical quality, and evaluation of biological relevance. Importantly, since worm genome annotations are improved frequently12, we updated previous protein-protein interaction data according to recent gene models. Finally, we provide an empirical estimate of the full size of the C. elegans interactome, through the implementation of a novel interaction mapping framework based exclusively on protein-protein interaction data13.
To extend the use of WI8 beyond protein-protein interaction analysis and to place WI8 into broader biological context, we integrated the resulting protein-protein interactions with complementary datasets such as physical and genetic interactions from curated literature, our new interolog dataset, phenotypic profiling data and a co-expression compendium. We also identified tissue localizations and developmental stages in which interacting pairs are most likely to be physiologically relevant whenever ‘anatomical annotation’14 or ‘spatiotemporal expression patterns’15 were available for both proteins.
Our new dataset, WI-2007, provides 1,816 high confidence binary protein-protein interactions. Previously published high-quality C. elegans binary protein-protein interactions were integrated with WI-2007 into the updated WI8 version of the worm interactome, providing 3,816 high-quality binary physical interactions between 2528 proteins. We demonstrated that WI8 is significantly enriched for functionally linked protein pairs, confirming its high biological relevance and demonstrating the value of unbiased large-scale Y2H screens in inferring protein function.
RESULTS
A new HT-Y2H dataset
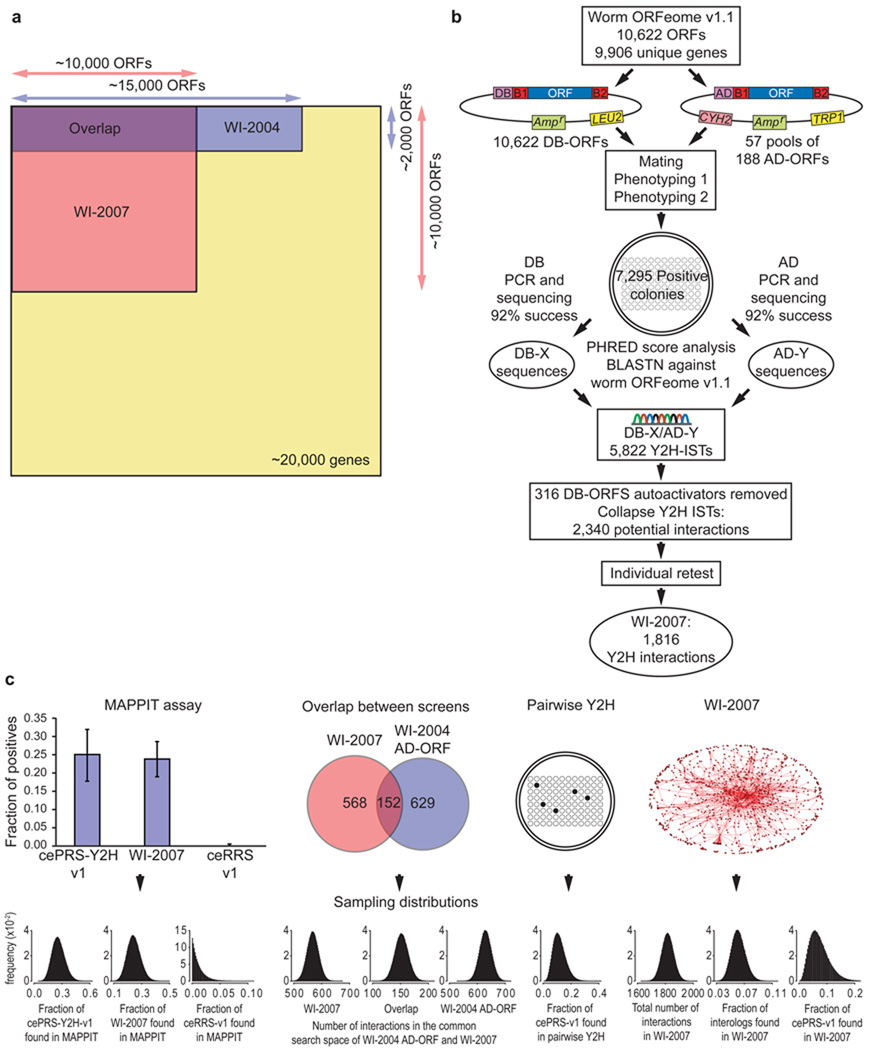
For this iteration of worm interactome mapping, we implemented a HT-Y2H strategy previously used for human interactome mapping5. We tested all ORFs in the worm ORFeome v1.18 against each other (a ~ 10,000 × ~ 10,000 matrix), a search space corresponding to ~ 24% of the total search space for a comprehensive C. elegans interactome map, excluding variants due to polymorphism, alternative transcription or splicing (Fig. 1a). We also ensured the quality of the new dataset by using stringent conditions and controls described previously5, including low expression of DNA-binding and activation-domain fusion proteins (DB-X and AD-Y), multiple reporter genes to ensure high precision, removal of all a priori and de novo DB-X auto-activators, and individual retesting of each positive protein-protein interaction. The resulting set of 1,816 protein-protein interactions between 1,496 proteins (Fig. 1b) is called “WI-2007”.
Figure 1.
Construction and characterization of WI-2007. (a) Search spaces of WI-2007 and WI-2004 relative to the whole proteome. WI-2007 results from a 10,000 ORFs matrix screen, which is three times larger than WI-2004, corresponding to one fourth of the entire theoretical search space (screening completeness ~ 24%). (b) Pipeline used for WI-2007. ORFs from ORFeome v1.1 were transferred into DB and AD vectors by recombinational cloning, then transformed into yeast cells. Each bait was then mated with pools of 188 AD-ORFs. Two steps of phenotyping were performed to isolate positive colonies, which were used to PCR-amplify DB-ORFs and AD-ORFs for sequencing, leading to the identification of 5,822 Interaction Sequence Tags (ISTs). After exclusion of autoactivators and collapsing of all ISTs corresponding to the same, non oriented pair, an individual retest was performed to generate the final WI-2007 dataset. (c) WI-2007 characterization. Ten measurements were used (left to right) : proportions of cePRS-Y2H-v1, a random sample of WI-2007 and ceRRS-v1 observed in MAPPIT; number of interactions detected in the common search space of WI-2007 and WI-2004: in WI-2007, in both screens, and in WI-2004 AD-ORF; proportion of cePRS-v1 detected in an independent pairwise Y2H experiment; total number of interactions in WI-2007, proportion of ultra-conserved interologs and cePRS-v1 recovered in WI-2007. The Sampling errors on the 10 measurements are modeled with Beta distributions. Precision, sampling sensitivity, assay sensitivity and the total number of interactions in C. elegans are computed using a Monte-Carlo simulation. Y axis label (frequency) applies to all ten sampling distributions…
Characterization of WI-2007
To further assess the quality of our new dataset and estimate the size of the complete worm interactome, we used a novel framework recently developed13 with a slightly different implementation relevant to the available data in C. elegans. This framework empirically measures several parameters to characterize a high-throughput binary protein-protein interactions dataset: screening completeness, the fraction of the proteome-wide space tested in the experiment; precision, the proportion of true biophysical interactions in the dataset; sampling sensitivity, the fraction of all detectable interactions for a particular assay found under given sampling conditions, which corresponds here to the saturation of a single screen; and assay sensitivity, the proportion of all biophysical interactions that can be identified by an assay at saturation.
To estimate these parameters we performed the following experiments. First, we used the Mammalian Protein-Protein Interaction Trap (MAPPIT) to measure how a random sample of WI-2007 performs in an independent protein interaction detection assay compared to a Positive Reference Set (cePRS-v1) and a Random Reference Set (ceRRS-v1). Second, the overlap between WI-2007 and our previous Y2H study in their common search space was used to quantify the level of saturation of our screen. Third, to evaluate the proportion of interactions that can be captured by our Y2H assay, the fraction of cePRS-v1 pairs recovered in a pairwise Y2H experiment and in WI-2007, as well as the proportion of widely conserved interologs found in WI-2007, were used. Introducing these measurements into a Monte-Carlo simulation, we computed the four parameters in our framework, as well as the expected size of the worm interactome. According to this model, the screening completeness equals 23.6%; the precision is estimated as 86% ± 16% (mean and standard deviation), the sampling sensitivity 31% ± 8%, the assay sensitivity 16% ± 3% and the size of the worm interactome 115,600 ± 26,400 (Fig. 1c).
Given the potential bias in cePRS-v1 and in the set of ultra-conserved interologs towards interactions that are easy to detect, the associated assay sensitivity may be an overestimate. Thus the predicted interactome size is likely to be a conservative estimate. The strength of this approach is that these calculations rely solely on protein-protein interactions, without depending on functional annotation or other types of genomic or proteomic data. Our estimate provides an endpoint for the worm interactome mapping project and can be used as a reference for evolutionary comparisons between interactome networks from different species.
A combined dataset of high-quality binary interactions
To provide an integrated high-quality binary protein-protein interaction dataset for C. elegans, we reprocessed the raw data from multiple smaller scale Y2H screens, encompassing proteins involved in vulval development1, protein degradation6, DNA damage response7, germline formation9, TGF-β signaling pathway11 and RNA interference10, along with unpublished Y2H interactions. This “biological processes” subset (BPmaps) contains 554 protein-protein interactions.
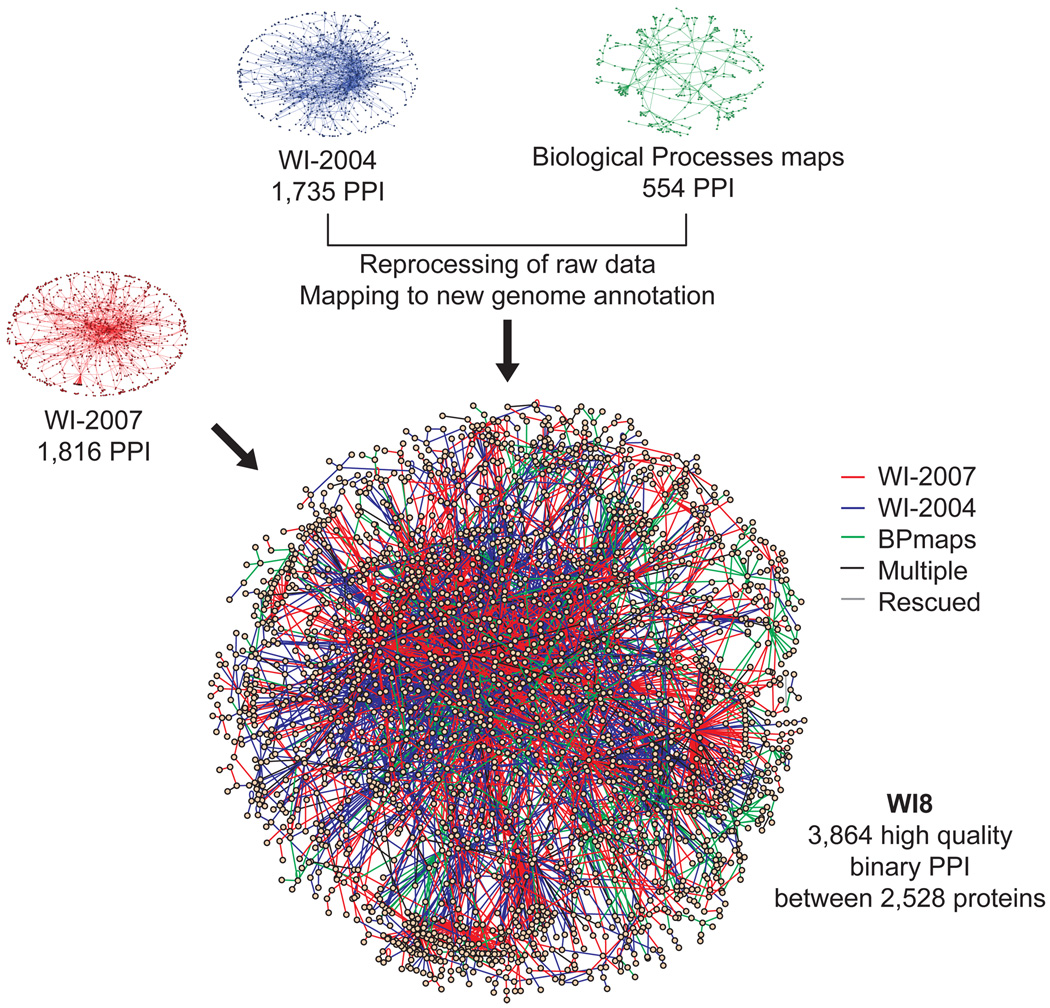
WI8 is the union of WI-2004, WI-2007 and BPmaps. The consolidated WI8 network (Fig. 2, Supplementary Table 1 online) contains 3,864 high-quality protein-protein interactions among 2,528 proteins. Approximately 40% of the interactions are novel, and the set excludes any lower confidence interactions from previous studies3. The WI8 physical interaction network can be visualized on our website (http://interactome.dfci.harvard.edu/C_elegans/) using N-Browse16 or VisAnt17.
Figure 2.
WI8 - an extended high-quality protein-protein interaction network. High-quality Y2H protein-protein interactions (PPI) from WI-2007 (1,816 interactions), WI-2004 (1,735 interactions), and diverse medium throughput biological processes based Y2H maps1,6–11 (554 interactions) were updated, filtered, assembled, and integrated into WI8. Two interactions, detected in both WI-2004 and WI-2007 with lower confidence were also “rescued”. The final dataset contains 3,864 high-confidence binary protein-protein interactions. In the network representation of WI8, the color of the edge indicates the dataset of origin: WI-2007 in red, WI-2004 in blue and biological process maps in green. Edges corresponding to several of these evidence types are shown in black, and edges corresponding to “rescued” interactions, i.e., supported by at least two lower confidence evidences, in grey. Only the main component of the network is shown.
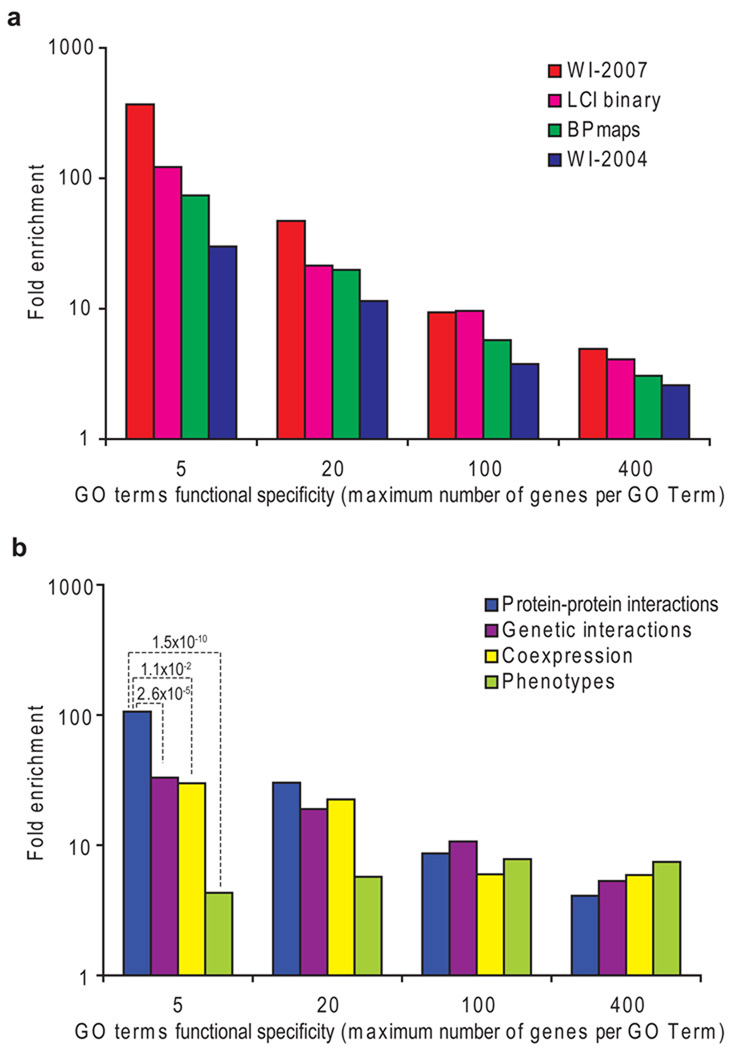
Confirmed Y2H interactions may be “biophysically” true interactions that do not actually occur in vivo if the involved proteins are not present at the same time and place within a multi-cellular organism, or with the proper post-translational modifications. We evaluated the overall biological relevance of WI8 by assessing the degree to which interacting pairs share Gene Ontology (GO) annotation terms, i.e., can be considered as functionally linked. A GO term may be specific or broad depending on the number of genes to which it is assigned. We therefore defined four different thresholds of functional specificity: less than 5, 20, 100 and 400 annotated genes per GO term. For all three component subsets of WI8 we compared the degree of functional linkage with that of binary interactions derived from the literature (LCI binary, Supplementary Table 2 online), normalizing for protein composition bias of each of these subsets. All data subsets showed a high enrichment for both broad and specific functional linkage (Fig. 3a), suggesting high biological relevance. The degree of functional linkage among WI-2007 was similar to or exceeded the literature enrichment at each functional specificity limit tested.
Figure 3.
Biological relevance. The Fold enrichment represents the frequency of functional linkage of protein pairs compared to random. The maximum number of genes associated to a particular GO term was used as an estimate of the functional specificity (the smaller the maximum number of genes per GO term, the higher the functional specificity), and the fold enrichments were calculated for several functional specificity levels (5, 20,100 and 400). (a) Enrichment of different components of WI8 and LCI binary for functional relationships. (b) Functional relationship enrichments for distinct types of experimental evidence. P-values assessing the difference between protein-protein interactions (PPI) and other types of evidence are shown for very specific GO terms (maximum 5 genes per GO term).
Various interactions in WI8 provide valuable new biological information. For example, EBP-1, a microtubule binding protein whose homologs are involved in a variety of microtubule mediated processes18, interacts with several proteins involved in microtubule dynamics, including UNC-14, a protein required for axon growth and sex myoblast migration19, VAB-8, a kinesin-like protein required for axon outgrowth and cell migrations20, and RSA-2, a protein specifically required for microtubule outgrowth from centrosomes and for spindle assembly21.
Integrated functional network
Integration of diverse large-scale datasets was previously used to demonstrate the coordination of interconnected yet distinct molecular machines involved in worm early embryogenesis22. Another recent publication23 describes Bayesian integration of functional linkages into a single network, which can be a valuable resource leading to interesting hypotheses. However, such a scoring scheme is highly dependent on the benchmark, which can strongly bias the predictions (Supplementary Discussion online). In contrast, we chose to provide an unweighted dataset that (i) does not artificially bias the network towards highly studied proteins, (ii) allows the user to select their own threshold when relevant, (iii) separates each type of experimental evidence and (iv) does not rely on an inevitably biased benchmark.
We integrated WI8 with five different sources of evidence for functional relationships: (i) mRNA co-expression data available in WormBase12 (Supplementary Table 3 online); (ii) RNAi phenotypes from RNAiDB24 (Supplementary Table 4 online); (iii) genetic interactions curated in WormBase12; (iv) interolog interactions; and (v) all binary and non-binary protein-protein interactions from the literature curated dataset (LCI). This integrated network involves 178,151 links between 6,176 genes and can be visualized online using N-Browse (http://interactome.dfci.harvard.edu/C_elegans/).
We compared the biological relevance of each type of data from the integrated network by calculating the enrichment for functional linkage, as described before for protein-protein interaction datasets (Fig. 3b). All the analyzed datasets showed highly significant enrichment for functional linkage (all P < 2.5 × 10−3). Interestingly, amongst the analyzed datasets, physical interactions appeared to be the best predictors of highly specific shared GO terms, whereas pairs sharing phenotypes showed the highest enrichment for less specific functional linkages. The phenotypic profiles used in this study were “gross” phenotypes, and more precise phenotypic observations would likely be better predictors for more precise functions but worse at predicting more global functions. Similarly, linkages from expression data were derived from a wide range of experimental conditions and could be better predictors of more specific linkages if a set of experimental conditions targeting a particular process was used. This observation reflects how these different datasets address biological questions at different levels, in the same way sequence and structure similarity better predict which proteins exert the same enzymatic activity than proteins belonging to the same pathways25.
Next we examined the overlap between component networks of each type. We observed significant overlap for almost all combinations of component networks (Table 1). WI8, LCI, interologs and genetic interactions exhibited higher overlap with one another than co-expression or phenotypic correlation with any other dataset. The strong association between the two physical interaction datasets and interologs (LCI and WI8 confirmed 56 and 194 predicted interologs, including 49 and 147 heterodimers, respectively) was expected and confirmed that many interactions are conserved through evolution. LCI shared higher overlaps with phenotypically correlated pairs, genetic interactions and interologs than WI8, most likely because lower-throughput assays often test physical interactions that are enriched a priori for a common phenotype, or are known to have interacting orthologs. Still, WI8 substantiated 57 pairs of genes with high co-expression amongst a wide range of experimental conditions, 9 pairs of genes with similar RNAi phenotypic profiles, and 14 pairs of genetically interacting genes (“Shared edges” section at http://interactome.dfci.harvard.edu/C_elegans/).
Table 1.
Overlap between datasets from the integrated functional network. Fold enrichment (FE) and significance (P) of the overlaps between distinct functional datasets. The fold enrichment is defined as the number of pairs shared between two datasets divided by the expected random number of shared pairs, and the significance is assessed by Fisher’s exact test.
| WI8 | LCI | Interologs | Genetic interactions | Phenotypes | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FE | P | FE | P | FE | P | FE | P | FE | P | |
| WI8 | ||||||||||
| LCI | 182.3 | 1.01E-37 | ||||||||
| Interologs | 91.4 | 1.13E-212 | 145.6 | 5.89E-75 | ||||||
| Genetic interactions | 23.9 | 1.59E-14 | 66.9 | 1.17E-72 | 24.1 | 6.58E-58 | ||||
| Phenotypes | 3.0 | 5.33E-03 | 4.6 | 1.02E-03 | 3.0 | 1.27E-16 | 3.3 | 3.83E-06 | ||
| Coexpression | 2.5 | 1.20E-08 | 2.6 | 3.20E-03 | 3.2 | 5.01E-103 | 1.6 | 1.61E-01 | 1.6 | 1.09E-21 |
Although significant and informative, these overlaps remain relatively low (Supplementary Table 5 online). This can be explained by lack of “screening completeness” of most datasets, i.e., most of these datasets are not genome or proteome wide. Indeed, more than 60% of genes-proteins in the network are present in one dataset only while less than 5% are present in four datasets or more. Furthermore, most of the screens that have led to the generation of these datasets (including our Y2H screens) are far from saturation and are probably limited by low sampling sensitivity in addition to inherent limitations of each assay, i.e., precision and assay-sensitivity. Finally, a perfect overlap is not expected due to intrinsic differences in the nature of the biological attributes measured in those datasets.
Module-scale biological networks
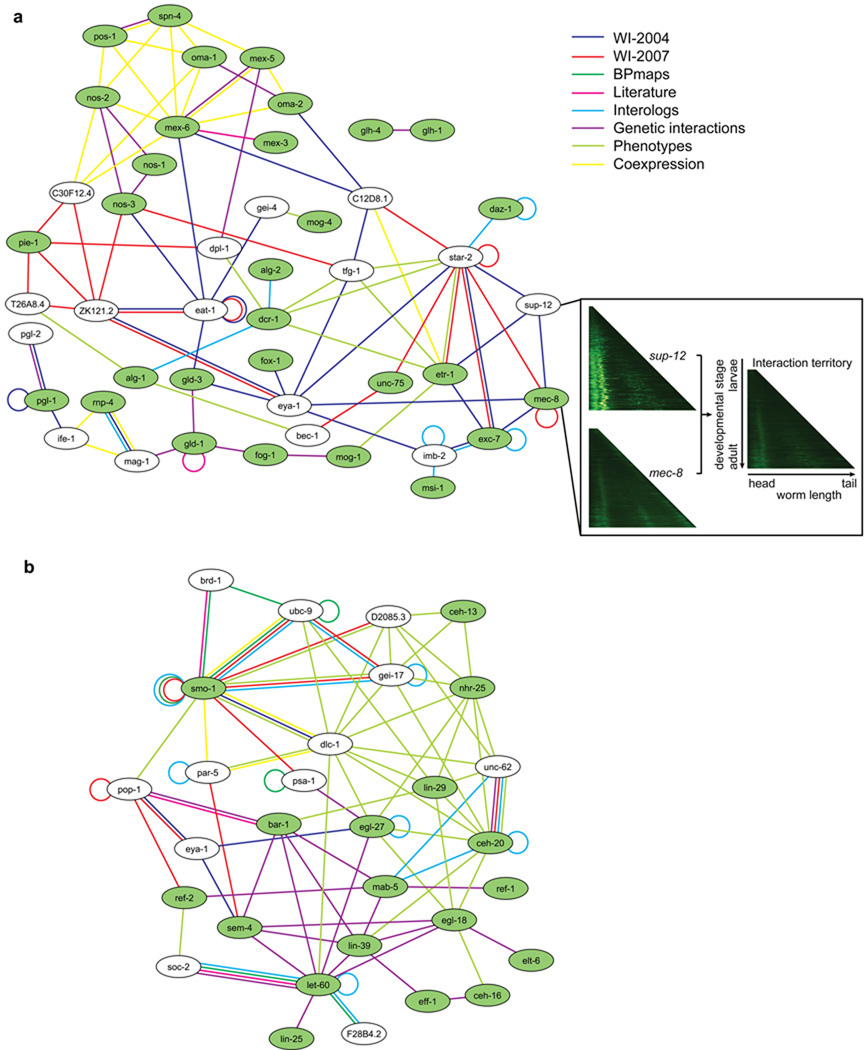
Module-scale biological sub-networks can be extracted from the integrated network by selecting ‘seed genes-proteins’ known to be associated with a specific process and then expanding by selecting neighboring genes-proteins. For example, with genes-proteins implicated in RNA-binding processes as seeds (Fig. 4a), nearly all genes-proteins in the expanded set were linked to several RNA-binding genes-proteins and at least two types of relationships. Most of these linked genes-proteins were thus predicted as functionally related to RNA-binding, and several (for example sup-12) were already annotated or predicted by sequence similarity to be associated with RNA-binding within WormBase. Other genes-proteins have annotations consistent with RNA-binding. For example, T26A8.4 codes for a protein predicted to be part of the CPSF sub-complex of the Polyadenylation factor I complex through clusters of eukaryotic Orthologous Groups (KOGs)26, and orthologous to yeast Caf120, which is part of the conserved Ccr4-Not transcriptional regulatory complex involved in mRNA initiation, elongation, and degradation.
Figure 4.
Examples of multiple evidence sub-networks. The networks represent genes-proteins (ellipses) relationships from several evidence sources: protein-protein interactions (PPI) from WI8 (separated according to origin): PPI from WI-2004 (dark blue), PPI from WI-2007 (red), PPI from biological processes maps (green), PPI from literature curation databases (magenta). The other types of functional links are: interologs (light blue), genetic interactions (violet), gross phenotypic similarity (light green), co-expression from a compendium of microarray experiments. Genes and their products are labeled using an unitalicized lower case version of the standard C. elegans three-letter system to reflect that the diagram combines links between both proteins and genes. (a) Genes related to RNA-binding. The genes annotated as ‘RNA-binding’ according to Wormbook are green ellipses in the network. White ellipses are genes-proteins linked to RNA-binding genes-proteins by at least one protein-protein interaction from WI8 and one additional piece of evidence. The inset shows the chronograms of sup-12 and mec-8 (left), and their predicted spatiotemporal pattern of interaction (right), corresponding to the stages and tissues where the interaction can take place. The color code of the chronograms represents the absolute GFP intensity measured (increasing values as black-green-yellow-white) for promoter(s) of the considered genes, along the worm length (x axis) and development stage (y axis)15. (b) Genes related to cell fusion. The genes-proteins annotated as ‘cell fusion’ according to Wormbook are green ellipses and genes-proteins linked to cell fusion genes-proteins by a protein-protein interaction from WI8 and one additional type of evidence are white ellipses.
When expanding from a seed set of genes-proteins involved in cell fusion (Fig. 4b), almost all added genes-proteins were linked to multiple nodes in the seed set, with many links supported by multiple evidence sources. For example, unc-62 had phenotypic correlation with seed genes-proteins nhr-25, lin-29 and ceh-20, physical, genetic and interolog links with ceh-20, and interolog links with mab-5. In contrast to the RNA-binding sub-network, where most links were physical interactions with few pairs being supported by multiple evidence types, in this example most links were either phenotypic or genetic interactions, and many physical interactions were supported by additional evidence.
Interestingly, WI-2007 provides new physical interactions between proteins not previously linked to one another, but at a network distance of two in the integrated network (Supplementary Fig. 1 online). In the RNA binding network, for example, star-2 and mec-8, which are known to be indirectly linked through sup-12, were found to directly interact. We found 1,157 new “triangle closures” of this kind (viewable within the “intersections” and “display” sections of http://interactome.dfci.harvard.edu/C_elegans/).
From “static” map to spatiotemporal interactome
Spatiotemporal expression patterns for ~ 2,000 worm genes have recently become available through large-scale studies of worms carrying endogenous promoters driving expression of Green Fluorescent Protein (GFP)14,15. Examination of the resulting GFP intensity patterns informs about where (tissue) and when (developmental stage) promoters are activated. The GFP profiles can be sorted according to developmental stage by length and aligned, forming a “chronogram” representation”15.
We performed computational “chronogram intersection” of the spatiotemporal expression patterns corresponding to two interacting proteins, and used these to infer a potential “interaction territory” (Supplementary Figs. 2 and 3 online). Interaction territories were also inferred based on explicit anatomical annotations14 for interacting proteins. We identified 111 common anatomical annotations and generated 69 chronogram intersections for protein-protein interactions from WI8 (viewable within the Localization section of http://interactome.dfci.harvard.edu/C_elegans/). Examples from the RNA-binding sub-network (Fig. 4a) include common interaction territories for SUP-12 and MEC-8 (Fig. 4a inset), MEC-8 and EXC-7, and MEP-1 and MOG-4 through chronogram intersections, and for an additional 21 interactions through anatomical annotations. Although this GFP-based technique has limitations related to resolution and coverage, these examples provide a glimpse of how integrating spatiotemporal expression information will eventually allow extraction of tissue-specific sub-networks corresponding to pathways, functional modules or protein complexes, once the technology improves and more data become available.
DISCUSSION
We describe the implementation of a novel integrated strategy for generating high confidence networks based on a highly stringent HT-Y2H assay combined with a quality control framework13, thus achieving an important step along the path to completion of the C. elegans interactome. Our estimated size of the complete C. elegans biophysical interactome is approximately 116,000 interactions, considering only a single protein isoform per gene. While WI8 provides 3,816 interactions, lack of screening completeness as well as incomplete sampling and assay sensitivity are key explanations for why 96–97% of the interactome remains untouched. From the overlap of two independent HT-Y2H screens, we estimate that a single high-throughput screen can capture ~ 30% of the detectable interactions and thus would need to be repeated multiple times to reach saturation. Even at saturation some interactions may not be detectable by Y2H because of intrinsic limitations of the assay, e.g., proteins may not be imported into the nucleus, proper folding may not occur because of the fusion with the DNA-binding or activation domains, interactions may require post-translational modifications or co-factors not present within S. cerevisiae. The proportion of interactions detectable with our HT-Y2H system is estimated at approximately 16%.
Several approaches under development, involving optimization of the experimental setup27 or systematic ORF fragmentation28, should improve the assay sensitivity in future interactome mapping projects. However, achieving comprehensive mapping of the interactome will require use of multiple assays with complementary assay sensitivities. For example, experiments conducted in mammalian cells may uncover some interactions missed by Y2H, but fail to find others because some interactions do not occur under the conditions tested27. In addition to solving sensitivity issues, additional cloning efforts will have to be undertaken to increase the screening completeness of the search spaces of future interactome mapping projects. WI8 represents an early milestone towards uncovering the complete interactome network, yet it is the most comprehensive and reliable protein-protein interactions dataset available today for C. elegans.
METHODS
LR cloning
10,622 worm ORFs corresponding to 9,906 genes from the worm ORFeome v1.1 resource were transferred into both pDB-dest and pAD-dest-CYH destination vectors to generate DB-ORF and AD-ORF fusions, respectively8. We used the products of the recombinational cloning reactions directly to transform E. coli via a selection for ampicillin resistance. After overnight growth in liquid culture, we prepared plasmid DNAs in 96-well format using a Qiagen 8000 Biorobot.
Yeast transformation
We transformed DB-ORF and AD-ORF individually into MATα MaV203 or MATa MaV103 yeast strains, respectively, in a 96-well format. DB-ORF transformed cells were spotted in a 96-well layout on solid synthetic complete (Sc) media lacking leucine (Sc-L). We then replica-plated transformant spots onto Sc-L plates containing uracil and 5-fluoro-orotic acid (Sc-L+5FOA) to suppress growth of auto-activators. Growing colonies were then cultured in liquid Sc-L medium and stored in glycerol for subsequent use.
We selected yeast cells transformed with AD-ORF plasmids in a 96-well layout on solid Sc medium lacking tryptophan (Sc-T). Growing colonies were then cultured in liquid Sc-T medium and stored in glycerol. Subsequently, we thawed and pooled aliquots of AD-ORF transformed yeast cells to generate 57 mini-libraries, each containing 188 individual AD-ORF transformants, referred to as “AD-188ORFs”.
Y2H screening
We mated 94 individual MATα MaV203 DB-ORF yeast strains, in a 96-well format, with the same MATa MaV103 AD-188ORFs mini-library on solid medium containing yeast extract, peptone and dextrose (YPD). Each DB-ORF 96-well plate was individually mated to all AD-ORFs compiled into 57 AD-188ORFs pools. After overnight growth at 30°C,we transferred the colonies to Sc-L-T plates lacking histidine and containing 20 mM 3-AT (Sc-L-T+3AT) to select for diploids that exhibited elevated expression levels of the GAL1::HIS3 Y2H marker. The same cells were transferred in parallel onto Sc-L+3AT plates containing tryptophan and cycloheximide (Sc-L+3AT+CYH). The pAD-dest-CYH vector contains the CYH2 negative selection marker that allows plasmid shuffling on cycloheximide-containing media. This step is crucial to eliminate auto-activators that can arise during Y2H selections. Auto-activators exhibit a 3AT+/3AT-CYH+ phenotype, while genuine positives exhibit a 3AT+/3AT-CYH− phenotype in this assay. We picked approximately 180,000 positive colonies from 3AT+/3AT-CYH− spots into a second-generation set of 96-well plates for further phenotypic screening.
Scoring Y2H assays
Consolidated and re-grown 3AT+/3AT-CYH− colonies were transferred to both Sc-L+3AT and Sc-L+3AT+CYH plates to confirm GAL1::HIS3 transcriptional activity, and to YPD to determine GAL1::lacZ transcriptional activity using a β-galactosidase filter assay. We selected colonies that retested 3AT+/3AT-CYH− and tested positive at levels equal or higher to that of the control DB-RB/AD-E2F interaction pair in our Y2H control set. Of the original ~ 180,000 3AT+/3AT-CYH− colonies, 7,295 passed this double phenotypic test and represent Y2H positives. We also systematically tested all DB-ORFs for auto-activation by growth on solid SC-L medium containing 20mM 3-amino-triazole (3-AT), identifying all strong auto-activators and removing them from further consideration as baits in Y2H.
Yeast PCR and IST sequencing
We performed PCR amplifications on all Y2H positive colonies to individually amplify DB-ORFs and AD-ORFs. The products from the PCR were purified and used as templates in a cycle-sequencing reaction to obtain two interaction sequence tags (ISTs) per Y2H positive.
WI-2007 IST analysis
The quality of the ISTs obtained by sequencing was measured by moving a sliding window of 10 base pairs to define the portion of the IST that had an average PHRED score of 10 or higher over at least 10% of their length. We aligned all sequences against the worm ORFeome v1.1 database (http://worfdb.dfci.harvard.edu/), and remapped them to WormBase version WS150. We retained only those 5,822 showing a BLASTN e-value ≤ 10−20. We collapsed all IST pairs corresponding to the same unordered gene locus pair.
Pairwise Y2H verification
We verified all Y2H interactions by mating fresh individual MATα MaV203 DB-ORF yeast cells with their corresponding individual MATa MaV103 AD-ORF yeast cells. For genes with multiple clones in the worm ORFeome v1.1, we used the clone with the highest similarity to the IST sequenced in the high-throughput screen for the retest. We tested the resulting diploids for their ability to activate two out of the three Y2H reporter genes. Of the 2,340 potential interactions, 78% (1,816) successfully passed this Y2H retest.
MAPPIT assay
In this system, the bait is fused to a STAT recruitment-deficient, homodimeric cytokine receptor and the prey protein to functional STAT recruitment sites (gp130). An interaction between bait and prey allows the activation of a ligand-dependent signaling transduction pathway, which controls the activation of a luciferase marker. MAPPIT was performed as described29 with minor changes. We transfected plasmids into human 293T cells in 96-well plates using a calcium phosphate protocol30. Transfected cells were cultured for 24 hours in DMEM medium supplemented with 10% fetal bovine serum and then stimulated with ligand (Epo) or left untreated for an additional 24 hours, followed by measurement of luciferase activity in triplicate.
Functional linkage estimation
The fold enrichment of a particular dataset is the number of distinct pairs (excluding homomeric interactions) sharing at least one GO term at a given functional specificity threshold, divided by the number of pairs expected at random, using a one-sided Fisher’s exact test (fold enrichment = odds ratio). We estimated the space of possible gene pairs by all unordered pairs between the genes in the input dataset to account for specific biases of each dataset, and then restricted this space to pairs in which both genes have one or more annotations at the considered functional specificity level. The number of genes associated with a particular GO term was used as an estimate of the functional specificity, and we calculated the fold enrichments for several functional specificity levels (5, 20,100 and 400). Differences between fold enrichments were assessed using an independent two-sample t-test. Supplementary Figs. 4 and 5 online detail the separate branches of the Gene Ontology.
Additional methods
Detailed descriptions of the MAPPIT scoring, WI-2007 characterization through Monte-Carlo simulation, overlap between component networks, module-scale sub-networks extraction, chronogram intersections and all datasets are available in Supplementary Methods online. WI8 is provided with MIMIX specifications as Supplementary Data 1 online. The integrated functional network is available as Supplementary Data 2 online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fabio Piano, members of Cancer Center for System Biology (CCSB) and the Vidal laboratory for helpful discussions; Andrei Petcherski from WormBase for the communication of worm genetic interactions; Zhenjun Hu from Boston University for VisAnt assistance and anonymous reviewers for their valuable criticisms; the worm interactome project was supported by grants from the National Institutes of Health— R01 HG001715 (M.V. and F.P.R.), R01 HG003224 (F.P.R.), F32 HG004098 (M.T.), T32 CA09361 (K.V.)—, an University of Ghent grant GOA12051401 (J.T.), and the “Fonds Wetenschappelijk Onderzoek – Vlanderen” (FWO-V) G.0031.06 (J.T.). I.L. had a postdoctoral fellowship with the FWO-V. K.C.G. and H.L.K. were supported by Department of the Army Award W81XWH-04-1-0307 and New York’s Science and Tech Ressources (NYSTAR) Contract #C040066. M.V. is a "Chercheur Qualifié Honoraire" from the “Fonds de la Recherche Scientifique” (FRS-FNRS, French Community of Belgium).
Footnotes
AUTHOR CONTRIBUTIONS
J.F.R., N.S. and A.R.C. coordinated experiments and data analysis. J.F.R., T.H.-K., J.M.S., F.G., S.C., P.B., N.L., N.A.-G., E.D., D.S., A.D, C.S., M.V., H.Y., M.B., S.M., M.D., M.T. and J.S.A. performed the high-throughput ORF cloning and Y2H screens. I.L., A.-S.D., P.B. and J.T. conducted the MAPPIT experiments. N.S., A.R.C., M.T., T.H., N.K., K.V, C.F., N.B., M.Y., C.L., A.S., H.-L.K. and K.C.G. performed the computational analyses. M.T., N.S., C.F., A.R.C., H.-L.K. and K.C.G. adapted or built the website and visualization tools. N. S., A.R.C., J.F.R., M.C., F.P.R. and M.V. wrote the manuscript. M.V. conceived the project. D.E.H., J.T., F.P.R. and M.V. co-directed the project.
REFERENCES
- 1.Walhout AJ, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 2.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 3.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 6.Davy A, et al. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–828. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulton SJ, et al. Combined functional genomic maps of the C. elegans DNA damage response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 8.Reboul J, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 9.Walhout AJ, et al. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 2002;12:1952–1958. doi: 10.1016/s0960-9822(02)01279-4. [DOI] [PubMed] [Google Scholar]
- 10.Kim JK, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 11.Tewari M, et al. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-β signaling network. Mol. Cell. 2004;13:469–482. doi: 10.1016/s1097-2765(04)00033-4. [DOI] [PubMed] [Google Scholar]
- 12.Bieri T, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat. Meth. 2008 doi: 10.1038/nmeth.1280. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt-Newbury R, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupuy D, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 16.Kao HL, Gunsalus KC. Browsing multidimensional molecular networks with the generic network browser (N-Browse) Curr Protoc Bioinformatics. 2008:11. doi: 10.1002/0471250953.bi0911s23. Chapter 9, Unit 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motegi F, Velarde NV, Piano F, Sugimoto A. Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Dev. Cell. 2006;10:509–520. doi: 10.1016/j.devcel.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Branda CS, Stern MJ. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. 2000;226:137–151. doi: 10.1006/dbio.2000.9853. [DOI] [PubMed] [Google Scholar]
- 20.Wolf FW, Hung MS, Wightman B, Way J, Garriga G. vab-8 is a key regulator of posteriorly directed migrations in C. elegans and encodes a novel protein with kinesin motor similarity. Neuron. 1998;20:655–666. doi: 10.1016/s0896-6273(00)81006-5. [DOI] [PubMed] [Google Scholar]
- 21.Schlaitz AL, et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunsalus KC, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 23.Lee I, et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat. Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 24.Gunsalus KC, Yueh WC, MacMenamin P, Piano F. RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 2004;32:D406–D410. doi: 10.1093/nar/gkh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol. 2000;297:233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- 26.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Meth. 2008 doi: 10.1038/nmeth.1281. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyckerman S, et al. Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 30.Lemmens I, Lievens S, Eyckerman S, Tavernier J. Reverse MAPPIT detects disruptors of protein-protein interactions in human cells. Nat. Protoc. 2006;1:92–97. doi: 10.1038/nprot.2006.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.