Abstract
Systems pharmacology involves the application of systems biology approaches, combining large-scale experimental studies with computational analyses, to the study of drugs, drug targets, and drug effects. Many of these initial studies have focused on identifying new drug targets, new uses of known drugs, and systems-level properties of existing drugs. This review focuses on systems pharmacology studies that aim to better understand drug side effects and adverse events. By studying the drugs in the context of cellular networks, these studies provide insights into adverse events caused by off-targets of drugs as well as adverse events-mediated complex network responses. This allows rapid identification of biomarkers for side effect susceptibility. In this way, systems pharmacology will lead to not only newer and more effective therapies, but safer medications with fewer side effects.
The clinical usefulness of a medication is related to its efficacy in treating specified diseases as well as its safety and tolerability in patients. It is important to understand not only the intended effects of a drug, but also the side effects and potential adverse events associated with treatment. While initial studies of drugs aim to elucidate mechanism of action and therapeutic effects, concerns over potential adverse events can prevent drugs from reaching the market and has led to several high-profile post-market drug failures. Systems pharmacology studies, which integrate large data sets such as protein–protein interaction networks and the FDA adverse event reports with computational analyses, can enhance the understanding of drug adverse events by looking at the effects of a drug in the context of cellular networks as well as exploring relationships between drugs. This can allow better understanding of adverse event pathogenesis and individual patient susceptibility to side effects, as well as identification of new targets. Identification of additional targets (off-targets) for drugs can subsequently lead to improved drug development to decrease off-target effects and potential repurposing of existing drugs for different diseases. We have previously surveyed the network-based computational techniques used in systems pharmacology studies1 and reviewed how systems pharmacology approaches are helping direct the evolution toward personalized medicine2,3 and directing drug discovery.3,4 This review discusses different ways in which drug side effects and adverse events arise in the context of cellular networks and recent advances in understanding these pathological processes arising from systems pharmacology studies of drug structures, targets, and their relationships to signaling networks that regulate both therapeutic and adverse biological responses (Figure 1).
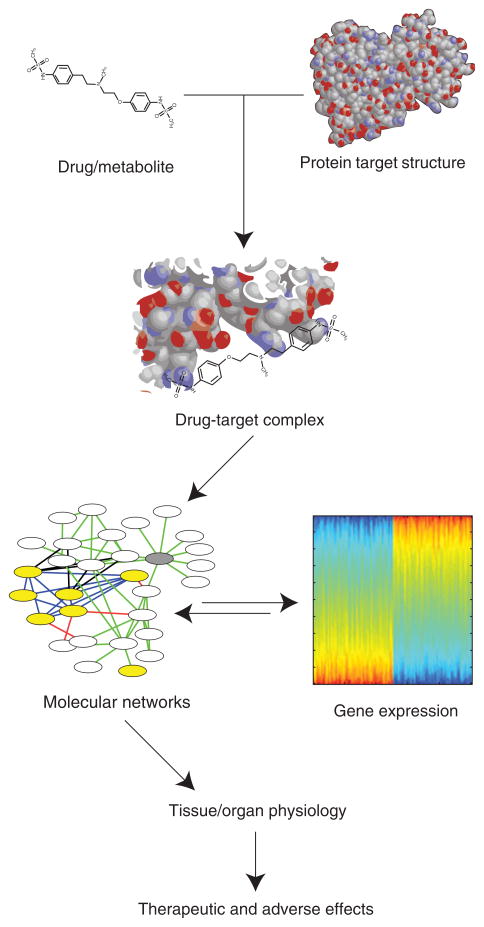
FIGURE 1.
Drug effects are mediated by a drug or metabolite binding a protein target and affecting the function of the target in the context of a cellular network. The response of the cellular network can cause changes in gene expression, enzymatic activation, and ultimately tissue or organ physiology. These give rise to the observed effects of drugs. Measurements or information about any step in the information flow from drug to effects can be informative about any of the preceding steps. Systems pharmacology studies can integrate various forms of data about drug responses to identify off-targets of drugs and predict potential adverse drug effects.
MECHANISMS OF DRUG EFFECTS, SIDE EFFECTS, AND ADVERSE EVENTS
Drugs exert their effects, both therapeutic and adverse, by interacting with molecular targets. These molecular targets, often receptors or enzymes, propagate the signal from the drug through effector pathways which ultimately cause the observable drug effects at the level of altered organ or whole body function. For many drugs, the therapeutic effect is mediated by a known protein target. Frequently, this target is located in the same tissue where a disease or pathological process occurs. These drugs are designed to interfere with the disease process or suppress the symptoms. Drug side effects are additional, usually undesirable, effects of medications. They can arise through the same mechanism of action as the therapeutic effect or through a distinct mechanism. Drug side effects and adverse events can be grouped based on their relationship to the desired therapeutic action of the drug.5 In this review, we focus on the following four classes of drug side effects: drug interacting with primary target in primary tissue, drug interacting with primary target in different tissue, drug or drug metabolite interacting with off-targets in either primary or different tissue, and drug adverse events arising through complex network interactions between drug primary or off-targets within or between tissues (Table 1).
TABLE 1.
Types of Drug-Induced Side Effects and Adverse Events
| Effector Compound | Effector Target | Tissue Affected | Example |
|---|---|---|---|
| Drug | Therapeutic target | Target tissue | Warfarin (anticoagulant) → inhibit VKOR → inhibit clotting → hemorrhagic stroke |
| Drug | Therapeutic target | Off-target tissue | Morphine (opioid analgesic) → μ-opiate receptor → inhibit myenteric plexus in intestine → constipation |
| Drug or metabolite | Off-target | Either target or off-target tissue | Delavirdine (HIV reverse transcriptase inhibitor) → histamine H4 receptor → severe rash |
| Drug, metabolite, or both | Multiple | Target, off-target, or multiple tissues | Domperidone (dopamine antagonist antiemetic) → HERG K+ channels and α-adrenergic receptors in heart → cardiac arrhythmias |
VKOR, vitamin K epoxide reductase.
ADVERSE EVENTS RELATED TO THE THERAPEUTIC EFFECT OF A DRUG
The first category of drug side effects occur when desired therapeutic effects can have additional negative consequences. Frequently, these side effects occur at doses of the medication higher than those required for the beneficial drug effects. For example, most medications designed to lower blood pressure can cause dizziness and fainting if blood pressure is lowered too much.6 Similarly, the most dangerous side effect of anticoagulants is bleeding, which can lead to hemorrhagic strokes when bleeding occurs in the brain.7 To avoid this type of side effect, appropriate dosing of the drug to maintain concentrations in blood within a narrow therapeutic window is required. Research on pharmacogenetics, reviewed elsewhere, has identified genetic variation in metabolic enzymes, transporters, and drug targets.8 These genomic variations affect metabolism and potency of drugs and thus relate to defining a patient-specific therapeutic window. Thus the drug-metabolizing capability of the individual becomes the important criterion when adverse event occurs due to drug-induced overcompensation of the pathophysiology. This class of effects provides a compelling rationale for pharmacogenetics and pharmacogenomics: identifying and classifying genetic changes and genomic variations in individuals for the design of personalized therapy.
ADVERSE EFFECTS MEDIATED BY THE THERAPEUTIC TARGET OF A DRUG
The second group of adverse events occurs when a drug’s target serves multiple purposes in different tissues of the body. Various types of receptors exist in many tissues. As such, pain medications such as morphine which achieves its analgesic effects by targeting μ-opioid receptors in the brain will also cause constipation by targeting the same type of opiate receptors in the gut thereby inhibiting peristalsis.9 Similarly, many chemotherapeutics will kill any fast growing cells in the body. While this does achieve the desired effect of cancer cell destruction, this will also suppress the immune system by killing dividing precursors to immune cells and hair loss by targeting growing cells in hair roots.10
Side effects due to the therapeutic drug target’s presence in multiple cell and tissue types as well as pathways can lead to attempts to develop better-targeted therapeutic agents that more specifically target the desired pathway or site of disease. Angiotensin-converting enzyme (ACE) inhibitors are used to decrease blood pressure in hypertension and prevent kidney damage in diabetes. Many patients taking ACE inhibitors develop a dry cough which is often severe enough to warrant discontinuing the medication.11 While ACE inhibition has the desired effect of decreasing levels of angiotensin II, and thus controlling hypertension, ACE is also responsible for the metabolism of bradykinin. ACE inhibition therefore has the undesired effect of causing bradykinin accumulation, which is thought in some individuals to mediate cough through proinflammatory mechanisms.12 As such, newer pharmaceuticals have been developed that block the angiotensin II pathway at the receptor level, thereby blocking the effects of angiotensin II without affecting bradykinin levels. Therefore angiotensin II receptor blocking drugs do not have this side effect and can be used when ACE inhibitors are not tolerated by a patient.13
DRUG SIDE EFFECTS MEDIATED BY OFF-TARGETS OF A DRUG
Adverse events and side effects can also be mediated by targets and pathways which are not intended to be perturbed by the drug. These are known as off-targets and are frequently not known or discovered until after observing an adverse event or side effect. For example, the HIV drug Rescriptor, which has its therapeutic effect by inhibiting HIV’s reverse transcriptase, can cause severe rashes by interacting with the histamine H4 receptor.14 This class of side effect could not be predicted based on the knowledge of the intended targets and functions of a drug. Therefore, during drug development, many drugs are screened for potential side effects by checking for interactions with common off-targets. Inhibition of the HERG ion channel, whose blockade by drugs is responsible for many severe drug-induced cardiac arrhythmias, is frequently checked during drug development.15 In other cases, the specific target responsible for a side effect is not clear. This thus requires understanding additional information about the drug such as its structure and metabolism. In the case of drug-induced agranulocytosis, multiple mechanisms may account for the side effect.16 In some of these hypothesized mechanisms, drugs are converted to highly reactive ions that can react with other cellular compounds in neutrophils, deplete the cell of ATP, and thereby lead to cell death.
SYSTEMS PHARMACOLOGY ALLOWS FOR THE PREDICTION AND IDENTIFICATION OF OFF-TARGETS
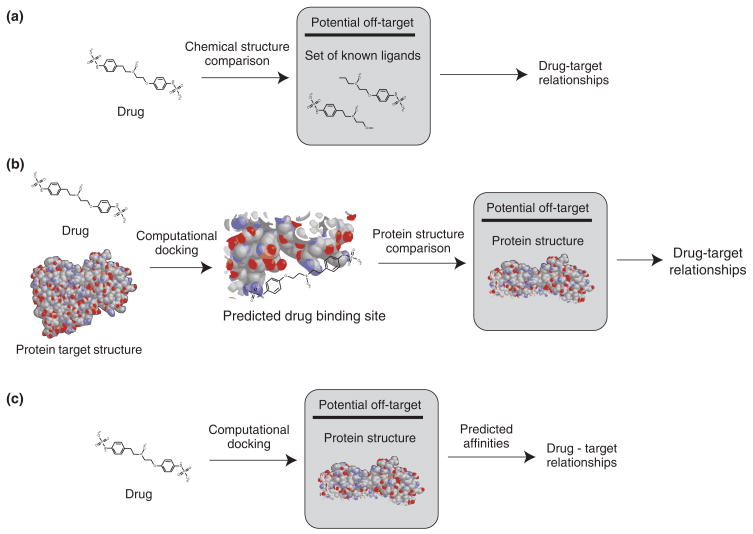
By providing methods for integration of multiple type of information about drugs and their effects, systems pharmacology studies facilitate the prediction and identification of drug off-targets.17 Prediction of drug off-targets can utilize many different sources and types of information about drugs to make their predictions (Figure 2). For example, there exist multiple databases containing known drug target information or ligand affinities.18 They have been used to construct ligand-based descriptions of drug targets which can be used to predict additional ligands that may bind. Keiser et al.14 used known ligand protein relationships and drug chemical structure information to make predictions using their similarity ensemble approach to predict drug targets. Drug targets were represented as the set of all ligands known to bind the protein. The structures of all drugs were compared to these ligand sets. As such, they were able to augment a network connecting drugs to their targets with new predictions of drug and drug target associations. They then analyzed how these predicted off-targets could explain the various side effects of different drugs (Figure 2(a)).
FIGURE 2.
Different strategies can be used to predict drug–target relationships. (a) Drug chemical structure can be compared to known ligands of a potential target. (b) Drug chemical structure and known target protein structure can be combined through computational docking to predict a binding site. This predicted binding site can be compared to protein structures of potential off-targets. (c) Drugs can be computationally docked to the protein structures of potential off-targets to generate predicted affinities and drug–target relationships.
Xie et al.19 studied the cholesteryl ester transfer protein (CETP) inhibitor, Torcetrapib. This medication was developed to treat high cholesterol, but has the side effect of inducing dangerous hypertension through an unknown mechanism. They used computational methods to dock the torcetrapib to its target CETP. After identification of putative binding sites, they searched for other proteins with known or predicted structures that were structurally similar to the drug-binding site (Figure 2(b)). The drug was then docked back to these predicted off-targets. This allowed the creation of predicted protein–ligand networks that demonstrated differences between CETP inhibitors which induce hypertension and those not known to have this side effect. Incorporating these off-target binding networks with known biological pathways allows further explanation of the pathophysiology of torcetrapib-induced hypertension by the binding of various nuclear receptors to dys-regulate the renin-angiotensin-aldosterone system, a major component of blood pressure regulation in humans.
Yang et al.20 computationally docked sets of drugs into binding pockets of 845 proteins to predict binding affinities. They applied this to sets of drugs known to cause two dangerous dermatological adverse events, Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN), as well as a set of controls. They reported that the drugs with the side effects had a distinct binding profile compared to the controls. From this analysis, combined with a gene co-citation network, which connects genes to genes discussed together in papers about the side effects, they were able to propose that MHC I protein heavy chain Cw*4 (1QQD) is the mediator of adverse event: drug-induced TEN. They report that using their docking-based approach, they can distinguish between different alleles of HLA-B for risk of abacavir-induced SJS (Figure 2(c)). Using a similar approach, they predicted candidate drug targets related to other severe drug adverse events including deafness, rhabdomyolysis, and cholestasis. Their methods have been implemented in SePreSA, a Server for the Prediction of Population Susceptible to Serious Adverse Drug Reactions, and SADR Gengle, which are internet-accessible programs available online.21,22
Campillos et al.23 also utilized the knowledge of drug chemical structures for the prediction of drug off-targets. However, they used additional knowledge provided by the known adverse reactions associated with each drug. Using drug package inserts, they assigned each drug a profile of adverse events associated with it. Combining drug structural similarity with effect similarity, measured by comparing the side effect profiles of drugs, they created a network that connected drugs to each other if they had a sufficiently similar side effect profile and chemical structure. In several cases, the drugs connected in the network were not known to share a protein target. They were then able to validate their approach by demonstrating that one of the drugs could in fact bind at least one of the targets of the other drug. This approach was thus able to predict previously unknown off-targets of drugs.
DRUG SIDE EFFECTS MEDIATED BY COMPLEX CELLULAR NETWORKS
Each of these grouping of adverse events discussed thus far has assumed that a single target or molecular perturbation leads to the adverse event. Many drugs have been shown to target multiple different proteins that can contribute to an adverse outcome.14 For example, HERG blockade alone is often not enough to induce arrhythmias, but HERG blockade in combination with α-adrenergic signals might be more dangerous in susceptible patients.24 Molecular networks are known to be robust to such perturbations and capable of compensating to minimize extreme effects.25 Variations in cellular networks in response to a drug can unmask silent phenotypes. The causes of this type of adverse drug events are inherently patient specific. Although a majority of patients taking medication will experience the therapeutic effect of most drugs, only few patients will experience a specific side effect or adverse event. However, these side effects can be devastating for the patient, ranging from hepatotoxicity and rhabdomyolysis to sudden cardiac death. Systems pharmacology approaches can help identify unknown targets of drugs, describe how drug signals propagate through cellular networks, increase the understanding of what patient-specific factors lead to adverse responses, and help predict adverse event associations for new drugs.
SYSTEMS PHARMACOLOGY ALLOWS PREDICTION AND EXPLANATION OF DRUG SIDE EFFECTS
Given the multiple different ways in which a drug can cause a side effect, it is unlikely that a single approach will be able to predict all types of side effects for all drugs. However, systems biology approaches allow integration of multiple different types of data sets and thus can provide a framework to explore the different types of side effects that a drug can produce. These approaches can provide more information about a drug through off-target identification and information about drug actions by exploring the known targets in the context of their interactions.
Using phosphoproteomic profiling in hepatocytes treated with different combinations of compounds, Cosgrove et al.26,27 reported patterns of signaling pathway activation that predicted drug-induced hepatotoxicity. They report that the integration of survival, stress, and apoptosis signals are integrated in the cellular network through the MEK–ERK, mTOR–p70 S6K, Akt, and p38–HSP27 pathways to determine the hepatotoxic side effects of drugs.
Wagner et al.28 profiled gene expression changes of genes associated with mitochondrial function as well as ATP level changes associated with treatment with various compounds and drugs. They found that they were able to identify expression profiles associated with drugs associated with drug-induced myopathy and myalgias. An additional surprising result suggested that propranolol might share this side effect which could also increase the risk of rhabdomyolysis in combination therapies with statins.
Scheiber et al.29 integrated the knowledge of drug structures and known adverse reactions to generate a network connecting adverse drug reactions that are caused by sets of drugs with shared characteristic chemical structures. By not focusing their study on the protein targets involved in the pathogenesis of adverse drug events, they were able to identify structural motifs of compounds that relate to the adverse drug events. They were able to use this approach to identify relationships between various adverse drug events as well.
In our work, we studied the systems pharmacology underlying specific forms of drug-induced arrhythmias.30 We focused on extracting a disease-centered network in an integrated human protein–protein interaction network. The gene products with interactions surrounding the products of genes related to congenital long-QT syndromes (LQTS) were used to construct an LQTS-selective neighborhood. We then used this to classify drugs based on their targets for risk of drug-induced QT prolongation and torsades de pointes, a dangerous cardiac arrhythmia characterized by polymorphic ventricular tachycardia. We found that we were able to implicate multiple proteins, including several receptors and kinases, in the pathogenesis of LQTS. We validated our results using the US Food and Drug Administration’s Adverse Event Reporting System database to identify drugs not previously known to have these side effects. This demonstrated how systems-level analysis of basic science data sets can enhance signal detection in clinical and epidemiological data sets.
CONCLUSION: PERSONALIZED MEDICINE AND PHARMACOLOGY
A transition to personalized medicine will require detailed case-by-case understanding of the pathogenesis of adverse drug events and side effects. Systems pharmacology approaches will help identify off-targets of drugs, thereby explain unintended effects. Other unintended effects will be explainable through exploration of the cross talk between signaling pathways affected by drug exposure. Patient-specific variation will need to be taken into account in order to correctly choose between drug options and decide on appropriate dosing (Figure 3). While most of the systems pharmacology studies discussed in this review have focused on acute or short-term adverse drug effects, future studies will be able to use techniques similar to those we described to understand more long-term drug side effects such as those related to epigenetic changes. It has been hypothesized that drug side effects such as tardive dyskinesia and drug-induced systemic lupus erythematosus are due to epigenetic changes arising from chronic drug exposure.31 The cardiovascular risks associated with the use of certain NSAIDs take many months to 1–2 years to be manifested. Understanding the mechanisms leading to these slowly developing adverse events will also require systems-level analyses.
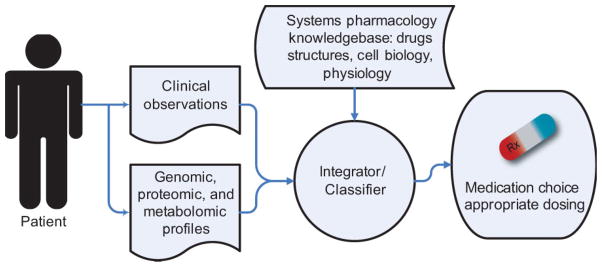
FIGURE 3.
Systems pharmacology approaches will allow clinical observations coupled with high dimensional data obtained from a patient’s genomic, proteomic, and metabolomic profiles to be integrated with a systems pharmacology knowledgebase to assist clinical decision making in choosing the correct medication and appropriate dosing.
The application of systems pharmacology approaches to personalize medicine will be enhanced by two emerging technologies, genomic sequencing and electronic medical records. As individual genome sequence becomes affordable and ubiquitous, patient-specific genomic data will be available to inform medical decision making. Tying this information to electronic medical records will allow population-based cross-sectional studies of genomic variations related to adverse drug events. Systems pharmacology will allow rapid integration of basic cell biology and physiology knowledge to more rapidly identify testable hypotheses for the etiology of drug side effects.
References
- 1.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25:2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wist AD, Berger SI, Iyengar R. Systems pharmacology and genome medicine: a future perspective. Genome Med. 2009;1:11. doi: 10.1186/gm11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boran AD, Iyengar R. Systems pharmacology. Mt Sinai J Med. 2010;77(4):333–44. doi: 10.1002/msj.20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boran AD, Iyengar R. Systems approaches to polypharmacology and drug discovery. Curr Opin Drug Discov Devel. 2010;13(3):297–309. [PMC free article] [PubMed] [Google Scholar]
- 5.Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt Sinai J Med. 2007;74:27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronow WS. Treating hypertension in older adults: safety considerations. Drug Saf. 2009;32:111–118. doi: 10.2165/00002018-200932020-00004. [DOI] [PubMed] [Google Scholar]
- 7.Kamali F. Genetic influences on the response to war-farin. Curr Opin Hematol. 2006;13:357–361. doi: 10.1097/01.moh.0000239708.70792.4f. [DOI] [PubMed] [Google Scholar]
- 8.Becquemont L. Pharmacogenomics of adverse drug reactions: practical applications and perspectives. Pharmacogenomics. 2009;10:961–969. doi: 10.2217/pgs.09.37. [DOI] [PubMed] [Google Scholar]
- 9.Holzer P. Opioid receptors in the gastrointestinal tract. Regul Pept. 2009;155:11–17. doi: 10.1016/j.regpep.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botchkarev VA. Molecular mechanisms of chemotherapy-induced hair loss. J Investig Dermatol Symp Proc. 2003;8:72–75. doi: 10.1046/j.1523-1747.2003.12175.x. [DOI] [PubMed] [Google Scholar]
- 11.Sica DA, Brath L. Angiotensin-converting enzyme inhibition-emerging pulmonary issues relating to cough. Congest Heart Fail. 2006;12:223–226. doi: 10.1111/j.1527-5299.2006.05746.x. [DOI] [PubMed] [Google Scholar]
- 12.Israili ZH, Hall WD. Cough and angioneurotic edema associated with angiotensin-converting enzyme inhibitor therapy. A review of the literature and pathophysiology. Ann Intern Med. 1992;117:234–242. doi: 10.7326/0003-4819-117-3-234. [DOI] [PubMed] [Google Scholar]
- 13.Cheung BM. Therapeutic potential of angiotensin receptor blockers in hypertension. Expert Opin Investig Drugs. 2006;15:625–635. doi: 10.1517/13543784.15.6.625. [DOI] [PubMed] [Google Scholar]
- 14.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KL, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi A, Dimino T, Vohra Y, Cui C, Yan GX. Preclinical strategies to assess QT liability and torsadogenic potential of new drugs: the role of experimental models. J Electrocardiol. 2004;37(suppl):7–14. doi: 10.1016/j.jelectrocard.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Tesfa D, Keisu M, Palmblad J. Idiosyncratic drug-induced agranulocytosis: possible mechanisms and management. Am J Hematol. 2009;84:428–434. doi: 10.1002/ajh.21433. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn M, Campillos M, Gonzàlez P, Jensen LJ, Bork P. Large-scale prediction of drug–target relationships. FEBS Lett. 2008;582:1283–1290. doi: 10.1016/j.febslet.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 18.Günther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, Schneider R, Skoblo R, Russell RB, Bourne PE, Bork P, Preissner R. SuperTarget and Matador: resources for exploring drug–target relationships. Nucleic Acids Res. 2008;36(Database issue):D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L, Li J, Bourne PE. Drug discovery using chemical systems biology: identification of the protein–ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Chen J, He L. Harvesting candidate genes responsible for serious adverse drug reactions from a chemical-protein interactome. PLoS Comput Biol. 2009;5:e1000441. doi: 10.1371/journal.pcbi.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Luo H, Chen J, Xing Q, He L. SePreSA: a server for the prediction of populations susceptible to serious adverse drug reactions implementing the methodology of a chemical-protein interactome. Nucleic Acids Res. 2009;37(Web Server issue):W406–W412. doi: 10.1093/nar/gkp312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, Xu L, He L. A CitationRank algorithm inheriting Google technology designed to highlight genes responsible for serious adverse drug reaction. Bioinformatics. 2009;25:2244–2250. doi: 10.1093/bioinformatics/btp369. [DOI] [PubMed] [Google Scholar]
- 23.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann P, Warner B. Are hERG channel inhibition and QT interval prolongation all there is in drug-induced torsadogenesis? A review of emerging trends. J Pharmacol Toxicol Methods. 2006;53:87–105. doi: 10.1016/j.vascn.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Alberghina L, Hofer T, Vanoni M. Molecular networks and system-level properties. J Biotechnol. 2009;144:224–233. doi: 10.1016/j.jbiotec.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Cosgrove BD, Alexopoulos LG, Saez-Rodriguez J, Griffith LG, Lauffenburger DA. A multipathway phosphoproteomic signaling network model of idiosyncratic drug- and inflammatory cytokine-induced toxicity in human hepatocytes. Conf Proc IEEE Eng Med Biol Soc. 2009;1:5452–5455. doi: 10.1109/IEMBS.2009.5334019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosgrove BD, King BM, Hasan MA, Alexopoulos LG, Farazi PA, Hendriks BS, Griffith LG, Sorger PK, Tidor B, Xu JJ, Lauffenburger DA. Synergistic drug-cytokine induction of hepatocellular death as an in vitro approach for the study of inflammation-associated idiosyncratic drug hepatotoxicity. Toxicol Appl Pharmacol. 2009;237:317–330. doi: 10.1016/j.taap.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner BK, Kitami T, Gilbert TJ, Peck D, Ramanathan A, Schreiber SL, Golub TR, Mootha VK. Large-scale chemical dissection of mitochondrial function. Nat Biotechnol. 2008;26:343–351. doi: 10.1038/nbt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheiber J, Jenkins JL, Sukuru SC, Bender A, Mikhailov D, Milik M, Azzaoui K, Whitebread S, Hamon J, Urban L, Glick M, Davies JW. Mapping adverse drug reactions in chemical space. J Med Chem. 2009;52:3103–3107. doi: 10.1021/jm801546k. [DOI] [PubMed] [Google Scholar]
- 30.Berger SI, Ma’ayan A, Iyengar R. Systems pharmacology of arrhythmias. Sci Signal. 2010;3:ra30. doi: 10.1126/scisignal.2000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csoka AB, Szyf M. Epigenetic side-effects of common pharmaceuticals: a potential new field in medicine and pharmacology. Med Hypotheses. 2009;73:770–780. doi: 10.1016/j.mehy.2008.10.039. [DOI] [PubMed] [Google Scholar]