Abstract
In bacteria, cytokinesis is dependent on lytic enzymes that facilitate remodeling of the cell wall during constriction. In this work, we identify a thus far uncharacterized periplasmic protein, DipM, that is required for cell division and polarity in Caulobacter crescentus. DipM is composed of four peptidoglycan-binding (LysM) domains and a C-terminal lysostaphin-like (LytM) peptidase domain. It binds to isolated murein sacculi in vitro, and is recruited to the site of constriction through interaction with the cell division protein FtsN. Mutational analyses showed that the LysM domains are necessary and sufficient for localization of DipM, while its peptidase domain is essential for function. Consistent with a role in cell wall hydrolysis, DipM was found to interact with purified murein sacculi in vitro and to induce cell lysis upon overproduction. Its inactivation causes severe defects in outer-membrane invagination, resulting in a significant delay between cytoplasmic compartmentalization and final separation of the daughter cells. Overall, these findings indicate that DipM is a periplasmic component of the C. crescentus divisome that facilitates remodeling of the peptidoglycan layer and, thus, coordinated constriction of the cell envelope during the division process.
INTRODUCTION
Most bacteria possess a cell wall that protects them against mechanical stresses and allows them to withstand their high internal osmotic pressure. In addition, it serves as a structural component required for maintaining proper cell shape and as a scaffold for the attachment of extracellular proteins (Vollmer et al., 2008a). The cell wall is constituted by a single bag-shaped macromolecule, the murein ‘sacculus’. It consists of peptidoglycan (murein), a dense meshwork of glycan strands (Gan et al., 2008), composed of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) subunits that are crosslinked by short peptide bridges (Holtje, 1998). Cell growth and division necessitate continuous remodeling of the murein sacculus, involving the synthesis of new cell wall material as well as the cleavage of existing bonds. Due to the turgor pressure exerted on the sacculus, these antagonistic activities need to be tightly synchronized to prevent generation of lesions in the peptidoglycan meshwork and, thus, rupture of the cell. Although the underlying regulatory mechanisms are still unclear, there is evidence for the assembly of peptidoglycan synthases and hydrolases into multi-protein complexes, thus facilitating their coordinate action (Holtje, 1998).
In recent years, two types of cell wall biosynthetic machineries have been identified, mediating longitudinal growth and cell division, respectively (den Blaauwen et al., 2008, Margolin, 2009). The function of the latter is critically dependent on the cytoskeletal protein FtsZ, a tubulin homologue that polymerizes into a ring-shaped structure at the future division site (Lutkenhaus, 2007, Li et al., 2007). This so-called Z ring then recruits, directly and indirectly, all other components of the division apparatus (Goehring and Beckwith, 2005). At a first stage, it associates with factors that stabilize the FtsZ polymers and tether them to the cytoplasmic membrane, including the actin homologue FtsA (Dai and Lutkenhaus, 1992), ZapA (Gueiros-Filho and Losick, 2002), and ZipA (Hale and de Boer, 1997). After a marked delay, a second set of proteins is recruited to the division site (Aarsman et al., 2005), among them the poorly characterized FtsQLB (Buddelmeijer and Beckwith, 2004) complex and FtsK, a factor with multiple functions in divisome assembly and chromosome segregation (Liu et al., 1998). The assembly process continues with the recruitment of various proteins implicated or involved in constriction of the peptidoglycan layer, such as PBP3 (Wang et al., 1998, Weiss et al., 1997), the putative membrane transporter FtsW, and the murein-binding protein FtsN (Addinall et al., 1997, Dai et al., 1993). Divisome maturation then finishes with the addition of several non-essential proteins, including several FtsN-like proteins (Arends et al., 2009, Gerding et al., 2009), the Tol/Pal complex (Gerding et al., 2007), which promotes invagination of the outer membrane, and various peptidoglycan hydrolases (Bernhardt and de Boer, 2003, Bernhardt and de Boer, 2004, Gerding et al., 2007, Uehara et al., 2009).
Bacteria contain a battery of lytic enzymes that target the peptidoglycan layer (Vollmer et al., 2008b). In E. coli, cells lacking multiple peptidoglycan hydrolases frequently display a chaining phenotype, indicating that the activity of these enzymes is required for splitting of the division septum during the final stages of cell division (Heidrich et al., 2001, Heidrich et al., 2002, Uehara et al., 2009). Recent work has identified two major groups of proteins involved in daughter cell separation, namely N-acetylmuramyl-L-alanine amidases and LytM-domain containing endopeptidases (Bernhardt and de Boer, 2004, Uehara et al., 2009). E. coli mutants lacking individual amidases only show a mild chaining phenotype, but combined inactivation of the three isoenzymes AmiA, AmiB and AmiC results in severe division defects (Heidrich et al., 2002, Priyadarshini et al., 2007). Likewise, single mutations in either of the four LytM factors produced by E. coli barely affect cell division, whereas a strain deficient in all of these proteins phenocopies the amidase triple mutant (Uehara et al., 2009). Interestingly, both EnvC and NlpD were shown to lack intrinsic hydrolase activity and instead serve as septum-specific activators of AmiA/B and AmiC, respectively (Uehara et al., 2010). Among the various factors involved in peptidoglycan hydrolysis, only AmiC and the LytM-domain factors EnvC and NlpD specifically accumulate at the septum during constriction (Bernhardt and de Boer, 2003, Bernhardt and de Boer, 2004, Uehara et al., 2009). In the case of AmiC, recruitment to midcell was shown to be dependent on the cell division protein FtsN (Bernhardt and de Boer, 2003).
Whereas divisome function and assembly are well studied in E. coli, the situation is less clear for other organisms. In recent years, the alpha-proteobacterium Caulobacter crescentus has evolved as an alternative model for the analysis of cell division. A prominent feature of C. crescentus is its asymmetric cell division, which generates two morphologically and physiologically distinct daughter cells (Poindexter, 1964, Brown et al., 2009). One of them is characterized by a long protrusion, called the stalk, the tip of which bears an adhesive organelle mediating surface attachment. The other sibling, by contrast, lacks a stalk and possesses a single, polar flagellum responsible for swimming motility. Whereas a new-born stalked cell can immediately start a new round of cell division, the swarmer cell has to differentiate into a stalked cell before it can resume its cell cycle. Immediately after birth, the C. crescentus cell elongates in an MreB-dependent manner by uniform insertion of new material throughout the entire murein sacculus. On assembly of the Z ring, but before the onset of constriction, an additional, band-like growth zone is established around the cell center (Aaron et al., 2007). The rate of localized peptidoglycan synthesis then further increases during cytokinesis, resulting from PBP3-dependent formation of the new cell poles. Apart from that, in every cell cycle, new peptidoglycan is inserted at the base of the stalk, thereby driving stalk elongation (Aaron et al., 2007, Schmidt and Stanier, 1966).
Sequence analyses indicate that C. crescentus contains homologues of all essential cell division proteins identified in E. coli as well as some of the accessory factors, such as the Tol/Pal complex (Nierman et al., 2001). While several of the core divisome components have been analyzed in detail (Costa et al., 2008, Martin et al., 2004, Thanbichler and Shapiro, 2006, Wang et al., 2006, Quardokus et al., 2001), no information is available on division-related cell wall hydrolases in this organism. Overall, C. crescentus produces a relatively small range of lytic enzymes involved in peptidoglycan remodeling. It lacks LD- and DD-carboxypeptidase activity (Markiewicz et al., 1983) and only contains a single N-acetylmuramyl-L-alanine amidase, related to E. coli AmiC (Nierman et al., 2001). However, analysis of its genome revealed at least seven genes that code for putative LytM-domain containing endopeptidases, some of which are predicted to be localized in the cell envelope (Nierman et al., 2001).
In the present work, we identify one of these proteins, now designated DipM, as a critical component of the C. crescentus cell division apparatus. We show that DipM interacts with the murein sacculus and localizes to the site of constriction in an FtsN-dependent manner. In its absence, cells show severe division and polarity defects, resulting from delayed invagination of the cell wall and outer membrane during cytokinesis. These findings suggest that DipM is required for proper peptidoglycan remodeling during cell division, thus contributing to coordinated constriction of the different cell envelope layers.
RESULTS
DipM is a peptidoglycan-binding protein localizing to the cell division site
To identify factors involved in peptidoglycan remodeling during cell division, we examined the subcellular localization of C. crescentus proteins carrying predicted peptidoglycan-binding domains. This screen turned our attention to CC1996 (Nierman et al., 2001), a putative 609-amino acid protein with a predicted molecular mass of 63 kDa. Based on the findings described in this study, CC1996 was named DipM (division and polarity-related metallopeptidase). Bioinformatic analyses using the PSORTb algorithm (Gardy et al., 2005) suggest that DipM carries an N-terminal signal sequence with a predicted signal peptidase I cleavage site between positions 24 and 25. The processed, periplasmic form of the protein contains four lysin-motif (LysM) domains and a C-terminal peptidase M23 (LytM) domain, connected by largely unstructured linker regions. LysM domains are conserved in a number of proteins involved in cell wall degradation and were proposed to have a general peptidoglycan-binding function (Joris et al., 1992, Bateman and Bycroft, 2000, Steen et al., 2003). Peptidase family M23, on the other hand, comprises various zinc metallopeptidases, many of which are involved in peptidoglycan remodeling (Iversen and Grov, 1973, Bernhardt and de Boer, 2004).
In order to analyze the localization pattern of DipM over the course of the C. crescentus cell cycle, the native dipM gene of wild-type strain CB15N was replaced with a dipM-mCherry fusion. Swarmer cells of the resulting strain (MT261) were transferred onto an agarose pad and observed while they progressed through their developmental program (Fig. 1A). In new-born cells, the fusion protein was largely dispersed, although it occasionally appeared to be excluded from the pole-proximal regions of the cytoplasm. During transition to the stalked phase, foci were briefly observed at the nascent stalked pole. The protein then started to concentrate at the future division site, forming a broad band that gradually condensed into a tight focus as constriction proceeded. Immediately after cytokinesis, the protein was once again dispersed uniformly within the cell. The concentration of DipM remained constant throughout the cell cycle (Fig. 1B), excluding the possibility that the observed localization pattern was a result of fluctuating protein levels. Thus, DipM appears to be actively relocated to midcell at the onset of cell division.
Figure 1. Subcellular localization and function of DipM.
(A) Cell cycle-dependent localization pattern of DipM. Swarmer cells of strain MT261 (dipM-mCherry) were immobilized on an M2G-agarose pad (t = 0 min) and visualized at the indicated time points by DIC and fluorescence microscopy (bar: 2 μm). Polar localization is indicated by arrows. The generation time was ~250 min.
(B) Abundance of DipM over the course of the cell cycle. M2G medium was inoculated with swarmer cells of wild-type strain CB15N (t = 0 min). Samples were collected in 20-min intervals and analyzed by immunoblotting with anti-DipM and anti-CtrA antiserum. The response regulator CtrA is differentially expressed during the cell cycle (Domian et al., 1997) and serves as a control for the synchrony of the culture. Asterisks indicate non-specific immunoreactive bands.
(C) Subcellular localization of DipM. Whole-cell lysate of CB15N was fractionated by ultracentrifugation. Samples of the cell lysate (L), the supernatant (S) and the sedimented membrane fraction (M) were analyzed by immunoblotting with anti-DipM, anti-CtrA, and anti-SpmX antiserum. The soluble response regulator CtrA (Domian et al., 1997) and the membrane-integral polarity factor SpmX (Radhakrishnan et al., 2008) were used as controls.
(D) Peptidoglycan-binding capacity of DipM. 6 μg DipMAA26-609 or MalE were mixed with 100 μg of isolated CB15N murein sacculi. Murein was collected by ultracentrifugation and washed once in binding buffer. The supernatant of the first centrifugation step (S), the supernatant of the washing step (W), and the resuspended pellet (P) were analyzed by SDS-PAGE, and proteins were visualized by Coomassie blue staining. For both proteins, control reactions were performed in the absence of murein sacculi.
(E) Cells of strain MT258 (ΔdipM) were grown to exponential phase in PYE medium and analyzed by DIC microscopy (bar: 5 μm).
To validate the predicted periplasmic localization of DipM, cell fractionation studies were performed. The protein was indeed detected in the soluble fraction (Fig. 1C), indicating that it is processed at the suggested cleavage site. In addition, synthesis of a DipM-β-lactamase fusion was found to confer ampicillin resistance to a β-lactam-sensitive reporter strain, confirming export of DipM to the periplasmic space (Fig. S1). As a test for peptidoglycan-binding activity, CB15N sacculi were mixed with purified DipM and sedimented by ultracentrifugation. DipM was exclusively recovered in the pellet fraction, whereas it remained in the supernatant without the addition of sacculi (Fig. 1D). By contrast, a periplasmic protein without peptidoglycan-binding capacity (MalE) was not sedimented under the same conditions. Collectively, these findings identify DipM as a periplasmic protein that might be involved in peptidoglycan remodeling at the cell division site. In support of this hypothesis, a ΔdipM mutant (MT258) displays a severe cell division defect (Fig. 1E). Its cell length was, on average, significantly increased and highly variable (13.4 ± 9.8 μm). In addition, 13.4% of the cells showed more than one constriction and 9.5% of them formed branches (Table S1).
Localization of DipM is dependent on its interaction with FtsN
The localization pattern observed for DipM is characteristic of proteins involved in cell division (Wang et al., 2006, Möll and Thanbichler, 2009, Costa et al., 2008, Thanbichler and Shapiro, 2006). In order to clarify whether DipM is in fact a component of the cell division apparatus, we introduced DipM-mCherry into a conditional ftsZ mutant (AM214). When depleted of FtsZ, the cells became filamentous and DipM was evenly distributed over the cell (Fig. 2A). On re-induction of FtsZ synthesis, foci were rapidly restored within the filaments, followed by the establishment of constrictions at the sites marked by these foci. To examine if DipM is part of the late cell division complex, the localization behavior of DipM-mCherry was further studied in a conditional ftsN mutant (AM128). After depletion of FtsN (Fig. S2), DipM-mCherry was again dispersed over the cell (Fig. 2B), although other divisome components, such as FtsZ, FtsA, FtsK, and FtsI, are known to assemble properly in this condition (Möll and Thanbichler, 2009). Restoration of FtsN synthesis was accompanied by concentration of DipM-mCherry in broad fluorescent patches, which gradually condensed into distinct foci. There was a notable lag between the accumulation of the fusion protein and the onset of constriction, and division proceeded more slowly than usual, suggesting that division complexes assembled in the absence of FtsN are trapped in an unfavorable state, necessitating longer recovery periods.
Figure 2. Localization of DipM is dependent on FtsZ and FtsN.
(A) Dependence of DipM localization on FtsZ. Strain AM214 (Pvan::Pvan-dipM-mCherry ftsZ::Pxyl-ftsZ) was grown to exponential phase in PYE medium supplemented with 0.3% xylose. The cells were washed and depleted of FtsZ by incubation for another 6 h in the absence of inducer. Two hours before analysis, expression of dipM-mCherry was induced by addition of 0.5 mM vanillate. Cells were transferred onto an M2G-agarose pad containing 0.3% xylose and visualized at the indicated timepoints by DIC and fluorescence microscopy (bar: 5 μm).
(B) Dependence of DipM localization on FtsN. Strain AM128 (Pxyl::Pxyl-dipM-mCherry ΔftsN Pvan::Pvan-ftsN) was grown to exponential phase in PYE medium containing 0.5 mM vanillate. The cells were washed and depleted of FtsN by cultivation for another 14 h in the absence of inducer. Two hours before analysis, expression of dipM-mCherry was induced by addition of 0.3% xylose. Cells were placed on an M2G-agarose pad containing 0.5 mM vanillate and visualized at the indicated timepoints by DIC and fluorescence microscopy (bar: 5 μm).
(C) Timing of DipM localization. Strains AM206 (Pxyl::Pxyl-dipM-mCherry Pvan::Pvan-ftsZ-ecfp) and AM216 (dipM-mCherry Pxyl::Pxyl-ftsK-ecfp) were grown to exponential phase in M2G medium. Two hours before harvest, expression of the fluorescent protein fusions was induced by addition of 0.3% xylose and/or 0.5 mM vanillate. Swarmer cells were isolated from the cultures, resuspended in M2G medium, and visualized at 15-min intervals using DIC and fluorescence microscopy. The graph shows the frequency of cells that display a noticeable midcell focus or constriction as a function of the cell cycle. At least 100 cells were analyzed per timepoint. The generation time was ~120 min.
(D) Quantitative analysis of the interaction between DipM and FtsN. Reporter strain E. coli BTH101 was transformed with combinations of plasmids encoding fusions of the T25 and T18 fragments of Bordetella pertussis adenylate cyclase to the yeast GCN4 leucin-zipper region (zip), the transmembrane linker MalGAA1-77 (tm), FtsN, and MalGAA1-77-DipMAA26-609 (tm-DipM) (see Supplemental Material). β-galactosidase activities were determined to assess the interaction between the different hybrids. Each value represents the average of three independent measurements, performed in triplicate.
Our depletion studies suggest that DipM is a late recruit to the cell division apparatus, requiring FtsN for proper localization. To determine the precise position of DipM in the assembly hierarchy of the divisome, we analyzed the localization patterns of FtsZ, FtsK and DipM in a time-course experiment (Fig. 2C). Surprisingly, DipM was detectable at midcell earlier than FtsK, a protein known to accumulate at the division site before FtsN (Möll and Thanbichler, 2009). This seeming discrepancy might be explained by the previous finding that FtsN is likely to be recruited to midcell in a gradual process, regulated by a positive feedback loop (Möll and Thanbichler, 2009, Gerding et al., 2009). Thus, a small number of FtsN molecules might already localize to midcell early during cell division and thus be in place to interact with DipM, while the majority of the protein follows at a later stage.
To further analyze the dependence of DipM localization on FtsN, we chose to examine whether the two proteins could interact in a Bacterial Adenylate Cyclase Two-Hybrid (BACTH) assay (Karimova et al., 1998). For this purpose, the periplasmic portion of DipM was fused to a transmembrane anchor, comprising the first 70 residues of the E. coli maltose transporter subunit MalG. The N-terminal, cytoplasmic tails of the resulting construct and of full-length FtsN were then fused to the T18 or T25 fragment of Bordetella pertussis adenylate cyclase. In the BACTH system, interaction between two proteins triggers the synthesis of cyclic AMP, which in turn induces expression of a lacZ reporter gene carried by the test strains. The combination of hybrids containing full-length FtsN and DipM indeed resulted in high β-galactosidase activities (Fig. 2D), supporting the idea that the two proteins bind to each other, directly or indirectly, during divisome assembly. Consistent with this hypothesis, FtsN and DipM can be co-purified from cell extracts by co-immunoprecipitation (Fig. S3A and S3B). A closer analysis of the interaction determinants suggests contacts of FtsN with both the N-terminal region and the C-terminal peptidase domain of DipM (Fig. S3C). Further studies indicate that FtsN can also interact with the late cell division proteins TolR (CC3232) and AmiC (CC1876) as well as with the polarity determinant TipN (Fig. S4), consistent with a general role of FtsN in the organization of outer envelope constriction and polar morphogenesis. Collectively, these findings identify DipM as a periplasmic component of the cell division apparatus, possibly involved in peptidoglycan remodeling at the cell division site.
The LysM domains of DipM are required for proper localization
The N-terminal part of DipM contains four LysM domains, organized in two tandem repeats. LysM domains are conserved in various proteins involved in cell wall degradation and might have a general peptidoglycan-binding function (Bateman and Bycroft, 2000, Steen et al., 2003). To examine how they contribute to the function of DipM, we generated derivatives of DipM lacking between one and four of these domains (Figs. 3A and S5). The mutant proteins were fused to the red fluorescent protein mCherry and synthesized in ΔdipM background to analyze their functionality and localization patterns. Full-length DipM-mCherry was able to restore wild-type morphology and showed the normal subcellular distribution, indicating that the fusion was fully functional under the conditions used (Fig. 3B). Similarly, derivatives lacking one or two LysM domains could functionally replace the wild-type protein. However, whereas the loss of one domain (DipMΔ123-166-mCherry) still allowed for proper localization, deletion of the first pair of LysM domains (DipMΔ123-216-mCherry, DipMΔ123-291-mCherry) markedly impaired the recruitment of DipM to midcell, leading to more diffuse fluorescent signals. Even more severe localization defects were observed for DipM derivatives lacking three or all four LysM domains (Fig. 3C). When synthesized in a ΔdipM mutant, these proteins were largely dispersed throughout the periplasm, forming only faint (DipMΔ123-340-mCherry) or barely visible (DipMΔ123-390-mCherry) foci. Given that both proteins largely failed to localize to the cell division plane in the wild-type background, these foci might not result from interaction with the division apparatus but rather from an enlargement of the periplasmic space at the site of constriction, caused by delayed invagination of the outer cell envelope. Irrespective of their highly aberrant localization pattern, both DipM derivatives were still able to support cell division, although the cells showed minor morphological defects such as slight elongation (Table S2) and enlarged poles. Thus, the LysM domains are critical for condensation of DipM at the cell division site. However, proper localization of DipM appears to be largely dispensable for cell division, although it improves the robustness of the division process.
Figure 3. The LysM domains of DipM are required for localization.
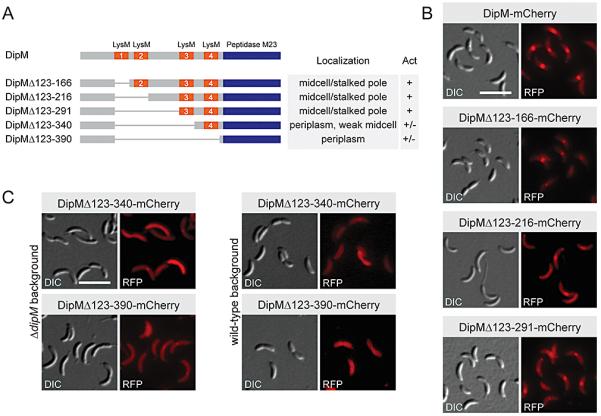
(A) Schematic representation of DipM and the different derivatives analyzed. The four LysM domains (residues 123-166, 173-216, 292-340, and 347-390) are shown in orange. The C-terminal LytM/Peptidase M23 domain (residues 501-609) is depicted in blue. The boxes on the right give the localization pattern and the activity (Act) of the proteins. Symbols: wild-type (+), partially impaired (+/−).
(B) Localization and functionality of DipM derivatives with lesions in the first pair of LysM domains. Strains AM205 (ΔdipM Pxyl::Pxyl-dipM-mCherry), AM222 (ΔdipM Pxyl::Pxyl-dipMΔAA123-166-mCherry), AM241 (ΔdipM Pxyl::Pxyl-dipMΔAA123-216-mCherry) and SS187 (ΔdipM Pxyl::Pxyl-dipMΔAA123-291-mCherry) were grown to exponential phase in PYE medium containing 0.3% xylose and visualized by DIC and fluorescence microscopy (bar: 5 μm).
(C) Localization and functionality of DipM derivatives lacking three or four LysM domains. Strains AM234 (ΔdipM Pxyl::Pxyl-dipMΔAA123-340-mCherry) and AM242 (ΔdipM Pxyl::Pxyl-dipMΔAA123-390-mCherry) were grown to exponential phase in PYE medium containing 0.3% xylose. Strains AM233 (Pxyl::Pxyl-dipMΔAA123-340-mCherry) and AM240 (Pxyl::Pxyl-dipMΔAA123-390-mCherry) were grown in PYE medium and induced with 0.3% xylose for 2 h. Subsequently, the cells were visualized by DIC and fluorescence microscopy (bar: 5 μm).
The peptidase domain of DipM is essential for function
To further dissect the determinants of DipM function, we generated a series of C-terminally truncated DipM derivatives, tagged with the red fluorescent protein mCherry (Figs. 4A and S5). The shortest deletion (DipMΔ501-609-mCherry) removed the entire C-terminal peptidase (LytM) domain, while other constructs additionally lacked the second pair (DipMΔ292-609-mCherry) or all four (DipMΔ123-609-mCherry) of the LysM domains. When synthesized in a ΔdipM background, none of these derivatives was able to functionally replace the wild-type protein, resulting in cells that displayed the characteristic ΔdipM mutant phenotype (Figs. 4B and 4C). Thus, the peptidase domain is absolutely required for DipM activity. However, it appears to be dispensable for localization, because the fusion proteins were still able to condense at the sites of constriction in a ΔdipM mutant (Fig. 4B) and to localize normally in the wild-type background (Fig. S6) as long as they contained one pair of LysM domains. The construct lacking both the peptidase and all four LysM domains was, by contrast, largely dispersed throughout the periplasm (Fig. 4C). As observed for other DipM derivatives defective in localization (compare Fig. 3C), the protein occasionally formed faint foci at the division sites or cell poles when synthesized in the ΔdipM strain, whereas it was evenly distributed in the wild-type background, again pointing to a local expansion of the periplasmic space in the absence of DipM activity.
Figure 4. The peptidase domain of DipM is required for function.
(A) Schematic representation of DipM and the derivatives analyzed. The four LysM domains are depicted in orange, the C-terminal LytM/Peptidase M23 domain in blue. The boxes on the right give the localization pattern and the activity (Act) of the proteins. Symbol: inactive (−).
(B) Localization and functionality of C-terminally truncated DipM derivatives. Strains AM225 (ΔdipM Pxyl::Pxyl-dipMΔAA501-609-mCherry) and AM231 (ΔdipM Pxyl::Pxyl-dipMΔAA292-609-mCherry) were grown to exponential phase in PYE medium containing 0.3% xylose and visualized by DIC and fluorescence microscopy (bar: 5 μm).
(C) Localization and function of a DipM derivative lacking all conserved domains. Strains AM232 (ΔdipM Pxyl::Pxyl-dipMΔAA123-609-mCherry) was grown in PYE medium containing 0.3% xylose. Strain AM200 (Pxyl::Pxyl-dipMΔAA123-609-mCherry) was grown in PYE medium and induced for 2 h with 0.3% xylose. Subsequently, the cells were visualized by DIC and fluorescence microscopy (bar: 5 μm).
(D) Peptidoglycan binding properties of the LysM and peptidase domains of DipM. 6 μg of the indicated DipM fragment were mixed with 100 μg isolated CB15N murein sacculi. Murein was collected by ultracentrifugation and washed once in binding buffer. The supernatant of the first centrifugation step (S), the supernatant of the washing step (W), and the resuspended pellet (P) were analyzed by SDS-PAGE and subsequent Coomassie blue staining. For both fragments, control reactions were performed in the absence of murein sacculi.
Together, these analyses indicate that the LysM domains are responsible for recruiting DipM to the division site, whereas the peptidase domain could provide a hydrolytic activity that promotes remodeling of the cell wall during constriction. To analyze the interaction of these two functional modules with the peptidoglycan layer, fragments of DipM comprising either the four LysM domains (DipMAA26-500) or the peptidase domain (DipMAA501-609) were purified and tested for their ability to co-sediment with isolated C. crescentus sacculi. The N-terminal fragment of DipM was recovered in the cell wall pellet after centrifugation, whereas it remained in solution in the absence of sacculi (Fig. 4D). By contrast, the isolated LytM domain failed to sediment with murein sacculi, indicating that it has no or rather weak peptidoglycan-binding activity. Thus, attachment of DipM to the peptidoglycan layer appears to rely mainly on its LysM domains.
DipM primarily acts in cell division
The ΔdipM mutant is characterized by inefficient cell division, enlarged cell poles and branching, suggesting that defects in DipM activity affect both cytokinesis and cell polarity. To differentiate between direct and indirect effects, we generated a strain (SW59) that expressed the dipM gene under the control of a vanillate-inducible promoter. Upon removal of the inducer, DipM levels started to decrease, reaching the detection limit within eight hours of further incubation (Fig. 5A). By twelve hours of depletion, we observed the first cases of cell elongation (Fig. 5B), concomitant with the development of rounded, enlarged poles (compare Fig. 5C). Although these morphological defects became more pronounced over time, the cells required at least sixteen hours of cultivation in the absence of vanillate to initiate branching, and more than twenty hours to reach the same branching frequency as a ΔdipM mutant strain. These findings suggest that inactivation of DipM primarily affects cell division and polar morphogenesis, whereas the effects on cell polarity might be indirect.
Figure 5. Mutation of DipM affects polar morphogenesis and cell polarity.

(A) Depletion of DipM. Strain SW59 (ΔdipM Pxyl::Pxyl-dipM) was grown in PYE medium supplemented with 0.3% xylose, washed twice, and resuspended in PYE medium containing 0.2% glucose. Samples were taken from the culture at two-hour intervals and analyzed by immunoblotting using anti-DipM antiserum. Asterisks indicate non-specific immunoreactive bands.
(B) Phenotypic consequences of DipM depletion. The cells from the samples described in (A) were analyzed by DIC microscopy (bar: 5 μm). Arrows indicate branches.
(C) Rounding of the poles in a ΔdipM mutant. Cells of strains MT258 (ΔdipM) and CB15N (WT) were grown in M2G medium and analyzed by electron cryo-tomography. The image shows a 19-nm and 13-nm slice, respectively, through the polar region of a reconstructed cell (bar: 100 nm). The asterisk indicates an abnormal bulge in the S-layer.
(D) Localization of TipN in the ΔdipM background. Strain AM263 (ΔdipM tipN::tipN-egfp) was grown in PYE medium and analyzed by DIC and fluorescence microscopy (bar: 5 μm). Arrowheads highlight TipN complexes with aberrant subcellular localization.
(E) Branching of cells in the absence of DipM. Strain MT258 (ΔdipM) was grown in M2G medium and analyzed by electron cryo-tomography. Shown are 19-nm slices through two reconstructed cells (bars: 50 nm). Stalked poles are indicated by arrows. The arrow head points to a chemoreceptor array, which is typically only found at a flagellated pole (Briegel et al., 2008).
In C. crescentus, the divisome-associated protein TipN contributes to the establishment of cell polarity by marking the newly generated poles after cell division (Huitema et al., 2006, Lam et al., 2006). Moreover, it is required for proper function of MreB, a key regulator of cell wall biosynthesis, and its ectopic localization was reported to induce the formation of branches (Lam et al., 2006). When synthesized in a ΔdipM background, a TipN-GFP fusion formed foci at the division sites and new cell poles, recapitulating the pattern observed in the wild-type situation. However, it was also found at the poles of lateral branches, at the constrictions of chained cells and, occasionally, at random positions within filamentous cells (Figs. 5D and S7). The branching phenotype of the ΔdipM mutant could thus originate from the establishment of ectopic polar growth zones at the positions marked by these TipN complexes. The tips of branches frequently displayed stalks (Figs. 5E, S8A and S8B), supporting the notion that they represent fully developed cell poles carrying the normal complement of polar proteins.
Deletion of DipM impairs invagination of the outer cell envelope
To analyze the cell division defects induced in the absence of DipM in more detail, we determined the subcellular distribution of the pole-organizing protein PopZ in the ΔdipM background. PopZ forms a polymeric scaffold that is responsible for polar attachment of the chromosomal origin regions and, in addition, interacts with signaling proteins involved in cell cycle regulation (Bowman et al., 2008, Ebersbach et al., 2008). Normally, newborn cells display a single PopZ complex at their old, flagellated pole. Soon after initiation of chromosome replication, the protein is then redistributed to both poles to capture the two segregating sister origin regions (Fig. S9). In the absence of DipM, however, PopZ predominantly localizes to both ends of the cell as well as to the division sites (Fig 6A). This finding suggests that in the ΔdipM mutant, division of the outer layers of the cell envelope lags significantly behind compartmentalization of the cytoplasm, allowing the two daughter cells to start the next cell cycle while cytokinesis is still in progress.
Figure 6. Cell division defects of DipM-deficient cells.
(A) Localization of PopZ in the ΔdipM background. Strain AM264 (ΔdipM Pvan::Pvan-popZ-eyfp) was grown in PYE medium, induced for 1 h with 0.5 mM vanillate, and analyzed by DIC and fluorescence microscopy (bar: 5 μm).
(B) Analysis of cellular compartmentalization using fluorescence loss in photobleaching (FLIP). Strain AM296 (ΔdipM Pxyl::Pxyl-egfp + pEJ216 [Pxyl-torAss-tdimer2]) was grown in PYE medium and induced for 2 h with 0.3% xylose. Cells were transferred onto an agarose pad, exposed to a series of 30-ms laser pulses at 50% laser intensity, and visualized by DIC and fluorescence microscopy before and after photobleaching. The region bleached is indicated by a rectangle. The arrow points to a constriction within the filament (bar: 2.5 μm).
(C) Ultrastructure of the division site in wild-type cells. Wild-type strain CB15N was grown in M2G medium and analyzed by electron cryo-tomography (11-nm slice, bar: 100 nm). Abbreviations: surface layer (SL), peptidoglycan (PG), cytoplasmic membrane (IM), and outer membrane (OM).
(D) Defects in peptidoglycan and outer membrane invagination in DipM-deficient cells. Strain MT258 (ΔdipM) was grown in M2G medium and analyzed by electron cryotomography. Shown are representative 29-nm (a, b) or 19-nm (c, d) slices through reconstructed cells at different stages of cell division (bars: a, c, d: 100 nm; b: 50 nm). Image b was median-filtered to reduce noise. Arrowheads indicate possible layers of peptidoglycan.
(E) Defects in polar morphology arising from cell division in the absence of DipM. Shown is a cryo-electron tomogram of a stalked MT258 (ΔdipM) cell (median filtered, 29-nm slice, bar: 100 nm).
In order to test this hypothesis further, filamentous ΔdipM cells were analyzed for the existence of multiple cytoplasmic compartments using the fluorescence-loss-in-photobleaching (FLIP) technique. For this purpose, we generated a dipM-deficient strain (AM296) that accumulated eGFP in the cytoplasm and, concurrently, exported the red fluorescent protein tDimer2 as a soluble protein to the periplasm. In untreated cells, diffuse green and red fluorescence was detectable along the entire cell body (Fig. 6B). After application of a laser pulse to one of the cell poles, eGFP was only bleached in a defined segment of the filament, usually discernable as a morphologically distinct compartment, while other regions of the cell remained unchanged. No recovery of fluorescence was observed after a 10-min interval, indicating discontinuity of the cytoplasmic space. By contrast, tDimer2 fluorescence decreased significantly along the entire length of cell, albeit to a lesser extent in the daughter cell compartments not exposed to the laser beam. The signal equilibrated completely over the course of the following ten minutes, suggesting that the different cytoplasmic compartments are surrounded by a common periplasm, allowing unrestrained diffusion of proteins within the cell envelope. Similar results were obtained for 80% of the cells investigated (n=26). In a wild-type culture, by contrast, only a small fraction of constricted cells (7.5 %) show cytoplasmic compartmentalization before division of the outer membrane (Judd et al., 2005), supporting involvement of DipM in the late stages of cytokinesis.
To visualize the division defects induced by the absence of DipM, the ultrastructure of a ΔdipM mutant was analyzed by electron cryo-tomography. Normally, the surface layer, the outer membrane and the cell wall dent inwards with the cytoplasmic membrane during constriction (Fig. 6C; Judd et al., 2005). In the mutant cells, however, a clear separation between the cytoplasmic membrane and the outer layers of the cell envelope was visible throughout all stages of cell division, resulting in an extensive widening of the periplasmic space at the division plane (Figs. 6D and S8D-G). Consistent with this finding, dipM-deficient cells exporting tDimer2 to the periplasm frequently display fluorescent foci at the sites of constriction (Fig. S10), indicating a local increase in the periplasmic volume (compare also Fig. 3C). Given the relatively mild filamentous or chaining phenotype of the ΔdipM strain, these structural abnormalities appear to still allow for cell division, albeit at a reduced rate, with some of the defects being passed on to the offspring (Figs. 6E and S8C). It was difficult to trace the cell wall at the division site, both in wild-type and mutant cells, although it could be readily detected throughout the remaining parts of the cell envelope. In some instances, we observed structures suggestive of peptidoglycan that lined both the outer membrane and the cytoplasmic membrane at the interface of the two daughter cell compartments (Fig. 6D, panels c and d). The observed defects in outer membrane invagination could thus be caused by the accumulation of supernumerary peptidoglycan layers that intercalate between the inner and outer cell envelope and thus disrupt the function of the Tol/Pal complex. However, current imaging technology does not provide sufficient resolving power to analyze these putative structural changes in detail.
DipM is involved in peptidoglycan hydrolysis
DipM contains a C-terminal peptidase (LytM) domain that is conserved among cell wall hydrolases. Given its localization pattern and the phenotype of the ΔdipM mutant, the protein is likely to play a role in peptidoglycan remodeling during cell division. To further investigate this possibility, we determined the effects of DipM overproduction, using a strain that expresses a plasmid-borne copy of dipM under the control of a xylose-inducible promoter. Upon induction, the protein level increased significantly, reaching a plateau by two hours of further incubation (Fig. 7A). Soon afterwards, the cells started to become spherical and lyse, with the frequency of ghost cells increasing dramatically over the course of the following hours (Fig. 7B). Thus, excess DipM strongly destabilizes the cell wall, supporting the notion that its peptidase domain has catalytic activity and acts on the peptidoglycan layer.
Figure 7. DipM displays peptidoglycan hydrolase activity in vivo and in vitro.
(A) Overproduction of DipM. Wild-type strain CB15N carrying plasmid pMT808 (Pxyl-dipM) was grown in PYE medium, and expression of dipM was induced by addition of 0.3% xylose. Samples were collected at the indicated timepoints and analyzed by immunoblotting using anti-DipM antiserum. The culture was maintained in exponential growth phase throughout the experiment. A lysate of strain MT258 (ΔdipM) was analyzed to control for non-specific immunoreactive bands (indicated by asterisks).
(B) Cell lysis upon overproduction of DipM. Cells treated as described in (A) were visualized by DIC microscopy (bar: 5 μm).
(C-E) Zymogram analysis of the peptidoglycan hydrolase activity of DipM. The indicated amounts of bovine serum albumin (BSA), lysozyme (Lys), DipM, and DipM fragments (in μg) were applied to SDS gels containing purified CB15N murein sacculi. The proteins in one of the gels were stained with Coomassie blue. The other gel was incubated in renaturation buffer to allow for refolding of the proteins (M: molecular mass standard). Subsequently, areas of lysis/binding were detected by staining of sacculi with methylene blue.
To determine whether DipM can in fact hydrolyze cell wall material, the mature form of the protein was isolated and tested for activity in a zymogram assay (Fig. 7C). For this purpose, DipM as well as the control proteins bovine serum albumin (BSA) and lysozyme were applied to a denaturing gel containing purified C. crescentus sacculi and refolded to their native state. Subsequently, the gel was treated with a peptidoglycan-binding dye, which is supposed to produce clear zones in all areas in which the sacculi have been degraded due to the presence of murein hydrolase activity. Whereas BSA, as expected, was inactive in this assay, no staining was observed in the vicinity of the lysozyme and DipM bands, suggesting that DipM may be able to cleave peptidoglycan in vitro. However, a recent study suggested that strong peptidoglycan-binding activity may be sufficient for a protein to yield a positive result in the zymogram assay (Uehara et al., 2010). To address this issue, we additionally tested fragments of DipM comprising only the N-terminal peptidoglycan-binding (LysM) domains (DipMAA-26-500) or the C-terminal peptidase domain (DipMAA501-609), respectively. Clear zones were indeed obtained for both of the truncated proteins, with the N-terminal fragment (Fig. 7D) producing a stronger signal than the peptidase domain (Fig. 7E). The latter does not co-sediment with murein sacculi, indicating that it lacks significant affinity for peptidoglycan (Fig. 4D). Its positive reaction in the zymogram assay is thus likely to reflect genuine, weak hydrolase activity. Nevertheless, the overall signal observed for the full-length protein may largely stem from the peptidoglycan-binding capacity of its four LysM domains.
DISCUSSION
The mechanisms that ensure the coordinated constriction of the Gram-negative cell envelope during cytokinesis are still unclear. Current data suggest that dynamic polymerization of FtsZ generates a pulling force that promotes invagination of the cytoplasmic membrane (Li et al., 2007, Osawa et al., 2008, Osawa et al., 2009). As a consequence, membrane-integral components of the cell wall biosynthetic apparatus, such as PBP3 and FtsN, may gradually move toward the cell center, ensuring that the peptidoglycan layer remains closely associated with the constricting membrane. The Tol/Pal complex might then mediate invagination of the outer membrane by interconnecting the inner and outer layers of the cell envelope (Gerding et al., 2007).
Localization of DipM
Despite many parallels in the composition of their division apparatus, C. crescentus and E. coli display marked differences in the progression of cell division with respect to the temporal and spatial regulation of events. In case of C. crescentus, division occurs concurrently with cell elongation, resulting in a conical constriction zone. We have now identified and characterized the first cell wall hydrolase involved in this process. DipM is produced throughout the cell cycle and localizes dynamically within the cell. After a short phase of uniform distribution, it accumulates transiently at the stalked pole and then starts to concentrate at the division plane, consistent with results from a previous large-scale analysis of protein localization in C. crescentus (Werner et al., 2009). The significance of the polar signals is unclear. Given that stalk elongation occurs by insertion of new peptidoglycan in a narrow polar growth zone (Aaron et al., 2007, Schmidt and Stanier, 1966), DipM might play a role in stalk biogenesis. However, we could not detect any noticeable defects in stalk morphology upon deletion of the dipM gene (data not shown), possibly indicating functional redundancy with other lytic enzymes. Localization of DipM to the division site is dependent on FtsN, consistent with the finding that the two proteins interact in two-hybrid and co-immunoprecipitation assays. However, while FtsN is thought to be a late recruit to the division site, DipM already starts to accumulate at midcell during early stages of the cell cycle, significantly before the onset of constriction. This seeming discrepancy may be resolved by the finding that even minute levels of FtsN are sufficient for divisome function (Möll and Thanbichler, 2009). Thus, it is possible that a small number of FtsN molecules already localize to midcell early during cell division, ready to initiate the recruitment of DipM. The bulk of the protein might then follow at a later stage, driven by a positive feedback loop that involves recognition of the nascent septal cell wall by the nonessential peptidoglycan-binding (SPOR) domain of FtsN (Gerding et al., 2009, Möll and Thanbichler, 2009). In agreement with an early role in cell division, overproduction of FtsN in E. coli can compensate for the lack of FtsK and suppress temperature-sensitive mutations in FtsA, FtsK, FtsQ and FtsI, proteins that are thought to be upstream of FtsN in the hierarchy of divisome assembly (Dai et al., 1993, Draper et al., 1998, Goehring et al., 2007).
We showed that the presence of LysM domains is essential for localization of DipM to the division plane. These modules are found in a variety of prokaryotic and eukaryotic proteins that bind to murein or chitin, possibly recognizing the N-acetylglucosamine or N-acetyl-muramic acid moiety (Buist et al., 2008, Ohnuma et al., 2008). In support of an interaction with the conserved glycan backbone of murein, LysM domains can interact efficiently with both A- and B-type peptidoglycan (Steen et al., 2003). In some cases, binding was shown to be restricted to certain regions of the cell wall, depending on the presence of inhibitory peptidoglycan modifications (Steen et al., 2003, Yamamoto et al., 2008). Consistent with these findings, an N-terminal fragment of DipM containing all four LysM domains binds to purified murein sacculi in vitro. It is conceivable that their peptidoglycan-binding activity mostly serves to tether the neighboring peptidase domain to the cell wall, thus enhancing its catalytic efficiency, whereas localization of DipM is achieved through its interaction with FtsN. However, the majority of DipM already localizes to the division site at a cell cycle stage at which FtsN is still largely dispersed throughout the cell. Its recruitment to midcell may thus occur in two discrete steps, similar to the situation seen for FtsN (Gerding et al., 2009, Möll and Thanbichler, 2009). Initially, a small number of molecules could be tethered to the divisome through direct interaction with FtsN, thereby allowing initiation of the constriction process. As cytokinesis proceeds, invagination of the peptidoglycan layer might be accompanied by the formation of structural intermediates that are specifically recognized by the LysM domains of DipM, thereby promoting further accumulation of DipM at the division site. In this way, the amount of DipM, and thus the level of endopeptidase activity present at midcell, could be dynamically adjusted over the course of the division process. Interaction of DipM with a conserved structure such as peptidoglycan is also supported by the fact that the protein is still targeted to midcell after heterologous expression in E. coli (Fig. S11). The nature of the putative DipM binding sites, however, still remains to be determined.
Previous work in E. coli has shown that FtsN interacts with a number of factors implicated in septum formation, including the amidase AmiC (Bernhardt and de Boer, 2003) as well as the transpeptidase PBP3 (Di Lallo et al., 2003, Karimova et al., 2005, Wissel and Weiss, 2004), the bifunctional transpeptidase/transglycosylase PBP1B (Müller et al., 2007) and the glycosyl transferase MtgA (Derouaux et al., 2008). The interaction network of the C. crescentus FtsN orthologue is much less well-defined. We have now revealed AmiC as a likely binding partner using BACTH analysis and provided clear evidence for an interaction between FtsN and the endopeptidase DipM. These findings further extend the range of enzymatic activities coordinated by FtsN, supporting a key role for this protein in the organization of peptidoglycan remodeling during cell division. Interestingly, FtsN and DipM show striking parallels in their domain composition and localization mechanisms. Both proteins contain peptidoglycan-binding domains that are recruited to the division site in isolated form (Gerding et al., 2009, Möll and Thanbichler, 2009). Although these domains are required for proper protein localization, their activities appear to be largely dispensable for function, suggesting that they mainly serve to improve the robustness of the constriction process. In each case, this goal might be achieved by a self-reinforcing mechanism whereby a small number of molecules is recruited to the divisome to initiate cell division, promoting the formation of new cell wall material that then provides additional binding sites for the protein. With FtsN serving as a central hub for peptidoglycan biosynthetic enzymes, these localization dynamics are likely to be inflicted on other proteins as well. Thus, factors involved in the late stages of cytokinesis might often be active at the division site significantly before they accumulate to detectable levels at this location, complicating the analysis of divisome assembly.
Function of DipM
Ultrastructural analysis of ΔdipM mutant cells revealed severe defects in cell wall and outer membrane constriction, leading to a considerable delay between compartmentalization of the cytoplasm and cell separation. Previous work has shown that division of the cytoplasm is sufficient to trigger entry into the next cell cycle (Thanbichler, 2009). Accordingly, the two daughter cells can initiate their developmental program although they are still connected by a common periplasm, explaining the high frequency of predivisional cells with both polar and medial PopZ complexes in the ΔdipM background. In cases where cell separation lags significantly behind the onset of polar development and growth in the daughter cell compartments, branches might emerge from the division sites. The polarity determinant TipN is indeed localized to constricted regions within elongated and filamentous ΔdipM cells, suggesting that the poles of the mutant cytoplasmic compartments have the same differentiation potential as regular cell poles.
The precise changes in cell wall architecture caused by lesions in dipM are difficult to assess. Even in the wild type, the peptidoglycan layer was barely detectable at the site of constriction, although it could be clearly resolved in the remaining parts of the cell, indicating transition of the invaginating cell wall to a less compact state. In E. coli, septal peptidoglycan has been proposed to have a three-ply structure, comprising two outer layers that form the new cell poles and an intermediate one that is degraded to facilitate daughter cell separation (Labischinski et al., 1991, Uehara and Park, 2008). Similarly, gradual constriction of the C. crescentus cell wall might involve a multi-layered intermediate. DipM, supported by other lytic enzymes, could function to remove the outer shells of this structure, thereby unlinking the daughter cells and maintaining a constant distance between the inner and outer membrane. In its absence, dissociation of the two membranes may disrupt the Tol/Pal complex and thus further impede outer membrane and S-layer invagination. Interestingly, the C. crescentus dipM mutant has some features in common with E. coli strains deficient in N-acetylmuramy-L-alanine amidase activity. Deletion of the amiABC genes induces chains of cells with compartmentalized cytoplasm that still adhere to each other via a common cell wall (Priyadarshini et al., 2007, Heidrich et al., 2001). The constrictions separating the individual compartments display conspicuous rings of inert peptidoglycan, resulting from excessive accumulation of septal cell wall material. Analysis of ΔdipM cells by electron cryotomography did not reveal thickening of the cell wall at the division sites. However, we cannot exclude the possibility that cells lacking DipM deposit multiple, loosely packed layers of peptidoglycan during constriction, with a density too low for resolution by low-contrast imaging techniques.
Although overexpression and zymogram analyses support a role for DipM in peptidoglycan degradation, its mechanism of action still remains to be determined. Previous work in E. coli has shown that the LytM-domain containing proteins EnvC and NlpD lack intrinsic hydrolase activity and rather serve as division site-specific activation factors for the amidases AmiA/B and AmiC, respectively (Uehara et al., 2010). The C. crescentus genome only encodes a single putative amidase, related to AmiC, which localizes to the constriction site during the late stages of cytokinesis (data not shown). Its inactivation has essentially no effect on cell division and does not prevent cell lysis upon overproduction of DipM (Fig. S12), excluding the possibility that DipM has a similar role as its E. coli homologues. Given that amidase activity is dispensable for the cleavage of septal peptidoglycan in C. crescentus, other cell wall hydrolases such as LytM-like endopeptidases might be required for this process. It is conceivable that the weak hydrolytic activity of DipM is stimulated through interaction with the cell division apparatus. Alternatively, DipM could contribute to the activation of other LytM-domain containing proteins.
LytM-like endopeptidases can target various bonds within the peptide side chains of murein. Several enzymes, such as the prototypic autolysin LytM from Staphylococcus aureus (Ramadurai et al., 1999) or the tail protein gp13 from Bacillus subtilis bacteriophage Φ29 (Cohen et al., 2009), were shown to cleave in-between two cross-linked peptides. The B. subtilis endopeptidase LytH, by contrast, hydrolyzes the L-Ala–D-Glu bond within individual disaccharide-tetrapeptide units (Horsburgh et al., 2003). C. crescentus lacks LD- and DD-carboxypeptidase activity, consistent with the observation that its peptidoglycan contains an unusually high percentage of pentapeptides (> 50%). Nevertheless, 44% of the monomeric peptide side chains are tetramers, indicating the existence of an enzymatic activity that releases tetrapeptides from cross-linked side chains via hydrolysis of the meso-A2pm–D-Ala bond (Markiewicz et al., 1983). Our results suggest DipM as a possible candidate for such an endopeptidase. However, although DipM undoubtedly has a major role in cell division, a ΔdipM mutant still manages to divide, albeit at reduced rate, indicating redundancy with other cell wall hydrolases.
There are parallels between DipM and the LytM-type endopeptidases of E. coli (Bernhardt and de Boer, 2004, Uehara et al., 2009). All of these proteins share the same conserved peptidase domain and support peptidoglycan remodeling during cell division. In addition, lesions in dipM and envC both result in enlargement of the periplasmic space at the constriction site (Bernhardt and de Boer, 2004). However, whereas deletion of dipM alone is sufficient to produce severe cell division defects, at least two of the E. coli paralogues have to be inactivated to achieve a significant chaining phenotype in standard growth conditions (Uehara et al., 2009). Moreover, there are clear differences in domain composition. DipM contains four LysM domains, at least two of which are required for localization to midcell. EnvC, by contrast, lacks apparent peptidoglycan-binding domains and bears an N-terminal coiled-coil region instead, while its paralogues only display a single LysM domain (Uehara et al., 2009). Thus, LytM factors are required for proper cell division in evolutionarily distinct bacteria, but the precise nature, number and function of the proteins involved appears to vary considerably.
MATERIALS AND METHODS
Media and growth conditions
The C. crescentus strains described in this study were derived from synchronizable wild-type strain CB15N (NA1000) (Agabian and Unger, 1978) and grown at 28°C in peptone-yeast-extract (PYE) medium (Poindexter, 1964) or M2-glucose (M2G) minimal medium. To induce the xylX promoter (Meisenzahl et al., 1997) or the vanA promoter (Thanbichler et al., 2007), media were supplemented with 0.3% xylose or 0.5 mM vanillate, respectively. For fluorescence microscopy, cells were grown to exponential phase in PYE medium and induced with vanillate (for 1 h) or xylose (for 2 h). Generalized transduction was performed using phage ΦCr30 (Ely and Johnson, 1977). Plasmids were introduced by electroporation (Ely, 1991). E. coli strains TOP10 (Invitrogen) or XL1-Blue (Stratagene) were used for general cloning purposes, whereas E. coli Rosetta(DE3)/pLysS (Merck) was used for protein overproduction. In both cases, cells were grown at 37°C in Super Broth (SB) (Botstein et al., 1975). E. coli BTH101 (Euromedex) was used for Bacterial Adenylate Cyclase Two-Hybrid (BACTH) analysis and grown at 28°C on MacConkey plates supplemented with 1% glucose-free maltose, kanamycin and ampicillin. For β-galactosidase assays, strains were grown in LB broth (Karl Roth, Germany) at 30°C. Synchronization of C. crescentus for microscopy and protein expression analysis was achieved by density gradient centrifugation, using Percoll (Tsai and Alley, 2001) or Ludox AS-40 (SigmaAldrich) (Ely, 1991), respectively. Antibiotics were added at the following concentrations (μg/ml; liquid/solid medium): spectinomycin (25/50), streptomycin (-/5), gentamicin (0.5/5), kanamycin (5/25) for C. crescentus and spectinomycin (50/100), gentamicin (15/20), kanamycin (30/50), chloramphenicol (20/30), ampicillin (100/200) for E. coli.
Construction of plasmids and bacterial strains
Plasmids and strains as well as details on their construction are described in Supplemental Tables S3, S4, S5 and S6. Gene replacement was performed with the help of sacB-containing suicide plasmids, using sucrose counter-selection to identify clones that have undergone double homologous recombination (Stephens et al., 1996). Genes of interest were expressed under the control of the vanA or xylX promoter using a previously described set of integration vectors (Thanbichler et al., 2007). All plasmids constructed were validated by DNA sequence analysis.
Immunoblot analysis
Peptide ‘IQAQPGEAESLPRPTP’, which is part of the N-terminal portion of DipM (AA 50-65), and ‘MRYAPTVKDKAKPVDP’, which is part of the C-terminal peptidase domain (AA 588-603), were used to immunize rabbits for the production of a polyclonal anti-DipM antibody (Eurogentec). Immunoblot analysis was performed as described previously (Thanbichler and Shapiro, 2006), using anti-CtrA (Domian et al., 1997), anti-DipM, or anti-SpmX (Radhakrishnan et al., 2008) rabbit antiserum at dilutions of 1:10,000 (CtrA, DipM) or 1:50,000 (SpmX).
Protein purification and murein pull-down assay
‘MAS’-DipM(AA26-610)-His6 (pAM072), ‘MAS’-DipM(AA501-609) (pAM091), ‘MAS’-DipM(AA26-500) (pAM092) and MalE-His6 (pAM142) were overproduced in E. coli Rosetta (DE3)/pLysS (Invitrogen). Proteins were enriched by nickel affinity chromatography and further purified by ion exchange or size exclusion chromatography, as described in the Supplemental Methods. Sacculi of C. crescentus CB15N were purified as described previously (Zahrl et al., 2005), starting with cells from three liters of an exponentially growing culture. The murein binding activity of DipM was examined using a modified murein pull-down assay (Ursinus et al., 2004). Murein sacculi were resuspended in binding buffer (20 mM Tris-maleate, 1 mM MgCl2, 30 mM NaCl, 0.05% Triton X-100, pH 6.8) to a final concentration of 100 μg/μl in a total volume of 400 μl. The solution was centrifuged for 30 min at 90.000 rpm and 4°C in a Beckman TLS-120.1 rotor to collect murein. Sedimented sacculi were resuspended in 400 μl binding buffer containing either 6 μg or no purified DipM. The mixture was incubated on ice for 30 min, centrifuged, and washed in 400 μl binding buffer. The supernatant of the first centrifugation step, the supernatant of the washing step, and the resuspended pellet were analyzed by SDS-PAGE followed by Coomassie blue staining.
Bacterial adenylate cyclase two-hybrid analysis
BACTH analysis was performed essentially as described (Karimova et al., 1998). The adenylate cyclase-deficient strain E. coli BTH101 was co-transformed with plasmids that encode hybrid proteins comprising the protein of interest fused to either the T25 or the T18 fragment of Bordetalla pertussis adenylate cyclase. The resulting strains were plated on MacConkey agar supplemented 1% maltose. Interaction was indicated by red colonies. β-galactosidase activity assays were used to quantify interaction between the hybrid proteins (Miller, 1972). For this purpose, cells were grown to exponential phase in LB medium containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. After recording the OD600 of the culture, 1-ml samples were sedimented by centrifugation and resuspended in 1 ml Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7). The cells were permeabilized by addition of 50 μl chloroform and 25 μl 0.1% SDS, followed by vigorous shaking for 10 s. The lysate was incubated at room temperature for 30 min. 500 μl aliquots of the samples were added to 500 μl Z-buffer supplemented with 50 mM 2-mercaptoethanol. The reaction was started by addition of 200 μl of o-nitrophenyl-β-D-galactoside (ONPG; 4 mg/ml), and the solution was incubated at room temperature. The reaction was stopped by addition of 500 μl 1M Na2CO3, and the reaction time was recorded. After centrifugation of the samples for 10 min at 14000 rpm, the A420 of the supernatant was measured against a cell-free blank. Enzymatic activities, in Miller Units (MU), were calculated using the following formula: MU = 1000*A420/ (incubation time in min)*(culture volume in ml)*OD600.
Light microscopy and photobleaching
For light microscopic analyses, exponentially growing cells were transferred onto pads made of 1% agarose (for still images) or 1% agarose in M2G medium, supplemented with inducer if necessary (for time-lapse analyses). When appropriate, the cover slides were sealed with VLAP (vaseline, lanolin and paraffin at a 1:1:1 ratio). Cells were visualized using a Zeiss AxioImager.M1 microscope equipped with a Plan Apochromat 100x/1.40 Oil DIC objective and a Cascade:1K CCD camera (Photometrics). Images were processed with Metamorph 7.1.2 (Universal Imaging Group) and Adobe Photoshop CS2 (Adobe Systems). Photo-bleaching experiments were performed with a 405 nm-solid state laser and a 2D-VisiFRAP Galvo System multi-point FRAP module (Visitron Systems, Germany), applying 30-ms pulses at a laser power of 50%.
Electron microscopy
For electron cryo-tomography, 2 ml of cell suspension were centrifuged for 5 min at 1500 g, and the pellet was resuspended in 30–50 μl of the supernatant. A solution of 10-nm colloidal gold (Ted Pella, Redlands, CA) was added to the cells immediately before plunge freezing and after treatment with BSA to avoid aggregation of the gold particles (Iancu et al., 2006). A 4 μl droplet of the sample solution was transferred to a glow-discharged R2/2 copper/rhodium grid, then automatically blotted and plunge-frozen in liquid ethane or a liquid ethane/propane mixture (Tivol et al., 2008) using a Vitrobot (FEI Company, Hillsboro, OR). The grids were stored under liquid nitrogen until data collection. Images were acquired using the FEI Polara ™ (FEI Company, Hillsboro, OR, USA) 300 kV FEG transmission electron microscope, equipped with a Gatan energy filter (slit width 20 eV) on a lens-coupled, cooled 4k x 4k Ultracam (Gatan, Pleasanton, CA). The pixel size on the specimen plane was 0.961 nm. Single-axis tilt series were recorded from −60 ° to 60° with an increment of 1° and an underfocus of 12 μm, using Leginon (Suloway et al., 2009). The cumulative dose was limited to 200 e/A2. Three-dimensional reconstructions were obtained using the IMOD software package (Mastronarde, 1997).
Zymography
The peptidoglycan hydrolase activity of DipM was analyzed by zymography essentially as described previously (Bernhardt and de Boer, 2004). SDS-polyacrylamide gels containing 0.04% (w/v) purified murein sacculi were loaded with 5 μg BSA (negative control), 5 μg lysozyme (positive control) and 5 μg, 2.5 μg and 1.25 μg purified DipM, and developed at a constant current of 20 mA at room temperature. The gels were incubated overnight in renaturation buffer (25 mM Tris/HCl, 1% TritonX-100, pH 8.0), shaken for 3 h in staining solution (0,1% methylene blue, 0.01% KOH), and destained in deionized water.
Cell fractionation
Cells of wild-type strain CB15N were grown in 80 ml PYE medium to an OD600 of 0.6, harvested by centrifugation, and washed in 80 ml0.2 M Tris/HCl (pH 8.0). The pellet was resuspended in 8 ml 60 mM Tris-HCl (pH 8.0) containing 0.2 M sucrose, 0.2 M EDTA, 10 mg/ml lysozyme, 100 μg/ml PMSF and 5 U/ml DNaseI. After 10 min incubation at room temperature, the sample was frozen in liquid nitrogen. The cells were thawed on ice and lysed by sonication. Cell debris was removed by centrifugation for 10 min at 4000 x g. Subsequently, membranes were sedimented by centrifugation for 1 h at 133,000 x g (4°C). After withdrawal of the supernatant, the pellet was washed in 0.2 M Tris/HCl (pH 8.0) and resuspended to a volume equivalent to that of the supernatant. Subsequently, samples from the supernatant and pellet fraction were analyzed by immunoblotting.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephanie Wick for excellent technical assistance and Grant R. Bowman, Lucy Shapiro, Yves V. Brun, and Patrick H. Viollier for providing plasmids, strains and antisera. We further thank Sebastian Poggio, Christine Jacobs-Wagner, Erin Goley and Lucy Shapiro for communicating unpublished results. This work was supported by funds from the Max Planck Society, National Institutes of Health (NIH) grant R01 AI067548 to G.J.J., and a gift to Caltech from the Gordon and Betty Moore Foundation.
REFERENCES
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- Agabian N, Unger B. Caulobacter crescentus cell envelope: effect of growth conditions on murein and outer membrane protein composition. J Bacteriol. 1978;133:987–994. doi: 10.1128/jb.133.2.987-994.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends SJ, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol. 2009;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein K, Lew KK, Jarvik V, Swanson CA. Role of antirepressor in the bipartite control of repression and immunity by bacteriophage P22. J Mol Biol. 1975;91:439–462. doi: 10.1016/0022-2836(75)90271-5. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Hardy GG, Trimble MJ, Brun YV. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol. 2009;54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddelmeijer N, Beckwith J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol Microbiol. 2004;52:1315–1327. doi: 10.1111/j.1365-2958.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Cohen DN, Sham YY, Haugstad GD, Xiang Y, Rossmann MG, Anderson DL, Popham DL. Shared catalysis in virus entry and bacterial cell wall depolymerization. J Mol Biol. 2009;387:607–618. doi: 10.1016/j.jmb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T, Priyadarshini R, Jacobs-Wagner C. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol Microbiol. 2008;70:634–651. doi: 10.1111/j.1365-2958.2008.06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- Derouaux A, Wolf B, Fraipont C, Breukink E, Nguyen-Disteche M, Terrak M. The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J Bacteriol. 2008;190:1831–1834. doi: 10.1128/JB.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lallo G, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology. 2003;149:3353–3359. doi: 10.1099/mic.0.26580-0. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Draper GC, McLennan N, Begg K, Masters M, Donachie WD. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Ely B, Johnson RC. Generalized transduction in Caulobacter crescentus. Genetics. 1977;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci U S A. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. Self-enhanced accumulation of FtsN at division sites, and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009 doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Robichon C, Beckwith J. Role for the nonessential N terminus of FtsN in divisome assembly. J Bacteriol. 2007;189:646–649. doi: 10.1128/JB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Holtje JV. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh GJ, Atrih A, Foster SJ. Characterization of LytH, a differentiation-associated peptidoglycan hydrolase of Bacillus subtilis involved in endospore cortex maturation. J Bacteriol. 2003;185:3813–3820. doi: 10.1128/JB.185.13.3813-3820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, Gan L, He Y, Jensen GJ. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2006;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Iversen OJ, Grov A. Studies on lysostaphin. Separation and characterization of three enzymes. Eur J Biochem. 1973;38:293–300. doi: 10.1111/j.1432-1033.1973.tb03061.x. [DOI] [PubMed] [Google Scholar]
- Joris B, Englebert S, Chu CP, Kariyama R, Daneo-Moore L, Shockman GD, Ghuysen JM. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett. 1992;70:257–264. doi: 10.1016/0378-1097(92)90707-u. [DOI] [PubMed] [Google Scholar]
- Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labischinski H, Goodell EW, Goodell A, Hochberg ML. Direct proof of a “more-than-single-layered” peptidoglycan architecture of Escherichia coli W7: a neutron small-angle scattering study. J Bacteriol. 1991;173:751–756. doi: 10.1128/jb.173.2.751-756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Draper GC, Donachie WD. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz Z, Glauner B, Schwarz U. Murein structure and lack of DD- and LD-carboxypeptidase activities in Caulobacter crescentus. J Bacteriol. 1983;156:649–655. doi: 10.1128/jb.156.2.649-655.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ME, Trimble MJ, Brun YV. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol. 2004;54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Möll A, Thanbichler M. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol Microbiol. 2009;72:1037–1053. doi: 10.1111/j.1365-2958.2009.06706.x. [DOI] [PubMed] [Google Scholar]
- Müller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Disteche M, Vollmer W. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma T, Onaga S, Murata K, Taira T, Katoh E. LysM domains from Pteris ryukyuensis chitinase-A: a stability study and characterization of the chitin-binding site. J Biol Chem. 2008;283:5178–5187. doi: 10.1074/jbc.M707156200. [DOI] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Din N, Brun YV. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol Microbiol. 2001;39:949–959. doi: 10.1046/j.1365-2958.2001.02287.x. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadurai L, Lockwood KJ, Nadakavukaren MJ, Jayaswal RK. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology. 1999;145(Pt 4):801–808. doi: 10.1099/13500872-145-4-801. [DOI] [PubMed] [Google Scholar]
- Schmidt JM, Stanier RY. The development of cellular stalks in bacteria. J Cell Biol. 1966;28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci U S A. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M. Spatial regulation in Caulobacter crescentus. Curr Opin Microbiol. 2009;12:715–721. doi: 10.1016/j.mib.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc. Microanal. 2008 doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Park JT. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursinus A, van den Ent F, Brechtel S, de Pedro M, Holtje JV, Löwe J, Vollmer W. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J Bacteriol. 2004;186:6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008a;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008b;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Khattar MK, Donachie WD, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, West L, Shapiro L. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J Bacteriol. 2006;188:1497–1508. doi: 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Pogliano K, Carson M, Guzman LM, Fraipont C, Nguyen-Disteche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissel MC, Weiss DS. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J Bacteriol. 2004;186:490–502. doi: 10.1128/JB.186.2.490-502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J. The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol Microbiol. 2008;70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology. 2005;151:3455–3467. doi: 10.1099/mic.0.28141-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







