Abstract
Fifty-nine single Cys-replacement mutants in helix VII and helix X of the lactose permease of Escherichia coli were subjected to site-directed fluorescence labeling in right-side-out membrane vesicles to complete the testing of Cys accessibility or reactivity. For both helices, accessibility/reactivity is relatively low at the level of the sugar-binding site where the helices are tightly packed. However, labeling of Cys substitutions in helix VII with tetramethylrhodamine-5-maleimide decreases from the middle toward the cytoplasmic end and increases toward the periplasmic end. Helix X is labeled mainly on the side facing the central hydrophilic cavity with relatively small or no changes in the presence of ligand. In contrast, sugar binding causes a significant increase in accessibility/reactivity at the periplasmic end of helix VII. When considered with similar findings from N-ethylmaleimide alkylation studies, the results confirm and extend support for the alternating access model.
Keywords: lactose, permease, symport, transport, symport, membranes, membrane proteins
As one of the most extensively studied members of the Major Facilitator Superfamily of membrane transport proteins, the lactose permease of Escherichia coli (LacY) catalyzes the coupled translocation of an H+ and a galactopyranoside (lactose/H+ symport). Since translocation is obligatorily coupled, sugar accumulation against a concentration gradient is achieved by using the free energy released from the downhill movement of H+ with the electrochemical H+ gradient (Δµ̃H+; interior negative and/or alkaline). Conversely, downhill sugar translocation by LacY drives uphill translocation of H+ with the generation of Δµ̃H+, the polarity of which depends on the direction of the sugar concentration gradient (1, 2).
X-ray structures of a conformationally restricted mutant (C154G) (3, 4) and wild type LacY (5) reveal 12 mostly irregular transmembrane helices arranged in 2 pseudo-symmetrical 6-helix bundles surrounding a large central water-filled cavity open to the cytoplasm only (an inward-facing conformation). However, multiple independent biochemical and biophysical studies, which include thiol cross-linking (6–13), site-directed alkylation (reviewed in 13, 14, 15–19), single molecule fluorescence resonance energy transfer (20), double electron-electron resonance (21) and Trp fluorescence quenching (22), not only demonstrate that an outward-facing conformation exists, but that a periplasmic cavity must open and close for transport to occur (13). By this means, the cytoplasmic cavity closes with opening of a wide hydrophilic cavity on the periplasmic side of LacY, which allows exposure of the sugar-binding site to either side of the membrane (the alternating access mechanism).
Helix VII contains Asp237 and Asp240, which are salt bridged to Lys358 (helix XI) and Lys319 (helix X), respectively (23–26). Although there are no irreplaceable residues with respect to transport activity in helix VII (27, 28), this transmembrane helix along with helix I is important for gating access to the sugar-binding site (17, 19). Furthermore, when three paired double-Cys mutants located in the interface between helices I/II and VII on the periplasmic side of LacY are cross-linked with relatively short homobifunctional reagents (<15 Å), all three mutants lose the ability to catalyze lactose transport (13). Strikingly, however, full activity is observed when cross-linking is mediated by a flexible reagent approximately 17 Å in length, a distance that agrees well with results from double electron-electron resonance (21). Molecular dynamics studies (29) are also consistent with the conclusion that a wide periplasmic cavity opens, as well as closes, during turnover of LacY.
Helix X also plays an essential role in active transport, containing His322 and Glu325, which are irreplaceable for H+ translocation and coupling (30, 31). Helix X neighbors helices VIII and IX, as well as helices VII and XI. Gly-scanning mutagenesis shows that the transport activity of mutant E325D can be rescued if Val316 is replaced with Gly, implying that the distance and/or flexibility between Glu325 and Arg302 (helix IX) is critical for deprotonation of Glu325 (32). Lys319 forms a charge pair with Asp240 (helix VII) (23, 25, 33), and there are also 6 positions in helix X that exhibit low transport activity if they are replaced with Cys and alkylated with NEM (34). Insertion of 2–6 continuous His residues between Leu313 and Glu314 also severely impacts activity (35), as do insertions of single or tandem factor Xa sites in various positions in periplasmic loop IX/X (X. Jiang, and H.R. Kaback, unpublished data).
Site-directed alkylation in situ with tetramethylrhodamine-5-maleimide (TMRM) is a more rapid and sensitive method compared to sulfhydryl alkylation with radiolabeled N-ethylmaleimide and autoradiography (15). Alkylation reflects the accessibility and/or reactivity of a Cys replacement at a given position, and any change in labeling induced by sugar binding or other perturbations indicates a change in the local environment (14). Site-directed TMRM labeling is applied here to single-Cys LacY mutants at each position in helices VII and X, and the overall findings from site-directed alkylation with TMRM or NEM are shown to be comparable. The results are consistent with the conclusion that the alternating access model is compatible with the global conformational change that occurs during turnover of LacY.
Materials and Methods
Materials
Tetramethylrhodamine-5-maleimide (TMRM, T-6027) was obtained from Molecular Probes, Invitrogen Corp. (Carlsbad, CA). 4-Nitrophenyl α-d-galactopyranoside was obtained from Sigma (St. Louis, MO). ImmunoPure immobilized monomeric avidin (Cat #20228) was obtained from Pierce (Rockford, IL). All other materials were reagent grade and obtained from commercial sources.
Plasmid construction
All single-Cys LacY replacements were obtained from the library of single-Cys LacY mutants encoded in plasmid pT7-5 (27, 34). DNA fragments encoding a given mutant were isolated by using restriction sites EcoR I and Sty I and then inserted into plasmid pT7-5 encoding C-less LacY with a biotin acceptor domain (BAD) from a Klebsiella pneumoniae oxaloacetate decarboxylase and a 6-His tag at the C terminus (15). Each mutant was verified by DNA sequencing of the entire lacY gene.
Growth of bacteria
E. coli T184 (lacY−Z−) transformed with plasmid pT7-5 encoding a given mutant was grown overnight at 37 °C in 50 mL Luria-Bertani broth containing ampicillin at a concentration of 100 µg/mL. The overnight cultures were then diluted 10-fold and induced with 1 mM isopropyl-1-thio-β-D-galactopyranoside when the OD600 reached 0.6. After additional growth for 2 h, cells were harvested and used for the preparation of right-side-out (RSO) membrane vesicles.
Preparation of RSO membrane vesicles
RSO membrane vesicles were prepared from 500 mL cultures of E. coli T184 expressing a given single-Cys mutant by lysozyme-ethylenediaminetetraacetic acid treatment and osmotic lysis (36, 37). The vesicles were resuspended to a protein concentration of 10–20 mg/mL in 100 mM potassium phosphate (KPi; pH 7.5) containing 10 mM MgSO4, frozen in liquid nitrogen and stored at −80 °C until use.
Labeling with TMRM
TMRM labeling was carried out following a protocol described previously (15, 16) with minor modifications. For each single-Cys mutant, 50 µL RSO membrane vesicles (0.1 mg of total protein) were incubated with 40 µM TMRM in the absence or presence of 1 mM 4-nitrophenyl α-d-galactopyranoside (αNPG) at 0 °C or 25 °C for 30 min or 30 s, as indicated. Reactions were terminated by adding dithiothreitol (DTT) to a final concentration of 10 mM. The membranes were solubilized with 2% n-dodecyl β-d-maltopyranoside (DDM) and mixed with 50 µL of 5% (vol/vol) immobilized monomeric avidin, which was pre-equilibrated in column buffer [50 mM NaPi (pH 7.5)/100 mM NaCl/0.02% DDM]. The mixture was incubated at 25 °C for 10 min with shaking, and then loaded onto Wizard columns (Promega, Madison, WI) on a vacuum manifold. After washing the avidin beads with column buffer, biotinylated LacY was eluted with 40 µL of column buffer containing 5 mM d-biotin. 10 µL of 5 × sodium dodecyl sulfate (SDS) loading buffer was added to each sample and then 10 µL of each sample was subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gel was imaged directly on an Amersham Typhoon™ 9410 Workstation (Amersham/GE Healthcare, Piscataway, NJ) as described. Finally, the SDS-PAGE gel was silver-stained and scanned.
To quantitatively measure the effect of NPG on TMRM labeling, the density of each band was estimated by using ImageQuant (Molecular Dynamics, GE Healthcare Bio-SciencesCorp., Piscataway, NJ). For each mutant, TMRM labeling in the absence of NPG was estimated by dividing the density of the protein band with TMRM signal, which was then normalized to 100% (equation 1):
Results
Helix VII
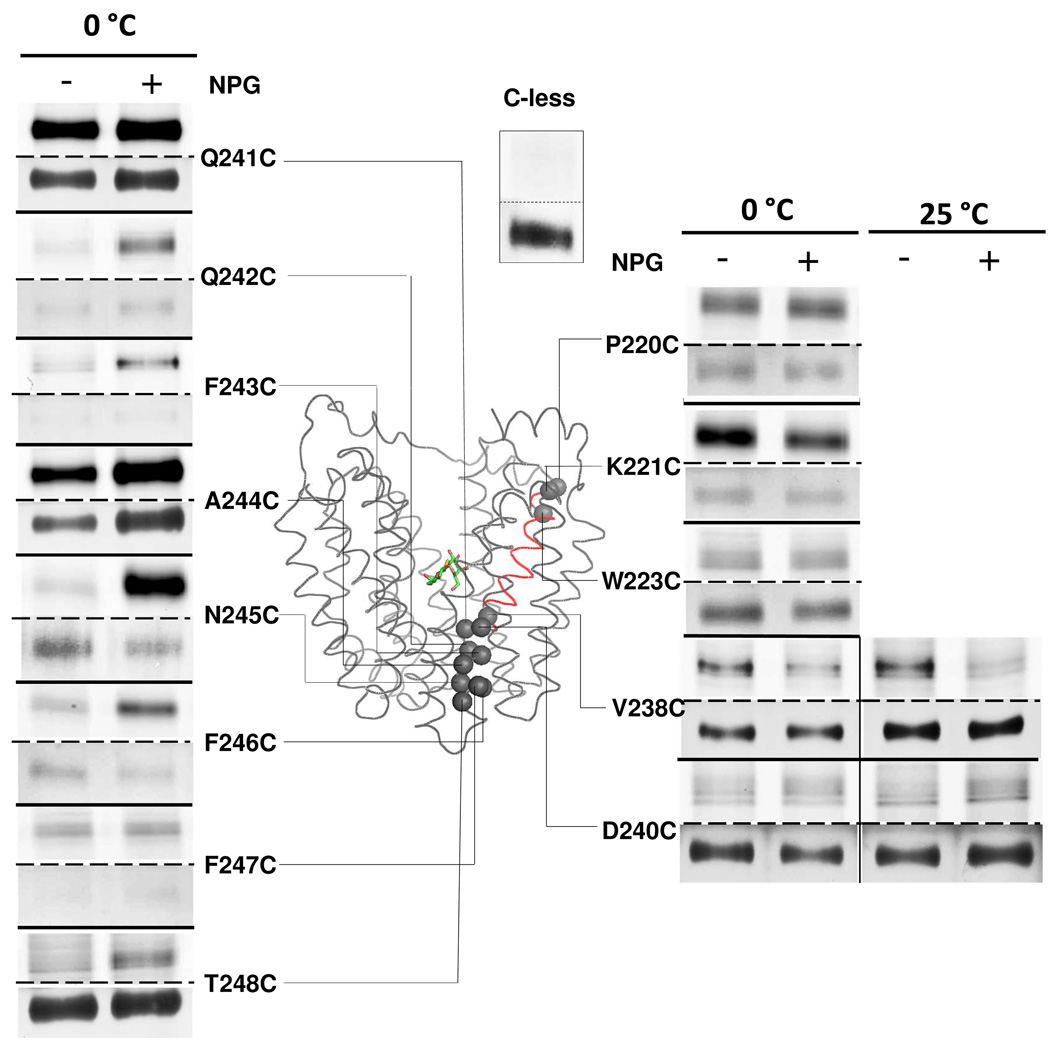
TMRM labeling of 29 single-Cys replacements in helix VII (from P220C to T248C) was carried out in the absence or presence of the high affinity galactopyranoside αNPG. Results for 13 such mutants are shown in Figure 1. With the exception of D237C, which is not expressed (24), each single-Cys mutant is expressed, albeit to different levels.
Figure 1. Labeling of single-Cys replacements in helix VII.
RSO vesicles containing given single-Cys LacY mutants (0.1 mg of total protein) were incubated with 40 µM TMRM for 30 min at 0 °C or 25 °C in the absence or presence of 1 mM αNPG as indicated. Reactions were terminated by addition of 10 mM DTT. The membranes were solubilized with 2% DDM and biotinylated LacY was purified with monomeric avidin, and then subjected to SDS-PAGE. LacY bands labeled with TMRM (upper panels) and silver-stained (lower panels) were imaged. The Cα atoms of labeled single-Cys mutants are shown as gray spheres superimposed on the backbone of LacY [PDB ID: 1PV7]. LacY is viewed perpendicular to the membrane with the N-terminal helix bundle on the left and the C-terminal helix bundle on the right. αNPG is shown as a stick model at the apex of the inward-facing cavity. Helix VII is highlighted in red. TMRM labeling of C-less LacY for 30 min at 0°C is shown as the negative control.
On the cytoplasmic side of the sugar-binding site, mutants P220C, K221C and W223C react with TMRM. Mutant K221C exhibits decreased TMRM labeling in the presence of αNPG, while mutants P220C and W223C exhibit no significant changes upon sugar binding.
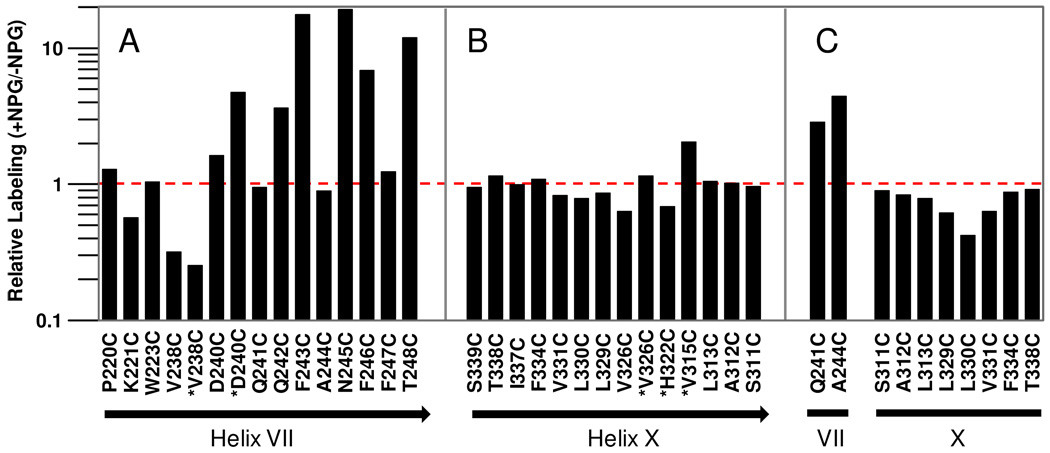
On the periplasmic side, αNPG binding causes decreased labeling with mutant V238C, and the effect is more pronounced at 25 °C relative to 0 °C. Mutant D240C barely reacts with TMRM at 0 °C, but labeling increases in the presence of αNPG at 25 °C. αNPG addition markedly increases labeling of mutants F243C and T248C, and strongly stimulates labeling of mutants Q242C, N245C and F246C at 0 °C. With mutants Q241C and A244C, the effect of αNPG is obscured by signal saturation, but when labeling is carried out at 0 °C for 30 s, αNPG induces a clear increase in labeling (compare Figs. 1 & 3). Single-Cys replacements at positions 222, 224–236 and 239 do not label in the absence or presence of αNPG at either 0 °C or 25 °C (data not shown).
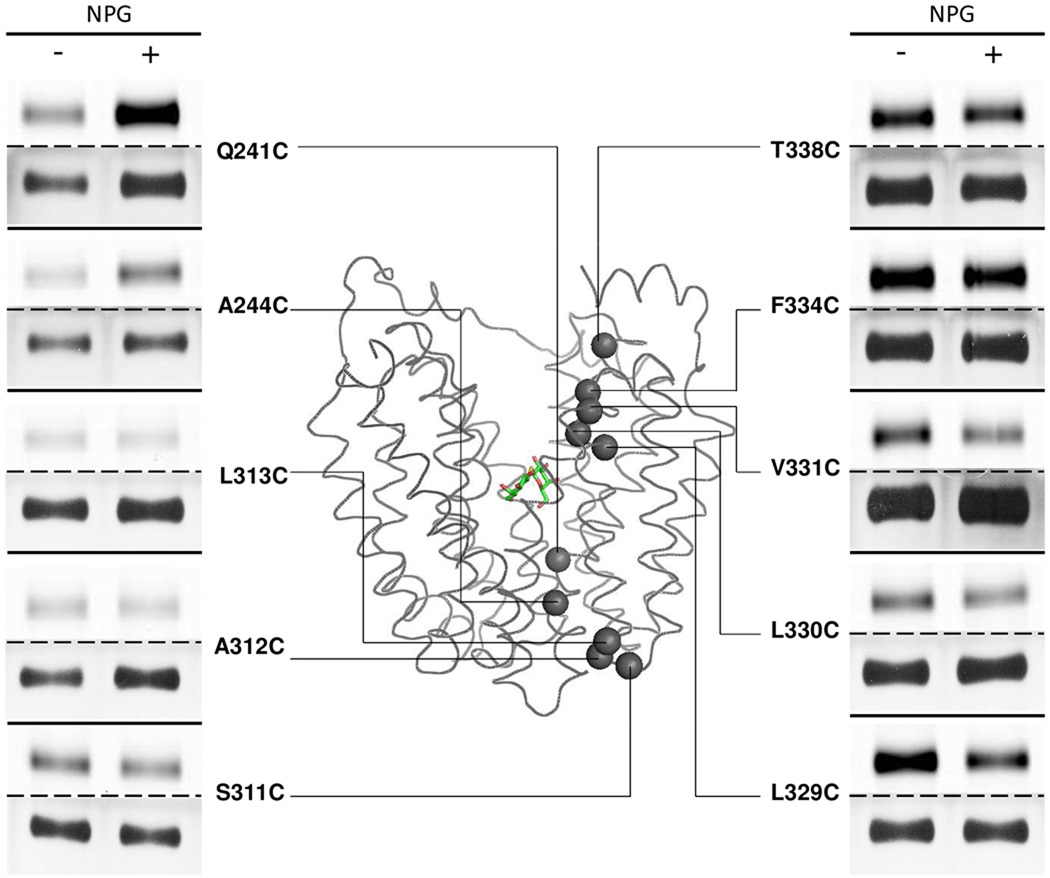
Figure 3. Labeling of given single-Cys replacements in helix VII or helix X.
Single-Cys mutants Q241C, A244C, S311C, A312C, L313C, L329C, L330C, V331C, F334C and T338C were labeled with TMRM for 30 s at 0 °C. TMRM-labeled bands (upper panels) and silver-stained bands (lower panels) corresponding to LacY were imaged. Experiments were carried out as described in Materials and Methods and in the legend to Fig. 1.
Results of TMRM labeling of helix VII at 0 °C are consistent with previous NEM labeling data (38). Due to enhanced sensitivity of fluorescence scanning with TMRM compared to autoradiography with NEM, three additional mutants on the periplasmic side of LacY (D240C, F243C and F247C) are identified that exhibit increased labeling in the presence of αNPG.
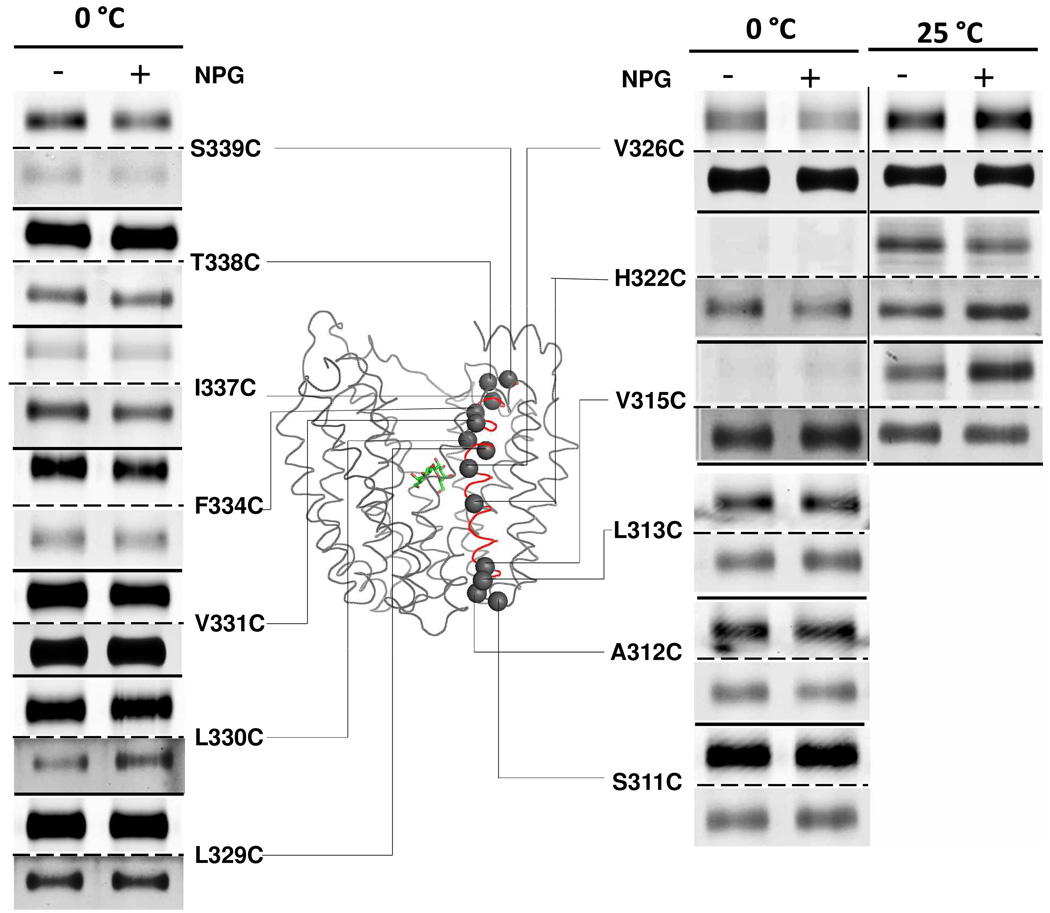
Helix X
Thirty mutants with single-Cys replacements in helix X (from S311C to Q340C) were subjected to TMRM labeling in the absence or presence of αNPG, and 13 of the mutants are shown (Fig. 2). Except for mutant Y336C, which is not expressed, each of the other mutants is expressed, although to different levels.
Figure 2. Labeling of single-Cys replacements in helix X.
RSO vesicles containing given single-Cys LacY mutants were labeled with TMRM for 30 min at 0 °C or 25 °C in the absence or presence of αNPG as indicated. Experiments were carried out as described in Materials and Methods and in the legend to Fig. 1. TMRM-labeled (upper panels) and silver-stained (lower panels) bands corresponding to LacY in SDS-PAGE gels were imaged. αNPG is shown as a stick model. Helix X is highlighted in red.
On the cytoplasmic side, mutant V326C exhibits decreased labeling in the presence of αNPG at 0 °C, but the effect is undetectable at 25°C due to signal saturation. With mutants L329C, L330C and V331C, signals are saturated after 30 min incubation with TMRM at 0 °C, but labeling for 30 s reveals decreased labeling upon αNPG binding (Fig. 3). Mutants F334C, I337C, T338C and S339C are labeled readily at 0°C, but NPG has no effect (Figs. 2 & 3).
On the periplasmic side, H322C labels at 25 °C but not at 0 °C, and addition of αNPG mildly decreases reactivity. Similarly, mutant V315 labels exclusively at 25 °C; however, addition of αNPG increases labeling. Mutants S311C, A312C and L313C at the periplasmic end of helix X label readily at 0 °C, but no significant changes are induced by αNPG (Figs. 2 & 3).
No labeling is observed with single-Cys mutants at positions 314, 316–321, 323–325, 327, 328, 332, 333, 335 or 340 at 0 °C, and positions 314, 316–321, 323–325 and 327 do not react with TMRM at 25 °C either (data not shown).
TMRM labeling of single-Cys replacements in helix X is also consistent with the alternating-access model. Thus, increased labeling is observed in the presence of αNPG on periplasmic side of the sugar-binding site (V315C) with decreased labeling on the cytoplasmic side (L329C, L330C and V331C). Except for specified positions at the very ends of helix X, most positions that label with TMRM are located on the side facing the central hydrophilic cavity.
Discussion
Both x-ray crystal structures of mutant C154G and wild type LacY exhibit an inward-facing conformation in which a large hydrophilic cavity faces the cytoplasm (3–5). In marked contrast, the periplasmic side is tightly packed and the single sugar-binding site at the apex of the hydrophilic cavity is completely blocked from the periplasmic side. Thus, in order to transport sugar across the membrane, there must be at least two conformations, one open to the cytoplasm and another open to the periplasm. Moreover, in order to prevent collapse of Δµ̃H+ from influx of H+, LacY can never be open to both sides. In order to accommodate these properties, alternating access models were proposed (39–41). Most recently, this type of model has received direct support from X-ray crystal structures (3–5, 42–44), as well as biochemical and biophysical studies (reviewed in 45).
Of the battery of biochemical/biophysical methods used, one of the most simple and straightforward is site-directed alkylation. By this means, the accessibility/reactivity of single-Cys mutants in a background devoid of native Cys residues is tested for reactivity with either radiolabeled NEM or fluorescent TMRM in the absence and presence of a high-affinity lactose analogue. The studies reveal that for the most part, Cys replacements on the periplasmic side of the sugar-binding site exhibit increased reactivity upon sugar binding, whereas decreased reactivity is observed on the cytoplasmic side (reviewed in 45). The results presented here focus on helices VII and X and represent completion of the study on almost all of the 417 positions in LacY except for the 17-residue C-terminal tail, which can be deleted without altering expression or activity (46, 47)
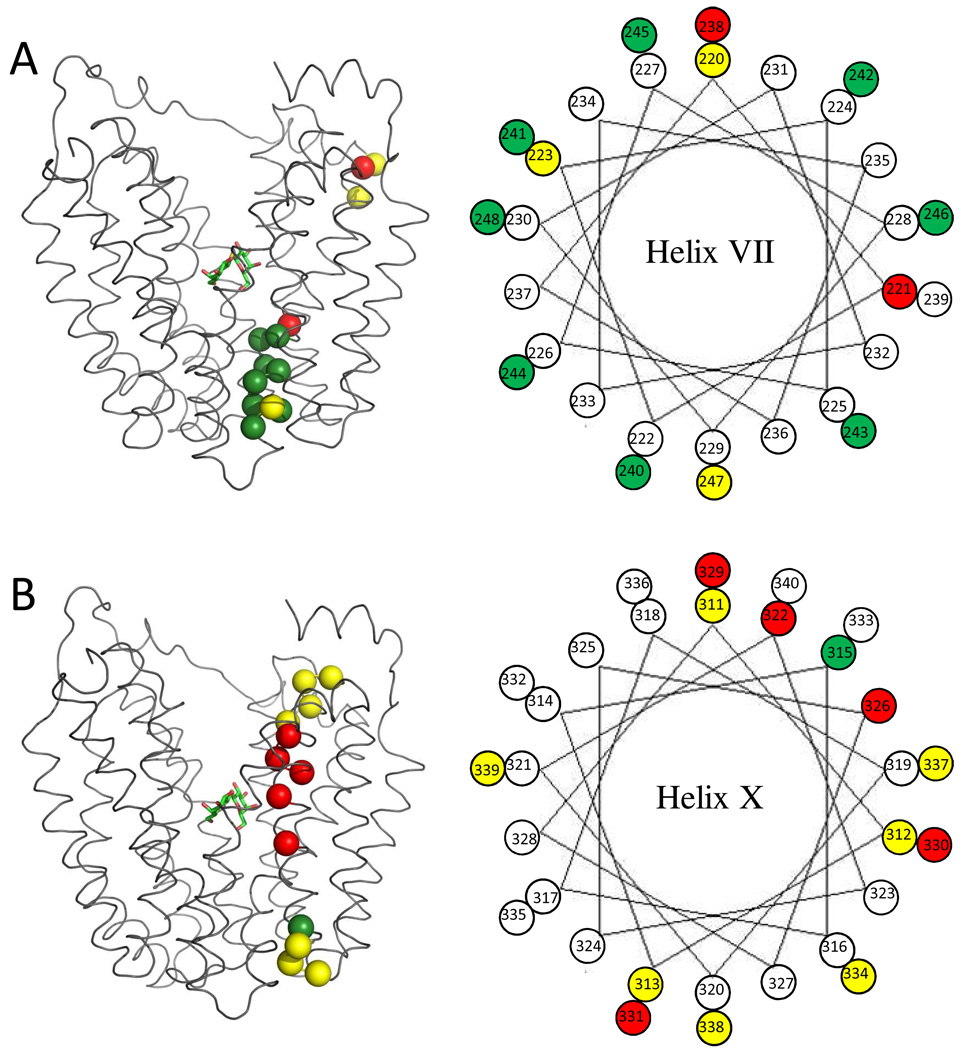
As shown here (Fig. 1), 13 out of the 29 single-Cys mutants in helix VII label with TMRM at 0°C. The reactivity of K221C at the cytoplasmic end of helix VII clearly decreases in the presence of αNPG, implying that the local environment changes upon sugar binding. W223C is not labeled by NEM (38), but reacts with TMRM, which is likely due to increased sensitivity of the latter method. Towards the periplasmic end, the thirteen mutants from L225C to T237C are not labeled at either 0 °C and 25 °C. This may be explained by the nature of helix packing in this region. Thus, helix X and helix XI may block TMRM access from the central hydrophilic cavity. Mutant V238C is located in the middle of the helix VII and at the same level as the sugar-binding site, and αNPG attenuates labeling, consistent with decreased accessibility. In contrast, most of the single-Cys mutants at the periplasmic end of helix VII exhibit enhanced labeling in the presence of αNPG (Figs. 1, 3 & 4), as previously reported for NEM labeling (38). The observations strongly suggest that this region of helix VII plays an important role in opening LacY on the periplasmic side. Notably, the effect of αNPG on TMRM labeling reverses between positions 238 and 240, consistent with the notion that apex of the hydrophilic cavity with the sugar-binding site is the fulcrum around which opening and closing occurs (3).
Figure 4. Effect of NPG binding on TMRM labeling of single-Cys replacements in helix VII and X.
Relative TMRM labeling of single-Cys replacements was calculated according to eq 1(see Materials and Methods). The red broken line indicates the labeling level in the absence of NPG, which is normalized to 1. (A) and (B), relative labeling of single-Cys replacements shown in Fig. 1 & 2, respectively. Black arrow beneath the bar graph indicates the periplasmic side of the helix. Labeling marked by “✱” was done at 25°C. (C), relative labeling of given single-Cys replacements shown in Fig. 3.
Unlike helix VII, helix X directly faces the cavity on the cytoplasmic side and tilts away from the center of the protein towards the periplasmic side. At 0 °C, mutants S311C, A312C and L313C at the periplasmic end are strongly labeled in the absence or presence of NPG. Intermolecular thiol cross-linking also shows that single-Cys replacements at these positions readily form LacY homodimers (48). In contrast, none of the mutants from E314C to F328C label at 0 °C. Tight packing of helix X with helix VII, helix VIII and helix IX in this region and/or blockade by the lipid bilayer likely prevent accessibility to TMRM. Mutant V315C appears to be buried by the abutting helices, but labels at 25°C in a manner that is increased by αNPG (Figure. 2). Thus, increased thermal motion probably causes the environment at position 315 to become more accessible to TMRM, and the effect is enhanced by substrate. His322, an irreplaceable residue involved in H+ translocation and coupling (1), is near the apex of the central cavity. Like V315C, H322C is labeled at 25 °C only, and αNPG weakly decreases labeling. αNPG also decreases labeling of mutants V326C, L329C, L330C, V331C (Figure. 2–4). These positions are on the cytoplasmic side of the sugar-binding site facing the central cavity. Decreased reactivity/accessibility at these positions is consistent with the idea that ligand binding induces closure of the cavity.
Notably, αNPG causes little or no change in labeling of Cys replacements in helix X. Furthermore, most positions that label, such as 315, 322, 326, 330, 334 and 338, are located on the same side of helix X that faces the central cavity. In contrast, αNPG significantly enhances labeling of Cys replacements in the periplasmic half of helix VII, and the labeled residues are distributed around the periplasmic end (Fig. 5). These observations may reflect different roles of the two helices in lactose/H+ symport. Since H322 and Glu325 are involved in H+ translocation, helix X may undergo relatively small structural changes during the turnover so that a constant distance between the two residues is maintained. In this regard, it may also be important that Lys319 is charge paired with Asp240 in helix VII (3–5, 23, 25, 49). In contrast, the periplasmic half of helix VII requires a much larger structural alteration to close and open the periplasmic side as LacY alternates between inward-facing and outward-facing conformations. Therefore helix VII might be more flexible and dynamic.
Figure 5. Distribution of labeled single-Cys replacements in helices VII and X.
Right: helical wheel diagrams are used to visualize the distribution of the TMRM-labeled residues on helix VII (A) and helix X (B). Positions labeled with TMRM are represented as colored spheres: Yellow, no change in the presence of αNPG; Red, decreased TMRM reactivity in the presence of NPG; Green, increased TMRM reactivity in the presence of NPG. Positions that do not label are shown as open circles. Left: the Cα atoms of the labeled residues on each helix are shown superimposed on LacY backbone as colored spheres and the color representation is same as that used in the helical diagrams. αNPG is shown as a stick model.
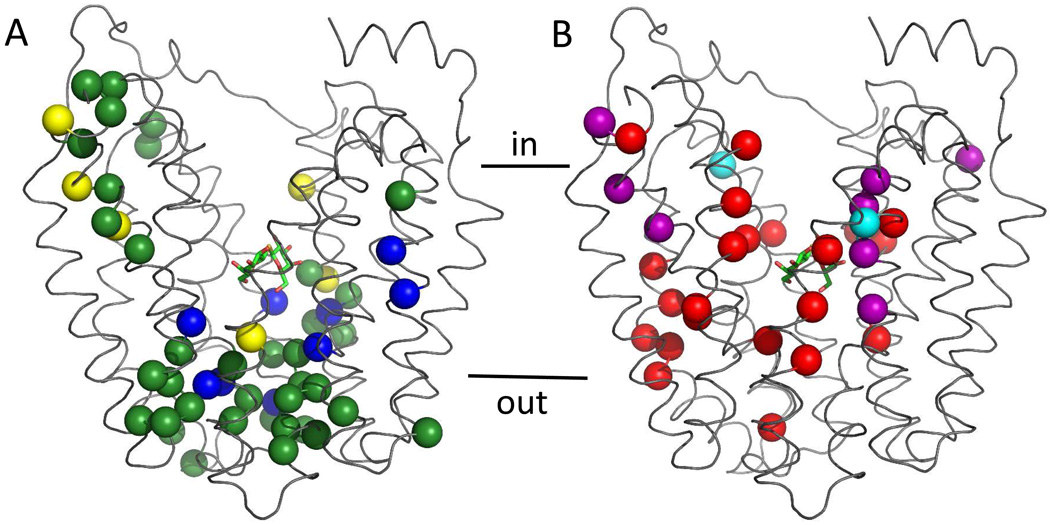
In previous studies from this laboratory, almost every residue of LacY was individually replaced with Cys and tested for reactivity with radiolabeled NEM (38, 50–55). Among these mutants, 65 replacements exhibit changes in reactivity in the presence of ligand. Taken together with the results presented here, more than 100 single-Cys replacements, covering all the positions that show changes in NEM labeling, were also tested with TMRM (15, 16). The results are highly consistent with each other. Most positions exhibiting increased labeling are located primarily on the periplasmic side of the sugar-binding site, while those exhibiting decreased labeling are located mainly on the cytoplasmic side (Fig. 6). Thus, the changes in reactivity observed with both labeling reagents provide strong support for the contention that alternating access provides a plausible model for the global conformational change that occurs during sugar translocation by LacY.
Figure 6. Distribution of single-Cys replacements exhibiting ligand-induced changes in TMRM or NEM reactivity.
Positions that exhibit ligand-induced changes in TMRM or NEM reactivity are superimposed on the backbone of LacY. LacY is viewed perpendicular to the membrane with the N-terminal helix bundle on the left, the C-terminal bundle on the right and TDG shown as a stick model. (A) Green spheres, increased reactivity with TMRM and NEM at positions 2, 3, 8,12, 14, 17, 25, 28, 31, 32, 42, 44, 49, 70, 71, 96, 100, 136, 157, 158, 159, 160, 161, 241, 242, 244, 245, 246, 248, 291, 308, 315, 359, 361, 363 and 364; blue spheres, increased reactivity with TMRM, but not NEM at positions 24, 29, 30, 45, 53, 240, 295, 298 and 362; yellow spheres, increased reactivity with NEM, but not TMRM, at positions 5, 11, 15, 265, 351 and 360. (B) Red spheres, decreased reactivity with both TMRM and NEM at positions 4, 21, 22, 27, 34, 60, 81, 84, 86, 87, 88, 122, 141, 145, 148, 238, 264, 268, 272, 329, 356 and 357; purple spheres, decreased reactivity with TMRM but not NEM at positions 5, 11, 15, 221, 322, 326, 330 and 331; cyan spheres, decreased reactivity with NEM, but not TMRM at positions 67 and 327.
Acknowledgement
We are indebted to Vladimir Kasho and Phillip Klebba for help with the preparation of Figs. 4 and 5. This work was supported by NIH grants DK051131, DK069463, GM073210 and GM074929, and NSF grant 0450970 to H.R.K.
The abbreviations
- LacY
the lactose permease of Escherichia coli
- Cys-less LacY
functional LacY devoid of native Cys residues
- RSO
right-side-out
- NEM
N-[ethyl-1-14C]ethylmaleimide
- NPG
4-Nitrophenyl α-D-galactopyranoside
- TDG
β-D-galactopyranosyl 1-thio-β-D-galactopyranoside
- KPi
potassium phosphate buffer
- DTT
1, 4-dithiothreitol
- EDTA
ethylenediamine tetraacetic acid
- TMRM
tetramethylrhodamine-5-maleimide
- BAD
biotin acceptor domain
- DDM
n-dodecyl β-D-maltopyranoside
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Reference
- 1.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaback HR. The Passion of the Permease. In: Wikström M, editor. Biophysical and Structural Aspects of Bioenergetics. Cambridge, UK: Royal Society of Chemistry; 2005. pp. 359–373. [Google Scholar]
- 3.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 4.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H(+) symport in LacY. Embo J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Kaback HR. A general method for determining helix packing in membrane proteins in situ: helices I and II are close to helix VII in the lactose permease of Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:14498–14502. doi: 10.1073/pnas.93.25.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Kaback HR. Helix proximity and ligand-induced conformational changes in the lactose permease of Escherichia coli determined by site-directed chemical crosslinking. J Mol Biol. 1997;270:285–293. doi: 10.1006/jmbi.1997.1099. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Hardy D, Kaback HR. Tilting of helix I and ligand-induced changes in the lactose permease determined by site-directed chemical crosslinking in situ. Biochemistry. 1998;37:15785–15790. doi: 10.1021/bi981501o. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Hardy D, Kaback HR. Transmembrane helix tilting and ligand-induced conformational changes in the lactose permease determined by site-directed chemical crosslinking in situ. J Mol Biol. 1998;282:959–967. doi: 10.1006/jmbi.1998.2065. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Hardy D, Kaback HR. Site-directed chemical cross-linking demonstrates that helix IV is close to helices VII and XI in the lactose permease. Biochemistry. 1999;38:1715–1720. doi: 10.1021/bi982342b. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Hardy D, Kaback HR. Tertiary contacts of helix V in the lactose permease determined by site-directed chemical cross-linking in situ. Biochemistry. 1999;38:2320–2325. doi: 10.1021/bi982288z. [DOI] [PubMed] [Google Scholar]
- 12.Sorgen PL, Hu Y, Guan L, Kaback HR, Girvin ME. An approach to membrane protein structure without crystals. Proc Natl Acad Sci U S A. 2002;99:14037–14040. doi: 10.1073/pnas.182552199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Guan L, Freites JA, Kaback HR. Opening and closing of the periplasmic gate in lactose permease. Proc Natl Acad Sci U S A. 2008;105:3774–3778. doi: 10.1073/pnas.0800825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaback HR, Dunten R, Frillingos S, Venkatesan P, Kwaw I, Zhang W, Ermolova N. Site-directed alkylation and the alternating access model for LacY. Proc Natl Acad Sci U S A. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Y, Ermolova N, Kaback HR. Site-directed Alkylation of LacY: Effect of the Proton Electrochemical Gradient. J Mol Biol. 2007;374:356–364. doi: 10.1016/j.jmb.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie Y, Sabetfard FE, Kaback HR. The Cys154-->Gly mutation in LacY causes constitutive opening of the hydrophilic periplasmic pathway. J Mol Biol. 2008;379:695–703. doi: 10.1016/j.jmb.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie Y, Zhou Y, Kaback HR. Clogging the periplasmic pathway in LacY. Biochemistry. 2009;48:738–743. doi: 10.1021/bi801976r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie Y, Kaback HR. Sugar binding induces the same global conformational change in purified LacY as in the native bacterial membrane. Proc Natl Acad Sci U S A. 2010;107:9903–9908. doi: 10.1073/pnas.1004515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Nie Y, Kaback HR. Residues Gating the Periplasmic Pathway of LacY. J Mol Biol. 2009;394:219–225. doi: 10.1016/j.jmb.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumdar DS, Smirnova I, Kasho V, Nir E, Kong X, Weiss S, Kaback HR. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc Natl Acad Sci U S A. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smirnova I, Kasho V, Choe JY, Altenbach C, Hubbell WL, Kaback HR. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci U S A. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smirnova I, Kasho V, Sugihara J, Kaback HR. Probing of the rates of alternating access in LacY with Trp fluorescence. Proc Natl Acad Sci U S A. 2009;106:21561–21566. doi: 10.1073/pnas.0911434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin-Toth M, Dunten RL, Gonzalez A, Kaback HR. Functional interactions between putative intramembrane charged residues in the lactose permease of Escherichia coli. Proc Natl Acad Sci U S A. 1992;89:10547–10551. doi: 10.1073/pnas.89.21.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunten RL, Sahin-Toth M, Kaback HR. Role of the charge pair aspartic acid-237-lysine-358 in the lactose permease of Escherichia coli. Biochemistry. 1993;32:3139–3145. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 25.Sahin-Toth M, Kaback HR. Properties of interacting aspartic acid and lysine residues in the lactose permease of Escherichia coli. Biochemistry. 1993;32:10027–10035. doi: 10.1021/bi00089a019. [DOI] [PubMed] [Google Scholar]
- 26.Voss J, Sun J, Venkatesan P, Kaback HR. Sulfhydryl oxidation of mutants with cysteine in place of acidic residues in the lactose permease. Biochemistry. 1998;37:8191–8196. doi: 10.1021/bi9802667. [DOI] [PubMed] [Google Scholar]
- 27.Frillingos S, Sahin-Tóth M, Persson B, Kaback HR. Cysteine-scanning mutagenesis of putative helix VII in the lactose permease of Escherichia coli. Biochemistry. 1994;33:8074–8081. doi: 10.1021/bi00192a012. [DOI] [PubMed] [Google Scholar]
- 28.Frillingos S, Sahin-Toth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. Faseb J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 29.Holyoake J, Sansom MS. Conformational change in an MFS protein: MD simulations of LacY. Structure. 2007;15:873–884. doi: 10.1016/j.str.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Kaback HR, Wu J. What to do while awaiting crystals of a membrane transport protein and thereafter. Acc Chem Res. 1999;32:805–813. [Google Scholar]
- 31.Kaback HR, Wu J. From membrane to molecule to the third amino acid from the left with the lactose permease of Escherichia coli. Quart Rev Biophys. 1997;30:333–364. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- 32.Weinglass AB, Smirnova IN, Kaback HR. Engineering conformational flexibility in the lactose permease of Escherichia coli: use of glycine-scanning mutagenesis to rescue mutant Glu325-->Asp. Biochemistry. 2001;40:769–776. doi: 10.1021/bi002171m. [DOI] [PubMed] [Google Scholar]
- 33.Voss J, Sun J, Kaback HR. Sulfhydryl oxidation of mutants with cysteine in place of acidic residues in the lactose permease. Biochemistry. 1998;37:8191–8196. doi: 10.1021/bi9802667. [DOI] [PubMed] [Google Scholar]
- 34.Sahin-Tóth M, Kaback HR. Cysteine scanning mutagenesis of putative transmembrane helices IX and X in the lactose permease of Escherichia coli. Protein Sci. 1993;2:1024–1033. doi: 10.1002/pro.5560020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna E, Hardy D, Kaback HR. Insertional mutagenesis of hydrophilic domains in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:11954–11958. doi: 10.1073/pnas.89.24.11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Short SA, Kaback HR, Kohn LD. Localization of D-lactate dehydrogenase in native and reconstituted Escherichia coli membrane vesicles. J Biol Chem. 1975;250:4291–4296. [PubMed] [Google Scholar]
- 37.Kaback HR. Bacterial Membranes. In: Kaplan NP, Jakoby WB, Colowick NP, editors. Methods in Enzymol. New York: Elsevier; 1971. pp. 99–120. [Google Scholar]
- 38.Venkatesan P, Kwaw I, Hu Y, Kaback HR. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: helix VII. Biochemistry. 2000;39:10641–10648. doi: 10.1021/bi000438b. [DOI] [PubMed] [Google Scholar]
- 39.Widdas WF. Inability of diffusion to account for placental glucose transfer in the sheep and consideration of the kinetics of a possible carrier transfer. J Physiol. 1952;118:23–39. doi: 10.1113/jphysiol.1952.sp004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell P. Molecule, group and electron transport through natural membranes. Biochem Soc Symp. 1963;22:142–168. [Google Scholar]
- 41.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 45.Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The Alternating Access Transport Mechanism in LacY. J Membr Biol. 2010;239:85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna E, Hardy D, Pastore JC, Kaback HR. Sequential truncation of the lactose permease over a three-amino acid sequence near the carboxyl terminus leads to progressive loss of activity and stability. Proc Natl Acad Sci USA. 1991;88:2969–2973. doi: 10.1073/pnas.88.8.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna E, Hardy D, Kaback HR. Evidence that the final turn of the last transmembrane helix in the lactose permease is required for folding. J Biol Chem. 1992;267:6471–6474. [PubMed] [Google Scholar]
- 48.Ermolova N, Guan L, Kaback HR. Intermolecular thiol cross-linking via loops in the lactose permease of Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:10187–10192. doi: 10.1073/pnas.1434239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JI, Hwang PP, Wilson TH. Lysine 319 interacts with both glutamic acid 269 and aspartic acid 240 in the lactose carrier of Escherichia coli. J Biol Chem. 1993;268:20007–20015. [PubMed] [Google Scholar]
- 50.Kwaw I, Zen KC, Hu Y, Kaback HR. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: helices IV and V that contain the major determinants for substrate binding. Biochemistry. 2001;40:10491–10499. doi: 10.1021/bi010866x. [DOI] [PubMed] [Google Scholar]
- 51.Frillingos S, Kaback HR. Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry. 1996;35:3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesan P, Hu Y, Kaback HR. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: helix X. Biochemistry. 2000;39:10656–10661. doi: 10.1021/bi0004403. [DOI] [PubMed] [Google Scholar]
- 53.Venkatesan P, Liu Z, Hu Y, Kaback HR. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: helix II. Biochemistry. 2000;39:10649–10655. doi: 10.1021/bi0004394. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Hu Y, Kaback HR. Site-directed sulfhydryl labeling of helix IX in the lactose permease of Escherichia coli. Biochemistry. 2003;42:4904–4908. doi: 10.1021/bi034078e. [DOI] [PubMed] [Google Scholar]
- 55.Ermolova N, Madhvani RV, Kaback HR. Site-directed alkylation of cysteine replacements in the lactose permease of Escherichia coli: helices I, III, VI, and XI. Biochemistry. 2006;45:4182–4189. doi: 10.1021/bi052631h. [DOI] [PubMed] [Google Scholar]