Abstract
Background
No commercially licensed vaccine or treatment is available for dengue fever, a potentially lethal infection that impacts millions of lives annually. New tools that target mosquito control may reduce vector populations and break the cycle of dengue transmission. Male mosquito seminal fluid proteins (Sfps) are one such target since these proteins, in aggregate, modulate the reproduction and feeding patterns of the dengue vector, Aedes aegypti. As an initial step in identifying new targets for dengue vector control, we sought to identify the suite of proteins that comprise the Ae. aegypti ejaculate and determine which are transferred to females during mating.
Methodology and Principal Findings
Using a stable-isotope labeling method coupled with proteomics to distinguish male- and female-derived proteins, we identified Sfps and sperm proteins transferred from males to females. Sfps were distinguished from sperm proteins by comparing the transferred proteins to sperm-enriched samples derived from testes and seminal vesicles. We identified 93 male-derived Sfps and 52 predicted sperm proteins that are transferred to females during mating. The Sfp protein classes we detected suggest roles in protein activation/inactivation, sperm utilization, and ecdysteroidogenesis. We also discovered that several predicted membrane-bound and intracellular proteins are transferred to females in the seminal fluids, supporting the hypothesis that Ae. aegypti Sfps are released from the accessory gland cells through apocrine secretion, as occurs in mammals. Many of the Ae. aegypti predicted sperm proteins were homologous to Drosophila melanogaster sperm proteins, suggesting conservation of their sperm-related function across Diptera.
Conclusion and Significance
This is the first study to directly identify Sfps transferred from male Ae. aegypti to females. Our data lay the groundwork for future functional analyses to identify individual seminal proteins that may trigger female post-mating changes (e.g., in feeding patterns and egg production). Therefore, identification of these proteins may lead to new approaches for manipulating the reproductive output and vectorial capacity of Ae. aegypti.
Author Summary
Dengue is a potentially lethal infection that impacts millions of humans annually. This disease is caused by viruses transmitted by infected female Aedes aegypti mosquitoes during blood feeding. No commercial vaccine or treatment is available for dengue infection. One way to break the disease transmission cycle is to develop new tools to reduce dengue vector populations. Seminal fluid proteins (Sfps) produced in the reproductive glands of male mosquitoes and transferred to females in the ejaculate during mating could be the target of such a tool. In related insects, Sfps modulate female reproduction and feeding patterns. Here we report 145 proteins that are transferred to females in the Ae. aegypti ejaculate. The proteins, which include Sfps and sperm proteins, fall into biochemical classes that suggest important potential roles in mated females. Of particular interest are proteins that could play roles in fertility and hormonal activity (including pathways involved in egg development and utilization of the blood meal). Our results lay important groundwork for new control strategies by identifying candidate proteins that may alter the reproductive biology or blood-feeding patterns of female Ae. aegypti and ultimately reduce the global burden of dengue.
Introduction
Male seminal fluid proteins (Sfps) influence female reproductive and feeding behaviors in a range of insects studied to date (reviewed in [1],[2]). Therefore, these proteins may provide targets or pathways that can be manipulated to reduce pathogen transmission by blood-feeding arthropods. The Aedes aegypti mosquito transmits several pathogens of concern to human health, including the viruses that cause dengue and dengue hemorrhagic fever (DHF) ([3]). Dengue, the most important mosquito-borne virus impacting human health, is a re-emerging disease in the tropical regions of the world. There is currently no vaccine against, or cure for, dengue, although research in this area is ongoing ([4]–[6]). Therefore, prevention of dengue infection depends heavily on control of its mosquito vector.
Understanding mosquito reproductive biology is critical to developing effective vector control methods. Previous research on Ae. aegypti suggests that mating and, specifically, male-derived proteins may play an important role in modulating female reproduction and feeding behavior. Upon mating, female Ae. aegypti undergo a series of time-dependent behavioral and physiological changes. Relative to virgin females, mated females have increased egg development and oviposition rates ([7],[8]), blood digestion rates ([9],[10]), and blood meal size ([10]). Mated females also have a lower likelihood of being inseminated by another male ([11]), of flying ([12],[13]), and of responding to host cues ([14]–[17]), and they have a reduced daily blood-feeding frequency ([18]). These changes in mated females appear to be induced by molecules produced in males' accessory glands (AG) and transferred to the female during mating ([9],[19]–[28]). In two other Dipteran species, individual AG-derived Sfps have been associated with functions in mated females. In Drosophila melanogaster, experimental studies have demonstrated that specific AG-derived Sfps influence a wide range of female post-mating behaviors including oogenesis (sex peptide), ovulation (ovulin), sperm storage and release from storage (Acp36DE, Acp29AB, CG1652/CG1656, CG17575, CG9997), propensity to re-mate (sex peptide), activity level (sex peptide), and feeding (sex peptide; reviewed in [2],[29]–[32]). In Anopheles gambiae, transglutaminase derived from male reproductive glands is necessary for the formation of the mating plug which is required for proper sperm storage ([33]). In both Ae. aegypti and D. melanogaster, secretions from the male AGs are necessary for fertility ([19],[34]). Identification of individual bioactive Sfps causing post-mating changes in female Ae. aegypti has not yet been accomplished and is a long-term goal of our research.
Previously, we identified over 250 proteins from male Ae. aegypti reproductive glands (AGs and seminal vesicles; [35]; L. Sirot, M. Wolfner, L. Harrington, unpubl. data). Fifty-three of those proteins were considered to be putative Sfps based on the criteria that they contained predicted secretion signal sequences and were not known to be housekeeping or structural proteins ([35]). However, we did not have direct evidence that those proteins were transferred to females during mating. In the current study, we identify a suite of proteins that are transferred from males to females during mating, and are thus candidate regulators of female behavior and physiology. Male- and female-derived proteins in the female reproductive tract were distinguished using an approach adapted from a study in D. melanogaster ([36]) that combined proteomics with a stable-isotope labeling technique (using 15N). We adapted this method to blood-feeding mosquitoes and discovered a set of proteins transferred from male to female Ae. aegypti. Among the Sfps we identified are potential modulators of protein activation/inactivation, sperm utilization, and ecdysteroidogenesis. Few of the Ae. aegypti Sfps we detected are homologs of known or predicted Sfps in other insect species, although many are in protein classes that are conserved across seminal fluid of a wide range of taxa ([37],[38]). Furthermore, our finding of intracellular and membrane-bound proteins in the transferred Sfps supports the hypothesis that Ae. aegypti Sfps are secreted, at least in part, through apocrine processes (pinching off of the apical portion of the cells into vesicles containing Sfps; [39]) in the accessory glands.
In the process of identifying the Sfps, we also identified a subset of 52 putative Ae. aegypti sperm proteins. The D. melanogaster homologs of many of the predicted Ae. aegypti sperm proteins are also sperm proteins ([40]), suggesting conservation of sperm-related function across Diptera. Some of the proteins we have identified may be useful targets for control of Ae. aegypti and may be applicable to other mosquito vectors.
Methods
15N-Labeling of female Ae. aegypti proteins
Overview
In order to distinguish male- and female-derived proteins in the mated female reproductive tract, we adapted a stable-isotope labeling technique that had been developed for D. melanogaster ([36]). Stable-isotope labeling of proteins makes their peptides unidentifiable by standard mass spectrometry analysis. Therefore, we mated labeled females to unlabeled males in order to identify male-derived proteins in the female reproductive tract. The steps involved in this process (described in detail below) are: stable-isotope labeling of yeast as food for mosquito larvae, rearing of mosquitoes on labeled yeast, mating of labeled females to unlabeled males, extraction of proteins from mated female reproductive tracts, and mass spectrometric analysis of the extracted proteins.
Yeast labeling
Saccharomyces cerevisiae strain D273-10B was used for all experiments. Two batches of yeast were prepared: one labeled with 15N and one unlabeled. Isotopic labeling was performed following the methods of Findlay et al. [36] with adjustments for incorporating the label into mosquitoes. Briefly, yeast was grown to saturation (∼16 h at 30°C) in 200 mL of minimal medium consisting of 20 g of glucose, 1.7 g of yeast nitrogen base without amino acids and either 5 g of 15N labeled ammonium sulfate (>99% 15N-enrichment; Cambridge Stable Isotopes, Andover MA, USA) or 5 g of unlabeled ammonium sulfate, in sterile water. The following day, 800 mL of additional medium was added and the yeast was grown for another 24 h. Yeast was harvested by centrifugation at 5,000 rpm at 4°C for 10 min, and the pellet was re-suspended in 30 mL of sterile water and centrifuged such that 15 mL of excess water was removed to yield a final volume of 15 mL of yeast slurry. The yeast slurry was stored at 4°C for less than two weeks before use.
Mosquito rearing
The Liverpool strain of Aedes aegypti L. was used for all experiments. Initially, eggs were hatched under vacuum and 200 first instar larvae were transferred to shallow pans containing 1 L of sterile water. Approximately 1 mL of the yeast slurry was added to each larval rearing pan every 1–2 days until pupation occurred. Preliminary data showed that females from the 15N-labeled treatment required additional nutrition beyond the yeast slurry (as assessed by their inability to fly and, thus, mate). Therefore, we supplemented the rearing pans with 200 mL of inoculum of rearing-water from a previous cohort of larvae grown under the same treatment (i.e., unlabeled or 15N-labeled yeast). The resulting larvae produced females that were able to fly and mated readily. Unlabeled males from the same mosquito strain were reared in pans of 200 larvae/1 L water and fed a 1∶1 mixture of Brewer's yeast and lactalbumin until pupation. All pupae were placed individually into vials and held for emergence. All adult mosquitoes were maintained on 20% sucrose solution on soaked cotton wicks. Reproductive tract samples were dissected from unlabeled and 15N-labeled virgin females to test the effectiveness of the labeling technique.
Mating
15N-labeled virgin females (3–5 days after emergence) were individually introduced into a 5 L bucket container that contained 20–40 unlabeled males (4–6 days after emergence). If a successful mating event was observed, both the female and male were removed from the bucket at the termination of mating as the pair began to separate. After mating, each female was placed individually in a test-tube and stored on ice for dissection. Females that did not mate within approximately 5 min were removed from the male cage and discarded; a new female was introduced into the male cage for mating. Dissections of the 15N-labeled females mated to unlabeled males were conducted within 30 min of mating.
Dissections
Female lower reproductive tracts (i.e., spermathecae, bursa, common oviduct) were dissected in MOPS buffer (80 mM NaCl, 10 mM KCl, 1 mM CaCl2, 0.2 mM MgCl2,10 mM MOPS) on ice and homogenized in 20 µl Dulbecco's PBS (DPBS) with protease inhibitors (Roche Complete Protease Inhibitor Tablets, Indianapolis, IN). Reproductive tracts from 20 females were pooled for each biological replicate. Samples were centrifuged at 12,000 rpm for 30 min at 4°C. The supernatant was transferred to a separate tube, the pellet was resuspended in 20 µl DPBS with protease inhibitors, and 20 µl of 2× SDS sample buffer (125 mM Tris–HCl pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.001% bromophenol blue) was added to each sample. The samples were boiled for 4 min and stored at −80°C.
Identification of proteins in sperm derived from seminal vesicle and testes
In order to distinguish Sfps from sperm proteins, we conducted proteomic analyses of sperm-enriched samples derived from the seminal vesicles or the testes. Seminal vesicles or testes from 40 males were dissected in DPBS with protease inhibitors, on ice. The tissues were then placed in a fresh droplet of DPBS, teased apart with a needle, swirled in the buffer to release sperm, and then removed from the droplet. The buffer droplet containing the sperm was transferred to a microcentrifuge tube containing 500 µl DPBST (DPBS with 0.1% Tween-20). The samples were spun at 20,800×g for 5 min at 4°C. The supernatant was discarded and the pellet was washed twice in DPBST. After the final wash, the pellet was resuspended in 2× SDS sample buffer. We considered the resulting samples as “sperm-enriched” as they contained not only sperm, but likely also some tissue, tissue secretions, and Sfps.
Protein separation and identification
Both the supernatant and the pellet samples were analyzed using two independent biological replicates of each sample type, except for the virgin females for which only one sample each was used for verification of our techniques. Protein separation and identification was conducted as previously described [35]. Briefly, proteins from each sample were separated by electrophoresis on a one-dimensional 5–15% gradient SDS polyacrylamide gel and visualized using Simply-Blue SafeStain (Invitrogen, Carlsbad, CA). The Cornell University Life Sciences Core Proteomics and Mass Spectrometry facility conducted in-gel digestion, tryptic peptide extractions, and Nano-LC-MS/MS. The nanoLC was carried out using an LC Packings Ultimate integrated capillary HPLC system equipped with a Switchos valve switching unit (Dionex, Sunnyvale, CA), which was connected in-line to a hybrid triple quadropole linear ion trap mass spectrometer, 4000 Q Trap (ABI/MDS Sciex, Framingham, MA) equipped with Micro Ion Spray Head ion source. The resulting MS and MS/MS data were submitted for database searching using the MASCOT search engine version 2.3 (Matrix Science, Inc., Boston, MA) or ProteinPilot software 1.0 (Applied Biosystems, Foster City, CA) against three databases (see “Databases” section below): the Ae. aegypti predicted peptide (AaegL1.2 Gene Build; hereafter “Vectorbase”) database (including supplemental peptides from AaegL 1.1 Gene Build, http://aaegypti.vectorbase.org/index.php; MASCOT program, Matrix Science, Boston, MA), a 6-frame translation of the Aedes genome (versionAaegL1.2; MASCOT program; hereafter “6-frame translation”), and a database of small (<150 amino acid) predicted peptides (ProteinPilot; hereafter “small peptides”). The default search settings used for protein identification were: one mis-cleavage for full trypsin with variable carbamidomethyl modification of cysteine, and a methionine oxidation. For the searches using MASCOT, the false discovery rate (FDR) was estimated for a measure of random identification from the same database. To estimate the FDR, an automatic decoy database search was performed in which a database of random sequences was generated and tested for raw spectra along with the real database.
Protein identifications were based on a significance threshold of <0.05. Additionally, we only considered proteins to be high confidence hits if either two different peptides from the same sample exceeded the significance threshold or if one peptide hit exceeded the significance threshold in two independent biological replicates (single or multiple peptide hits are reported in Tables S1 and S2). Hits from the 6-frame translation and the small peptides database were compared to the Vectorbase database using BLASTP, and queries with a significant match were removed. Any remaining hits from the 6-frame translation were searched for a predicted gene using the GeneID 1.2 prediction program (http://genome.crg.es/geneid.html) and only these hits were used in further analyses. We tested for predicted secretion signal sequences using SignalP 3.0 (www.cbs.dtu.dk/services/SignalP/, [41]) and for predicted protein domains using SMART (http://smart.embl-heidelberg.de/, [42]) and Pfam (http://pfam.sanger.ac.uk/, [43]). The relative quantitation of identified proteins in each biological sample was estimated using the exponentially modified protein abundance index (emPAI, [44]) and are reported in Tables S1 and S2.
Databases
All of the databases are derived from sequencing of the Liverpool strain. The Vectorbase database is based on 8× coverage and includes 4,758 supercontigs, 1.3 gigabases, 15,988 genes, and 17, 402 predicted peptides (http://www.vectorbase.org/Aedes_aegypti/Info/Index). To obtain the 6-frame translation database, the Ae. aegypti contigs were downloaded from Vectorbase (version AaegL1.2, September 2009). A program written in Visual Basic 6.0 by one of the authors (JMCR) obtained the 6-frame translation using the eukaryotic codon table. Sequences between stop codons were written to a fasta file retaining the frame and contig coordinates. This database includes 81,550,487 sequences. The small peptides database was generated by one of the authors (JMCR) through ab initio gene prediction based on the AaegL1.1 release using the GeneID prediction program (http://genome.crg.es/software/geneid/, [45]) and includes 24,092 sequences.
Identification of homologs to Ae. aegypti Sfp and sperm protein-encoding genes
By using BLASTP, we identified homologs of the Ae. aegypti proteins in the genomes of three other Diptera (Culex quinquefasciatus, Anopheles gambiae, and D. melanogaster) based on amino acid similarity. The divergence time between Aedes and Drosophila is predicted to be 250 million years ago ([46]). The Anopheles and Aedes genera are predicted to have diverged approximately 226 million years ago ([47]). Divergence time for Aedes and Culex is more recent than the divergence between Aedes and Anopheles ([47]). For the proteins identified from the Ae. aegypti Gene Build database, we defined homologs as reciprocal best BLASTP hits with ≥30% identity and E values≤10−3. For proteins identified from the Supplemental predicted peptides, the 6-frame translation and the small peptides databases, we used unidirectional BLASTP searches of the Ae. aegypti hits against the databases from the other three species.
Results and Discussion
Isotope labeling technique
In order to identify male-derived proteins that are transferred to females during mating, we used a whole-organism isotope labeling method. The principle of this method is to mate males to females whose proteins are labeled with the stable isotopes so as to exclude the female proteins from proteomic identification. Specifically, female proteins are labeled with 15N, which shifts their masses upward such that the masses of female-derived peptides do not match those expected in a standard search (uncorrected for 15N) of a predicted protein database. The method was developed by Krijgsveld et al. [48] and first used to identify Sfps by Findlay et al. [36] in D. melanogaster. We adapted this method to label female-derived proteins in Ae. aegypti. As with D. melanogaster, we reared larvae on yeast whose only nitrogen source was 15N. However, in order to generate females that could fly and mate, we had to supplement the larvae with an inoculum of rearing-water from larvae previously reared on 15N-labeled yeast (see Methods for more detail). To verify that the 15N-labeling technique sufficiently labeled female-derived proteins, we conducted nanoLC-MS/MS on protein samples from the reproductive tracts of labeled and unlabeled virgin females. We initially analyzed protein samples from two arbitrarily-chosen molecular weight ranges (∼30 kD to 50 kD and ∼98 kD to 120 kD) of both types of females. Using the Vectorbase database, we identified 115 proteins from the unlabeled female samples (Table S3) and no proteins from the labeled female samples. To further verify the labeling technique, we conducted nanoLC-MS/MS on the remaining gel sections from the labeled virgin female sample. We did not identify proteins from any of these samples. These results demonstrate that any proteins we identify in the reproductive tracts of labeled females mated to unlabeled males are highly likely to be male-derived. Furthermore, the technique we developed for in vivo stable-isotope labeling of Ae. aegypti proteins could be applied to other studies (e.g., to quantify proteomic changes in females in response to mating and/or blood-feeding or to distinguish mosquito-derived proteins from those of their pathogens, parasites, and/or endosymbionts; [49]).
Proteins transferred to females during mating
In our search against the Vectorbase database, we identified 128 proteins in the reproductive tracts of labeled females after mating with unlabeled males (Tables 1 and 2). Since the Ae. aegypti genome sequence is relatively new and thus the current annotation might still be missing actual genes, we also searched our mass spectrometry results against a 6- frame translation database of the Ae. aegypti contigs (Version AaegL1.2) and against a database of predicted small peptides (<150 amino acids). In our search against the 6-frame translation, we identified 12 novel predicted semen proteins (Tables 1 and 2). We identified 5 novel predicted small peptides from the small peptides database (Tables 1 and 2). The sequences of the unannotated predicted proteins and peptides are provided in Table S4. Thus, in total, we identified 145 male-derived predicted proteins that are transferred during mating to females. These proteins include Sfps and sperm proteins. For all searches, the FDR was ≤1%.
Table 1. Predicted seminal fluid proteins transferred in Aedes aegypti ejaculate.
| Molecular function | Predicted protein class | Aa a | Molecular function (cont.) | Predicted protein class | Aa |
| Binding | Annexin | 11302 | Proteolysis/ Catalysis (cont.) | Protease | 01588 |
| Calcyphosine | 08489 | 02000 | |||
| Dipeptidase | 08893 | 06403 | |||
| Fibrinogen | 01713 | 06414 | |||
| Kakapo | 02829 | 06421 | |||
| Lectin | Supp4872 | 06429 | |||
| 04679 | 10725 | ||||
| Mitochondrial brown fat uncoupling protein | 07046 | 11558 | |||
| Moesin | 07915 | 12217 | |||
| Mucin | 00718 | 15386 | |||
| Odorant-binding | AaegSfp1 | Protease inhibitor | 02715 | ||
| Phosphatidylethanol- binding protein | 11263 | AaegSfp2 | |||
| AaegSfp3 | |||||
| Tubulin β-chain | 02848 | Thioesterase | 03569 | ||
| None | 00479 | Transferase | 03746 | ||
| 08274 | None | 02793 | |||
| 09201 | 13559 | ||||
| Oxidoreductase | Catalase | 13407 | 17451 | ||
| Decarboxylase | 05790 | 17460 | |||
| Dehydrogenase | 04338 | Structural | Actin | 01928 | |
| 05308 | 05964 | ||||
| 06928 | Vitellogenin | 05815 | |||
| 10464 | None | 03348 | |||
| 12014 | Transport | Cytochrome c oxidase subunit | 00929 | ||
| NADH-ubiquinone oxidoreductase subunit | 05946 | 13751 | |||
| Peroxidase | 04112 | Glutamate receptor | 09813 | ||
| Proteolysis/ Catalysis | Aminopeptidase | 02399 | Mitochondrial glutamate carrier | 11276 | |
| 02978 | Sodium/calcium exchanger | 12480 | |||
| 07201 | Other | Niemman-Pick Type C-2 | 09760 | ||
| Asparaginase | 02796 | Rab GDP-dissociation inhibitor | 12904 | ||
| ATP synthase subunit | 06516 | Venom allergen | 09239 | ||
| 07777 | None | 04944 | |||
| 11025 | 05219 | ||||
| 12035 | 10824 | ||||
| Supp3543 | |||||
| 12819 | Supp4095 | ||||
| ATPase | 14053 | AaegSfp5 | |||
| Dehydrogenase | 04294 | AaegSfp6 | |||
| Dynein | 11478 | AaegSfp7 | |||
| Gamma glutamyl transpeptidase | 10935 | AaegSfp8 | |||
| 14580 | AaegSfp9 | ||||
| Glutathione transferase | 11741 | AaegSfp10 | |||
| Hydrolase | 03666 | AaegSfp11 | |||
| 06485 | AaegSfp12 | ||||
| Kinase | 12359 | AaegSfp13 | |||
| 12731 | AaegSfp14 | ||||
| Lipase | 07063 | ||||
| Mannosidase | 05763 |
5-digit numbers are the Vectorbase database identification numbers without the proceeding “AAEL0”. Numbers with “Supp” prefix refer to proteins from the Supplementary predicted peptide database from AaegL1.1 Gene Build. Numbers with the prefix “AaegSfp” refer to proteins from either the 6-frame translation or the small peptides databases. The amino acid sequences for all of the “Supp” and “AaegSfp” predicted proteins are given in Table S4.
Table 2. Predicted sperm proteins transferred in Aedes aegypti ejaculate.
| Molecular function | Predicted protein class | Aa a | Molecular function (cont.) | Predicted protein class | Aa |
| Binding | Aminopeptidase | 06975 | Proteolysis/ Catalysis (cont.) | Protease inhibitor | AaegSp1 |
| Heat shock protein 70 | Supp4130 | None | 06509 | ||
| Histone | 00490 | 10754 | |||
| 15674 | 17349 | ||||
| Reticulocalbin | 14589 | Structural | Actin | 01673 | |
| Tubulin α-chain | 06642 | 11197 | |||
| 13229 | Myosin | 12543 | |||
| None | 00637 | Tubulin β-chain | 02851 | ||
| 08779 | 05052 | ||||
| 10149 | Transport | ADP, ATP carrier | 04855 | ||
| 10882 | Cytochrome c | 04457 | |||
| 14231 | Cytochrome c oxidase subunit | 05170 | |||
| Oxido-reductase | Dehydrogenase | 00454 | Ubiquinol-cytochrome c reductase unit | 03675 | |
| 02881 | 05269 | ||||
| 03757 | Voltage-dependent anion-selective channel | 01872 | |||
| 08166 | None | 17508 | |||
| NADH-ubiquinone oxidoreductase | 12552 | Other | Netrin receptor | 07195 | |
| Proteolysis/Catalysis | Aconitase | 03734 | None | 09707 | |
| Aminopeptidase | 00108 | 12282 | |||
| ATP synthase subunit | 02827 | 17096 | |||
| 05173 | Supp4104 | ||||
| 05610 | Supp7141 | ||||
| 05798 | AaegSp2 | ||||
| 08787 | AaegSp3 | ||||
| 08848 | |||||
| 12175 | |||||
| Kinase | 06042 | ||||
| Protease | 03308 |
5-digit numbers are the Vectorbase database identification numbers without the proceeding “AAEL0”. Numbers with “Supp” prefix refer to proteins from the Supplementary predicted peptide database from AaegL1.1 Gene Build. Numbers with the prefix “AaegSp” refer to proteins from either the 6-frame translation or the small peptides databases. The amino acid sequences for all of the “Supp” and “AaegSp” predicted proteins are given in Table S4.
Nine of the identified proteins share identical amino acid sequence with other Ae. aegypti predicted proteins in the regions that were identified by our mass spectrometry analysis. As a result, we cannot distinguish amongst these proteins in our samples. For simplicity, we have listed just one protein from each of these pairs or groups of indistinguishable proteins in Tables 1 and 2. However, we list the identities of all proteins in these pairs or groups in Table S5.
Comparison to previously identified Ae. aegypti reproductive gland proteins
Of the 145 transferred proteins we identified using the whole-organism isotope labeling method, 123 are newly-recognized components of Ae. aegypti semen. The remaining 22 were previously identified as putative Sfps ([35]), and we demonstrate here that they are transferred to females during mating. Thirty-one additional proteins were identified as putative Sfps in our previous study ([35]), but we did not detect them as transferred in our current study. Those proteins may not be transferred to females, or may be transferred at quantities below our detection threshold or with post-translational modifications that render them unidentifiable by standard mass spectrometry. Of the 123 newly-recognized seminal proteins, 84 were previously identified from the reproductive glands of Ae. aegypti, however they were not designated as putative Sfps because they lacked predicted secretion signal sequences ([35]; L. Sirot, M. Wolfner, L. Harrington, unpubl. data).
Distinguishing seminal fluid proteins from sperm proteins
In order to distinguish Sfps from sperm proteins among those transferred to females, we conducted a proteomics analysis of sperm-enriched samples from the seminal vesicles (SVs) and testes of virgin males. Sperm-enriched samples were obtained by releasing sperm from these organs, pelleting the sperm by centrifugation, and washing them repeatedly, as in Dorus et al. [40] (see Methods for details). We found 101 proteins that overlapped between our sperm-enriched samples from seminal vesicles and our sperm-enriched samples from testes. Of these 101 putative sperm proteins, 52 were detected as transferred to females during mating, providing a high-confidence subset of putative Ae. aegypti sperm proteins (Table 2).
Of the 145 total transferred proteins (see section “Proteins transferred to females during mating”), 16 were isolated from only one of the sperm-enriched tissues (SV: 5; testes: 11) and therefore were considered SV- or testes-derived Sfps, respectively, although we recognize that these could be sperm proteins. Additionally, 77 of the transferred proteins did not overlap with either sperm-enriched sample. Together, the 5 SV-derived Sfps, 11 testes-derived Sfps, and 77 of the 145 total transferred proteins that did not overlap with the sperm-enriched samples comprise a total of 93 proteins assigned with high-confidence as Ae. aegypti Sfps (Table 1).
Seminal fluid proteins
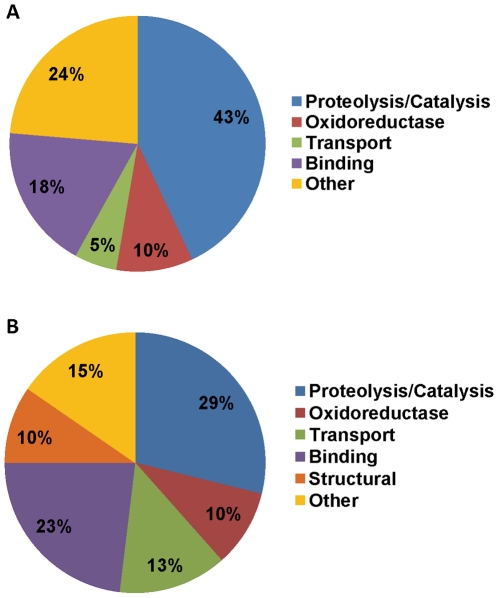
The Ae. aegypti Sfps identified represent a wide-range of predicted protein classes including proteolysis regulators, lectins, lipases, oxidoreductases, a cysteine-rich secretory protein (CRISP) and a venom allergen, and fall into a variety of Gene Ontology predicted molecular function classes (Table 1; Fig. 1A). Unlike Sfps in Drosophila ([31],[36],[50]) and An. gambiae ([33],[51]), in which some groups of Sfps tend to be spatially clustered, we found little evidence for spatial clustering of the 93 Sfp genes in the Ae. aegypti genome, with one exception. Four Sfp genes (AAEL006403; AAEL006414; AAEL006421; AAEL006429) clustered within a 23 kB region on supercontig 1.204. One gene in this region (AAEL006430) encodes a protein that was not detected in the present study and may either not be transferred or may be transferred at a level that was undetectable by our methods. The proteins encoded by all five of the genes in this region have predicted trypsin domains and their shared amino acid sequence identities range from 36 to 64%.
Figure 1. Gene Ontology molecular function categories of Aedes aegypti seminal fluid and putative sperm proteins.
A. Seminal fluid proteins; B. Putative sperm proteins.
As might be expected, the genes encoding the Sfps identified in this study tend to have highly male-biased expression when gene expression of whole males is compared to gene expression of whole females ([52]). Half (26 of 51) of the genes for which microarray data are available have significantly higher expression (at P≤0.001) in males than in non-blood-fed females (O. Marinotti, pers. comm.), as compared to a genome average of 16% (Chi-square test; X21 = 47.7; P<0.001; [52]). By comparison, 63% of the D. melanogaster Sfps identified by [36] have significantly higher expression (at P≤0.01) in males than in females as compared to a genome average of 12% (X21 = 345.9; P<0.001; [53]). Surprisingly, transcript levels of two Ae. aegypti Sfp-encoding genes (encoding a predicted kinase, AAEL012359, and a predicted zinc metalloprotease, AAEL012217) are significantly higher in non-blood-fed females than in males.
Sequence comparisons to other Diptera
Table S1 shows the extent to which homologs of the 93 putative Sfps can be detected in three other Dipteran genomes (Cx. quinquefasciatus, An. gambiae, and D. melanogaster). In comparison to known or predicted Sfp- or male AG-encoding genes from An. gambiae or D. melanogaster, Ae. aegypti Sfp genes generally had little sequence similarity. None of the An. gambiae homologs to Ae. aegypti Sfps is among the previously-identified An. gambiae AG genes ([33],[51]) or mating plug protein-encoding genes ([33]). Only two D. melanogaster (CG31704, and CG5162) homologs to Ae. aegypti Sfps (AaegSfp3 and AAEL005815, respectively; range of percent identity: 41–59%) are among the known D. melanogaster Sfps or AG genes (Table S1; [31],[31], [36],[54]). Additionally, three Ae. aegypti Sfps (AAEL005815; AAEL004112; AAEL000718) share sequence similarity (range: 30–84% identity) with four of the 13 AG genes identified from the Mediterranean fruit fly, Ceratitis capitata (clones 11c, 18a, 30c, 33a; [55]).
Below, we discuss (i) the unannotated, newly-identified predicted proteins from the 6-frame translation and small peptides database, (ii) new insights into the mode of Ae. aegypti Sfp secretion from Vectorbase identified proteins, and (iii) the potential biological functions of a subset of Sfps in modulating reproductive and physiological processes within the mated female. In our previous report on Ae. aegypti Sfps ([35]), we discussed the potential roles of these proteins in a number of processes including protein folding, antimicrobial activity, and sperm utilization by females. Although the proteins we have identified in the current report include many in the same classes and predicted functions as our previous report (and include 22 of the same proteins), we have specifically chosen to highlight proteins that suggest functions in the mated female that were not discussed in our previous report ([35]). These protein classes are not necessarily the most highly represented of the predicted Sfps.
Unannotated seminal fluid proteins
Based on comparisons to the 6-frame translation and small peptides databases, we discovered 14 previously unannotated predicted Sfps (Table 1). Of the 9 predicted proteins from the 6-frame translation, only one (AaegSfp2) had a significant hit to the SMART or PFAM database. This protein includes a predicted secretion signal sequence and a Kazal-type serine protease inhibitor domain. Interestingly, another predicted secreted protein of the 9 hits (AaegSfp8) shared a high degree of sequence similarity (70% identity; E value = 2e−74) with one of the Vectorbase predicted proteins, AAEL010824. AAEL010824 and AaegSfp8 are located on the same contig within 34 Kb of each other. Two of the other hits from the 6-frame database also had predicted secretion signal sequences. Of the 5 hits to the small peptides database, we found one predicted secreted Kazal-type serine protease inhibitor (AaegSfp3), one predicted secreted odorant-binding protein (AaegSfp1), and three hits with no predicted secretion signal sequence and no predicted protein domains.
Mode of secretion
Sixty-two of the Sfps that we identified are predicted intracellular or membrane-bound proteins (e.g., ATPases, dipeptidyl peptidase, gamma glutamyl transpeptidase, glutathione S-transferase, angiotensin converting enzyme). Predicted intracellular proteins also have been reported in the seminal fluid of other organisms including bed bugs ([56]), honey bees ([57]), and humans ([58], [59]), and in the AGs and seminal fluids of D. melanogaster ([60]; G. Findlay & W. Swanson, pers. comm.). In bed bugs, Sfps of predicted intracellular origin have been considered potential cell contaminants ([56]); whereas, in honeybees, it has been suggested that these proteins may be secreted through non-standard secretion routes ([57]). In Ae. aegypti, our finding of intracellular and membrane-bound Sfps is consistent with the hypothesized modes of secretion in the AGs of this species: based on cytological studies using light and electron microscopy, cells in the anterior portion of the glands are thought to secrete proteins by pinching off apically (“apocrine secretion”; [39],[61]), whereas cells in the posterior portion of the glands are thought to secrete proteins through granules and/or via rupture of the cell membrane (“holocrine secretion”; [61],[62]; but see [39]). Inside the female reproductive tract, the ejaculate contains “vacuoles” that grow over time after mating and then disappear by 24 h post-mating ([62]). Four of the Sfps we identified are subunits of the membrane-bound vacuolar-type proton ATPase and thus may be part of these vacuoles. Future studies could investigate whether the male-derived vacuolar ATPase is functional in the ejaculate vacuole membrane in the female reproductive tract and therefore may play a role in regulating the release of the contents of the vacuoles.
Importantly, our results emphasize that studies of Sfps in other species should not exclude proteins whose sequence (alone) suggest that they are intracellular and membrane-bound proteins. For species in which intracellular and/or membrane-bound proteins are found in the seminal fluid (e.g., bed bugs and honey bees), further research should be conducted on the mode of secretion of the male reproductive gland cells. Furthermore, secretion by Ae. aegypti accessory glands could serve as a model for secretion in systems such as the male reproductive glands of mammals, since mammalian prostasomes, exosomes, and epididymisomes contain many of the same classes as found among Ae. aegypti Sfps ([63]).
Proteolysis
Across a wide range of organisms, seminal fluid is rich in proteolysis regulators (e.g., [37],[38],[64]–[67]). Ae. aegypti is no exception. Thirteen of the 93 Sfps we identified in Ae.aegypti are predicted regulators of proteolysis. These include predicted trypsins, a zinc carboxypeptidase, a metalloprotease, a serine protease inhibitor (serpin), an angiotensin converting enzyme, and a proprotein convertase in the subtilisin/kexin type 4 (PCSK4) family (Tables 1 and S1). The predicted or known functions of seminal fluid proteolysis regulators include regulating the liquefaction of semen and/or mucus in the female reproductive tract ([68],[69]); protection of sperm from premature acrosome reactions ([67]); and activation and/or degradation of other reproductive proteins ([70]). We have previously discussed the potential role of predicted proteolysis regulators in Ae. aegypti seminal fluid ([35]). Here, we highlight the novel finding of a predicted proprotein convertase subtilisin/kexin type 4 (PCSK4) protein in insect seminal fluid.
PCSK4s can activate precursors of membrane receptors, peptide hormones, antibacterial peptides and neuropeptides through proteolytic processing ([71],[72]). Interestingly, the predicted PCSK4 (AAEL010725) found in this study shares close sequence similarity (56% identity; E-value: 2e-158) with the Ae. aegypti protein that processes vitellogenin (AAEL003652; [72]). To our knowledge, proprotein convertases have not been reported previously in insect seminal fluid. Transcripts of the D. melanogaster gene encoding one protein in this class (CG10702) are enriched (relative to whole body) in the AGs ([73]). The protein they encode contains a predicted secretion signal sequence (SignalP; [41]), but it has not yet been detected in the seminal fluid in this species ([36]).
Steroidogenesis
One of the Sfps we identified (AAEL009760) is a predicted sterol carrier in the Niemann-Pick type C-2 (NPC2) family. In insects, sterol carriers are essential for the production of ecdysteroids (ECDs) (e.g., [74]). ECDs are hormones that influence molting, gametogenesis, vitellogenesis, and other reproductive processes (reviewed in [75],[76]). In Ae. aegypti, ECDs are essential components of a signaling cascade linking blood meal intake with vitellogenesis (e.g., [77]; reviewed in [78], [79]). Although a role for the male-derived NCP2-like protein has not yet been determined in Ae. aegypti, findings in other Dipteran species suggest that male AGs can not only synthesize ECD (An. gambiae, [80]) but also stimulate ECD production in females (D. melanogaster, [81]). We suggest that AAEL009760 could potentially contribute to the regulation of ECD biosynthesis in Ae. aegypti male reproductive tracts and/or in mated females.
Sperm and sperm-associated proteins
From Ae. aegypti sperm-enriched tissue samples, we identified 101 sperm or sperm-associated proteins. Fifty-two were found among proteins transferred to females during mating (Table 2); the remaining 49 proteins were not detected as transferred (Table S6). These latter 49 proteins included 7 that have homologs found in the D. melanogaster sperm proteome ([40]). It is possible that some of these 49 Ae. aegypti proteins are components of somatic cells of the testis and/or SV tissues and play a role in spermatogenesis or sperm maintenance, whereas others could be sperm proteins whose abundance was too low for us to detect in the transferred samples or whose post-translational modifications rendered them unidentifiable by standard mass spectrometry. In the remainder of this section, we will only discuss the 52 proteins detected as transferred (hereafter referred to as “putative sperm proteins”).
Sequence comparisons to other Diptera
Table S2 shows the extent to which homologs of the 52 putative sperm proteins can be detected in three other Dipteran genomes (Cx. quinquefasciatus, An. gambiae, and D. melanogaster). Of the 30 proteins with D. melanogaster homologs, 17 (57%) of the homologs were found among the identified sperm proteins of D. melanogaster ([40]). This level of homology between Ae. aegypti putative sperm proteins and D. melanogaster sperm proteins suggests that the sperm-related functions of these proteins are conserved between the two species. Non-sperm D. melanogaster homologs of Ae. aegypti putative sperm proteins may also serve reproductive functions, as suggested by gene expression patterns. Of the 13 D. melanogaster homologs not found in the D. melanogaster sperm proteome (Table S2), transcripts of 5 are enriched in male and female reproductive tissues, transcripts of 1 are enriched only in male reproductive tissues, and transcripts of 6 are enriched only in female reproductive tissues ([73]).
Unnannotated sperm proteins
We discovered three previously unannotated predicted sperm proteins from the 6-frame translation database (Table 2). One predicted protein from the 6-frame translation, AaegSp1, contained a predicted secretion signal sequence and a Kazal serine protease inhibitor domain. The other two hits contained no predicted secretion signal sequence and no conserved protein domains.
Potential functions
The likely biological functions of putative Ae. aegypti sperm proteins include spermatogenesis, serving as structural components of mature sperm, and sperm locomotion (Fig. 1B). Spermatogenesis-related proteins that we found among Ae. aegypti putative sperm proteins include the heat shock protein 70, actin, and tubulin (α- and β-chains). Ae. aegypti putative sperm proteins that might contribute to sperm structure or motility include actin, tubulins, dyneins, ATP synthases, protein kinases and kinesin motor proteins ([82]–[86]). Ae. aegypti putative sperm proteins that are predicted mitochondrial enzymes include malate dehydrogenase, aconitase, cytochrome c oxidase, and ubiquinol-cytochrome c reductase. In other animals, these proteins generate the energy necessary for sperm locomotion via oxidative phosphorylation and the citric acid cycle ([87]–[92]).
Summary and conclusion
Secretions of the reproductive glands of male Ae. aegypti have previously been shown to induce post-mating changes in female reproductive and feeding behavior ([22],[23],[26]). In order to lay the groundwork for identifying specific proteins causing these effects, we report here 145 male-derived proteins that are transferred to females during mating in Ae. aegypti. We distinguished 93 seminal fluid proteins from 52 predicted sperm proteins, thus contributing to the growing understanding of insect ejaculate proteomes ([33],[35],[36],[40],[57],[88],[93]–[95]). Twenty-two of these proteins were previously identified as male reproductive gland proteins ([35]), and we demonstrate here that they are transferred to the female.
The Sfps identified in this study suggest roles in protein activation/inactivation, ecdysteroidogenesis, and sperm utilization. Furthermore, our discovery that many predicted intracellular and membrane-bound proteins are transferred to females in the seminal fluid indicates that findings of such proteins in the seminal fluid of other species (e.g., [56],[57]) may also result from apocrine and/or holocrine secretion from the male reproductive glands ([39],[61]). The putative sperm proteins of Ae. aegypti show sequence conservation within Diptera and 17 of their D. melanogaster homologs are sperm proteins in that species ([40]) indicating potential conservation of sperm-related functions.
Genes encoding Sfps showed higher male-biased expression than the genome average. On the one hand, this is not unexpected because Sfps are made in the male reproductive tract and are then transferred to females. On the other hand, it is not necessarily predicted a priori that Sfp-encoding genes will be male-biased in their expression, and the way we identified the proteins was without bias regarding their genes' expression. That 49% of the Ae. aegypti Sfp-encoding genes for which there are microarray data are not male-biased in expression will be important to bear in mind in designing future screens for Sfps.
Together, our results provide a foundation for functional analyses to associate individual Sfps with their function in the mated female. Once functions are identified for individual proteins, investigations of the pathways by which they induce effects on male and female reproductive biology could identify novel targets for control of Ae. aegypti and dengue transmission. Of particular interest is to determine how specific Sfps modulate female behavior and physiology (e.g., egg production and blood feeding) and to investigate candidate genes which increase the reproductive success of male Ae. aegypti that are to be used in genetic control strategies.
Supporting Information
Predicted seminal fluid proteins transferred in Aedes aegypti ejaculate
(0.18 MB DOC)
Predicted sperm proteins transferred in Aedes aegypti ejaculate
(0.11 MB DOC)
Proteins identified from unlabeled unmated Aedes aegypti female samplea
(0.07 MB DOC)
Amino acid sequences of unannotated predicted sperm and seminal fluid proteins from Aedes aegypti
(0.07 MB DOC)
Aedes aegypti seminal fluid proteins and sperm proteins indistinguishable by peptides identified through mass spectrometry
(0.05 MB DOC)
Putative Aedes aegypti sperm proteins that were not detected as transferred to females during mating
(0.08 MB DOC)
Acknowledgments
We thank D. Severson, University of Notre Dame, for providing us with material to start the Liverpool colony. We thank O. Marinotti for providing insights and sharing data regarding gene expression patterns of the predicted proteins we report. We thank G. Findlay and W. Swanson for helpful discussions regarding experimental design and data interpretation. L. Cator, G. Findlay, E. Kelleher, M. Sirot, and three anonymous reviewers provided insightful feedback on an earlier draft of this manuscript. We thank S. Zhang, S. Baumgart, R. Sherwood, W. Chen, and A. Ptak of the Cornell University Life Sciences Core Laboratories Center, and S. Pitcher for technical help.
Footnotes
The authors have declared that no competing interests exist.
This work was funded in part by grant R21AI0176828 from National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by NYC-139437 HATCH project to LCH and MFW. LKS was supported in part by a National Institutes of Health National Research Service Award fellowship F32GM074361. JMCR was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gillott C. Male accessory gland secretions: Modulators of female reproductive physiology and behavior. Annu Rev Entomol. 2003;48:163–184. doi: 10.1146/annurev.ento.48.091801.112657. [DOI] [PubMed] [Google Scholar]
- 2.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annu Rev Entomol. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 4.Clements DE, Coller BAG, Lieberman MM, Ogata S, Wang G, et al. Development of a recombinant tetravalent dengue virus vaccine: Immunogenicity and efficacy studies in mice and monkeys. Vaccine. 2010;28:2705–2715. doi: 10.1016/j.vaccine.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble CG, Chen YL, Dong HP, Gu F, Lim SP, et al. Strategies for development of dengue virus inhibitors. Antiviral Res. 2010;85:450–462. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillett JD. Behaviour differences in 2 strains of Aedes aegypti. Nature. 1955;176:124–125. doi: 10.1038/176124a0. [DOI] [PubMed] [Google Scholar]
- 8.Wallis RC, Lang CA. Egg formation and oviposition in blood-fed Aedes aegypti L. Mosq News. 1956;16:283–286. [Google Scholar]
- 9.Downe AER. Internal regulation of rate of digestion of blood meals in mosquito, Aedes aegypti. J Insect Physiol. 1975;21:1835–1839. doi: 10.1016/0022-1910(75)90250-4. [DOI] [PubMed] [Google Scholar]
- 10.Houseman JG, Downe AER. Methods of measuring blood meal size and proteinase activity for determining effects of mated state on digestive processes of female Aedes aegypti (L) (Diptera, Culicidae). Can Entomol. 1986;118:241–248. [Google Scholar]
- 11.Gwadz RW, Craig GB, Jr, Hickey WA. Female sexual behavior as the mechanism rendering Aedes aegypti refractory to insemination. Biol Bull Mar Biol Lab Woods Hole. 1971;140:210–214. doi: 10.2307/1540069. [DOI] [PubMed] [Google Scholar]
- 12.Taylor B, Jones MDR. Circadian rhythm of flight activity in mosquito Aedes aegypti (L) - phase-setting effects of light-on and light-off. J Exp Biol. 1969;51:59–70. doi: 10.1242/jeb.51.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Jones MDR. The programming of circadian flight-activity in relation to mating and the gonotrophic cycle in the mosquito, Aedes aegypti. Physiol Entomol. 1981;6:307–313. [Google Scholar]
- 14.Fernandez NM, Klowden MJ. Male accessory-gland substances modify the host-seeking behavior of gravid Aedes aegypti mosquitoes. J Insect Physiol. 1995;41:965–970. [Google Scholar]
- 15.Klowden MJ, Lea AO. Humoral inhibition of host-seeking in Aedes aegypti during oocyte maturation. J Insect Physiol. 1979;25:231–235. doi: 10.1016/0022-1910(79)90048-9. [DOI] [PubMed] [Google Scholar]
- 16.Lavoipierre MMJ. Biting behaviour of mated and unmated females of an African strain of Aedes aegypti. Nature. 1958;181:1781–1782. doi: 10.1038/1811781a0. [DOI] [PubMed] [Google Scholar]
- 17.Lavoipierre MMJ. Presence of a factor inhibiting biting in Aedes aegypti. Nature. 1958;182:1567–1568. doi: 10.1038/1821567a0. [DOI] [PubMed] [Google Scholar]
- 18.Judson CL. Feeding and oviposition behavior in mosquito Aedes aegypti (L) .i. Preliminary studies of physiological control mechanisms. Biol Bull. 1967;133:369–377. doi: 10.2307/1539832. [DOI] [PubMed] [Google Scholar]
- 19.Adlakha V, Pillai MKK. Involvement of male accessory gland substance in fertility of mosquitoes. J Insect Physiol. 1975;21:1453–1455. doi: 10.1016/0022-1910(75)90207-3. [DOI] [PubMed] [Google Scholar]
- 20.Borovsky D, Carlson DA, Hancock RG, Rembold H, Vanhandel E. De-novo biosynthesis of juvenile hormone-III and hormone-I by the accessory glands of the male mosquito. Insect Biochem Molec. 1994;24:437–444. doi: 10.1016/0965-1748(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 21.Craig GB. Mosquitoes - female monogamy induced by male accessory gland substance. Science. 1967;156:1499–1501. doi: 10.1126/science.156.3781.1499. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs MS, Craig GB, Despommi DD. Protein nature of substance inducing female monogamy in Aedes aegypti. J Insect Physiol. 1969;15:701–709. [Google Scholar]
- 23.Hiss EA, Fuchs MS. Effect of matrone on oviposition in mosquito, Aedes aegypti. J Insect Physiol. 1972;18:2217–2227. doi: 10.1016/0022-1910(72)90250-8. [DOI] [PubMed] [Google Scholar]
- 24.Klowden MJ, Chambers GM. Male accessory-gland substances activate egg development in nutritionally stressed Aedes aegypti mosquitoes. J Insect Physiol. 1991;37:721–726. [Google Scholar]
- 25.Leahy MG, Craig GB. Accessory gland substance as a stimulant for oviposition in Aedes aegypti and Aedes albopictus. Mosq News. 1965;25:448–452. [Google Scholar]
- 26.Lee JJ, Klowden MJ. A male accessory gland protein that modulates female mosquito (Diptera : Culicidae) host-seeking behavior. J Am Mosq Control Assoc. 1999;15:4–7. [PubMed] [Google Scholar]
- 27.Ramalingam S, Craig GB. Functions of male accessory-gland secretions of Aedes mosquitoes (Diptera-Culicidae) - transplantation studies. Can Entomol. 1976;108:955–960. [Google Scholar]
- 28.Shutt B, Stables L, Aboagye-Antwi F, Moran J, Tripet F. Male accessory gland proteins induce female monogamy in Anopheline mosquitoes. Med Vet Entomol. 2010;24:91–94. doi: 10.1111/j.1365-2915.2009.00849.x. [DOI] [PubMed] [Google Scholar]
- 29.Chapman T. The soup in my fly: Evolution, form and function of seminal fluid proteins. PLoS Biol. 2008;6:1379–1382. doi: 10.1371/journal.pbio.0060179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isaac RE, Li CX, Leedale AE, Shirras AD. Drosophila male sex peptide inhibits sleep and promotes locomotor activity in the post-mated female. P Roy Soc B-Biol Sci. 2010;277:65–70. doi: 10.1098/rspb.2009.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravi Ram K, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47:427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- 32.Sirot LK, LaFlamme BA, Sitnik J, Rubinstein CD, Avila FW, et al. Molecular social interactions: Drosophila seminal fluid proteins as a case study. Adv Genet. 2009;68:23–56. doi: 10.1016/S0065-2660(09)68002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers DW, Baldini F, Battaglia F, Panico M, Dell A, et al. Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PloS Biol. 2009;7 doi: 10.1371/journal.pbio.1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue L, Noll M. Drosophila female sexual behavior induced by sterile males showing copulation complementation. P Natl Acad Sci USA. 2000;97:3272–3275. doi: 10.1073/pnas.060018897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirot LK, Poulson RL, McKenna MC, Girnary H, Wolfner MF, et al. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: Potential tools for control of female feeding and reproduction. Insect Biochem Molec Biol. 2008;38:176–189. doi: 10.1016/j.ibmb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Findlay GD, Yi XH, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:1417–1426. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller JL, Ripoll DR, Aquadro CF, Wolfner MF. Comparative structural modeling and inference of conserved protein classes in Drosophila seminal fluid. P Natl Acad Sci USA. 2004;101:13542–13547. doi: 10.1073/pnas.0405579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poiani A. Complexity of seminal fluid: a review. Behav Ecol Sociobiol. 2006;60:289–310. [Google Scholar]
- 39.Dapples CC, Foster WA, Leaa AO. Ultrastructure of the accessory gland of the male mosquito, Aedes aegypti (L.) (Diptera: Culicidae). Int J Insect Morphol Embryol. 1974;3:279–291. [Google Scholar]
- 40.Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, et al. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–1445. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- 41.Emanuelsson SB, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 42.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. P Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D11–D22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, et al. Exponentially modified protein abundance index (emPAI) for estimates of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Blanco E, Parra G, Guigo R. Using GeneID to identify genes. In: Baxevanis A, editor. Current Protocols in Bioinformatics. New York: John Wiley & Sons Inc; 2002. [Google Scholar]
- 46.Severson DW, deBruyn B, Lovin DD, Brown SE, Knudson DL, et al. Comparative genome analysis of the yellow fever mosquito Aedes aegypti with Drosophila melanogaster and the malaria vector mosquito Anopheles gambiae. J Hered. 2004;95:103–113. doi: 10.1093/jhered/esh023. [DOI] [PubMed] [Google Scholar]
- 47.Reidenbach KR, Cook S, Betrone MA, Harbach RE, Wiegmann BM, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, et al. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- 49.Findlay GD, Swanson WJ. Proteomics enhances evolutionary and functional analysis of reproductive proteins. Bioessays. 2010;32:26–36. doi: 10.1002/bies.200900127. [DOI] [PubMed] [Google Scholar]
- 50.Holloway AK, Begun DJ. Molecular evolution and population genetics of duplicated accessory gland protein genes in Drosophila. Mol Biol Evol. 2004;21:1625–1628. doi: 10.1093/molbev/msh195. [DOI] [PubMed] [Google Scholar]
- 51.Dottorini T, Nicolaides L, Ranson H, Rogers DW, Crisanti A, et al. A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. P Natl Acad Sci USA. 2007;104:16215–16220. doi: 10.1073/pnas.0703904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dissanayake S, Ribeiro JMC, Wang MH, Dunn WA, Yan G, et al. aeGEPUCI: a database of gene experession in the dengue vector mosquito, Aedes aegypti. BMC Res Not. 2010;3:248. doi: 10.1186/1756-0500-3-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takemori N, Yamamoto MT. Proteome mapping of the Drosophila melanogaster male reproductive system. Proteomics. 2009;9:2484–2493. doi: 10.1002/pmic.200800795. [DOI] [PubMed] [Google Scholar]
- 55.Davies SJ, Chapman T. Identification of genes expressed in the accessory glands of male Mediterranean fruit flies (Ceratitis capitata). Insect Biochem Molec Biol. 2006;36:846–856. doi: 10.1016/j.ibmb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 56.Reinhardt K, Naylor RA, Siva-Jothy MT. Ejaculate components delay reproductive senescence while elevating female reproductive rate in an insect. P Natl Acad Sci USA. 2009;106:21743–21747. doi: 10.1073/pnas.0905347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baer B, Heazlewood JL, Taylor NL, Eubel H, Millar AH. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics. 2009;9:2085–2097. doi: 10.1002/pmic.200800708. [DOI] [PubMed] [Google Scholar]
- 58.Fung KYC, Glode LM, Green S, Duncan MW. A comprehensive characterization of the peptide and protein constituents of human seminal fluid. Prostate. 2004;61:171–181. doi: 10.1002/pros.20089. [DOI] [PubMed] [Google Scholar]
- 59.Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7 doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, et al. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a pro-cathepsin and a soluble gamma-glutamyl transpeptidase. Proteome Sci. 2006;4:9. doi: 10.1186/1477-5956-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramalingam S. Secretion in the male accessory-glands of Aedes aegypti (l) (Diptera, Culicidae). Int J Insect Morphol Embryol. 1983;12:87–96. [Google Scholar]
- 62.Jones JC, Wheeler RE. Studies on spermathecal filling in Aedes aegypti (Linnaeus) .I. Description. Biol Bull. 1965;129:134–150. doi: 10.2307/1539731. [DOI] [PubMed] [Google Scholar]
- 63.Hermo L, Jacks D. Nature's ingenuity: Bypassing the classical secretory route via apocrine secretion. Mol Reprod Dev. 2002;63:394–410. doi: 10.1002/mrd.90023. [DOI] [PubMed] [Google Scholar]
- 64.Dean MD, Clark NL, Findlay GD, Karn RC, Yi XH, et al. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus). Mol Biol Evol. 2009;26:1733–1743. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braswell WE, Andres JA, Maroja LS, Harrison RG, Howard DJ, et al. Identification and comparative analysis of accessory gland proteins in Orthoptera. Genome. 2006;49:1069–1080. doi: 10.1139/g06-061. [DOI] [PubMed] [Google Scholar]
- 66.Veselsky L, Cechova D. Distribution of acrosin inhibitors in bull reproductive tissues and spermatozoa. Hoppe Seylers Z Physiol Chem. 1980;361:715–722. doi: 10.1515/bchm2.1980.361.1.715. [DOI] [PubMed] [Google Scholar]
- 67.Wojtczak M, Dietrich GJ, Ciereszko A. Transferrin and antiproteases are major proteins of common carp seminal plasma. Fish Shellfish Immunol. 2005;19:387–391. doi: 10.1016/j.fsi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 68.Robert M, Gibbs BF, Jacobson E, Gagnon C. Characterization of prostate-specific antigen proteolytic activity on its major physiological substrate, the sperm motility inhibitor precursor semenogelin I. Biochemistry US. 1997;36:3811–3819. doi: 10.1021/bi9626158. [DOI] [PubMed] [Google Scholar]
- 69.Malm J, Hellman J, Hogg P, Lilja H. Enzymatic action of prostate-specific antigen (PSA or hK3): Substrate specificity and regulation by Zn2+, a tight-binding inhibitor. Prostate. 2000;45:132–139. doi: 10.1002/1097-0045(20001001)45:2<132::aid-pros7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 70.Ravi Ram K, Sirot LK, Wolfner MF. Predicted seminal astacin-like protease is required for processing of reproductive proteins in Drosophila melanogaster. P Natl Acad Sci USA. 2006;103:18674–18679. doi: 10.1073/pnas.0606228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barr PJ. Mammalian subtilisins - the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- 72.Chen JS, Raikhel AS. Subunit cleavage of mosquito pro-vitellogenin by a subtilisin-like convertase. P Natl Acad Sci USA. 1996;93:6186–6190. doi: 10.1073/pnas.93.12.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 74.Huang X, Warren JT, Buchanan JA, Gilbert LI, Scott MP. Drosophila Niemann-Pick Type C-2 genes control sterol homeostasis and steroid biosynthesis: a model of human neurodegenerative disease. Development. 2007;134:3733–3742. doi: 10.1242/dev.004572. [DOI] [PubMed] [Google Scholar]
- 75.Clements AN. The Biology of Mosquitoes. London: Chapman & Hall; 2000. 752 [Google Scholar]
- 76.Brown MR, Sieglaff DH, Rees HH. Gonadal ecdysteroidogenesis in Arthropoda: occurrence and regulation. Annu Rev Entomol. 2009;54:105–125. doi: 10.1146/annurev.ento.53.103106.093334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fallon AM, Hagedorn HH, Wyatt GR, Laufer H. Activation of vitellogenin synthesis in mosquito Aedes aegypti by ecydsone. J Insect Physiol. 1974;20:1815–1823. doi: 10.1016/0022-1910(74)90211-x. [DOI] [PubMed] [Google Scholar]
- 78.Klowden MJ. Endocrine aspects of mosquito reproduction. Arch Insect Biochem Physiol. 1997;35:491–512. [Google Scholar]
- 79.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem Molec. 2005;35:661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 80.Pondeville E, Maria A, Jacques JC, Bourgouin C, Dauphin-Villemant C. Anopheles gambiae males produce and transfer the vitellogenic steroid hormone 20-hydroxyecdysone to females during mating. P Natl Acad Sci USA. 2008;105:19631–19636. doi: 10.1073/pnas.0809264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harshman LG, Loeb AM, Johnson BA. Ecdysteroid titers in mated and unmated Drosophila melanogaster females. J Insect Physiol. 1999;45:571–577. doi: 10.1016/s0022-1910(99)00038-4. [DOI] [PubMed] [Google Scholar]
- 82.Baccetti B. Insect sperm cells. Adv Insect Physiol. 1972;9:315–397. [Google Scholar]
- 83.Cummins J. Sperm motility and energetics. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Burlington, MA: Elsevier Ltd; 2009. pp. 185–206. [Google Scholar]
- 84.Dallai R, Bellon PL, Lanzavecchia S, Afzelius BA. The Dipteran sperm tail: ultrastructural characteristics and phylogenetic considerations. Zool Scr. 1993;22:193–202. [Google Scholar]
- 85.Dorus S, Karr TL. Sperm proteomics and genomics. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Burlington, MA: Elsevier Ltd; 2009. pp. 435–469. [Google Scholar]
- 86.Werner M, Simmons LW. Insect sperm motility. Biol Rev. 2008;83:191–208. doi: 10.1111/j.1469-185X.2008.00039.x. [DOI] [PubMed] [Google Scholar]
- 87.Blum MS, Taber S. Chemistry of the drone honey bee reproductive system - III. Dehydrogenases in washed spermatozoa. J Insect Physiol. 1965;11:1489–1501. [Google Scholar]
- 88.Collins AM, Caperna TJ, Williams V, Garrett WM, Evans JD. Proteomic analyses of male contributions to honey bee sperm storage and mating. Insect Molec Biol. 2006;15:541–549. doi: 10.1111/j.1365-2583.2006.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karr TL. Fruit flies and the sperm proteome. Hum Mol Genet. 2007;16:R124–R133. doi: 10.1093/hmg/ddm252. [DOI] [PubMed] [Google Scholar]
- 90.Mann T. Studies on the metabolism of semen. 1. General aspects, occurrence and distribution of cytochrome, certain enzymes and coenzymes. Biochem J. 1945;39:451–458. [PMC free article] [PubMed] [Google Scholar]
- 91.Mann T. Studies on the metabolism of semen. 2. Glycolysis in spermatozoa. Biochem J. 1945b;39:458–465. [PMC free article] [PubMed] [Google Scholar]
- 92.Ruiz-Pesini E, Diez C, Lapen AC, Perez-Martos A, Montoya J, et al. Correlation of sperm motility with mitochondrial enzymatic activities. Clin Chem. 1998;44:1616–1620. [PubMed] [Google Scholar]
- 93.Andres JA, Maroja LS, Harrison RG. Searching for candidate speciation genes using a proteomic approach: seminal proteins in field crickets. P Roy Soc B-Biol Sci. 2008;275:1975–1983. doi: 10.1098/rspb.2008.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Findlay GD, MacCoss MJ, Swanson WJ. Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Res. 2009;19:886–896. doi: 10.1101/gr.089391.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walters JR, Harrison RG. Combined EST and proteomic analysis identifies rapidly evolving seminal fluid proteins in Heliconius butterflies. Molec Biol Evol. 2010;27:2000–2013. doi: 10.1093/molbev/msq092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted seminal fluid proteins transferred in Aedes aegypti ejaculate
(0.18 MB DOC)
Predicted sperm proteins transferred in Aedes aegypti ejaculate
(0.11 MB DOC)
Proteins identified from unlabeled unmated Aedes aegypti female samplea
(0.07 MB DOC)
Amino acid sequences of unannotated predicted sperm and seminal fluid proteins from Aedes aegypti
(0.07 MB DOC)
Aedes aegypti seminal fluid proteins and sperm proteins indistinguishable by peptides identified through mass spectrometry
(0.05 MB DOC)
Putative Aedes aegypti sperm proteins that were not detected as transferred to females during mating
(0.08 MB DOC)