Abstract
Adoptive immunotherapy is evolving to assume an increasing role in treating cancer. Most imaging studies in adoptive immunotherapy to date have focused primarily on locating tumor-specific T-cells rather than understanding their effector functions. In this study, we report the development of a non-invasive imaging strategy to monitor T-cell activation in living subjects by linking a reporter gene to the Granzyme B promoter (pGB), whose transcriptional activity is known to increase during T-cell activation. Because pGB is relatively weak and does not lead to sufficient reporter gene expression for non-invasive imaging, we specifically employed two signal amplification strategies, namely the Two Step Transcription Amplification (TSTA) strategy and the CMV enhancer (CMVe) strategy, to maximize firefly luciferase reporter gene expression. While both amplification strategies were capable of increasing pGB activity in activated primary murine splenocytes, only the level of bioluminescence activity achieved with the CMVe strategy was adequate for non-invasive imaging in mice. Using T-cells transduced with a reporter vector containing the hybrid pGB-CMVe promoter, we were able to optically image T-cell effector function longitudinally in response to tumor antigens in living mice. This methodology has the potential to accelerate the study of adoptive immunotherapy in preclinical cancer models.
Keywords: Bioluminescence imaging, Granzyme B, T-cell activation, Two step transcriptional amplification (TSTA), CMV enhancer
Introduction
Cellular immunity plays a key role in immunosurveillance of early cancers, prevention of relapse from minimal residual disease, and these roles are being exploited in the design of cancer vaccines (1) and adoptive immune cell therapies (2-4). To achieve full therapeutic potential for adoptively transferred T-cells, large numbers need to be injected into patients (5, 6). Current culture techniques needed to reach these numbers can cause negative effects on the functional characteristics of the modified T-cells, making them less effective in patients (5). Therefore, it is important to monitor the function of cytotoxic and/or helper T-cells prior to and after adoptive transfer.
Currently, cytotoxic T-cell function is measured through cell killing assays on target cells, whereas helper T-cells are analyzed for their cytokine production during exposure to target antigens. In living subjects, however, T-cell function is measured by therapeutic outcome (e.g., reduction in tumor volume). While tumor-specific T-cells may have great efficacy in CTL assays, they are often ineffective against target tumor cells when injected in living subjects (reviewed in(7)). Research into the cell culture conditions and re-targeting of T-cells using chimeric T-cell receptors has improved the efficacy of tumor-specific T-cells in living subjects. Recently Morgan et al.(8) found partial success in treating melanoma patients using re-targeted T-cells partly due to culturing retargeted T-cells for less than a week. Various strategies for improving the efficacy of immunoadoptive therapy such as modifications of cell culture conditions, alterations in T-cell repertoire, and identifying the cell population(s) responsible for tumor eradication would greatly benefit from an imaging tool to non-invasively visualize the function of transferred T-cells in living subjects.
In this study, we pursued this goal by linking the promoters of T-cell activation markers to reporter genes commonly employed for molecular imaging. Markers of T-cell activation such as interleukin-2 (IL-2) and Granzyme B are routinely used to gauge the activation status of T-cells in cell culture and ex vivo (9). The cytokine IL-2 is produced mainly by CD4+ T-cells in the early stages of activation. In contrast, the expression of Granzyme B in CD8+ T-cells signifies their full differentiation and acquisition of killing potential (10). The full-length Granzyme B promoter (~9kb) has been well characterized and contains in its distal end binding sites for the AP-1, CBF, and CRE transcription factors (11, 12) which are induced during T-cell activation (13) and can drive reporter gene expression during T-cell activation at a level sufficient for in vitro detection (12). By coupling this promoter to a bioluminescence reporter gene, we inferred that CTL function could be visualized in live small animals. However, our initial experiments showed the bioluminescence signal generated by this promoter to be too weak for detection in living animals.
Detection of reporter gene expression from tissue-specific promoters can indeed be difficult in living animals such as mice and rats (14, 15). Several methods have been reported to increase promoter activity in order to enhance the production of reporter or therapeutic proteins-- these include multimerizing promoters, using full length promoters, creating hybrid promoters, and using two step systems based on the yeast two hybrid system (14, 16). Hybrid promoters that use the CMV enhancer (CMVe), a strong transcriptional enhancer, in conjunction with the tissue-specific promoter (17, 18) and the Two Step Transcription Amplification strategy (TSTA) (see Supplementary Fig. S1 online) (14) have been used to increase reporter gene expression and enable visualization in small animals. These two methods are applied here to the Granzyme B promoter and we compare these two approaches for the ability to visualize T-cell effector function in small animals using bioluminescence imaging (BLI).
Materials and Methods
Animals
Mice were purchased from Jackson Laboratories (Bar Harbor, ME) and housed under pathogen free conditions at the Stanford Research Animal Facility. All animal procedures were approved by the Institutional Administrative Panel on Laboratory Animal Care at Stanford University (APLAC #9759).
Cells
The tumor cell line EL4 (C57BL/6, H2b, thymoma) and its derivative E.G7 (EL4 cells stably expressing chicken OVA cDNA) (19) purchased from ATCC (Manassas, VA).
Lentivirus production and transduction
High titer lentiviral vectors were produced using a modified version of the protocol presented in Zhang et al. (20) for details see Supplemental Methods.
Primary murine splenocytes from C57BL/6 and OT1 mice were first depleted of B-cells (only for cell culture studies) using Easy Sep Reagent (Stem Cell Tech, Vancouver, BC, Canada), then pre-stimulated for 36hrs with 2.5μg/ml Concanavalin A (Calbiochem, San Diego, CA) in cRPMI, 50μM βME, 10ng/ml IL-7 (R & D Systems, Minneapolis, MN). 5×106 cells were then transduced for 3-4hrs in 500ul OptiMEM + 8ug/ml polybrene (Sigma, St.Louis, Mo) in 12well tissue culture plates. Cells were then cultured in a total of 2.5ml/well cRPMI, 50uM βME, 10ng/ml IL-15 (R & D Systems, Minneapolis, MN), 10U/ml IL-2 (Chiron, Emeryville, CA), and 50mM α-MethylMannoside (Calbiochem, San Diego, CA) for 48hrs prior to further study. For in vitro studies, murine T-cells were activated with anti-CD3 and anti-CD28 antibodies (1μg/ml, BD Biosciences, San Jose, CA).
Adoptive transfer
48hrs post transduction cells were labeled with carboxyfluorescein succinimidyl ester (CFSE, Invitrogen) as previously described (21). 10×106 labeled T-cells in 200μl PBS were injected via the tail vein into congenic albino C57BL/6 mice bearing EL4 (non-target) and E.G7 (target) tumor cells (1×106 cells in the left and right shoulder respectively) inoculated subcutaneously six days prior to adoptive T-cell transfer.
Cell Isolation and Flow cytometry
E.G7 and EL4 tumors, spleens and lymph nodes were excised from mice. For E.G7 and EL4 tumors, single cell suspensions were created by dissociatiation of tumors with Liberase Blendzymes (Roche) as described (22) and the dissociated tumor cells were stained with various antibodies. Antibodies against CD4, CD8, CD69, Vα2 and Vβ5.1 were purchased from Biolegend (San Diego, CA). Samples were acquired on a FACSCalibur (BD Biosciences, San Jose CA). In Figures 2E and 2F Mean Fluorescence Intensity (MFI) for GFP was quantified by gating on either CD4+ or CD8+ cells. In Figures 3 and 4 cells were gated on CFSE and CD8 double positive cells. Data was analyzed using FlowJo Software (TreeStar, Ashland, OR).
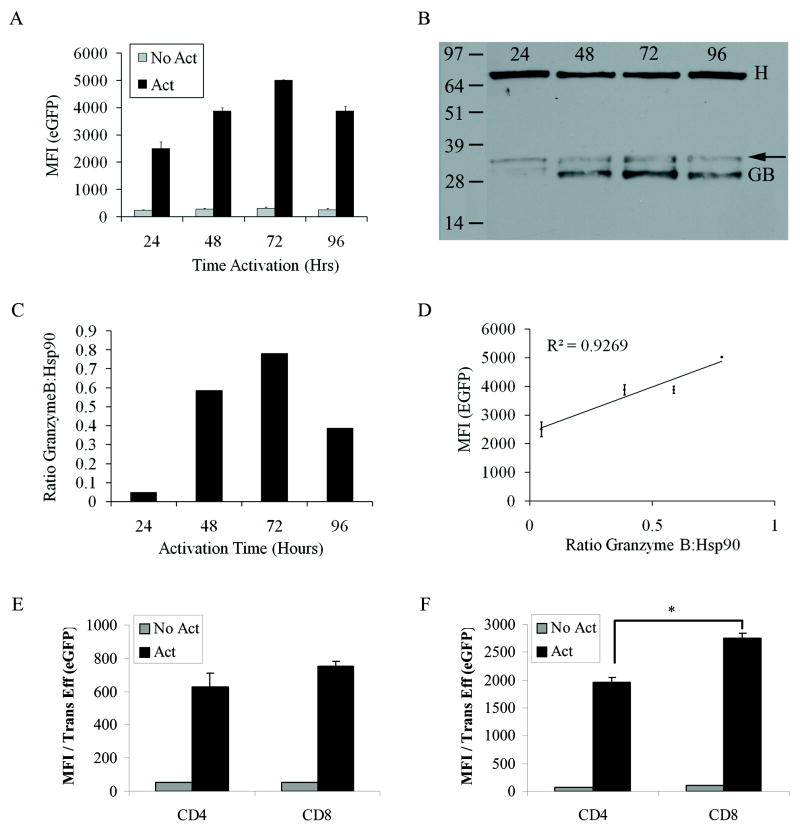
Figure 2.
Characterization of the hybrid pGBe promoter. A) eGFP mean fluorescence intensity (MFI) from activated or unactivated splenocytes transduced with a lentiviral vector containing pGBeG. B) Western blot of extracts from murine splenocytes transduced with pGBeG. T-cells were activated for the indicated period of time 48hrs after transduction. Cells were lysed and a total of 6ug of protein were loaded into lanes and then probed with antibodies to either Granzyme B (GB) or Hsp90 as a loading control (H). The arrow indicates other forms of GB which migrate at 31-33kDa due to differential glycosylation (52). C) Graphical analysis of the Western blot in (B). ROIs were drawn around each band and the signal intensities, calculated with ImageJ software, of each GB band was normalized to the intensities of the corresponding Hsp90 bands. D) MFIs are plotted against the protein ratios to correlate GB expression with reporter gene expression. E) and F) Expression of eGFP from splenocytes transduced with (E) pGBeG or (F) pGBTSTA-EGFP lentiviral vector (*p=0.005). Data was normalized to transduction efficiency (% positive GFP) of each vector in either CD4 or CD8 T-cells. Flowjo software was used to calculate MFI in A, D, E, and F. N=3, Student t test
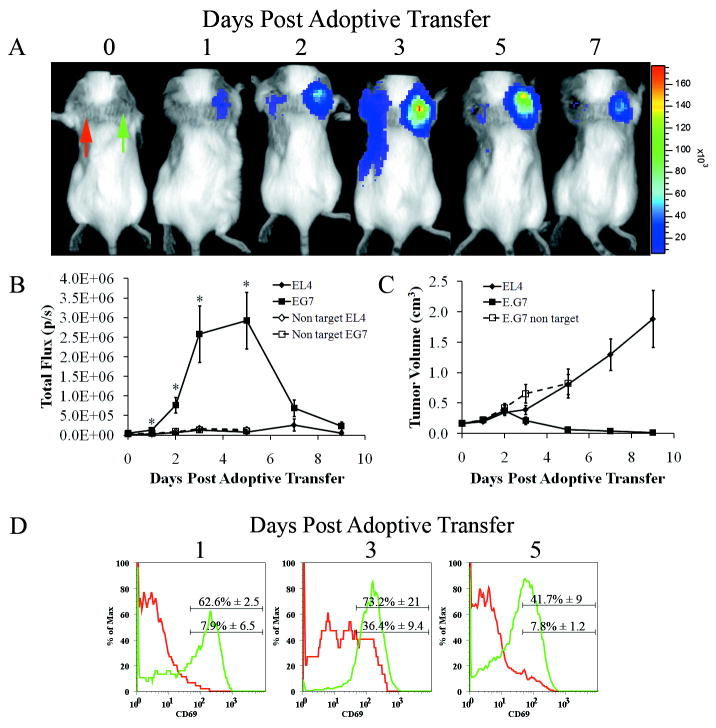
Figure 3.
Activation of pGB in T-cells occurs at the target site. A) Representative mouse after adoptive transfer with 10×106 OT1 CD8+ T cells transduced with a lentiviral vector containing pGBeLT imaged on the days indicated above the panel. The images are scaled to the reference color bar shown on the right, with red indicating the highest and purple the lowest signal. Red arrow (EL4) and green arrow (EG.7) B) Quantitative analysis of regions of interest (ROI) on EL4 and E.G7 tumors shown in (A) following transfer of OT1 T-cells (closed symbols) or non-targeted T-cells (open symbols) (N=5, Error Bars=SEM, *denotes statistical significance between EL4 and E.G7 using OT1 T-cells, p=0.009 (day 3) and 0.004 (day 5) using ANOVA with Bonferroni test. C) Tumor volumes of mice from (A). Tumor measurements started six days post tumor implantation, which corresponds to day 0 of adoptive transfer (N=5, Error Bars=SEM). D) At day 1, 3, and 5 days post adoptive transfer, tumors (EL4 (red line) and E.G7 (green line)) were removed and analyzed by flow cytometry for the presence of CD8+ T-cells and CD69 expression. In each graph the upper and lower numbers represent the percent CD69 positive cells in E.G7 and EL4 tumors respectively (N=2).
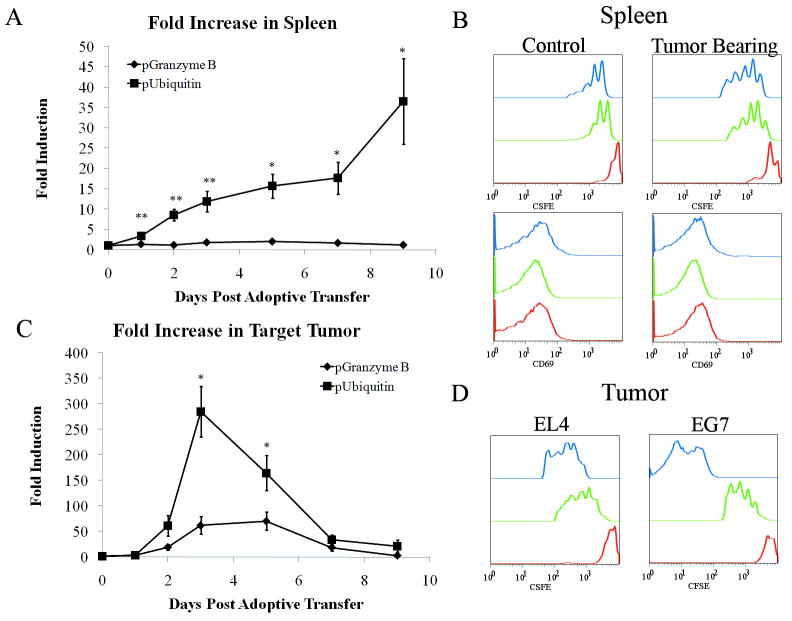
Figure 4.
Bioluminescence signal from the pGB is due to T-cell activation and not T-cell proliferation. A) ROI analysis of spleens from mice adoptively transferred with 10×106 OT1 CD8+ T-cells labeled with CFSE and transduced with lentiviral vectors containing pUbiLG (squares) or pGBeLT (diamonds). ROIs were drawn around the spleen and total flux calculated using Living Image Software. Flux measurements were then normalized by dividing with the average flux value from day 0 for each mouse to determine the fold induction and plotted on the graph. N=5 for each group, error bars=SEM, and *denotes statistical significance, p=0.002 (day 5), 0.004 (day 7) and 0.01 (day 9) using ANOVA with Bonferroni’s test. **Days 1, 2, and 3 were statistically different (p<0.005) when performing ANOVA, however multiple comparisons test did not show these time points to be statistically different. B) At 24hrs (red line), 72hrs (green line), and 120hrs (blue line) post adoptive transfer, spleens were analyzed by flow cytometry for the loss of CFSE (top panels) and CD69 expression (bottom panels). Non-tumor bearing mice served as controls (left panels) and received the same adoptive transfer. Data shown were gated on total CFSE labeled CD8+ T-cells and is representative of 2 animals at each time point. C) Quantitative analysis of bioluminescence signal from E.G7 tumors from adoptively transferred mice. ROI analysis was performed as described in (A) except ROIs were drawn over E.G7 tumors. N=5 for each group. Error bars=SEM, *denotes statistical significance, p=0.0028 (day 3) and p=0.04 (day 5) using ANOVA and Bonferroni tests. D) As in (B) tumors were removed, dispersed and analyzed by flow cytometry for the loss of CFSE in tumor infiltrating T-cells to determine T-cell division.
Statistical Analysis
Two-tailed student T tests were used to evaluate the significant difference or p values between samples in Figures 1 and 2. Standard deviations (SD) of mean values are depicted as error bars in Figures 1 and 2. ANOVA and the Bonferroni multiple comparison test (α=0.05) were used to evaluate significant difference in Figures 3 and 4. Standard error of mean (SEM) is depicted as error bars in Figures 3 and 4.
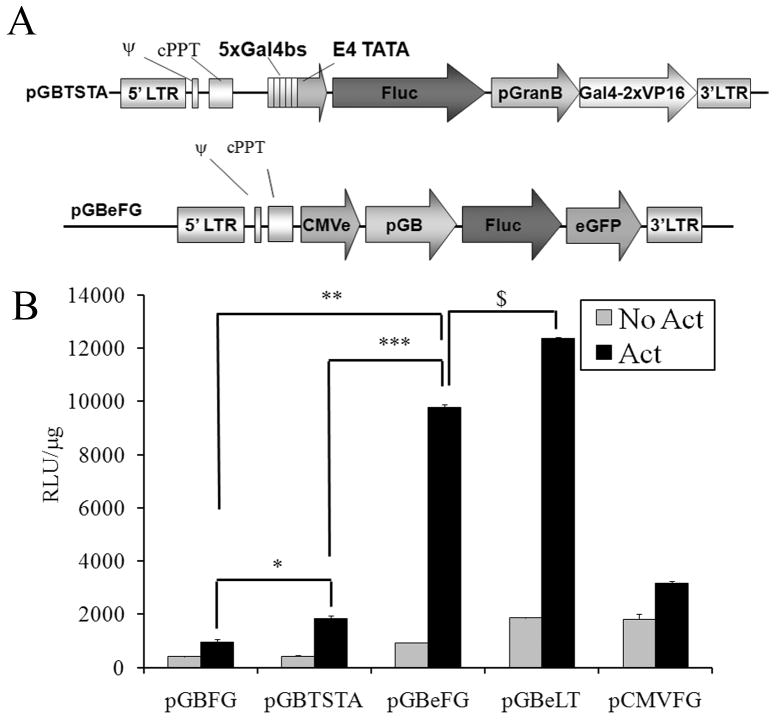
Figure 1.
CMVe coupled promoter significantly amplifies reporter gene expression compared to TSTA and full length CMV promoter in primary murine splenocytes. A) Map of pGBTSTA and pGBeFG lentiviral vectors. B) Splenocytes were transduced as described in Materials and Methods with lentiviral vectors carrying the 828bp Granzyme B promoter (pGB) driving expression of fluc-egfp fusion, pGB in the TSTA system (pGBTSTA) driving fluc expression, pGB with CMV enhancer (pGBe) driving expression of fluc-egfp (pGBeFG) or fluc2-tdtomato (pGBeLT), or pCMV driving expression of fluc-egfp (pCMVFG). 48hrs after transduction, cells were stimulated with anti-CD3 and anti-CD28 antibodies or left unstimulated. 24hrs later, cells were harvested and analyzed for luciferase activity using an in vitro luciferase assay. Data is normalized to total protein per sample. N=3, Students T-test. Symbols above bars denote statistical significance; *p<0.05
Results
Two transcription enhancer strategies increase the activity of the -828bp Granzyme B promoter
In vitro, the -828bp Granzyme B promoter (pGB) drives low firefly luciferase (fluc) reporter gene expression when transiently transfected into primary murine splenocytes and activated with anti-CD3 and anti-CD28 antibodies (1 μg/ml) as compared to the CMV promoter (pCMV) (250 Relative Light Units (RLU)/μg of protein for pGB compared to 9930 RLU/μg for pCMV, see Supplementary Fig. S2A). A similar comparison was observed in transiently transfected Jurkat cells after normalization to control vector (See Supplementary Figure S2B). A minimum of 10000 RLU/μg of protein is needed in cell culture in order to detect fLuc activity living animals (data not shown). To increase reporter gene expression driven by pGB, we created two lentiviral vectors that carried either 1) pGB in the TSTA system to drive fLuc expression (pGBTSTAF), or 2) a hybrid promoter, in which the CMV enhancer was cloned upstream of pGB (pGBe) to drive the expression of a fLuc-EGFP (FG) fusion protein (pGBeFG) (Fig. 1A). As controls, we also created vectors in which FG expression was under control of pGB only (pGBFG) or pCMV (pCMVFG). Splenocytes were transduced with lentiviral vectors carrying pGBeFG, pGBTSTAF, pGBFG or pCMVFG. Upon activation of splenocytes using anti-CD3 and anti-CD28 antibodies, fLuc activity from cells transduced with pGBeFG was 10-fold higher than cells transduced with pGBFG (p<0.05) and 5-fold higher than pGBTSTAF. Since all the lentiviral vectors we generated were from the same backbone and they transduced primary splenocytes with equal efficiency (35%, Supplementary Fig. 3) we concluded the difference in luciferase expression was due to differences in the promoters and not a change in transduction efficiency. To further elevate reporter activity, we replaced the FG reporter gene with a fusion gene, LT, composed of a synthetic firefly luciferase (luc2, see Materials and Methods) and the tandem tomato red fluorescent protein to create pGBeLT (Fig. 1B). In subsequent experiments we transduced splenocytes with the pGBeLT vector.
Hybrid promoter maintains characteristics of the endogenous Granzyme B promoter for driving Granzyme B expression
In primary murine splenocytes transduced with the pGBe-eGFP lentiviral vector (pGBeG), the mean fluorescence intensity (MFI) peaked at 72hrs of sustained activation with anti-CD3 and CD28 antibodies (Fig. 2A). Granzyme B protein expression in the same splenocytes transduced with pGBeG (Fig. 2B-C) directly correlated with fluorescence expressed from reporter gene (R2=0.92) (Fig. 2D). To further characterize the hybrid promoter by determining its expression in T-cell subsets, we transduced B cell-depleted murine splenocytes with pGBeG and analyzed cells by flow cytometry. eGFP expression was detected in both activated CD4+ and CD8+ T-cells (Fig. 2E); however, eGFP expression was significantly higher in CD8+ T-cells than in CD4+ T-cells (p=0.008). Also, splenocytes transduced with pGBeG had a 7 fold higher eGFP expression when compared to splenocytes transduced with the pGBTSTA-eGFP lentiviral vector (p=0.0004, Fig. 2E-F).
Activated T-cells are detected in target tumor using bioluminescence imaging
As proof of principle for the detection of T-cell activation in living animals, we used OT1 transgenic mice whose CD8+ T-cells express a T-cell receptor specific for a chicken ovalbumin peptide, residues 257-264, in the context of H2-Kb (23). Purified CD8+ T-cells from OT1 transgenic mice were transduced with the pGBeLT lentiviral vector. Transduced T-cells were then injected via the tail vein into congenic albino C57BL/6 mice bearing 6 day old target (E.G7) and non-target (EL4) tumors implanted behind the right and left shoulder respectively. These mice are congeneic to wild type C57BL/6 mice. 24hrs after adoptive transfer, bioluminescence signal was detected in the area of the E.G7 tumor but not where EL4 cells had been injected (Fig. 3A). Similar to the cell culture kinetics, this signal peaked 72hrs post adoptive transfer (Fig. 3B) and was significantly larger than the signal from the EL4 tumor (p=0.009). In tumor bearing animals in which syngeneic polyclonal, non-targeted T-cells labeled with pGBeLT were adoptively transferred, no signal could be detected from the sites of EL4 or E.G7 tumor grafts at any time during the study period (Fig. 3B), even though T-cells could be detected in these tumors when they were transduced with a lentiviral vector carrying a constitutively active promoter (see Supplementary Fig. S4, online). The peak signal intensity from the E.G7 tumor coincided with tumor regression (Fig. 3C). CD8+ T-cells isolated from E.G7 tumors had a higher percentage of cells expressing CD69 than CD8+ T-cells isolated from EL4 tumors as early as 24hrs post adoptive transfer and remained higher at 72hrs and 120hrs post adoptive therapy (Fig. 3D). In the draining lymph nodes we were not able to detect any fLuc expression until day 3 post adoptive therapy (Supplementary Fig. S5A online) even though we were able to detect OT1 T-cells 24hrs post adoptive therapy in draining lymph nodes from both tumors by flow cytometry (Supplementary Fig. S5B-D online).
Bioluminescent signal from target tumor is due to CTL effector function and not T-cell proliferation
To determine whether the signal in the E.G7 tumor was a result of CTL effector function or stemmed from a mere increase in the number of intra-tumoral T-cells, we compared the bioluminescence signal from pGBeLT transduced T-cells to the signal from T-cells transduced with a lentiviral vector carrying a fusion gene, LG, composed of luc2 and egfp driven by the constitutive human Ubiquitin C promoter (pUbiLG). The human Ubiquitin C promoter (pUbi) is not influenced by T-cell activation through the T-cell receptor (data not shown) and has been used to study T-cell proliferation in a tumor model (24, 25). Since the spleen serves as a site of T-cell filtration and is not a site of CTL effector function, we did not expect to detect bioluminescence from pGBeLT transduced OT1 T-cells in the spleens of tumor bearing mice. We observed an increase in the signal from the spleen in tumor bearing mice injected with pUbiLG transduced OT1 T-cells (Supplementary Fig. S6 online). Since both pGB and pUbi have different promoter strength, we represented the bioluminescent signal from both promoters as fold increase from day 0 to compare reporter gene expression from both promoters on the same graph (Fig. 4A). There was a significant difference between the bioluminescent signal from pGB and pUbi transduced OT1 T-cells in the spleen. The fold increase in bioluminescence from pUbiLG transduced T-cells in the spleen increased over the course of the experiment. In contrast, signal from pGBeLT transduced T-cells in the spleen did not change significantly (p>0.05) through the course of the experiment (Fig. 4B). This was confirmed with flow cytometric analysis on spleens from mice that received adoptively transferred OT1 T-cells that were transduced with the lentiviral vector and labeled with CFSE (Fig. 4B). CFSE-labeled splenocytes of non-tumor burdened mice showed only one round of cell division and no CD69 expression. CFSE labeled cells isolated from splenocytes of tumor carrying mice had undergone at least 4 rounds of cell division, as indicated by the decrease of CFSE label. However, they did not show any expression of CD69. In the tumor, we would expect increased pUbiLG signal before the peak of pGBeLT signal because of the dynamics T-cell response, in which proliferation precedes differentiation into effector cells (reviewed in [(26)). There was a statistically significant increase in the fold induction between day 3 and day 2 in T-cells transduced with pUbiLG in the target tumor (p=0.003). However, this occurred at a similar time as the peak fold induction in pGBeLT transduced T-cells in the target tumor (Fig. 4C). In the target tumor, by day 5 most of the T-cells had complete loss of CFSE (Fig. 4D), indicating that they had undergone several rounds of division. In addition, the signal from pGBeLT transduced T-cells displays similar kinetics as shown in Fig. 4 after normalizing the signal by the number of OT1 T-cells isolated from the spleen or tumor (Supplementary Fig. S7, online).
Discussion
In this study of T-cell receptor dependent T-cell activation in vivo, we used the minimal Granzyme B promoter (pGB) for expressing reporter genes for in vivo imaging because it is well characterized (11, 12), and there has also been success using the promoter’s human homologue to detect activated T-cells in transgenic mice via flow cytometry (27). Furthermore, it can be detected by flow cytometry by staining for CD107a expression, which has been used to detect antigen-specific CD8+ T-cells (28) and NK cell activity (29). However, when we transiently transfected primary T-cells with pGB-Fluc, the amount of reporter gene expression was weak compared to that of pCMV-Fluc and insufficient for long-term in vivo imaging (data not shown). We therefore chose two enhancer strategies, CMVe and TSTA, to increase pGB activity.
Both CMVe and TSTA elements enhanced pGB activity, with CMVe being superior. One of its drawbacks was the relatively high background bioluminescence emitted from cells transduced with luciferase reporters driven by pGBe; however, it responded best to activation of the T-cells with a 10-fold gain in signal. This strong response is most likely due to the presence of transcription factor binding sites for CREB (cAMP-response element-binding protein) and NF-κB in CMVe (30, 31). Both of these transcription factors are up-regulated during T-cell activation (32, 33). One concern may be that the addition of CMVe to a tissue specific promoter might alter the promoter’s properties, such as its kinetics of activation and cell specificity. In our hands, addition of CMVe did not alter activation kinetics of pGB-driven gene expression: peak gene expression by both the hybrid promoter and endogenous Granzyme B promoter occurred at 72hrs. Surprisingly, the reporter gene expression was detected not only in CD8+ T-cells but also in CD4+ T-cells. This could be due to lack of regulatory sequences that are located further upstream of the minimal promoter (34). However, cytotoxic CD4+ T-cells have been described before, and Hanson et al (35) also observed Granzyme B mRNA in some CD4+ T-cell clones isolated from their transgenic mice. Granzyme B mRNA was also found in CD4+ T-cells by other groups (36-38) including regulatory T (Treg) cells (39). Others have shown that the use of CMVe to increase weak promoter activity retains cell specificity (40, 41). Therefore, detection of reporter gene activity in CD4+ T-cells could be due to expression from the minimal Granzyme B promoter and not due to the addition of CMVe.
As proof of principle, we used purified CD8+ T-cells from OT1 transgenic mice to detect T-cell activation in vivo as measured by luciferase expression driven by pGB. Granzyme B promoter activity peaked 72hrs post T-cell transfer in the target tumor (E.G7), even when tumors were implanted 6 days after adoptive transfer of T-cells (data not shown). This correlated with our in vitro measurements of Granzyme B protein expression as assessed by Western blot. In addition, the peak signal for pGBeLT transduced OT1 T-cells coincided with tumor regression. A small signal could be detected in the parental, non-target tumor (EL4) and we were able to detect OT1 T-cells in these tumors by flow cytometry. However, the signal from pGBeLT transduced OT1 T-cells in the EL4 tumors did not increase significantly over the course of the study and the loss of CFSE label in these intra-tumoral T-cells could be due to activated/proliferating T-cells in the circulation that were trapped in the tumor at the time of harvest (42). This is supported by the observation that OT1 T-cells in the spleen and in EL4 tumors had undergone the same number of cell divisions.
Other groups have reported visualization of T-cell activation in living subjects (25, 43, 44). Ponomerev et al. (43) used a synthetic promoter consisting of the multiple repeats of the binding site for the transcription factor nuclear factor of activated T-cells (NFAT), which is a commonly found sequence in many promoters that are triggered during T-cell activation. This promoter was used to drive the expression of Herpes Simplex Virus Type 1 Thymidine Kinase (HSV1-tk) for micro positron emission tomography imaging (μPET). This study enabled non-invasive imaging of T-cell activation using tumors derived from transduced Jurkat T-cells and direct activation through the use of stimulatory antibodies against CD3 and CD28. However, the authors did not proceed to further non-invasively detect T-cell activation in response to an actual tumor or infection. Shu et al. (25) visualized T-cell activation by following the proliferative response of T-cells to actual tumors using reporter genes under the control of the constitutively expressed Ubiquitin C promoter (pUbi). Since TCR dependent T-cell activation leads to proliferation (26, 45), we wanted to ensure that the signal increase from pGBeLT transduced T-cells in the target tumor was in fact due to T-cell effector function and not to a simple increase in cell number. For this we determined if the kinetics of pUbiLG and pGBeLT transduced T-cells in the target tumor hold true to T-cell response dynamics. It is not clear if there is a significant amount of proliferation prior to T-cells developing effector function. In our study, both signals peaked at similar times in the E.G7 tumors. Other studies using the OT1 mouse model have shown that only cells that had lost CFSE label (cells that had undergone several rounds of proliferation) in the E.G7 tumors expressed Granzyme B (42, 46). Therefore in the target tumor the kinetics may be linked. On the single cell level, the events of TCR dependent activation and proliferation occur within the first 24hrs of antigen recognition (47), but in our study, proliferation as measured by an increase in signal from pUbiLG transduced T-cells was not visualized completely until 48hrs post adoptive transfer. This is most likely due to the limit of detection of bioluminescence imaging (48), in which a critical number of cells must be focalized in order to be detected by the CCD camera. These initial events of T-cell dynamics occur in the draining lymph nodes (26); however we were not able to detect signal in lymph nodes due to the limit of detection of bioluminescence imaging and at later time points due to overlapping signal from the tumors. However we were able to observe signal from the draining lymph nodes ex vivo and we were able to detect OT1 T-cells by flow cytometry. 24hrs post adoptive therapy, we were able to detect activated OT1 T-cells by flow cytometry in the draining lymph nodes of E.G7 tumors, however we did not observe signal from pGBeLT transduced T-cells in these lymph nodes until day 3. This finding is supported by the fact that Granzyme B is a marker for T-cell effector function (28) and in a previously published study, Granzyme B expressing T-cells were also only detected in lymph node T-cells that had undergone several rounds of division (46). The spleen is normally not a site where T-cells acquire effector function and in our model should not show an increase in signal from pGBeLT transduced T-cells and therefore serves as a control for visualizing T-cells as they acquire effector function. We did not observe any statistical increase in signal from pGBeLT transduced T-cells in tumor challenged mice and OT1 T-cells isolated from the spleens did not show expression of CD69. Since the spleen is a lymphoid filtration organ, we did observe an accumulation of OT1 T-cells transduced with pUbiLG and since these cells did have a loss of CFSE label, they could be proliferating cells that had migrated from the draining lymph nodes and entered the circulation (42, 46). This spatially and temporally defined bioluminescence signal, together with our flow cytometry data showing CD69 expression on the tumor-infiltrating T-cells, indicates that we were most likely visualizing T-cells as they acquired effector function.
In summary, we have created a hybrid promoter based on CMVe and pGB that significantly increases reporter gene expression over pGB alone, and also correlates with endogenous Granzyme B protein production. Using a bioluminescent reporter, the level of transcriptional activity from this hybrid promoter upon T-cell stimulation in vivo is sufficient to be detected in small living animals. We believe that this method will be applicable over a broad range of animal models that benefit from functional, non-invasive visualization of CTL function such as immune response to infection and adoptive immunotherapy. By replacing pGB with other promoters turned on during T-cell activation, such as the IL-2 promoter, IFN-γ promoter, or NFAT synthetic promoter (44) it should be feasible to visualize different stages of T-cell function and differentiation non-invasively in vivo and in vitro, however kinetics of activation are dependent on disease model (44). In addition, by replacing the bioluminescence reporter genes with reporter genes for PET imaging, it should be feasible to monitor T-cell activation in patients. Our lab has recently imaged T-cell localization using PET reporter gene imaging and a constitutive promoter in glioma patients undergoing T-cell therapy (49). Recently, Radu et al. developed a small molecule PET probe capable of imaging lymphoid organs and immune activation (50), which may be useful to image inflammation (51) and leukemic cells, but is somewhat limited due to tumor uptake of the PET probe and cannot discern tumor from activated T-cells. Therefore, imaging of tumor targeted T-cell activation may eventually be possible in patients by utilizing a T-cell activation promoter.
Supplementary Material
Acknowledgments
We thank Dr. Remi Creusot for critical reading of our manuscript.
Grant Support: NCI ICMIC P50CA114747 (S.S.G., C.H.C., M.H.B.), NCI RO1 CA082214 (S.S.G.), NIH P01 CA49605-19 (C.H.C.) and the Canary Foundation.
Footnotes
Conflicts of Interest: S.S.G. is founder of CellSight Inc which develops and markets strategies for imaging of cell trafficking; C.H.C. has financial interest in Caliper LifeSciences, an in vivo imaging company.
References
- 1.Finn OJ. Cancer vaccines: between the idea and the reality. Nature Reviews Immunology. 2003;3:630–41. doi: 10.1038/nri1150. [DOI] [PubMed] [Google Scholar]
- 2.Weiss DW. Reflections on tumor origin, immunogenicity, and immunotherapy. Cancer Immunology Immunotherapy. 1984;18:1–4. doi: 10.1007/BF00205391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leen AM, Rooney CM, Foster AE. Improving T Cell Therapy for Cancer. Annual Review of Immunology. 2007;25:243–65. doi: 10.1146/annurev.immunol.25.022106.141527. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauce D, Bodinier M, Garin M, et al. Retrovirus-mediated gene transfer in primary T lymphocytes impairs their anti-Epstein-Barr virus potential through both culture-dependent and selection process-dependent mechanisms. Blood. 2002;99:1165–73. doi: 10.1182/blood.v99.4.1165. [DOI] [PubMed] [Google Scholar]
- 6.Hunder NN, Wallen H, Cao J, et al. Treatment of Metastatic Melanoma with Autologous CD4+ T Cells against NY-ESO-1. New England Journal of Medicine. 2008;358:2698–703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley ME, Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nature Reviews Cancer. 2003;3:666–75. doi: 10.1038/nrc1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–9. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coligan J, Bierer B, Margulies D, Shevach E, Strober W. Short Protocols in Immunology: A Compendium of Methods from Current Protocols in Immunology. Hoboken, NJ: John Wiley & Sons; 2005. [Google Scholar]
- 10.Prendergast J, Helgason C, Bleackley R. Quantitative polymerase chain reaction analysis of cytotoxic cell proteinase gene transcripts in T cells. Pattern of expression is dependent on the nature of the stimulus. J Biol Chem. 1992;267:5090–5. [PubMed] [Google Scholar]
- 11.Babichuk CK, Duggan BL, Bleackley RC. In vivo regulation of murine granzyme B gene transcription in activated primary T cells. Journal of Biological Chemistry. 1996;271:16485–93. doi: 10.1074/jbc.271.28.16485. [DOI] [PubMed] [Google Scholar]
- 12.Babichuk CK, Bleackley RC. Mutational analysis of the murine granzyme B gene promoter in primary T cells and a T cell clone. Journal of Biological Chemistry. 1997;272:18564–71. doi: 10.1074/jbc.272.30.18564. [DOI] [PubMed] [Google Scholar]
- 13.Kuo CT, Leiden JM. Transcriptional regulation of T lymphocyte development and function. Annual Reviews Immunology. 1999;17:149–87. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- 14.Iyer M, Wu L, Carey M, et al. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. PNAS. 2001;98:14595–600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray S, Paulmurugan R, Hildebrandt I, et al. Novel Bidirectional Vector Strategy for Amplification of Therapeutic and Reporter Gene Expression. Human Gene Therapy. 2004;15:681–90. doi: 10.1089/1043034041361271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nettelbeck DM, Jerome V, Muller R. Gene therapy: designer promoters for tumour targeting. Trends in Genetics. 2000;16:174–81. doi: 10.1016/s0168-9525(99)01950-2. [DOI] [PubMed] [Google Scholar]
- 17.Chen IY, Gheysens O, Ray S, et al. Indirect imaging of cardiac-specific transgene expression using a bidirectional two-step transcriptional amplification strategy. Gene Therapy. 2010;17:827–38. doi: 10.1038/gt.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 19.Moore MW, Carbone FR, Bevan MJ. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54:777–85. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F, Wang L-P, Brauner M, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–9. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 21.Parish C, Warren H. Use of the Intracellular Fluorescent Dye CFSE to Monitor Lymphocyte Migration and Proliferation. In: Coligan J, Bierer B, Margulies D, et al., editors. Current Protocols in Immunology. Hoboken, NJ: John Wiley & Sons, Inc; 2001. pp. 4.9.1–4.9.10. [Google Scholar]
- 22.Cho RW, Wang X, Diehn M, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–71. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 23.Hogquist K, Jameson S, Heath W, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 24.Kim YJ, Dubey P, Ray P, Gambhir SS, Witte ON. Multimodality imaging of lymphocytic migration using lentiviral-based transduction of a tri-fusion reporter gene. Molecular Imaging & Biology. 2004;6:331–40. doi: 10.1016/j.mibio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Shu CJ, Guo S, Kim YJ, et al. Visualization of a primary anti-tumor immune response by positron emission tomography. Proc Natl Acad Sci U S A. 2005;102:17412–7. doi: 10.1073/pnas.0508698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanzavecchia A, Sallusto F. Dynamics of T Lymphocyte Responses: Intermediates, Effectors, and Memory Cells. Science. 2000;290:92–7. doi: 10.1126/science.290.5489.92. [DOI] [PubMed] [Google Scholar]
- 27.Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593–7. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- 28.Betts MR, Brenchley JM, Price DA, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of Immunological Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 29.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. Journal of Immunological Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Hunninghake GW, Monick MM, Liu B, Stinski MF. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. Journal of Virology. 1989;63:3026–33. doi: 10.1128/jvi.63.7.3026-3033.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambucetti LC, Cherrington JM, Wilkinson GW, Mocarski ES. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO Journal. 1989;8:4251–8. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niller HH, Hennighausen L. Phytohemagglutinin-induced activity of cyclic AMP (cAMP) response elements from cytomegalovirus is reduced by cyclosporine and synergistically enhanced by cAMP. Journal of Virology. 1990;64:2388–91. doi: 10.1128/jvi.64.5.2388-2391.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazan U, Thomas D, Alcami J, et al. Stimulation of a human T-cell clone with anti-CD3 or tumor necrosis factor induces NF-kappa B translocation but not human immunodeficiency virus 1 enhancer-dependent transcription. Proc Natl Acad Sci U S A. 1990;87:7861–5. doi: 10.1073/pnas.87.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson BA, John VA, Henschler R, et al. Upstream elements bestow T-cell and haemopoietic progenitor-specific activity on the granzyme B promoter. Gene. 1999;234:101–7. doi: 10.1016/s0378-1119(99)00173-0. [DOI] [PubMed] [Google Scholar]
- 35.Hanson RD, Sclar GM, Kanagawa O, Ley TJ. The 5’-flanking region of the human CGL-1/granzyme B gene targets expression of a reporter gene to activated T-lymphocytes in transgenic mice. Journal of Biological Chemistry. 1991;266:24433–8. [PubMed] [Google Scholar]
- 36.Ebnet K, Levelt CN, Tran TT, Eichmann K, Simon MM. Transcription of granzyme A and B genes is differentially regulated during lymphoid ontogeny. Journal of Experimental Medicine. 1995;181:755–63. doi: 10.1084/jem.181.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lancki DW, Hsieh CS, Fitch FW. Mechanisms of lysis by cytotoxic T lymphocyte clones. Lytic activity and gene expression in cloned antigen-specific CD4+ and CD8+ T lymphocytes. Journal of Immunology. 1991;146:3242–9. [PubMed] [Google Scholar]
- 38.Wang M, Windgassen D, Papoutsakis E. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-Cell activation. BMC Genomics. 2008;9:225. doi: 10.1186/1471-2164-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao X, Cai SF, Fehniger TA, et al. Granzyme B and Perforin Are Important for Regulatory T Cell-Mediated Suppression of Tumor Clearance. Immunity. 2007;27:635–46. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Liu BH, Wang X, Ma YX, Wang S. CMV enhancer//human PDGF-[beta] promoter for neuron-specific transgene expression. Gene Therapy. 2004;11:52–60. doi: 10.1038/sj.gt.3302126. [DOI] [PubMed] [Google Scholar]
- 41.Gruh I, Wunderlich Stephanie, Winkler Monica, Schwanke Kristin, Heinke Jennifer, Blomer Ulrike, Ruhparwar Arjang, Rohde Bettina, Li Ren-Ke, Haverich Axel, Martin Ulrich. Human CMV immediate-early enhancer: a useful tool to enhance cell-type-specific expression from lentiviral vectors. The Journal of Gene Medicine. 2007 doi: 10.1002/jgm.1122. ePub. [DOI] [PubMed] [Google Scholar]
- 42.Boissonnas A, Combadiere C, Lavergne E, et al. Antigen Distribution Drives Programmed Antitumor CD8 Cell Migration and Determines Its Efficiency. Journal of Immunology. 2004;173:222–9. doi: 10.4049/jimmunol.173.1.222. [DOI] [PubMed] [Google Scholar]
- 43.Ponomarev V, Doubrovin M, Lyddaney C, et al. Imaging TCR-Dependent NFAT-Mediated T-Cell Activation with Positron Emission Tomography In Vivo. Neoplasia. 2001;3:480–8. doi: 10.1038/sj.neo.7900204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Na I-K, Markley JC, Tsai JJ, et al. Concurrent visualization of trafficking, expansion and activation of T lymphocytes and T-cell precursors in vivo. Blood. 2010 doi: 10.1182/blood-2009-12-259432. blood-2009-12-259432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brehm M, Selin L, Welsh R. CD8 T cell responses to viral infections in sequence. Cellular Microbiology. 2004;6:411–21. doi: 10.1111/j.1462-5822.2004.00390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curtsinger JM, Gerner MY, Lins DC, Mescher MF. Signal 3 Availability Limits the CD8 T Cell Response to a Solid Tumor. Journal of Immunology. 2007;178:6752–60. doi: 10.4049/jimmunol.178.11.6752. [DOI] [PubMed] [Google Scholar]
- 47.Henrickson SE, von Andrian UH. Single-cell dynamics of T-cell priming. Current Opinion in Immunology. 2007;19:249–58. doi: 10.1016/j.coi.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Massoud T, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes & Development. 2003;17:545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 49.Yaghoubi SS, Jensen MC, Satyamurthy N, et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nature Clinical Practice Oncology. 2009;6:53–8. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radu CG, Shu CJ, Nair-Gill E, et al. Molecular imaging of lymphoid organs and immune activation by positron emission tomography with a new [18F]-labeled 2[prime]-deoxycytidine analog. Nature Methods. 2008;14:783–8. doi: 10.1038/nm1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brewer S, Nair-Gill E, Wei B, et al. Uptake of [18F]1-(2’-deoxy-2’-arabinofuranosyl) Cytosine Indicates Intestinal Inflammation in Mice. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.01.003. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dieter EJ, Jürg T. Granzymes, a Family of Serine Proteases Released from Granules of Cytolytic T Lymphocytes upon T Cell Receptor Stimulation. Immunological Reviews. 1988;103:53–71. doi: 10.1111/j.1600-065x.1988.tb00749.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.