Abstract
The N-terminal protein processing pathway is an essential mechanism found in all organisms. However, it is widely believed that deformylase, a key enzyme involved in this process in bacteria, does not exist in eukaryotes, thus making it a target for antibacterial agents such as actinonin. In an attempt to define this process in higher eukaryotes we have used Arabidopsis thaliana as a model organism. Two deformylase cDNAs, the first identified in any eukaryotic system, and six distinct methionine aminopeptidase cDNAs were cloned. The corresponding proteins were characterized in vivo and in vitro. Methionine aminopeptidases were found in the cytoplasm and in the organelles, while deformylases were localized in the organelles only. Our work shows that higher plants have a much more complex machinery for methionine removal than previously suspected. We were also able to identify deformylase homologues from several animals and clone the corresponding cDNA from human cells. Our data provide the first evidence that lower and higher eukaryotes, as well as bacteria, share a similar N-terminal protein processing machinery, indicating universality of this system.
Keywords: Arabidopsis thaliana/homologues/methionine removal/peptide deformylase/protein processing
Introduction
In all organisms, a protein chain starts universally with the same amino acid, namely methionine. In some cases, an N-formyl group may modify this first amino acid as a result of the early addition of an N-formyl onto the amino group of the methionine bound to initiator tRNAMet. This latter process has been found in the organelles of all eukaryotic organisms examined and in the cytoplasm of prokaryotes, except Archaea (for a review see Meinnel et al., 1993b; see also Halbreich and Rabinowitz, 1971; Guillemaut and Weil, 1975; Crosti et al., 1977; Steinmetz and Weil, 1989; Takeuchi et al., 1998). However, the first (formyl)methionine is often not retained at the N-terminus of the protein as it is excised from the mature protein by a specialized hydrolytic machinery. More than 60% of all proteins lose their N-terminal methionine. Two enzyme families are involved in the co-translational process of methionine removal, methionine aminopeptidase (MAP) and peptide deformylase (PDF) (for reviews see Meinnel et al., 1993b; Bradshaw et al., 1998; Giglione et al., 2000). PDF removes all N-formyl groups, unmasking the amino group of the first methionine, a prerequisite for the subsequent action of MAP.
Although the two steps involved in methionine removal are essential in all organisms (for reviews see Meinnel et al., 1993b; Giglione et al., 2000; Lowther and Matthews, 2000), the role of this process is still poorly understood. Among the various hypotheses concerning the crucial role of methionine removal is the idea that it allows further post-translational modifications (Kendall et al., 1990; Krishna and Wold, 1993), including N-myristoylation of several important proteins involved in signal transduction and growth in eukaryotic cells (Johnson et al., 1994). It has also been suggested that the removal of the first methionine is linked to a molecular device, the so-called N-end rule, which governs the half-life of proteins (Arfin and Bradshaw, 1988). This pathway, which is the first step leading to the proteasome, depends strictly on the nature of the N-termini of proteins (Varshavsky, 1996).
Most of our knowledge about N-terminal protein processing comes from diverse work with model organisms including bacteria (for PDF and MAP), yeast and mammals (for MAP). The data have been used to generalize the conclusions for all cell types. For instance, although protein synthesis starts with an N-formylmethionine in the organelles of all eukaryotes, there does not appear to be a PDF in the yeast Saccharomyces cerevisiae or in the nematode Caenorhabditis elegans (see data quoted in Giglione et al., 2000). This led to the idea that eukaryotic cells do not have a PDF. One of the major consequences of this conclusion is that PDF is now considered to be a very good target for developing antibiotics (Giglione et al., 2000). Indeed, PDF is the target of the naturally occurring antibiotic actinonin and of other artificial bactericides (Apfel et al., 2000; Chen et al., 2000; Huntington et al., 2000). The activities of MAP proteins are also the targets for several specific inhibitory drugs that are thought to be promising therapeutic agents. Fumagillin and various derivatives such as TNP-470 were demonstrated to inhibit MAP2 activity and to be very potent anti-cancer drugs, acting by blocking angiogenesis (Sin et al., 1997; Lowther et al., 1999; Turk et al., 1999). Given that N-terminal processing has become the target for new drugs, a better characterization of this process in higher eukaryotes is required.
Using the complete Arabidopsis thaliana genome, we investigated N-terminal cleavage in higher eukaryotes. For the first time, we have identified two different peptide deformylases in a eukaryotic organism. In addition, we report the existence of six novel methionine aminopeptidases in A.thaliana. We characterized the activity of the corresponding proteins in vivo and in vitro, studied the mRNA expression pattern and showed a specific subcellular localization for each protein. Taken together, our results reveal the existence of an intricate machinery aimed at removing the first methionine of proteins in plants. We finally identified homologues of deformylase in Drosophila melanogaster, and cloned the corresponding human cDNA. Our data favour the idea of a universality of the N-terminal protein processing mechanism.
Results
Identification, cloning and characterization of cDNAs encoding seven enzymes involved in the removal of the N-terminal methionine from proteins in A.thaliana
Enzymes displaying MAP activity are classified into two groups, MAP1 and MAP2. Proteins of the MAP1 group were first identified in eubacteria. Yeast and mammalian cells also possess a MAP1; both show the same catalytic core as bacterial MAP1 and have N-terminal pre-sequences including two conserved zinc fingers (Chang et al., 1992). Eukaryotic MAP2 enzymes also have N-terminal extensions compared with those found in Archaea. Both MAP1 and MAP2 are located in the cytoplasm of eukaryotes. Nevertheless, it has been suggested that the N-terminal pre-sequences favour the interaction of the catalytic core with particular targets (Arfin et al., 1995; Li and Chang, 1995; Zuo et al., 1995). Although the sequence similarity between the two MAP types is very low, the 3D structures of the catalytic core are homologous (Liu et al., 1998; Tahirov et al., 1998). Bacterial PDF proteins are metalloproteases of ∼20 kDa with an unusual metal binding site (for a recent review on PDF see Giglione et al., 2000). There are two sub-classes of PDF, PDF1 and PDF2, both containing two signature sequences and the HEXXH motif of the ‘zinc’ metalloproteases superfamily (Meinnel et al., 1997).
Using BLAST analysis (Altschul et al., 1990), we searched the genome sequence of A.thaliana for sequences similar to PDF and/or MAP genes. Because of the variability of the pre-sequences, we chose to restrict our primary analysis to the catalytic domains of the two enzymes. Consequently, the sequences of Escherichia coli MAP1 and PDF1, Pyrococcus furiosus MAP2 and Bacillus stearothermophilus PDF2 were used as primary input inquiries. Four MAP1 sequences (named MAP1A, 1B, 1C and 1D, respectively), one MAP2 sequence (MAP2A) and one PDF1 sequence (PDF1A) were retrieved (Table I). This result was unexpected since PDF-like sequences have only previously been found in prokaryotic genomes. Moreover, such a large number of MAP genes was surprising in an organism believed to have a low gene redundancy. In comparison, similar analysis with the complete genome of C.elegans, which has approximately the same size and gene number as A.thaliana, and with S.cerevisiae revealed no PDF and only one MAP1 and one MAP2 sequence.
Table I. Various cDNAs used in this study.
| Enzyme | Organism | Cell localizationa | EST (location of the match)b (GenBank)c | cDNA (GenBank)c | Gene sequence (GenBank)c | Chromosome location |
|---|---|---|---|---|---|---|
| MAP1A | A.thaliana | cytoplasm | Z34850(3′) Z33915 (3′) ATTS3579 (3′) | AF250960 | AC002387 (AL021961 AL161584) | 2 (+ partial duplication on chr. 4) |
| MAP1B | A.thaliana | plastids | H37448 (5′) | AF250961 | AC011810 | 1 |
| AV442047(5′) | AC027134 | |||||
| AA720204(m) | ||||||
| AA394671(3′) | ||||||
| AV439915(3′) | ||||||
| AV520476(3′) | ||||||
| AA651355(3′) | ||||||
| MAP1C | A.thaliana | plastids | T42700(5′) | AF250962 | AB028607 | 3 |
| mitochondria | AI997694(3′) | |||||
| MAP1D | A.thaliana | plastids | AV556325(3′) | AF250963 | ATAP21 | 4 |
| mitochondria | AV542276(3′) | AL161590 | ||||
| MAP2A | A.thaliana | cytoplasm | AI996382(3′) | AF250964 | AC004005 | 2 |
| MAP2B | A.thaliana | cytoplasm | AI999464(3′) | AF300880 | AL138647 | 3 |
| Z18043(m) | ||||||
| T04113(5′) | ||||||
| F19846(3′) | ||||||
| PDF1A | A.thaliana | mitochondria | – | AF250959 | AC007591 | 1 |
| PDF1A | L.esculentum | ND | AW617250 (L.hirsutum) | AF271258 | – | – |
| PDF1A | H.sapiens | ND | AI859289(3′) | AF239156 | AC012040 | 16 |
| AL045195(3′) | AC026464 | |||||
| AI765656(3′) | ||||||
| PDF1B | A.thaliana | plastids | AV440729(3′) | AF269165 | AL163792 | 5 |
| mitochondria | AV442746(5′) | |||||
| AV534215(3′) | ||||||
| PDF1B | L.esculentum | plastids | AI772897 | AF250958 | – | – |
| mitochondria | AI775816 | |||||
| AW223746 | ||||||
| AW039294 |
Using rapid analysis of cDNA ends (RACE), we cloned the corresponding full-length cDNAs (see data in Table I). The catalytic cores of the MAP1 are very similar (>40–50% amino acid sequence identity) to the bacterial counterparts (Figure 1A). MAP1A closely resembles the yeast enzyme whereas MAP1B, MAP1C and MAP1D displayed unique N-terminal pre-sequences but were very similar to each other around amino acid positions 90–150 (Figure 1A). The homology extended through the catalytic domain (pink in Figure 1A, region 150–430). Most interestingly, these three MAP1s lack the zinc finger of MAP1A. Deletion of this N-terminal pre-sequence in yeast MAP1 yielded an enzyme with full MAP activity in vitro but unable to complement a map-conditional mutant in vivo (Zuo et al., 1995). Finally, PDF1A also carried an 80–90 residue N-terminal extension not found in bacterial PDF (Figure 1C).
Fig. 1. Alignment of the amino acid sequences of PDF and MAP from higher eukaryotes. The sequences of MAP and PDF (see text) were used as input probes with the help of the BLAST software (Altschul et al., 1990) to search data at the web site of the Arabidopsis Information Resource (http://www.arabidopsis.org/cgi-bin/blast/TAIRblast.pl) and at the NCBI. Matching EST sequences were compared with each other and assembled when overlapping. The genomic fragments were analysed with the various software predicting intron–exon splice sites (Hebsgaard et al., 1996) and, by using the amino acid sequence homologies, assembled and compared with the ESTs. Arabidopsis thaliana cDNAs were cloned and sequenced. The deduced amino acid sequence is shown. Translated sequences were aligned with Clustalx (Jeanmougin et al., 1998) and then manually. A series of question marks indicates that the corresponding nucleotide sequence is partial or missing. The numbering of each of the three amino acid sequence sets is shown below each block of 150 residues. Amino acids in italic at the N-terminus indicate putative alternative translation starts. Residues strictly conserved within the catalytic core are shown in red or in green for the ligands of the catalytic metal. (A) MAP1 alignments. The amino acid similarity between groups is shown in blue for cytoplasmic MAP (bottom) or in pink for organellar MAP (top). (B) MAP2 alignments. The deduced amino acid sequence at the 3′ end of the EST of the Bombyx mori sequence was modified because of an obvious frame-shift error. The poly-basic common amino acid stretch is shown in pink. The second poly-basic sequence found in animal sequences is in blue. (C) PDF alignments. The additional similarity shared by plant PDF1A (top) is coloured in pink. Specific features of PDF1B types (bottom) are shown in blue. Secondary structure elements are indicated on top.
The corresponding genes were mapped in the A.thaliana genome (Table I). Genome sequence analysis showed that (i) there is a short, partial and non-functional duplication of an internal sequence of MAP1A on chromosome 4 and (ii) the MAP2 gene is duplicated, with one copy on chromosome 2 (MAP2A) and another on chromosome 3 (MAP2B). The cDNA of MAP2B was cloned. The two MAP2 cDNAs encode near-identical proteins (Figure 1B).
Existence of homologues of A.thaliana MAP and PDF in other higher plants, and the cloning and characterization of a cDNA encoding an additional PDF, PDF1B
The seven cloned cDNA sequences (six MAP and one PDF1A) were used to screen the many plant genomic and expressed sequence tags (EST) sequences available at http://www.arabidopsis.org/cgi-bin/blast/TAIRblast.pl. Several partial sequences encoding homologues of MAP1A and MAP2 were identified in rice, alfalfa, soybean, tomato, sorghum, corn and pine (Figure 1A and B). The plant MAP1As closely resemble their animal and fungal counterparts, with similar N-terminal pre-sequences and conservation of the histidines and cysteines of the zinc finger motif. Likewise, MAP2 from plants is similar to enzymes in the other kingdoms (Figure 1B). Homologues of MAP1B, 1C and 1D were found in the same plants (see Figure 1A). Interestingly, at least three distinct MAP1 sequences related to A.thaliana MAP 1B, 1C and 1D were identified in rice (Oryza sativa) (Figure 1A). This suggests that the MAP sequence set obtained from A.thaliana is representative of the general situation in other plants, both dicots and monocots.
A similar analysis was performed with the sequence of A.thaliana PDF1A. Very similar sequences (>70%) were found in tomato, rice, poplar and corn (Figure 1C). The full-length PDF1A from tomato (Lycopersicon esculentum) was cloned by construction of an L.esculentum RACE library (Table I). The catalytic domain of this complete PDF1A sequence is 70% identical to A.thaliana PDF1A, and the N-terminal extensions are similar (Figure 1C). Two divergent, partial PDF1 sequences (which we called PDF1B) were found in a rice genomic fragment and in a tomato EST. These PDF1B sequences exhibited only ∼30–35% identity with the catalytic domain of PDF1A and do not display the long L2 insertion and the V1 variable loop found in PDF1A. Thus, both the rice (i.e. a monocot) and tomato (i.e. a dicot) genomes include two distinct PDFs, PDF1A and PDF1B. We were able to isolate the full-length cDNA encoding tomato PDF1B by screening our RACE tomato library. The resulting sequence information was used to identify and clone the A.thaliana PDF1B, which, at the time of our initial analyses, had not yet been covered in the Arabidopsis genome database. The full-length amino acid sequence of A.thaliana PDF1B is indeed similar to L.esculentum PDF1B (>75%; Figure 1C).
The products of the PDF and MAP genes display deformylase and methionine aminopeptidase activities, respectively
Our data above suggested the existence of unknown mRNAs encoding MAP (the forms MAP1B, 1C and 1D) and PDF (PDF1A and PDF1B) homologues in plants. Nevertheless, we had no evidence that these transcripts encoded functional proteins with MAP or PDF activity. As plant MAP1 and PDF resembled their eubacterial counterparts, we tested whether they complement map (the gene for MAP) or def (the gene for PDF) defects in E.coli. A def-conditional strain was already available (PAL421Tr-pMAKdef) and we constructed a similar map-conditional strain (PAL314Tr-pMap). The three MAP1 cDNAs isolated from A.thaliana under the control of the lac promoter indeed complemented the map defect (data not shown).
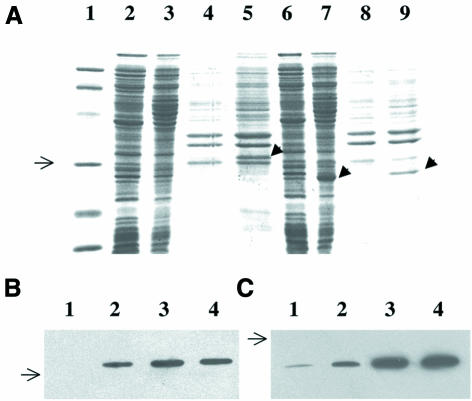
Expression of PDF1A in strain PAL421Tr did not allow bacterial growth at a non-permissive temperature. To check the production of the protein, the protein fused to the His6 tag was expressed in E.coli. This plasmid initially failed to complement the thermosensitivity of PAL421Tr. Western blot analysis with anti-His6 tag antibodies indicated that PDF1A was produced but remained in the insoluble fraction, as inclusion bodies (Figure 2B). Indeed, the N-terminal domain of the protein is hydrophobic. Therefore, we made a construct, pQdefAtΔ, encoding only the catalytic domain (i.e. residues 79–279). The protein produced was soluble (Figure 2A and C) and the plasmid complemented the def-conditional strain (Table II). A crude extract of the strain producing the catalytic domain of PDF1A was assayed for N-formyl group cleavage using two synthetic peptides, Fo-Met-Ala and Fo-Met-Ala-Ser. Its activity was >3000-fold higher than the control extract, and the addition of nickel improved the stability and linearity of the enzyme kinetics (data not shown). These characteristics are consistent with those of bacterial PDF (Ragusa et al., 1998) and show that PDF1A has peptide deformylase activity. Finally, sequences encoding both full-length protein and the truncated catalytic domain of PDF1B were fused to the His6 tag. Both constructs complemented the def-conditional strain PAL421Tr (Table II). A crude protein extract containing the catalytic domain showed a strong nickel-dependent stimulation of deformylation activity with a synthetic N-formylated peptide compared with the control (data not shown).
Fig. 2. Expression of PDF1A in E.coli. (A) PAGE analysis. Samples corresponding to 100 µg of protein of the indicated JM101Tr cell extract, expressing a His-tagged PDF1A from the plasmid indicated, were analysed by 12% SDS–PAGE and stained with Coomassie Blue. Lane 1 corresponds to the Bio-Rad low molecular weight markers (97, 66.2, 45, 31, 21.5 and 14.4 kDa). Lane 2, pQdef1a, soluble fraction, uninduced; lane 3, pQdef1a, soluble fraction, induced for 20 h with IPTG; lane 4, pQdef1a, insoluble fraction, uninduced; lane 5, pQdef1a, insoluble fraction induced for 20 h with IPTG; lane 6, pQdef1aΔ, soluble fraction, uninduced; lane 7, pQdef1aΔ, soluble fraction, induced for 20 h with IPTG; lane 8, pQdef1aΔ, insoluble fraction, un-induced, PDF (form 1); lane 9, pQdef1aΔ, insoluble fraction, induced for 20 h with IPTG. The black arrow indicates the induced proteins. (B and C) Time-course analysis of the expression of pQdef1a and pQdef1aΔ after induction with IPTG. The crude extracts were obtained and separated by SDS–PAGE as in (A). The gels were subjected to western blot analysis using anti-His tag mouse monoclonal antibodies. Exposure was for 1 min in both cases. Lane 1, uninduced (t = 0); lane 2, induced for 1 h with 0.1 mM IPTG; lane 3, induced for 5 h; lane 4, induced for 20 h. The arrow shows the location of the 31 kDa protein molecular weight marker. (B) pQdef1A, insoluble fraction (no signal was obtained with the soluble fraction). (C) pQdef1AΔN, soluble fraction.
Table II. Complementation of strain PAL421Tr at non-permissive temperature with various constructs.
| Plasmid used | PDF overproductiona | PDF in soluble fractionb | Complementation of strain PAL421Trc |
|---|---|---|---|
| pQE60 | – | – | – |
| pUCdef | +++ | ++ | + |
| pQdef1a | +++ | ND | – |
| pQdef1aΔN | +++ | +++ | + |
| pQdef1b | + | ND | + |
| pQdef1bΔN | ++ | ++ | + |
aProduction of the overproduced protein in JM101Tr, as assessed by PAGE and protein concentration measurement, was approximately 0 (–), 0.2 ± 0.1 (+), 1.0 ± 0.5 (++) or 4.0 ± 2.0 (+++) mg of PDF obtained from 20 ml of harvested bacteria.
bWhen PDF could not be shown by Coomassie staining (PDF1B) or western blot analysis (PDF1A) using anti-His-tag antibodies, ND (not detectable) is indicated.
cPAL421Tr strain was transformed at 30°C by the indicated plasmid. Ampicillin-resistant cells were then re-streaked at 42°C on pre-heated LB plates containing 100 µg/ml ampicillin and 0.5 mM IPTG (2 g/l glucose in the case of pUCdef).
+, a plasmid allowing normal cell growth after 12–24 h.
–, no growth after 24 h.
PDF and MAP genes are differently expressed in various A.thaliana tissues
Altogether, the above data demonstrated that the various cloned cDNAs expressed distinct proteins displaying either MAP or PDF activity similar to those found in bacterial cells. The unexpected occurrence of six MAPs and two PDFs raised the question concerning their relative expression in various tissues from A.thaliana. Poly(A)+ mRNAs were therefore prepared from fruits (2-week-old siliques), leaves, flowers and roots. Expression was systematically compared with that of elongation factor 1α, a housekeeping gene. Quantitative reverse transcription PCR (RT–PCR) experiments were performed to confirm the results. The sizes of seven of the eight mRNAs (except PDF1A) studied matched perfectly the length of the cloned cDNA. However, the PDF1A mRNA was 950 ± 50 bp and not 1050 ± 50 bp as expected. Primer extension analysis in 5′ and 3′ directions revealed that the 5′ extremity of the PDF1A cDNA, which was selected as the longest clone (see Materials and methods), was longer than that of the average mRNAs (data not shown). As a consequence, the first AUG codon of the PDF1A mRNAs may thus correspond to the second in-frame ATG codon of the cloned cDNA.
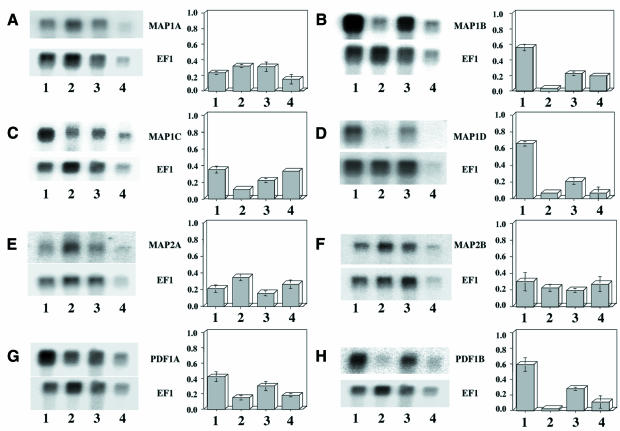
MAP1A, MAP1C, MAP2A, MAP2B and PDF1A mRNAs were all present and with a difference of abundance between 2- and 3-fold in the various tissues (Figure 3). In contrast, MAP1B, MAP1D and PDF1B mRNAs were strongly induced in green tissues, i.e. leaves and flowers (Figure 3). Finally, we noted that the amount of MAP2A was very low, <10% of that found for MAP2B.
Fig. 3. Expression of various MAP and PDF mRNAs in A.thaliana tissues. Two micrograms of poly(A)+ mRNAs were resolved by gel electrophoresis and analyzed by northern blotting as described in Materials and methods. The specific activity of the probe was in most cases ∼1 × 109 d.p.m./µg. Lane 1, leaves; lane 2, roots; lane 3, flowers; lane 4, siliques. Exposure was over 4 days. In each case, the blot was further re-hybridized with the elongation factor 1α probe (bottom) and exposed for 5 h. A fluorograph is shown for each specific transcript (PDF or MAP, top; EF1α, bottom). The table next to each panel is the average ratio of the intensity of the probe of interest to that of EF1α for three experiments. (A) MAP1A; (B) MAP1B; (C) MAP1C; (D) MAP1D; (E) MAP2A; (F) MAP2B; (G) PDF1A; (H) PDF1B.
The various MAPs and PDFs are localized in diverse cell compartments
Various prediction software, including chloroP (Emanuelsson et al., 1999), signalP (Nielsen et al., 1999), PSORT (Nakai and Horton, 1999) and Predotar (http://www.inra.fr/Internet/Produits/Predotar/), were used to analyse the function of the N-terminal domains. The PDF1A, PDF1B, MAP1B, MAP1C and MAP1D pre-sequences were predicted to contain targeting signals. In particular, they contain the high level of hydroxylated residues that is a feature of plant organellar targeting signals (Small et al., 1998). It has been suggested that the N-terminal extensions of MAP1A and MAP2 are involved in subcellular cytoplasmic targeting (Arfin et al., 1995; Li and Chang, 1995; Zuo et al., 1995). To determine the subcellular localization of the various proteins, their N-terminal extensions, as defined by comparison with the bacterial enzymes (i.e. E.coli MAP1 and PDF1 and P.furiosus MAP2; see Figure 1 and Table III), were fused to green fluorescent protein (GFP). The constructs were expressed transiently in epidermal onion cells after biolistic delivery. Onion cells possess low intrinsic red chlorophyll fluorescence of chloroplasts and have large and regular cells allowing the different compartments to be easily visualized by microscopy (Chiu et al., 1996). GFP fluorescence was monitored by epifluorescence microscopy and confocal laser scanning microscopy.
Table III. Escherichia coli strains and plasmids.
| Strain or plasmid | Genotype or markers | Reference |
|---|---|---|
| K37 | galK, rpsL | Miller and Friedman (1980) |
| JC10240 | Hfr(PO45) thr-3000 recA56 srl-300::Tn10 relA ilv-318 spoT1 thi-1 rpsE2300 | Csonka and Clark (1980) |
| PAL421Tr | fmsΔ1, galK, rpsL, recA56, srl-300::Tn10 | Meinnel and Blanquet (1994) |
| PAL314Tr | mapΔ1, galK, rpsL, recA56, srl-300::Tn10 | this work |
| JM101Tr | supE, thi, Δ(lac-pro), recA56, srl-300::Tn10, F′(traD36, proA+B+, lacIq, lacZΔM15) | Hirel et al. (1988) |
| pBluescriptSK | bla | Stratagene |
| pBEBNB | derivative of pBluescript SK with the BglII and NcoI sites inserted between the EcoRI and BamHI sites | Meinnel et al. (1993a) |
| pQE60 | bla, C-terminal His6-tag fusion vector | Qiagen |
| pMAK705 | cat, thermosensitive pSC101 replicon | Hamilton et al. (1989) |
| pUCdef | derivative of pUC18 with a chromosomal fragment from E.coli, fms, fmt′ | Meinnel and Blanquet (1993) |
| pMGDA-GFP | derivative of pSmRS-GFP with a SalII–NcoI fragment carrying the complete sequence of chloroplast membrane lipid MGDA synthetase from spinach fused in-frame to GFP | personal gift of E.Maréchal (Grenoble) |
| pXL1071 | bla, map | Mayaux et al. (1987) |
| pXΔ | bla, mapΔ1 | this work |
| pM1A | derivative of pBluescriptSK with the full-length cDNA encoding A.thaliana MAP1A | this work |
| pM1B | derivative of pBluescriptSK with the full-length cDNA encoding A.thaliana MAP1B | this work |
| pM1C | derivative of pBluescriptSK with the full-length cDNA encoding A.thaliana MAP1C | this work |
| pM1D | derivative of pBEBNB with the full-length cDNA encoding A.thaliana MAP1D | this work |
| pM2 | derivative of pBluescriptSK with the full-length cDNA encoding A.thaliana MAP2 | this work |
| pD1a | derivative of pBluescriptSK with the full-length cDNA encoding A.thaliana PDF1A | this work |
| pDLe1b | derivative of pBEBNB with the full-length cDNA encoding L.esculentum PDF1B | this work |
| pDAt1b | derivative of pBEBNB with the full-length cDNA encoding A.thaliana PDF1B | this work |
| pHuPD | derivative of pBluescriptSK with the full-length cDNA encoding the H.sapiens PDF homologue | this work |
| pMap | derivative of pMAK705, map | this work |
| pQdef1a | derivative of pQE60 and pD1a with an NcoI–BamHI insertion containing the whole A.thaliana PDF1A ORF (starting at third ATG) cloned between the NcoI and BglIII sites in-frame with the 6-His tag | this work |
| pQdef1b | derivative of pQE60 with an NcoI–BglIII insertion containing the whole L.esculentum PDF1B cDNA and the 3′ untranslated sequence cloned between the NcoI and BamHI sites | this work |
| pQdef1aΔN | same as pQdef1a but lacking codons 2–78 of the def gene | this work |
| pQdef1bΔN | same as pQdef1b but lacking codons 2–83 of the def gene | this work |
| pSmGFP | bla, CamV35S-smGFP-NosT | Davis and Vierstra (1998) |
| pSmRSGFP | bla, CamV35S-smRSGFP(S65T)-NosT | Davis and Vierstra (1998) |
| pDG1a | derivative of pSmGFP with an XbaI–BamHI fragment carrying the complete 5′ sequence of A.thaliana PDF1A (up to codon I83) inserted in-frame in between XbaI and BamHI | this work |
| pDG1aΔ1 | same as pDG1a1a but deletion of the first 18 bases (including ATG1) | this work |
| pDG1aΔ2 | same as pDG1a1a but deletion of the first 39 bases (including ATG1 and ATG2) | this work |
| pDRG1a | same as pDG1a, but derivative of pSmRS-GFP | this work |
| pDRG1aΔ1 | same as pDG1aΔ1, but derivative of pSmRS-GFP | this work |
| pDRG1aΔ2 | same as pDRG1aΔ2, but derivative of pSmRS-GFP | this work |
| pDG1b | derivative of pSmGFP with an XbaI–BamHI fragment carrying the complete 5′ sequence of A.thaliana PDF1B (up to codon I82) inserted in-frame in between XbaI and BamHI | this work |
| pDG2b | derivative of pSmGFP with an XbaI–BamHI fragment carrying the complete 5′ sequence of L.esculentum PDF1B (up to codon I89) inserted in-frame in between XbaI and BamHI | this work |
| pDRG1b | same as pDG1b, but derivative of pSmRS-GFP | this work |
| pMG1a | derivative of pSmGFP with an XbaI–BamHI fragment carrying the 5′ sequence of A.thaliana MAP1A (up to codon I141) inserted in-frame in between XbaI and BamHI | this work |
| pMG1b | derivative of pSmGFP with an XbaI–BamHI fragment carrying the 5′ sequence of A.thaliana MAP1A (up to codon I126) inserted in-frame in between XbaI and BamHI | this work |
| pMG1c | derivative of pSmGFP with an XbaI–BamHI fragment carrying the 5′ sequence of A.thaliana MAP1A (up to codon I99) inserted in-frame in between XbaI and BamHI | this work |
| pMG1d | derivative of pSmGFP with an XbaI–BamHI fragment carrying the 5′ sequence of A.thaliana MAP1A (up to codon L105) inserted in-frame in between XbaI and BamHI | this work |
| pMG2 | derivative of pSmGFP with an XbaI–BamHI fragment carrying the 5′ sequence of A.thaliana MAP2A (up to codon I126) inserted in-frame in between XbaI and BamHI | this work |
| pMRG2 | same as pMG2, but derivative of pSmRS-GFP | this work |
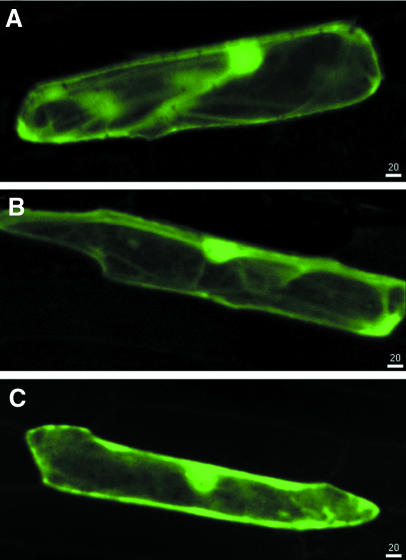
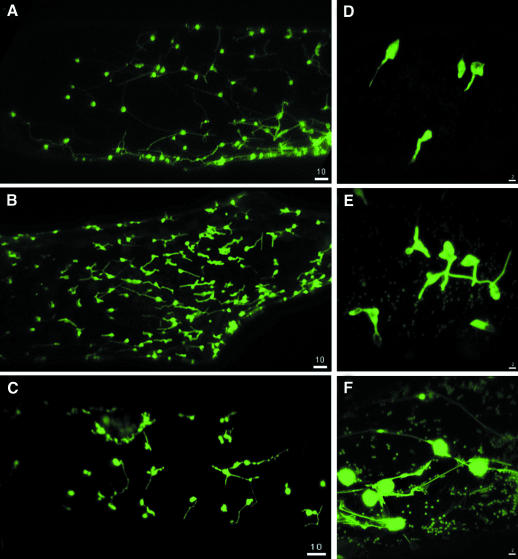
Production of the MAP1A–GFP or MAP2–GFP fusions resulted in GFP fluorescence throughout the cytoplasm, a pattern similar to that obtained with GFP alone (Figure 4). MAP1C–GFP and MAP1D–GFP were localized in both chloroplasts and mitochondria (Figure 5B and C). MAP1B–GFP was only found in chloroplasts (Figure 5A). As a control for plastid localization, we used a fusion of GFP to membrane lipid monogalactosyldiacylglycerol (MGDA) synthase from Spinacia oleracea, a nucleus-encoded protein shown to be located in chloroplasts by immunolocalization methods (Miege et al., 1999). In the onion cell system, MGDA–GFP fusions were efficiently targeted to plastids (data not shown) and the fluorescence pattern was indistinguishable from that of MAP1B–GFP fusions. This confirms the plastid localization of MAP1B, MAP1C and MAP1D. All three fusions were also found in stromules. Stromules are plastid connections that appeared as GFP-filled tubules emanating from the plastid surface (Figure 5). They are known to be abundant in tissues that contain chlorophyll-free plastids such as onion cells (Kohler and Hanson, 2000). In our experiments, GFP flowed through stromules (Figure 5), suggesting that MAP1 proteins are translocated between interconnected plastids. These observations are consistent with a system of coordination of plastid activities by inter-plastid communication (Kohler et al., 1997).
Fig. 4. The various pre-sequences of MAP1A and MAP2 target the protein to the cytoplasm. Confocal microscopy analysis of the expression of various GFP fusions in onion epidermal cells. Each image results from the horizontal projection of 20 different images. The white bar with a number indicates the scale in the two dimensions in micrometres. (A) GFP; (B) MAP1A–GFP fusion; (C) MAP2A–GFP fusion.
Fig. 5. The various pre-sequences of MAP1B, 1C and 1D target the protein to the organelles. Same analysis as Figure 4 but with the N-pre-sequences of MAP1B (A and D), MAP1C (B and E) and MAP1D (C and F).
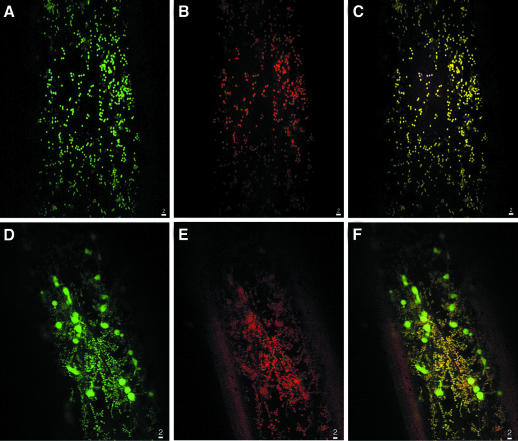
The PDF1A mRNAs include two putative alternative AUG start codons (see above). This phenomenon can be the basis for routing a protein to distinct cell compartments (Small et al., 1998). Three constructions in which the various ATG codons are the first AUG of the transcript were constructed. All three constructs resulted in a mitochondrial localization of GFP (Figure 6A). The mitochondrial localization was confirmed by labelling experiments in the presence of Mitotracker, a specific mitochondrial probe (Figure 6B and C). In the case of the construct with the most downstream start codon, which corresponds to the most abundant transcript (see above), many positive cells were observed and the fluorescent signal was stronger. Arabidopsis thaliana PDF1B, like MAP1C and MAP1D, was found in plastids and mitochondria (Figure 6D, E and F). This double localization in the case of PDF1B was further confirmed with a fusion of the pre-sequence of tomato PDF1B fused to GFP (data not shown).
Fig. 6. The various pre-sequences of plant PDF1A and PDF1B target the protein to the mitochondria and the plastids, respectively. Same analysis as Figure 4 but with the N-pre-sequences of PDF1B (A–C) and PDF1B (D–F). (B) and (E) were obtained after incubation of the slide with Mitotracker and analysis of its intrinsic fluorescence (see Materials and methods). (C) and (F) show the double labelling. The yellow colour results from the mixture of green and red.
From the above localization data (summarized in Table I), it thus appears that the process of removal of the first methionine occurs not only in the cytoplasm but also in the organelles of plants. In this context, we compared the protein sequences of several organelle gene products with the corresponding DNA genome data. A majority of the proteins indeed lacked the N-formyl or N-formylmethionine (Widger et al., 1984; Kamp et al., 1987; Hauska et al., 1988; Michel et al., 1988; Houtz et al., 1989; Yokoi et al., 1990, 1991; Schmidt et al., 1992; Braun and Schmitz, 1993; Gabler et al., 1994; Herz et al., 1994; Shanklin et al., 1995; Maffey et al., 1997). Some organellar membrane proteins apparently retained their N-formyl (Sebald and Wächter, 1980; Scheller et al., 1989), a situation also observed in eubacteria (von Heijne, 1989). Analysis of the specificity with which methionine removal occurs in plants fits perfectly with the known selectivity of eubacterial MAP and PDF (Meinnel et al., 1993b; Giglione et al., 2000).
Identification of MAP and PDF homologues in eukaryotic protists and Drosophila
The discovery of a novel pathway dedicated to methionine removal in higher plants raised the question of its existence in other eukaryotes. Interestingly, a gene encoding a PDF homologue was identified in the nuclear genome of Plasmodium falciparum, a eukaryotic protist related to green algae (Figure 1C; see also Meinnel, 2000). This PDF has an N-terminal pre-sequence and is very similar to PDF1B, the ‘plastid’ PDF form (see Figure 1C). Together with our results, this phylogeny strongly suggests that the P.falciparum PDF1B is routed to the apicoplast, the non-photosynthetic plastid of apicomplexan protists. Taking advantage of our data on MAP, we also found an organellar MAP1 on chromosome 5 of P.falciparum (Figure 1A). In addition, one organellar MAP1B gene on chromosome 2 (contig 16482) and one mitochondrial PDF1A sequence on chromosome 6 (contig 12497) could be identified in the genome of a protist devoid of plastid, the soil amoeba Dictyostelium discoideum (Figure 1A and C).
Very recently, the near complete genome of the fruit fly Drosophila was made available (Adams et al., 2000). One MAP1A and one MAP2 sequence were retrieved; indeed all eukaryotes have these two genes (Figure 1A and B). Unexpectedly, a search for MAP1B, MAP1C and MAP1D homologues identified one gene on chromosome 2L (DDBJ/EMBL/GenBank accession Nos AC007451 and AE003628). It is very similar to the three plant organellar MAP1s, including the sequence along the N-terminal domain (Figure 1A). In addition, it did not contain the Cys-His zinc finger that characterizes all known cytoplasmic MAP1As. Thus, the fruit fly appears to have an organellar-targeted MAP and consequently an N-terminal protein processing pathway in mitochondria. This suggests that the Drosophila genome may also have a gene encoding a PDF with an N-terminal targeting pre-sequence. Indeed, two adjacent PDF sequences were identified on chromosome 3R (DDBJ/EMBL/GenBank accession Nos AE003688 and AC007822). The annotated deduced protein (AAF54540), which is derived from the fusion of the two genes, appears however to be incorrectly processed. Re-analysis of the data using various tools available at http://www.fruitfly.org/seq_tools/splice.html (Reese et al., 2000) suggested that there are actually two genes encoding two PDFs with different extended N-terminal pre-sequences (Figure 1C). In addition, a TATA box and two poly-adenylation signals were found in between the two genes. The analysis of two ESTs from another insect of the dipter an order, the African malaria mosquito Anopheles gambiae, strengthened this prediction. These ESTs were overlapping, which allowed us to reconstitute one nearly complete cDNA. The deduced protein pre-sequence (Figure 1C) corresponded to that of one of the two PDF sequences found in the Drosophila genome. The amino acid homology continued upstream of the proposed splicing site within the sequence translated from the putative intron. We concluded that the fly genome encodes two PDF1s, with distinct N-terminal pre-sequences. Although this situation appears to be similar to that of higher plants, both fly sequences are most related to the mitochondrial form, i.e. PDF1A.
Cloning the human PDF1A homologue cDNA
The above data were highly suggestive that a machinery required for N-terminal methionine removal occurs not only in higher plant organelles but also in the mitochondria of insects. Such a result was completely unexpected and led us to wonder whether mammalian cells also possess such a system. PDF was a better marker for this machinery than MAP, since MAP1 sequences are found in all organisms (Figure 1A). Because PDF is thought to be a good target for new antibiotics (Giglione et al., 2000), and given that substantial human genome sequence is now available, we looked for PDF homologues in human. Using PDF1A and PDF1B as input sequences, we searched the complete human EST and genome data. Analysis of the ESTs produced a family of five candidates (AI859289, AL045195, AW501444, AI765656 and AW499510). Two clearly came from bacterial DNA contamination (AW499510 and AW501444), whereas the three others were associated with 18 additional related sequences in Unigene Hs.130849 (http://www.ncbi.nlm.nih.gov/UniGene/; Schuler, 1996, 1997). With the help of these ESTs, we identified the associated gene fragment composed of two exons on human chromosome 16 (Table I). We cloned the corresponding, full-length cDNA from human fetal tissues by RACE. The deduced PDF was very similar to insect and A.thaliana PDF1A, i.e. the mitochondrial PDF (Figure 1C). Data from Serial Analysis of Gene Expression (Velculescu et al., 1995) of Unigene Hs.130849 at http://www.ncbi.nlm.nih.gov/SAGE/ revealed that this sequence was expressed at the same level in all types of tissues in human, a situation similar to that of Arabidopsis PDF1A (see previous results). We also searched the mouse EST and identified four sequences (AA438042, AV113186, AA162512 and AA153405) related to human PDF1A (Unigene Mm.34708). The clone corresponding to the longest insertion was re-sequenced and the sequence extended (Figure 1C). Although the cDNA insertion was partial, it was quite similar to the human PDF1A homologue. Finally, homologues of the human PDF1A sequence were also found in the Rattus norvegicus EST library and in a partial genome sequence of the freshwater pufferfish Tetraodon nigroviridis (Figure 1C).
These data show that the mitochondrial PDF1A homologue is constitutively expressed in many animal cells. It is, therefore, likely that animal cells possess the machinery of N-terminal protein processing that we identified and characterized for the first time in higher plants.
Discussion
In this paper, we describe the identification of a multi-gene family encoding two enzyme types (PDF and MAP) that are both dedicated to the N-terminal processing pathway in higher plants. Taken together, our results show that the occurrence of the eight genes in A.thaliana encoding the various proteins reflects a special meaning. Indeed, most of these gene products fill specific functions in the cell through the combination of (i) their relative tissue-specific expression (Figure 3) and/or (ii) their capacity to be routed to different cell compartments where de novo protein synthesis occurs, i.e. the cytoplasm, the plastids and/or the mitochondria (Table I). For instance, MAP1A is the only MAP1 found in the cytoplasm, and PDF1A and MAP1C are the only PDF and MAP in the mitochondria of non-green tissues. Similarly, PDF1B is the only PDF in plastids. In contrast, MAP2A and MAP2B appear to be redundant, as both genes are constitutive and encode cytoplasmic MAP2. Nevertheless, >90% of the mRNAs come from the transcription of MAP2B. The family of MAP1 is very interesting, since the various sequences differ only in their N-terminal pre-sequence. As is the case for several such plant organellar proteins (for a review see Small et al., 1998), we were able to demonstrate that this pre-sequence by itself is enough to direct each protein product to its proper cell compartment. Nuclear-encoded organellar proteins are believed to be encoded by genes that have been transferred from the organelle genome (Martin et al., 1998). For instance, phylogenetic analysis suggests that yeast and human cytoplasmic MAP1 are derived from an organellar protein that itself was derived from an ancestral endosymbiotic eubacterium (Keeling and Doolittle, 1996; Bradshaw et al., 1998). Our work supports this hypothesis, as we were able to identify for the first time authentic organellar MAP1. The occurrence of organellar PDF genes that were previously believed to be found only in eubacteria is further evidence in favour of this theory.
We have found in animal genomes, including insects, fish and human, sequences homologous to mitochondrial PDF1A and MAP1D. It was previously believed that N-formylmethionine was not removed from proteins in eukaryotes and that, in turn, deformylase is a unique characteristic of eubacteria (Mazel et al., 1994). PDF has thus been considered to be an attractive target for antibacterial therapy (Giglione et al., 2000). This view was based on several indirect data including the absence of PDF genes in the S.cerevisiae and C.elegans genomes. Moreover, several authors have shown that various mammalian mitochondrial proteins are N-blocked by Edman sequencing and that this reaction was only possible after treatment of the proteins under mild acidic conditions. Although effective for N-deformylation (Sheehan and Young, 1958), this treatment also removes the block to Edman sequencing due to other N-substitutions. Note that such analysis has been carried out for only six bovine proteins of the 13 that are encoded in the mitochondrial genome, and that no similar data are available for insect, fish or human. There are two possible explanations for these data. First, if indeed there is no deformylation and further removal of the first methionine in animal mitochondria, then organellar PDF and MAP1 might be remnants of ancient functions with no current role. A second, more likely hypothesis is that N-terminal processing does occur in animal mitochondria but that the proteins undergoing the modification have not been identified. In the light of our present data, a re-evaluation of the N-terminal sequence data of human mitochondrially encoded proteins is required.
The demonstration of a PDF homologue in human cells might question deformylase as an ideal target for new antibiotics, as defined recently (see Giglione et al., 2000). Nevertheless, a number of antibiotics currently in use are known to act on both bacteria and mitochondria. Before any clinical trials, the role of the mammalian PDF homologue and the impact of its inhibition on eukaryotic cells should indeed be elucidated.
Materials and methods
Bacterial strains and plasmids
The bacterial strains and plasmids used are shown in Table III. Pal314Tr was constructed as follows. Plasmid pXL1071, which bears a 1.1 kb SmaI–PstI fragment expressing the whole E.coli map gene, was digested with HpaI, which cleaves within the beginning of the MAP open reading frame (ORF). It was then ligated to two complementary oligonucleotides (5′-TAAGGATCCTA), which introduce a unique BamHI restriction site, an in-frame TAA translation stop codon and a +1 base frame shift. This yielded plasmid pXLΔ, which expresses a truncated inactive MAP. The SacI–PstI fragment of pXLΔ was then sub-cloned between the same sites of pMAK705 to yield pMapΔ. Using the procedure described by Hamilton et al. (1989), we constructed, using strain K37 as starting material, strain Pal314-pMap. This strain was made recombination deficient by conjugation with the recA56 Hfr strain JC10240. This yielded Pal314Tr-pMap. At non-permissive temperature (42°C), this strain fails to grow on Luria–Bertani (LB) plates. At 30°C the strain grows.
Plant material and culture conditions
Arabidopsis thaliana (Wisconsin ecotype) seeds were sown after surface sterilization on Murashige and Skoog (ICN) medium (Anderson and Wilson, 2000) containing 1% agarose and incubated in a growth chamber (22°C/18°C, 14 h daylight) for up to 2 weeks. Alternatively, for long-term growth, seeds were directly sown on soil or sand and grown in a greenhouse. Lycopersicon esculentum (Money Marker) was grown in a greenhouse (24°C/18°C, 16 h daylight) for 1 month.
General molecular biology techniques
Restriction enzymes and T4 DNA ligase were purchased from New England BioLabs and Bethesda Research Laboratories. Buffers used were those indicated by the suppliers. Oligonucleotides were synthesized by MWG-AG-Biotech. A Perkin-Elmer 2400 GeneAmp PCR system was used for all PCR analyses. Nucleotide sequences were determined by the Big-Dye terminator method (Perkin-Elmer), with DNA templates purified with the Qiagen mini-prep. kit and analysed on a semi-automated sequencing system (model 373A; Applied-Biosystems). Nucleotide sequence data (11 cDNAs) are available at DDBJ/EMBL/GenBank under accession Nos AF239156, AF250958, AF250959, AF250960, AF250961, AF250962, AF250963 AF250964, AF269165, AF271258 and AF300880.
DNA was extracted from 2-week-old seedlings by using the DNeasy plant maxi kit (Qiagen). RNA was prepared from A.thaliana tissues as described previously (Kay et al., 1987). The RNAs were further purified with an RNeasy kit (Qiagen). Poly(A)+ RNA samples for electrophoresis were prepared with the Oligotex kit (Qiagen). RNA concentrations were quantified by measuring absorbance at 260 nm and equal amounts (1–2 µg) were size-fractionated by electrophoresis through 6% formaldehyde–1.2% agarose gels prior to transfer to nylon membranes (Hybond-N; Amersham-Pharmacia Biotech). Transcript sizes were assessed by comparison with two RNA ladders (Gibco-BRL, 1.77–0.16 kb; Promega Biotech, 6.58–0.28 kb) after staining with methylene blue (Sambrook et al., 1989).
An oligolabelling kit (Amersham-Pharmacia Biotech) was used to label probes with the various full-length cDNAs (Table I) as templates and [α-32P]dATP and [α-32P]dCTP (800 Ci/mmol). Probes were purified with the QIAquick nucleotide removal kit (Qiagen). Northern and Southern blotting were performed as described (Sambrook et al., 1989) with stringent hybridization and washing. Radioactivity was revealed with phosphor screens on a PhosphorImager (Storm 840; Molecular Dynamics). The systematic lower signal of the mRNAs in siliques is likely to be due to the accumulation of mRNAs encoding seed storage proteins. This was verified by using a seed-specific probe (encoding the seed-storage protein albumin 2S) that hybridized strongly with the silique mRNA lane only (data not shown). RT–PCR analyses were performed using the Superscript ONE-STEP kit (Gibco-BRL) with oligonucleotides the sequence of which overlapped two adjacent exon borders to avoid cross amplification with contaminant genomic DNA. After gel electrophoresis, blotting and hybridization, radioactivity associated with the amplified fragments was quantified on a PhosphorImager. Quantitative results were used to confirm the data obtained with northern blot experiments.
RACE involved libraries from A.thaliana seedlings (2 weeks) and L.esculentum young leaves (∼1 cm) constructed starting from 1 µg of poly(A)+ mRNA purified from total RNA with the Poly(A) Quick mRNA isolation kit (Stratagene). The Marathon™ cDNA Amplification kit (Clontech) and the recommendations of the manufacturer were used for the construction of both RACE libraries. The RACE library from human fetus was purchased from Clontech. Screening for the 5′ and 3′ extremities involved nested PCR using the Advantage cDNA PCR kit (Clontech) for the first amplification step and the Advantage-HF2 PCR kit (Clontech) or PfuTurbo DNA Polymerase (Stratagene) for the second amplification. Amplification experiments of MAP1C led to three PCR products. Random cloning showed that one of the PCR products was a consequence of mRNAs incorrectly spliced at the level of intron 4. About one half of the cloned cDNAs corresponded to fragments either containing the full-length intron 4 or with incorrect splicing at this level, the intron being shifted by 37 and 64 bases upstream of the authentic acceptor and donor sites, respectively. Both aberrant and wrongly spliced mRNAs encoded greatly truncated proteins that were expected to be inactive. Similarly, ∼50% of the PCR products of MAP2A showed a lack of cleavage of their intron 1. Finally, two different 3′ PCR products were found for A.thaliana PDF1B. The difference concerned the 3′ untranslated sequence: one of the two transcripts showed splicing of the terminal intron whereas the other did not. RT–PCR experiments indicated that the unspliced transcript accounted for ∼10–20% of the mRNAs (data not shown).
PCR products were further cloned in pBluescriptSK or pEBNB. In most cases, a dozen of the longest clones (at the 5′ and 3′ ends of the cDNA) were analysed and half were sequenced. The longest clones were used for the reconstruction of the full-length cDNAs. This was achieved in two steps, using an internal unique restriction site, except for MAP1B where three steps were necessary.
Protein analysis
JM101Tr cells expressing the given plasmid were grown at 23°C for 8–12 h in LB medium supplemented with 50 µg/ml ampicillin until the OD600 reached ∼0.9. Cells were induced with 0.1 mM isopropyl-β-d-thiogalactopyranlside (IPTG) and left for another 24 h under agitation. The cells were then re-suspended in 20 mM KH2PO4 pH 7.3, 150 mM NaCl, broken by ultra-sonication and centrifuged for 5 min in an Eppendorf microfuge (12 000 rpm) at 4°C. The supernatant was collected and 1 vol. mixed with 1 vol. of loading buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol). The pellet was re-suspended and solubilized in the same buffer. Protein concentrations were measured according to Bradford (1976) with the Bio-Rad protein assay kit. Immunoglobulin G was used as the protein standard. The Mini-PROTEAN III system (Bio-Rad) was used for polyacrylamide gel electrophoresis (PAGE) on SDS denaturing gels (12% gels, 0.75 mm thickness). Gels were stained with Bio-Safe Coomassie Stain (Bio-Rad). Proteins were electro-transferred on nitrocellulose BA85 membranes (Schleicher and Schüll) with a wet transfer unit (Hoefer, MiniTank Transphor Unit) in the cold room. Western blots with mouse anti-His-tag monoclonal antibodies (Clontech; dilution 1/2000) and peroxidase-linked anti-mouse antibodies from sheep (dilution 1/5000) were developed with the ECL detection reagents (Amersham-Pharmacia Biotech) and X-Ray films (Kodak).
Peptide deformylase activity assay was coupled to formate dehydrogenase essentially as described (Lazennec and Meinnel, 1997). PDF activity was assayed at 37°C as the increase of the absorbance at 340 nm of NADH (εM = 6300 M–1 cm–1). The standard assay was performed in a final volume of 200 µl with a 1 cm optical path. The time-course absorbance change was measured with an Ultrospec 4000 spectrophotometer (Amersham-Pharmacia Biotech). The reaction mixtures contained 50 mM HEPES pH 7.5, 1 mM Fo-Met-Ala-Ser (Bachem) or Fo-Met-Ala (Sigma), 12 mM NAD+ (Boehringer), 1 mM NiCl2 and 4.5 U/ml yeast formate dehydrogenase (Sigma). The reaction was started by the addition of 5 µl of crude bacterial extract diluted in 50 mM HEPES pH 7.5.
Plant cell GFP transient expression and particle bombardment
Two plasmids, pSmGFP and pSmRSGFP, expressing GFP under the control of the cauliflower mosaic virus 35S promoter, were used and expressed transiently in plant cells. Both encoded soluble highly fluorescent variants of GFP for use in higher plants (Haseloff et al., 1997; Davis and Vierstra, 1998). In addition to the Q80R, F99S, M153T and V163A substitutions borne by both constructs, smGFP and smRSGFP differed at amino acid position S65; RSGFP bearing the S65T substitution causing an increased fluorescence output and higher expression levels. Note that re-sequencing of the CamV35S–GFP region (data available upon request) of these plasmids allowed us to identify a unique XbaI restriction site just upstream from the BamHI site.
Biolistic bombardments were performed with a PDS-1000/He instrument (Bio-Rad). Acceleration of gold micro-carriers (1.0 µm) coated with 1.2 µg of pure plasmid DNA (purified with Qiagen mini-prep kits) spotted on macro-carriers was used to transform onion epidermal cells. Bombardment parameters were as follows: vacuum, 28 inches Hg; distance to target, 6 cm; helium pressure, 650 p.s.i. Onion scales were left for 12–24 h in the dark at 24°C, and epidermal tissues removed and layered in water on glass slides for microscopy. The tissues were examined under a Reichert epifluorescence microscope and then a confocal microscope.
GFP and Mitotracker visualization
For the Reichert epifluorescence microscope, GFP-specific fluorescence was observed by using the filters BP475–495 nm (excitation), DS510 nm (dichroic) and LP520–560 nm (barrier). Confocal microscopy was carried out using a confocal laser scanning microscope (Sarastro 2000; Molecular Dynamics) running on a SiliconGraphics Indy workstation. The confocal microscope was equipped with a 25 mW argon laser (488 nm), a 510 EFLP short pass filter for excitation and an upright Nikon microscope. A reflect short pass beam-splitter filter (565DRLP) was used to separate the emission beam in two. GFP fluorescence was collected through a bandpass filter (530DF30). In some experiments Mitotracker Red CMXRos (Molecular Probes, Leiden) was used to stain mitochondria. The dye was added to a final concentration of 1 µM. Mitotracker Red fluorescence was collected through the 565DRLP and a long pass filter (600ELFP). Images (512 × 512 pixels) were generated using two-pass Kalman averaging and 40×, 60× or 100× Nikon oil objectives with NA 1.3–1.4.
Acknowledgments
Acknowledgements
The authors wish to thank A.Kondorosi, F.Dardel, J.Bauly and J.Leung for critical review of the manuscript, J.-R.Prat and S.Brown for help with confocal microscopy experiments and M.Crespi for advice with biolistic bombardment. E.Maréchal and J.Joyard (CEA, Grenoble, France) are kindly acknowledged for the generous gift of plasmid pMGDA-GFP. Sequence data for D.discoideum chromosomes were obtained from The Sanger Centre website at http://www.sanger.ac.uk/Projects/D.discoideum/. Sequencing of D.discoideum chromosomes was accomplished as part of the EUDICT consortium with support from The European Union. This work was supported by ATIPE and PCV grants from the C.N.R.S. to T.M. and by the Fondation pour la Recherche Médicale. C.G. is supported by a post-doctoral fellowship from the Association pour la Recherche sur le Cancer (ARC, Villejuif, France).
References
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2196. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson M. and Wilson,F. (2000) Growth, maintenance and use of Arabidopsis genetic resources. In Hames,B.D. (ed.), Arabidopsis, A Practical Approach. Oxford University Press, Oxford, UK, pp. 1–26. [Google Scholar]
- Apfel C. et al. (2000) Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J. Med. Chem., 43, 2324–2331. [DOI] [PubMed] [Google Scholar]
- Arfin S.M. and Bradshaw,R.A. (1988) Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry, 27, 7979–7984. [DOI] [PubMed] [Google Scholar]
- Arfin S.M., Kendall,R.L., Hall,L., Weaver,L.H., Stewart,A.E., Matthews, B.W. and Bradshaw,R.A. (1995) Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc. Natl Acad. Sci. USA, 92, 7714–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bradshaw R.A., Brickey,W.W. and Walker,K.W. (1998) N-terminal processing: the methionine aminopeptidase and N α-acetyl transferase families. Trends Biochem. Sci., 23, 263–267. [DOI] [PubMed] [Google Scholar]
- Braun H.P. and Schmitz,U.K. (1993) Purification and sequencing of cytochrome b from potato reveals methionine cleavage of a mitochondrially encoded protein. FEBS Lett., 316, 128–132. [DOI] [PubMed] [Google Scholar]
- Chang Y.H., Teichert,U. and Smith,J.A. (1992) Molecular cloning, sequencing, deletion and overexpression of a methionine aminopeptidase gene from Saccharomyces cerevisiae. J. Biol. Chem., 267, 8007–8011. [PubMed] [Google Scholar]
- Chen D.Z. et al. (2000) Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry, 39, 1256–1262. [DOI] [PubMed] [Google Scholar]
- Chiu W., Niwa,Y., Zeng,W., Hirano,T., Kobayashi,H. and Sheen,J. (1996) Engineered GFP as a vital reporter in plants. Curr. Biol., 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Crosti P., Gambini,A., Lucchini,G. and Bianchetti,R. (1977) Eukaryotic N10-formyl-H4folate:methionyl-tRNAf transformylase. Some properties of the Euglena gracilis enzyme. Biochim. Biophys. Acta, 477, 356–370. [DOI] [PubMed] [Google Scholar]
- Csonka L.N. and Clark,A.J. (1980) Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J. Bacteriol., 143, 529–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J. and Vierstra,R.D. (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol., 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen,H. and von Heijne,G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci., 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabler L., Herz,U., Liddell,A., Leaver,C.J., Schroder,W., Brennicke,A. and Grohmann,L. (1994) The 42.5 kDa subunit of the NADH: ubiquinone oxidoreductase (complex I) in higher plants is encoded by the mitochondrial nad7 gene. Mol. Gen. Genet., 244, 33–40. [DOI] [PubMed] [Google Scholar]
- Giglione C., Pierre,M. and Meinnel,T. (2000) Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol. Microbiol., 36, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Guillemaut P. and Weil,J.H. (1975) Aminoacylation of Phaseolus vulgaris cytoplasmic, chloroplastic and mitochondrial tRNAsMet and of Escherichia coli tRNAsMet by homologous and heterologous enzymes. Biochim. Biophys. Acta, 407, 240–248. [DOI] [PubMed] [Google Scholar]
- Halbreich A. and Rabinowitz,M. (1971) Isolation of Saccharomyces cerevisiae mitochondrial formyltetrahydrofolic acid:methionyl-tRNA transformylase and the hybridization of mitochondrial fMet-tRNA with mitochondrial DNA. Proc. Natl Acad. Sci. USA, 68, 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C.M., Aldea,M., Washburn,B.K., Babitzke,P. and Kushner,S.R. (1989) New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol., 171, 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Siemering,K.R., Prasher,D.C. and Hodge,S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl Acad. Sci. USA, 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauska G., Nitschke,W. and Herrmann,R.G. (1988) Amino acid identities in the three redox center-carrying polypeptides of cytochrome bc1/b6f complexes. J. Bioenerg. Biomembr., 20, 211–228. [DOI] [PubMed] [Google Scholar]
- Hebsgaard S.M., Korning,P.G., Tolstrup,N., Engelbrecht,J., Rouze,P. and Brunak,S. (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res., 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz U., Schroder,W., Liddell,A., Leaver,C.J., Brennicke,A. and Grohmann,L. (1994) Purification of the NADH:ubiquinone oxidoreductase (complex I) of the respiratory chain from the inner mitochondrial membrane of Solanum tuberosum. J. Biol. Chem., 269, 2263–2269. [PubMed] [Google Scholar]
- Hirel P.H., Lévêque,F., Mellot,P., Dardel,F., Panvert,M., Mechulam,Y. and Fayat,G. (1988) Genetic engineering of methionyl-tRNA synthetase: in vitro regeneration of an active synthetase by proteolytic cleavage of a methionyl-tRNA synthetase-β-galactosidase chimeric protein. Biochimie, 70, 773–782. [DOI] [PubMed] [Google Scholar]
- Houtz R.L., Stults,J.T., Mulligan,R.M. and Tolbert,N.E. (1989) Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc. Natl Acad. Sci. USA, 86, 1855–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington K., Yi,T., Wei,Y. and Pei,D. (2000) Synthesis and antibacterial activity of peptide deformylase inhibitors. Biochemistry, 39, 4543–4551. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., Thompson,J.D., Gouy,M., Higgins,D.G. and Gibson,T.J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem. Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- Johnson D.R., Bhatnagar,R.S., Knoll,L.J. and Gordon,J.I. (1994) Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem., 63, 869–914. [DOI] [PubMed] [Google Scholar]
- Kamp R.M., Srinivasa,B.R., von Knoblauch,K. and Subramanian,A.R. (1987) Occurrence of a methylated protein in chloroplast ribosomes. Biochemistry, 26, 5866–5870. [Google Scholar]
- Kay R., Chan,A., Daly,M. and McPherson,J. (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science, 236, 1299–1302. [DOI] [PubMed] [Google Scholar]
- Keeling P.J. and Doolittle,W.F. (1996) Methionine aminopeptidase-1: the MAP of the mitochondrion? Trends Biochem. Sci., 21, 285–286. [PubMed] [Google Scholar]
- Kendall R.L., Yamada,R. and Bradshaw,R.A. (1990) Cotranslational amino-terminal processing. Methods Enzymol., 185, 398–407. [DOI] [PubMed] [Google Scholar]
- Kohler R. and Hanson,M.R. (2000) Plastid tubules of higher plants are tissue-specific and developmentally regulated. J. Cell Sci., 113, 81–89. [DOI] [PubMed] [Google Scholar]
- Kohler R.H., Cao,J., Zipfel,W.R., Webb,W.W. and Hanson,M.R. (1997) Exchange of protein molecules through connections between higher plant plastids. Science, 276, 2039–2042. [DOI] [PubMed] [Google Scholar]
- Krishna R.G. and Wold,F. (1993) Post-translational modification of proteins. Adv. Enzymol. Relat. Areas Mol. Biol., 67, 265–298. [DOI] [PubMed] [Google Scholar]
- Lazennec C. and Meinnel,T. (1997) Formate dehydrogenase-coupled spectrophotometric assay of peptide deformylase. Anal. Biochem., 244, 180–182. [DOI] [PubMed] [Google Scholar]
- Li X. and Chang,Y.H. (1995) Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl Acad. Sci. USA, 92, 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Widom,J., Kemp,C.W. and Clardy,J. (1998) Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science, 282, 1324–1327. [DOI] [PubMed] [Google Scholar]
- Lowther W.T. and Matthews,B.W. (2000) Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta, 1477, 157–167. [DOI] [PubMed] [Google Scholar]
- Lowther W.T., Orville,A.M., Madden,D.T., Lim,S., Rich,D.H. and Matthews,B.W. (1999) Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant and inhibited enzymes for the mechanism of catalysis. Biochemistry, 38, 7678–7688. [DOI] [PubMed] [Google Scholar]
- Maffey L., Degand,H. and Boutry,M. (1997) Partial purification of mitochondrial ribosomes from broad bean and identification of proteins encoded by the mitochondrial genome. Mol. Gen. Genet., 254, 365–371. [DOI] [PubMed] [Google Scholar]
- Martin W., Stoebe,B., Goremykin,V., Hansmann,S., Hasegawa,V. and Kowallik,K.V. (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature, 393, 162–165. [DOI] [PubMed] [Google Scholar]
- Mayaux J.F., Soubrier,F. and Latta,M. (1987) Cloning E.coli genes by oligonucleotide hybridization. Nucleic Acids Res., 15, 10593–10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D., Pochet,S. and Marliere,P. (1994) Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J., 13, 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T. (2000) Peptide deformylase of eukaryotic protists: a target for new antiparasitic agents? Parasitol. Today, 16, 165–168. [DOI] [PubMed] [Google Scholar]
- Meinnel T. and Blanquet,S. (1993) Evidence that peptide deformylase and methionyl-tRNA(fMet) formyltransferase are encoded within the same operon in Escherichia coli. J. Bacteriol., 175, 7737–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T. and Blanquet,S. (1994) Characterization of the Thermus thermophilus locus encoding peptide deformylase and methionyl-tRNA(fMet) formyltransferase. J. Bacteriol., 176, 7387–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Guillon,J.M., Mechulam,Y. and Blanquet,S. (1993a) The Escherichia coli fmt gene, encoding methionyl-tRNA(fMet) formyltransferase, escapes metabolic control. J. Bacteriol., 175, 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam,Y. and Blanquet,S. (1993b) Methionine as translation start signal: a review of the enzymes of the pathway in Escherichia coli. Biochimie, 75, 1061–1075. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Lazennec,C., Villoing,S. and Blanquet,S. (1997) Structure–function relationships within the peptide deformylase family. Evidence for a conserved architecture of the active site involving three conserved motifs and a metal ion. J. Mol. Biol., 267, 749–761. [DOI] [PubMed] [Google Scholar]
- Michel H., Hunt,D.F., Shabanowitz,J. and Benett,J. (1988) Tandem mass spectrometry reveals that three photosystem II proteins of spinach chloroplasts contain N-acetyl-O-phosphothreonine at their NH2 termini. J. Biol. Chem., 263, 1123–1130. [PubMed] [Google Scholar]
- Miege C., Marechal,E., Shimojima,M., Awai,K., Block,M.A., Ohta,H., Takamiya,K., Douce,R. and Joyard,J. (1999) Biochemical and topological properties of type A MGDG synthase, a spinach chloroplast envelope enzyme catalyzing the synthesis of both prokaryotic and eukaryotic MGDG. Eur. J. Biochem., 265, 990–1001. [DOI] [PubMed] [Google Scholar]
- Miller H.I. and Friedman,D.I. (1980) An E.coli gene product required for λ site-specific recombination. Cell, 20, 711–719. [DOI] [PubMed] [Google Scholar]
- Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Brunak,S. and von Heijne,G. (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng., 12, 3–9. [DOI] [PubMed] [Google Scholar]
- Ragusa S., Blanquet,S. and Meinnel,T. (1998) Control of peptide deformylase activity by metal cations. J. Mol. Biol., 280, 515–523. [DOI] [PubMed] [Google Scholar]
- Reese M.G., Kulp,D., Tammana,H. and Haussler,D. (2000) Genie—Gene finding in Drosophila melanogaster. Genome Res., 10, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Scheller H.V., Okkels,J.S., Hoj,P.B., Svendsen,I., Roepstorff,P. and Moller,B.L. (1989) The primary structure of a 4.0-kDa photosystem I polypeptide encoded by the chloroplast psaI gene. J. Biol. Chem., 264, 18402–18406. [PubMed] [Google Scholar]
- Schmidt J., Herfuth,E. and Subramanian,A.R. (1992) Purification and characterization of seven chloroplast ribosomal proteins: evidence that organelle ribosomal protein genes are functional and that NH2-terminal processing occurs via multiple pathways in chloroplasts. Plant Mol. Biol., 20, 459–465. [DOI] [PubMed] [Google Scholar]
- Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- Schuler G.D. et al. (1996) A gene map of the human genome. Science, 274, 540–546. [PubMed] [Google Scholar]
- Sebald W. and Wächter,E. (1980) Amino acid sequence of the proteolipid subunit of the ATP synthase from spinach chloroplasts. FEBS Lett., 122, 307–311. [Google Scholar]
- Shanklin J., DeWitt,N.D. and Flanagan,J.M. (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell, 7, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan J.C. and Young,D.H. (1958) The use of N-formylamino acids in peptide synthesis. J. Am. Chem. Soc., 80, 1154–1158. [Google Scholar]
- Sin N., Meng,L., Wang,M.Q., Wen,J.J., Bornmann,W.G. and Crews,C.M. (1997) The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl Acad. Sci. USA, 94, 6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I., Wintz,H., Akashi,K. and Mireau,H. (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol. Biol., 38, 265–277. [PubMed] [Google Scholar]
- Steinmetz A. and Weil,J.H. (1989) Protein synthesis in chloroplasts. In Marcus,A. (ed.), The Biochemistry of Plants. Academic Press, San Diego, CA, pp. 193–227. [Google Scholar]
- Tahirov T.H., Oki,H., Tsukihara,T., Ogasahara,K., Yutani,K., Ogata,K., Izu,Y., Tsunasawa,S. and Kato,I. (1998) Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J. Mol. Biol., 284, 101–124. [DOI] [PubMed] [Google Scholar]
- Takeuchi N., Kawakami,M., Omori,A., Ueda,T., Spremulli,L.L. and Watanabe,K. (1998) Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization and gene structure. J. Biol. Chem., 273, 15085–15090. [DOI] [PubMed] [Google Scholar]
- Turk B.E., Griffith,E.C., Wolf,S., Biemann,K., Chang,Y.H. and Liu,J.O. (1999) Selective inhibition of amino-terminal methionine processing by TNP-470 and ovalicin in endothelial cells. Chem. Biol., 6, 823–833. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc. Natl Acad. Sci. USA, 93, 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature, 341, 456–458. [DOI] [PubMed] [Google Scholar]
- Widger W.R., Cramer,W.A., Hermodson,M., Meyer,D. and Gullifor,M. (1984) Purification and partial amino acid sequence of the chloroplast cytochrome b-559. J. Biol. Chem., 259, 3870–3876. [PubMed] [Google Scholar]
- Yokoi F., Vassileva,A., Hayashida,N., Torazawa,K., Wakasugi,T. and Sugiura,M. (1990) Chloroplast ribosomal protein L32 is encoded in the chloroplast genome. FEBS Lett., 276, 88–90. [DOI] [PubMed] [Google Scholar]
- Yokoi F., Tanaka,M., Wakasugi,T. and Sugiura,M. (1991) The chloropast gene for ribosomal protein CL23 is functional in tobacco. FEBS Lett., 281, 64–66. [DOI] [PubMed] [Google Scholar]
- Zuo S., Guo,Q., Ling,C. and Chang,Y.H. (1995) Evidence that two zinc fingers in the methionine aminopeptidase from Saccharomyces cerevisiae are important for normal growth. Mol. Gen. Genet., 246, 247–253. [DOI] [PubMed] [Google Scholar]