Abstract
Background
Cytochrome P450 2D6 (CYP2D6) inhibition reduces the concentration of 4-hydroxylated tamoxifen metabolites, but the clinical relevance remains uncertain.
Methods
We conducted a large case–control study nested in the population of 11 251 women aged 35–69 years at diagnosis of stage I–III breast cancer between 1985 and 2001 on Denmark’s Jutland Peninsula and registered with the Danish Breast Cancer Cooperative Group. We identified 541 recurrent or contralateral breast cancers among women with estrogen receptor–positive (ER+) disease treated with tamoxifen for at least 1 year and 300 cancers in women with ER-negative (ER−) disease never treated with tamoxifen. We matched one control subject per case patient on ER status, menopausal status, stage, calendar time, and county, genotyped the CYP2D6*4 allele to assess genetic inhibition, and ascertained prescription history to assess drug–drug inhibition. We estimated the odds ratio (OR), associating CYP2D6 inhibition with breast cancer recurrence and adjusted for potential confounding with logistic regression. To address bias from incomplete information on CYP2D6 function, we used Monte Carlo simulation to complete a record-level probabilistic bias analysis. All statistical tests were two-sided.
Results
The frequency of the CYP2D6*4 minor allele was 24% in case patients with ER+ tumors, 23% in case patients with ER− tumors, and 22% each in control subjects with ER+ and ER− tumors. In women with ER+ tumors, the associations of one functional allele with recurrence (OR = 0.99; 95% confidence interval = 0.76 to 1.3) and no functional allele with recurrence (OR = 1.4; 95% confidence interval = 0.84 to 2.3) were near null, as were those for women with ER− tumors. The near-null associations persisted when evaluated by intake of medications, by combining genotype with medication history, in the probabilistic bias analysis, or by restricting the analysis to women with ER expression confirmed by re-assay.
Conclusion
The association between CYP2D6 inhibition and recurrence in tamoxifen-treated patients is likely null or small.
CONTEXTS AND CAVEATS
Prior knowledge
The cytochrome P450 2D6 (CYP2D6) enzyme, which metabolizes tamoxifen, is inhibited by the selective serotonin reuptake inhibitor paroxetine, but it is not known whether women with fewer than two functional CYP2D6*4 alleles or those who take selective serotonin reuptake inhibitors are poor candidates for tamoxifen therapy.
Study design
Five hundred and forty -one women in the Danish Breast Cancer Cooperative Group registry with recurrent or contralateral estrogen receptor–positive breast cancer who were treated with tamoxifen and 300 women with estrogen receptor–negative breast cancer who were never treated with tamoxifen were matched on clinical and tumor characteristics, CYP2D6 genotype, and selective serotonin reuptake inhibitor prescription history with control subjects from the same registry who had no recurrent or contralateral breast cancer.
Contribution
There was no statistically significant association between CYP2D6 inhibition and breast cancer recurrence in tamoxifen-treated women. The near-null association persisted regardless of whether CYP2D6 inhibition was assessed by genotype, by intake of medications that inhibit CYP2D6 function, or by a combination of genotype and medication history.
Implications
Tamoxifen treatment can be effective in women with estrogen receptor –positive breast cancer who have fewer than two functional CYP2D6 alleles or who take medications, such as selective serotonin reuptake inhibitors, that inhibit the CYP2D6 enzyme.
Limitations
Genotyping data for only one CYP2D6 allele were available, so the association between other CYP2D6 alleles and breast cancer recurrence was not ascertained. There was no information on tamoxifen adherence by case patients and control subjects, so the full extent of tamoxifen treatment was unknown.
From the Editors
Tamoxifen (TAM), a selective estrogen receptor (ER) modulator, halves the risk of breast cancer recurrence in patients with nonmetastatic ER-positive (ER+) disease and is also a potent therapy in women with metastatic ER+ disease (1). The effectiveness of tamoxifen therapy is, however, incomplete. Some women relapse and others do not respond at all. Mechanisms of resistance to tamoxifen therapy and predictive markers of susceptibility to resistance other than lack of ER expression have been widely researched (2–4). Accurate markers are clinically important, allowing prediction of tamoxifen response, adverse effects, and personalization of combined therapies (5,6). Tamoxifen and its primary metabolite (N-desmethyl tamoxifen) are metabolized mostly by the gene product of cytochrome P450 2D6 (CYP2D6) (7,8) to 4-hydroxytamoxifen (9) and 4-hydroxy-N-desmethyltamoxifen (10,11) (now often called endoxifen). These secondary metabolites bind the receptor about 100-fold more readily than tamoxifen and are thus the most important modulators of the ER in the tamoxifen pathway (12). Women who inherit two functional CYP2D6 alleles have higher steady-state concentrations of 4-hydroxytamoxifen (8,13) and 4-hydroxy-N-desmethyltamoxifen (7,13,14) than women who inherit no functional alleles when treated with tamoxifen. Women who inherit one functional allele have intermediate concentrations (13). Similarly, women who inherit two functional alleles and take the potent CYP2D6 inhibitor paroxetine (a selective serotonin reuptake inhibitor [SSRI]) have lower concentrations of 4-hydroxy-N-desmethyltamoxifen (7,14) when treated with tamoxifen. It has been suggested that ER+ breast cancer patients with nonfunctional alleles of CYP2D6 or those who take other medications that inhibit CYP2D6 function may be poor candidates for adjuvant tamoxifen therapy (15–17) because their lower concentrations of the potent metabolites may place them at higher risk for relapse or failure to respond.
Although the molecular and pharmacological bases for this hypothesis are compelling, earlier clinical epidemiology studies focusing on associations between CYP2D6 inhibition and breast cancer outcomes have had widely heterogeneous results (18). Thirteen published studies have examined the association between inheriting a variant CYP2D6 allele and risk of breast cancer recurrence or mortality (19–31), and several have been recently updated (32,33). Relative risks reported in these studies range from 0.52 to 6.7, with six reporting associations below the null and seven reporting associations above the null (Phomogeneity < .001). A similarly heterogeneous pattern has been found in studies of the association between drug-induced CYP2D6 inhibition and breast cancer recurrence (34–41). Some of these studies have been criticized on the grounds of small sample size, survivor and other selection biases, potential for uncontrolled confounding by prognostic markers, and information bias arising from retrospective or absent information on use of CYP2D6 inhibiting medications or from noncentralized testing of ER expression [see (18) for a review]. Genotype frequencies were not in Hardy–Weinberg equilibrium in many of the earlier studies (20,22,26,29,31,32). The inconsistent pattern of associations from earlier studies, combined with limitations offering only a partial explanation for the heterogeneity, cautions against any strong inference based on results available to date.
To address limitations of earlier research and to provide a precise estimate obtained from a large well-identified study population, we conducted a study of the association between genetic and pharmacological markers of CYP2D6 inhibition and breast cancer recurrence nested in a population-based clinical registry of breast cancer patients in Denmark (42).
Methods
Study Population
The source population consisted of 11 251 female residents of the Jutland Peninsula in Denmark aged 35–69 years between 1985 and 2001, who were diagnosed with stage I, II, or III breast cancer as defined by the Union for International Cancer Control (UICC) (43), and who were registered with the Danish Breast Cancer Cooperative Group (DBCG) (42). Since 1977, the DBCG has enrolled into its clinical database nearly all Danish breast cancer patients younger than 70 years at diagnosis. The same 10-year follow-up protocol is used for all patients registered with the DBCG (44), regardless of their participation in clinical trials. Approximately half of the DBCG patients are enrolled in clinical trials (44). Thus, studies nested in the DBCG registry combine the data quality advantages of a clinical trial with the generalizability benefits of a representative population. The study was approved by the Regional Committee on Biomedical Research Ethics of Aarhus County in Denmark and by the Boston University Medical Campus Institutional Review Board.
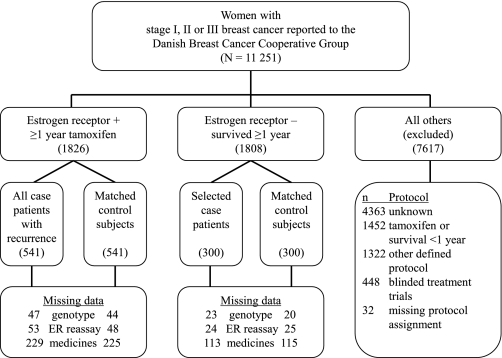
We divided the source population into three groups: women whose tumor expressed the ER and who were treated with tamoxifen for at least 1 year (ER+/TAM+, n = 1826); women whose tumor did not express the ER, who were not treated with tamoxifen, and who survived for at least 1 year (ER−/TAM−, n = 1808); and all other breast cancer patients (n = 7617, such as ER+ patients not treated with tamoxifen and ER− patients who were treated with tamoxifen) who were excluded (Figure 1). ER+/TAM+ women were assigned to tamoxifen- therapy protocols lasting 1, 2, or 5 years, depending on the guideline prevailing in Denmark at the time of diagnosis (44). As discussed below, most women assigned to 1- or 2-year protocols actually were likely to receive tamoxifen therapy for much longer. We included ER−/TAM− women to estimate the direct association between CYP2D6 inhibition and the breast cancer recurrence rate. Follow-up time began 1 year after the date of breast cancer diagnosis and continued until the date of the first breast cancer recurrence, death from any cause, loss to follow-up (eg, emigration), 10 years of follow-up, or until September 1, 2006.
Figure 1.
Design used to identify the study sample and to collect data. The source population consisted of 11 251 female residents of the Jutland Peninsula in Denmark aged 35–69 years who were diagnosed with stage I, II, or III breast cancer between 1985 and 2001. Most of the women (n = 4363) excluded because of unknown protocol had stage I breast cancer treated without a guideline protocol from the Danish Breast Cancer Cooperative Group. Missing data for medicines were because of breast cancer diagnosed before establishment of the Registry of Medicinal Products in 1995. Genotyping results and ER re-assay results were missing for a small proportion of subjects due to unavailable tumor blocks or indeterminate assay results. Case patients and control subjects with missing data were excluded from analyses that required the variable with a missing result (45). ER = estrogen receptor.
Case patients were women with local or distant breast cancer recurrence or contralateral breast cancer occurrence during their follow-up time. We designed the ER+/TAM+ sample size to achieve statistical power of 90% to detect an odds ratio (OR) of 1.5 associating reduced CYP2D6 function with recurrence risk. We therefore included all 541 ER+/TAM+ case patients. The number of ER−/TAM− case patients was greater than needed to achieve adequate statistical power. We therefore selected ER−/TAM− case patients at random (n = 300), after frequency matching as close as possible to the distribution of stage and calendar period of diagnosis among ER+/TAM+ case patients (Figure 1).
For each case patient, we selected without replacement one control subject from members of the source population who were alive and had no recurrence or contralateral breast cancer after the same amount of follow-up time. We matched control subjects to case patients according to group membership (ER+/TAM+ or ER−/TAM−), menopausal status at diagnosis (premenopausal or postmenopausal), date of breast cancer surgery (caliper matched ± 12 months), county of residence at time of diagnosis, and cancer stage at diagnosis (UICC stage I, II, or III).
Data Collection From Danish Registries
We used the Danish Civil Personal Registration number to link datasets. The number is a unique identifier assigned to all Danish residents who were alive on April 1, 1968, born thereafter, or upon immigration.
We collected data from the DBCG registry on demographic (age, menopausal status, and hospital of diagnosis), tumor (UICC stage, histological grade, and ER expression), and therapy characteristics (primary surgical tumor management, receipt of radiation therapy, receipt of chemotherapy, and receipt of tamoxifen therapy). We collected information from the Danish National Registry of Patients on the conditions included in the Charlson comorbidity index (46) that were present at the time of breast cancer diagnosis.
We obtained data on receipt of prescriptions for SSRIs and other potential CYP2D6 inhibitors by linking to the Registry of Medicinal Products, which is maintained by Statistics Denmark as part of the Danish national health-care system. Because this registry’s records are only complete from 1995 forward , analyses incorporating prescription information are limited to case patients and control subjects diagnosed with breast cancer that year or later (Figure 1).
Data Collection From Archived Tissue Samples
Laboratory personnel were blinded to all clinical information, including case or control status, ER status, and receipt or nonreceipt of tamoxifen therapy.
Tissue Processing.
We collected formalin-fixed paraffin-embedded tissue blocks from the pathology department archives of treating hospitals. We reviewed hematoxylin- and eosin-stained glass slides and the original pathology reports to identify those blocks to be processed. All tissue blocks were processed using the laboratory’s established sterile protocols designed to avoid case contamination. For DNA extractions, three to six 10-μm paraffin sections were cut from each sample and placed in a 1.5-mL microtube. Parallel whole sections were cut from tumor blocks for confirmatory ER expression assays.
DNA Extraction.
Before DNA extraction, tissue samples were deparaffinized by repeated treatment with xylene. After deparaffinization, DNA was extracted by repeated ethanol treatment, proteinase K digestion, and the QIAmp DNA FFPE Tissue kit (Qiagen AB, Dusseldorf, Germany) according to the manufacturer’s protocol.
Genotyping.
From each tissue sample, 50 ng of extracted DNA were amplified with 25 μL polymerase chain reaction with 50 denaturation cycles at 92°C for 15 seconds, followed by annealing and extension at 60°C for 90 seconds, using primers and reagents supplied with TaqMan genotyping kits (ABI kit: C-27102431-D0, Applied BioSystems [ABI], Foster City, CA; https://products.appliedbiosystems.com/ab/en/US/adirect/ab?cmd=ABAssayDetailDisplay&assayID=C__27102431_D0&Fs=y) to assay CYP2D6*4 (rs3892097, the minor allele with no function). All samples were assayed in duplicate using the MX3000P real-time polymerase chain reaction system (Stratagene, Cedar Creek, TX). Positive and negative control assays for the variant were identified by sequencing peripheral blood DNA from 30 healthy individuals and were included with each set of assays.
Validation Substudies
Laboratory Assays.
For 106 participants, we paired lymph node tissue blocks with tumor tissue blocks and processed them according to the protocols for sterile technique, DNA extraction, and genotyping described above.
ER expression at diagnosis was the basis for inclusion in the study and segregation into ER+/TAM+ or ER−/TAM− groups. During the study period, the DBCG recommended an evolving series of assays for ER expression for all Danish breast cancer patients (47). Because assay methods have improved over time, and to reduce the potential effect of variability across diagnosing hospitals, we re-assayed ER expression using whole sections from the original diagnostic paraffin-embedded tissues and primary antibody against ER alpha (clone 6F11; Novocastra, Newcastle Upon Tyne, UK). Heat-induced epitope retrieval for ER was achieved by incubation in a Tris–EDTA buffer, pH 9 (Target Retrieval Solution, pH 9; Dako, Glostrup, Denmark) using a microwave oven. Sections were stained on a Lab Vision Autostainer (Thermo Fisher Scientific, Fremont, CA) using the EnVision+ detection system (Dako) and visualized with horseradish peroxidase and diaminobenzadine. Slides were scored positive for ER when there was distinct nuclear staining of neoplastic cells. The percentage of positive cells was recorded and for the purposes of this study, a cutoff point of at least 10% positive tumor was chosen for ER- positivity in accordance with previous DBCG recommendations (47).
Registry Data.
We selected 20 ER+/TAM+ and 10 ER−/TAM− patients from one participating hospital. An investigator (J.P.G.) or a surgical colleague under his supervision reviewed their medical records blinded to the DBCG registry data. Half of the women in each group were diagnosed during the period 1985–1993 and half were diagnosed during the period 1994–2001. To guide the review of medical records, we adapted a standardized medical abstraction form and accompanying codebook from similar research tools used in earlier validation (48,49) and data collection (50) studies of breast cancer patients. The abstraction ascertained demographic information, tumor characteristics, therapy characteristics, recurrence status, and occurrence of a second primary breast cancer.
Analytic Variables
Recurrence.
We used the DBCG definition of breast cancer recurrence, that is, any type of breast cancer or distant metastases diagnosed subsequent to the initial course of therapy (51). Given the follow-up time in the source population, all recurrences occurred between 1 and 10 years after the primary breast cancer diagnosis.
Genotype Category.
We classified case patients and control subjects as having two functional CYP2D6*4 alleles, one functional CYP2D6*4 allele, or no functional CYP2D6*4 allele.
Prescription Status.
Prescriptions in the Registry of Medicinal Products are coded using the Anatomical Therapeutic Chemical (ATC) classification system of the World Health Organization. We defined SSRI antidepressants as all medications included in ATC group N06AB. This group comprises the following drugs: zimeldine, fluoxetine, citalopram, paroxetine, sertraline, alaproclate, fluvoxamine, etoperidone, and escitalopram.
We classified case patients and control subjects as those with no record of a SSRI prescription during their follow-up time (never SSRI) and those with any record of a prescription for a SSRI during their follow-up time (ever SSRI). We used a similar procedure to classify case patients and control subjects as ever or never users of another prescription medication that is a CYP2D6 inhibitor or substrate, as defined previously (37). In earlier investigations using this study population, we observed a near-null association between use of SSRIs or other CYP2D6 inhibitors and recurrence risk (35–37).
Index of CYP2D6 Inhibition.
To combine genetic and pharmacological information on CYP2D6 inhibition, we used the following index: 1) high inhibition: no functional allele or paroxetine or fluoxetine use during 30% or more of time on tamoxifen, 2) high intermediate inhibition: one functional allele and use of any CYP2D6 inhibitor (SSRI or other) during 30% or more of time on tamoxifen, 3) low intermediate inhibition: one functional allele and use of any CYP2D6 inhibitor during less than 30% of time on tamoxifen or no history of CYP2D6 inhibitor use, 4) low inhibition: two functional alleles and some history of CYP2D6 inhibitor use not previously classified, and 5) no inhibition: two functional alleles and no history of CYP2D6 inhibitor use. Women with missing genotype or medication history were excluded from this analysis.
Covariates.
We defined the following set of covariates: time of breast cancer diagnosis, age at diagnosis, Charlson comorbidity index score at diagnosis, menopausal status at diagnosis, county of residence at diagnosis, UICC stage at diagnosis, histological grade, surgery type, and receipt of systemic adjuvant chemotherapy.
Statistical Analysis
Conventional Analysis and Analysis of Validation Substudies.
Analyses were conducted within strata of the two groups (ER+/TAM+ or ER−/TAM−). We computed the frequency and proportion of case patients and control subjects within categories of assigned protocol of tamoxifen duration, genotype, SSRI use, use of other CYP2D6 inhibitors or substrates, the index of CYP2D6 inhibition, and the covariates. We tested whether the genotypes at the CYP2D6*4 locus observed among control subjects were in Hardy–Weinberg equilibrium by calculating the χ2 test statistic, with expected genotype frequencies based on the observed prevalence of major and minor alleles (52). We performed these calculations in all control subjects combined and within group strata.
We estimated the rate ratio associating CYP2D6 inhibition with breast cancer recurrence as the odds ratio in a conditional logistic regression, including genetic information, use of CYP2D6-inhibiting medications, or the index of CYP2D6 inhibition as the prognostic variable, conditioned on the matched factors. We adjusted for potential confounding in a logistic regression model, which included the marker of CYP2D6 inhibition, time to recurrence or control subject selection, menopausal status, stage, receipt of chemotherapy, receipt of radiation therapy, and type of surgery. We repeated this analysis restricting it to the ER+/TAM+ women who received no chemotherapy. All estimates of association are accompanied by a 95% confidence interval (95% CI) calculated by the profile likelihood method. We computed the two-sided P value for a test of homogeneity of the adjusted odds ratios in the ER+/TAM+ women vs the corresponding adjusted odds ratios in the ER−/TAM− women.
As noted above, prescription data were missing for women diagnosed before 1995. In addition, genotyping results and ER re-assay results were missing for a small proportion of subjects (Figure 1), due to unavailable tumor blocks or indeterminate assay results. Case patients and control subjects with missing data were excluded from analyses that required the variable with a missing result (45).
We examined the concordance of CYP2D6 genotypes in the 106 DNA samples extracted from normal tissue with the genotypes in the DNA samples extracted from the paired tumor tissues. We calculated the crude odds ratio associating failure to confirm the original ER status with recurrence risk. In a subanalysis, we calculated genotype associations with recurrence rate after excluding participants for whom re-assay of the tumor showed ER expression discordant with the assay result at diagnosis. We evaluated the concordance of the information from the DBCG registry with the information from the gold standard medical records. All analyses were performed using SAS version 9 (SAS Institute Inc., Cary, NC).
Quantitative Bias Analysis.
To address bias from incomplete information on CYP2D6 function, we conducted a quantitative bias analysis (53) informed by classifications based on comprehensive genotyping of the CYP2D6 gene in another study population of German breast cancer patients (54). We used this study to estimate the sensitivity (s = 187/303) and specificity (t = 186/189) of CYP2D6 functional classification based on genotyping only the CYP2D6*4 allele, as well as the positive predictive value (PPVco = 187/190) and negative predictive value (NPVco = 186/302) of reduced CYP2D6 function in control subjects based on genotyping only the CYP2D6*4 allele. We set the prevalence of reduced CYP2D6 function in control subjects (pco = 303/492) equal to the prevalence observed in the German study population (54) based on the comprehensive genotype. For the probabilistic bias analysis, we assigned beta distributions to all of these classification parameters using standard methods (α = numerator + 1, β = total − numerator + 1) (53). The prevalence of reduced CYP2D6 function in case patients (pca) is a function of the prevalence in control subjects and the association between reduced function and breast cancer recurrence (OR):
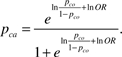 |
In the ER−/TAM− strata, we set OR = 1. In the ER+/TAM+ group, OR was the parameter we wished to estimate, so we substituted in its place a beta distribution with α = 1.83 and β = 4.54 scaled to the interval ln|1| to ln|2.77|. This distribution has minimum OR = 1 (null association), maximum OR = 2.77 [the strongest association reported in the study of German breast cancer patients (54)], and mode of OR = 1.22 [the result of a recent meta-analysis (55) of this topic among Caucasians (OR = 1.22, 95% CI = 0.88 to 1.68)]. Finally, we calculated the positive- predictive value in case patients (PPVca) and negative- predictive value in case patients (NPVca) as
where s is sensitivity and t is specificity.
Note that
so, when OR is greater than 1.0, NPVca is less than NPVco, which means that a case patient classified as having normal function based only on having no *4 allele is more likely to be reclassified as truly having reduced function than an analogous control subject. With this model and assigned probability distributions, we used Monte Carlo simulation to complete a record-level probabilistic bias analysis following established methods (53). The bias analysis was performed using SAS version 9.
Results
Conventional Analysis and Bias Analysis
More than 90% of the women in the ER+/TAM+ group were postmenopausal and had stage II or III disease (Table 1), a consequence of the DBCG criteria for assignment to standard tamoxifen protocols during the era when the study population was diagnosed with breast cancer (44). More than 80% of the breast cancer patients underwent mastectomy, which is consistent with treatment patterns in Denmark during this era (56), and a substantial proportion (34%–47%) received radiation therapy (Table 1). As expected, the prevalence of chemotherapy was much higher in the ER−/TAM− group (63% in control subjects and 83% in case patients) than in the ER+/TAM+ group (12% in control subjects and 13% in case patients) (Table 1).
Table 1.
Frequency and proportion of breast cancer recurrence case patients and matched control subjects within group strata*
| Patient characteristic | ER+/TAM+, No. (%) |
ER−/TAM−, No. (%) |
||
| Case patients | Control subjects | Case patients | Control subjects | |
| CYP2D6*4 genotype | ||||
| Two functional alleles | 299 (61) | 308 (62) | 167 (61) | 173 (62) |
| One functional allele | 154 (31) | 159 (32) | 91 (33) | 94 (34) |
| No functional allele | 41 (8.3) | 30 (6.0) | 19 (6.9) | 13 (4.6) |
| Missing† | 47 | 44 | 23 | 20 |
| SSRI prescription | ||||
| Ever | 47 (15) | 48 (15) | 17 (9.1) | 23 (12) |
| Never | 265 (85) | 268 (85) | 170 (90) | 162 (88) |
| Missing‡ | 229 | 225 | 113 | 115 |
| Other CYP2D6 inhibitors | ||||
| Ever | 70 (22) | 58 (18) | 38 (20) | 35 (19) |
| Never | 242 (78) | 258 (72) | 149 (80) | 150 (81) |
| Missing‡ | 229 | 225 | 113 | 115 |
| CYP2D6 inhibition index | ||||
| No inhibition | 123 (41) | 125 (41) | 80 (44) | 83 (46) |
| Low inhibition | 51 (17) | 56 (18) | 34 (19) | 28 (16) |
| Low intermediate inhibition | 85 (29) | 88 (29) | 48 (26) | 50 (28) |
| High intermediate inhibition | 14 (4.7) | 11 (3.6) | 5 (2.7) | 7 (3.9) |
| High inhibition | 25 (8.4) | 24 (7.9) | 16 (8.7) | 11 (6.1) |
| Missing†‡ | 243 | 237 | 117 | 121 |
| Diagnosis year§ | ||||
| 1985–1993 | 235 (43) | 234 (43) | 107 (36) | 100 (33) |
| 1994–1996 | 113 (21) | 112 (21) | 81 (27) | 83 (28) |
| 1997–2001 | 193 (36) | 195 (36) | 112 (37) | 117 (39) |
| Age category at diagnosis, y | ||||
| 35–44 | 16 (3.0) | 13 (2.4) | 68 (23) | 58 (19) |
| 45–54 | 116 (21) | 111 (21) | 120 (40) | 113 (38) |
| 55–64 | 286 (53) | 281 (52) | 82 (27) | 86 (29) |
| 65–69 | 123 (23) | 136 (25) | 30 (10) | 43 (14) |
| Charlson comorbidity index | ||||
| 0 | 450 (83) | 444 (82) | 271 (90) | 267 (89) |
| 1 or 2 | 81 (15) | 86 (16) | 27 (9.0) | 30 (10) |
| ≥3 | 10 (1.8) | 11 (2.0) | 2 (0.7) | 3 (1.0) |
| Menopausal status at diagnosis§ | ||||
| Premenopausal | 34 (6.3) | 34 (6.3) | 121 (40) | 121 (40) |
| Postmenopausal | 507 (94) | 507 (94) | 179 (60) | 179 (60) |
| UICC tumor stage at diagnosis§ | ||||
| I | 9 (1.7) | 9 (1.7) | 25 (8.3) | 25 (8.3) |
| II | 250 (46) | 250 (46) | 153 (51) | 153 (51) |
| III | 282 (52) | 282 (52) | 122 (41) | 122 (41) |
| Histological grade | ||||
| I | 108 (25) | 144 (35) | 27 (11) | 23 (10) |
| II | 234 (54) | 215 (52) | 125 (49) | 98 (43) |
| III | 92 (21) | 57 (14) | 103 (40) | 106 (47) |
| Missing | 107 | 125 | 45 | 73 |
| Surgery type | ||||
| Breast-conserving surgery | 58 (11) | 71 (13) | 47 (16) | 56 (19) |
| Mastectomy | 483 (89) | 470 (87) | 252 (84) | 244 (81) |
| Missing | 0 | 0 | 1 | 0 |
| Radiation therapy | ||||
| Yes | 183 (34) | 191 (35) | 128 (44) | 123 (47) |
| No | 358 (66) | 350 (65) | 166 (56) | 137 (53) |
| Missing | 0 | 0 | 6 | 40 |
| Tamoxifen protocol, y | ||||
| 1 | 247 (46) | 249 (46) | Not applicable | Not applicable |
| 2 | 98 (18) | 92 (17) | ||
| 5 | 196 (36) | 200 (37) | ||
| Systemic adjuvant chemotherapy | ||||
| Yes | 70 (13) | 65 (12) | 248 (83) | 188 (63) |
| No | 471 (87) | 476 (88) | 52 (17) | 112 (37) |
| Current ER expression | ||||
| Positive | 451 (92) | 474 (96) | 72 (26) | 70 (25) |
| Negative | 37 (7.6) | 19 (3.9) | 204 (74) | 205 (75) |
| Not available† | 53 | 48 | 24 | 25 |
| CYP2D6*4 genotype restricted to concordant ER | ||||
| Two functional alleles | 265 (61) | 287 (63) | 121 (62) | 121 (61) |
| One functional allele | 135 (31) | 140 (31) | 63 (32) | 67 (34) |
| No functional allele | 38 (8.7) | 28 (6.1) | 12 (6.1) | 9 (4.5) |
| Missing† | 13 | 19 | 8 | 8 |
The source population consisted of 11 251 female residents of the Jutland Peninsula in Denmark aged 35–69 years who were diagnosed with stage I, II, or III breast cancer between 1985 and 2001. Subjects were estrogen receptor (ER) positive and received at least 1 year of tamoxifen therapy (ER+/TAM+) or ER negative and never received tamoxifen therapy and survived at least 1 year after diagnosis (ER−/TAM−). CYP2D6 = cytochrome P450 2D6; SSRI = selective serotonin reuptake inhibitor; UICC = Union for International Cancer Control.
No tissue available for assay or assay results indeterminate.
Primarily missing because the breast cancer was diagnosed before establishment of the Registry of Medicinal Products in 1995.
Variable included in risk set sampling to match control subjects to case patients.
The frequency of the CYP2D6*4 minor allele was 24% in ER+ case patients, 23% in ER− case patients, and 22% among both the ER+/TAM+ control subjects and the ER−/TAM− control subjects. The minor allele frequency among control subjects compares well with the minor allele frequency reported in reference populations (57,58). The CYP2D6*4 allele was in Hardy–Weinberg equilibrium among all control subjects (P = .21 overall, P = .13 in ER+/TAM+ control subjects, and P = .97 in ER−/TAM− control subjects).
In the ER+/TAM+ group, the associations of one functional allele with breast cancer recurrence (adjusted OR = 0.99, 95% CI = 0.76 to 1.3) and no functional allele with breast cancer recurrence (adjusted OR = 1.4, 95% CI = 0.84 to 2.3) were near null (Table 2), as was the association of genetically reduced function (zero or one functional CYP2D6*4 allele) with breast cancer recurrence (adjusted OR = 1.1, 95% CI = 0.81 to 1.4). Similar near-null associations were found for the ER−/TAM− group (adjusted OR = 0.91, 95% CI = 0.62 to 1.3; adjusted OR = 1.3, 95% CI = 0.60 to 2.9; and adjusted OR = 0.95, 95% CI = 0.66 to 1.4, respectively; Table 2). When we restricted the ER+/TAM+ group to women who received no chemotherapy, the associations of one functional allele with breast cancer recurrence (adjusted OR = 1.0, 95% CI = 0.76 to 1.4) and no functional allele with breast cancer recurrence (OR = 1.5, 95% CI = 0.88 to 2.6) were similarly small or null. No estimate of association in the ER+/TAM+ group was statistically– significantly different from the corresponding estimate of association in the ER−/TAM− group (Table 2).
Table 2.
Associations between CYP2D6 inhibition and breast cancer recurrence within strata*
| CYP2D6 inhibition category | ER+/TAM+ |
ER−/TAM− |
P§ | ||||
| Case patients/control subjects | Matched OR (95% CI)† | Adjusted OR (95% CI)‡ | Case patients/control subjects | Matched OR (95% CI)† | Adjusted OR (95% CI)‡ | ||
| CYP2D6*4 genotype | |||||||
| Two functional alleles | 299/308 | 1 (reference) | 1 (reference) | 167/173 | 1 (reference) | 1 (reference) | |
| One functional allele | 154/159 | 0.99 (0.75 to 1.3) | 0.99 (0.76 to 1.3) | 91/94 | 1.0 (0.72 to 1.5) | 0.91 (0.61 to 1.3) | .72 |
| No functional allele | 41/30 | 1.4 (0.81 to 2.3) | 1.4 (0.84 to 2.3) | 19/13 | 1.5 (0.68 to 3.2) | 1.3 (0.60 to 2.9) | .88 |
| CYP2D6*4 genotype | |||||||
| Two functional alleles | 299/308 | 1 (reference) | 1 (reference) | 167/173 | 1 (reference) | 1 (reference) | |
| Less than two functional alleles | 195/189 | 1.0 (0.81 to 1.4) | 1.1 (0.81 to 1.4) | 110/107 | 1.1 (0.77 to 1.7) | 0.95 (0.66 to 1.4) | .54 |
| SSRI prescription | |||||||
| Never | 265/268 | 1 (reference) | 1 (reference) | 170/162 | 1 (reference) | 1 (reference) | |
| Ever | 47/48 | 0.95 (0.62 to 1.5) | 0.91 (0.58 to 1.4) | 17/23 | 0.71 (0.37 to 1.4) | 0.80 (0.38 to 1.7) | .77 |
| Other CYP2D6 inhibitor | |||||||
| Never | 242/258 | 1 (reference) | 1 (reference) | 149/150 | 1 (reference) | 1 (reference) | |
| Ever | 70/58 | 1.3 (0.86 to 1.9) | 1.4 (0.89 to 2.0) | 38/35 | 1.1 (0.66 to 1.8) | 1.2 (0.66 to 2.2) | .68 |
| CYP2D6 inhibition index | |||||||
| No inhibition | 123/125 | 1 (reference) | 1 (reference) | 80/83 | 1 (reference) | 1 (reference) | |
| Low inhibition | 51/56 | 0.88 (0.56 to 1.4) | 0.91 (0.58 to 1.4) | 34/28 | 1.2 (0.68 to 2.2) | 1.3 (0.68 to 2.5) | .37 |
| Low intermediate | 85/88 | 0.95 (0.64 to 1.4) | 0.91 (0.61 to 1.4) | 48/50 | 0.88 (0.54 to 1.4) | 0.74 (0.42 to 1.3) | .56 |
| High intermediate | 14/11 | 1.3 (0.56 to 2.9) | 1.3 (0.56 to 3.1) | 5/7 | 0.68 (0.19 to 2.5) | 0.70 (0.17 to 2.9) | .46 |
| High inhibition | 25/24 | 0.95 (0.50 to 1.8) | 0.95 (0.5 to 1.8) | 16/11 | 1.5 (0.61 to 3.5) | 1.2 (0.42 to 3.6) | .71 |
The source population consisted of 11 251 female residents of the Jutland Peninsula in Denmark aged 35–69 years who were diagnosed with stage I, II, or III breast cancer between 1985 and 2001. Associations between CYP2D6 inhibition and breast cancer recurrence within strata of women with tumors that expressed the estrogen receptor (ER) and who received at least 1 year of tamoxifen therapy (ER+/TAM+) or women with tumors that did not express the ER and who never received tamoxifen therapy and who survived at least 1 year after diagnosis (ERP−/TAM−). CI = confidence interval; CYP2D6 = cytochrome P450 2D6; OR = odds ratio; SSRI = selective serotonin reuptake inhibitor.
Estimated using conditional logistic regression with conditioning on the matched factors (time to recurrence or control selection, county, menopausal status, and stage).
Estimated using logistic regression with adjustment for time to recurrence or control selection, menopausal status, stage, receipt of chemotherapy, receipt of radiation therapy, and type of surgery.
Two-sided P for test of homogeneity of the adjusted odds ratio in ER+/TAM+ women vs the adjusted odds ratio in ER−/TAM− women.
As we reported previously (37), neither SSRI medications nor other CYP2D6 inhibitors increased the risk of breast cancer recurrence (Table 2). When we included information on medications that inhibit CYP2D6 function with the genetic information, the near-null results persisted in all categories of the index of CYP2D6 inhibition and in both groups (Table 2). These associations were less precisely measured because prescription information was not available throughout the study period.
The probabilistic bias analysis revealed that the results were unlikely to have been substantially biased by the absence of genetic information at alleles other than CYP2D6*4, assuming a valid bias model. The estimate of the association between reduced function and breast cancer recurrence in the ER+/TAM+ group changed from 1.1 (95% CI = 0.81 to 1.4) in the conventional analysis reported above to 1.3 (95% simulation interval 0.87 to 1.9) in the bias analysis. In the ER−/TAM− group, the estimate of association changed from 0.95 (95% CI = 0.66 to 1.4) in the conventional analysis reported above to 0.97 (95% CI = 0.71 to 1.3) in the bias analysis. Substituting genetic information from a second cohort of persons without breast cancer (57) yielded comparable results.
Validation Substudies
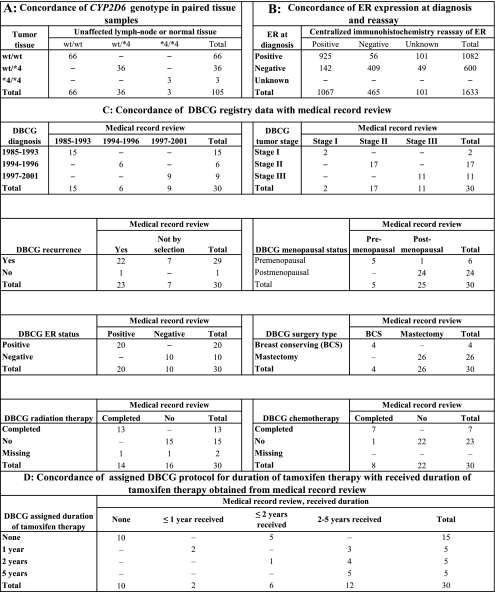
DNA extracted from normal tissue vs DNA extracted from paired tumor samples showed perfect concordance between CYP2D6 genotypes (Figure 2, A), so all DNA extractions used for genotyping in this study were from tumor samples. Concordance between positive ER expression at diagnosis and the centralized immunohistochemistry re-assay results (94%, Figure 2, B) was better than the concordance between negative ER expression at diagnosis and the re-assay results (74%, Figure 2, B). Further stratification of these results by case and control status was also informative. In the ER+/TAM+ group, 96% of tumors from control participants expressed the ER when re-assayed, whereas only 92% of tumors from case patients with breast cancer recurrence expressed the ER when re-assayed (Table 1). The crude odds ratio associating failure to express the ER in the re-assay with recurrence equaled 2.0 (95% CI = 1.2 to 3.6). In the ER−/TAM− group, the crude odds ratio associating failure to confirm the absence of ER expression with recurrence was null (OR = 1.0, 95% CI = 0.71 to 1.5). Limiting the genotype analyses to women whose ER expression at re-assay was concordant with ER status at diagnosis did not appreciably change the adjusted estimates of association in the ER+/TAM+ group (one functional allele OR = 1.0, 95% CI = 0.78 to 1.4 and no functional allele OR = 1.5, 95% CI = 0.87 to 2.4) or in the ER−/TAM− group (one functional allele OR = 0.86, 95% CI = 0.55 to 1.4 and no functional allele OR = 1.1, 95% CI = 0.40 to 3.0).
Figure 2.
Contingency tables showing frequencies of subjects in the validation substudies. The source population consisted of 11 251 female residents of the Jutland Peninsula in Denmark aged 35–69 years who were diagnosed with stage I, II, or III breast cancer between 1985 and 2001. A) Validation of DNA extracted from normal tissue vs DNA extracted from paired tumor samples. Total sums to 105, not the 106 paired samples assayed, because genotyping failed in one sample. B) Validation of Danish Breast Cancer Cooperative Group (DBCG) estrogen receptor (ER) status at diagnosis by centralized immunohistochemistry re-assay of ER expression. Unknown indicates that no malignant tissue was available for assay or assay results indeterminate. C) Validation of clinical and patient characteristics by medical record review. The 30 patients selected were all treated at Aalborg Hospital. D) Validation of assigned duration of tamoxifen therapy at treatment outset against received duration of tamoxifen therapy by medical record review. The 30 patients selected were all treated at Aalborg Hospital. CYP2D6 = cytochrome P450 2D6.
The 30 patients selected for the registry validation substudy were typical of the parent study population, except that they were all treated at Aalborg Hospital. Of the 22 patients categorized as having a recurrence in the DBCG registry, 19 had a recurrence and three had a second primary breast cancer according to the medical record review. One patient had a recurrence that was not registered in the DBCG (Figure 2, C).
All patients who were categorized as receiving tamoxifen by the DBCG had at least 1 year of treatment, and no patient categorized as not receiving tamoxifen by the DBCG had evidence in the medical record of receiving tamoxifen. Most importantly, medical record review showed that most patients assigned to 1- or 2-year treatment protocols at diagnosis actually received tamoxifen treatment for a longer duration (Figure 2, D).
Discussion
In this population-based study, we observed no substantial association between CYP2D6 inhibition and breast cancer recurrence. The adjusted odds ratio associating no functional CYP2D6*4 allele with breast cancer recurrence in the ER+/TAM+ group (OR = 1.4, 95% CI = 0.84 to 2.3) approximately equaled the corresponding adjusted odds ratio in the ER−/TAM− group (OR = 1.3, 95% CI = 0.60 to 2.9). Taken together, these results should caution against overinterpretation of the former association because no association via the tamoxifen pathway is plausible to explain the latter association. The near-null association persisted regardless of whether we assessed CYP2D6 inhibition only by *4 genotype, by intake of medications that inhibit CYP2D6 function, by combination of *4 genotype with medication history, by limiting the ER+/TAM+ group to women who received no chemotherapy, by limiting the dataset to women with confirmed ER expression, or by analysis of the potential bias arising from misclassification of CYP2D6 function because we genotyped only the *4 allele.
The study design took advantage of the long-standing high-quality clinical database maintained by the DBCG (42) and the tissue archives maintained by the pathology departments of Danish hospitals (59). Linking these resources yielded a study effectively immune to selection bias and, to our knowledge, the largest study to date of the association between CYP2D6 inhibition and breast cancer recurrence. The 184 recurrent cases with at least one *4 allele in the ER+/TAM+ group represent approximately one-third of the accumulated cases with genetically identified reduced function reported in studies thus far.
Data quality was high, with perfect concordance between genotypes in DNA extracted from tumor tissue and lymph node tissue in the validation subset and with alleles at the CYP2D6*4 locus in Hardy–Weinberg equilibrium among all control subjects and consistent with the expected allele frequencies. Concordance of re-assayed ER expression with ER status at diagnosis was consistent with earlier reports (60,61) and showed the expected associations with recurrence risk. The association between failure to express the ER at re-assay and recurrence risk in the ER+/TAM+ group provides the study design with face validity and demonstrates the study’s ability to detect associations between rare events and recurrence risk.
All recurrences in the validation subset were confirmed by medical record review, which is consistent with an earlier and much larger validation study that reported a positive predictive value of 99.4% for breast cancer recurrence recorded by the DBCG (62). We observed perfect agreement for all of the matched factors except for one patient’s menopausal status. Only duration of tamoxifen therapy differed frequently between the registry data and medical records, with many patients who were originally assigned to tamoxifen therapy of short duration receiving tamoxifen for longer durations. This discrepancy likely resulted from modifications to the tamoxifen protocol occurring after the patient’s diagnosis and initial assignment to tamoxifen durations of 1 or 2 years. The discrepancy actually strengthens our study’s results because it suggests that the null association did not arise from some patients receiving short-duration tamoxifen therapy.
The major limitation of this study was absence of genotyping data for CYP2D6 alleles other than *4. Complete genotyping of the DNA samples extracted from paraffin-embedded tissues required more resources than were available to us. To address this limitation, we implemented a quantitative bias analysis, which assumed that case patients with recurrence were more likely to carry alleles with reduced function than were control subjects. All of the parameters of the bias model were informed by published external data sources. Assuming an accurate bias model, this bias analysis showed that the near-null results would have changed very little with complete genotyping data. A second limitation was the absence of information on tamoxifen adherence by case patients and control subjects. About half of tamoxifen-treated patients fail to complete the intended duration of their tamoxifen therapy (63). Among the 20 ER+/TAM+ patients included in our medical record review, six did not complete their intended duration of tamoxifen therapy, two because of recurrent breast cancer. Tamoxifen adherence is related to recurrence risk (64) and may be predicted by CYP2D6 genotype (65), in which case, adherence to the intended duration would be a causal intermediate between CYP2D6 genotype and recurrence. Results adjusted for adherence would be more biased than without adjustment, usually toward the null (66).
Earlier studies of the association between CYP2D6 inhibition and risk of breast cancer recurrence or mortality in tamoxifen-treated women have reported widely heterogeneous results (55), and there is no adequate explanation for this heterogeneity (18). Although the heterogeneity presents an important barrier in interpreting a quantitative synthesis (18,67), a recent meta-analysis concluded that the effect of CYP2D6 inhibition on recurrence risk may be relatively small (55). This conclusion is consistent with our results, with reasonable bounds that can be placed on the expected association (18), and with the well-understood pharmacology of tamoxifen therapy (18,68), which requires that tamoxifen and its metabolites overwhelm estrogen in competition for binding to the ER. It is common for genetic research to initially show strong associations, sometimes in more than one study (69), only to find that the strength of association diminishes or disappears as evidence accumulates (70–72). Despite early enthusiasm for the potential to identify poor candidates for tamoxifen therapy by CYP2D6 genotyping (15), the true association between CYP2D6 inhibition and breast cancer recurrence risk in tamoxifen-treated patients is likely to be null or small.
Funding
US National Cancer Institute at the National Institutes of Health (R01 CA118708 to T.L.L.); Danish Cancer Society (DP06117 to S.H.-D.); Karen Elise Jensen Foundation (H.T.S.).
Footnotes
The authors thank the Danish Breast Cancer Cooperative Group for access to its registry data and for preparing the initial dataset.
The granting agencies had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the article; or the decision to submit the article for publication.
References
- 1.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Wolf DM, Gottardis MM, Jordan VC. Tamoxifen-resistant growth. In: Jordan VC, editor. Long-Term Tamoxifen Treatment for Breast Cancer. Madison, WI: University of Wisconsin Press; 1994. pp. 181–198. [Google Scholar]
- 3.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11(4):643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- 4.Jordan VC. Tamoxifen or raloxifene for breast cancer chemoprevention: a tale of two choices—point. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2207–2209. doi: 10.1158/1055-9965.EPI-07-0629. [DOI] [PubMed] [Google Scholar]
- 5.Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. Pathways to tamoxifen resistance. Cancer Lett. 2007;256(1):1–24. doi: 10.1016/j.canlet.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milano A, Dal LL, Sotiriou C, Piccart M, Cardoso F. What clinicians need to know about antioestrogen resistance in breast cancer therapy. Eur J Cancer. 2006;42(16):2692–2705. doi: 10.1016/j.ejca.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95(23):1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 8.Coller JK, Krebsfaenger N, Klein K, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol. 2002;54(2):157–167. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75(2):305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 10.Lien EA, Solheim E, Kvinnsland S, Ueland PM. Identification of 4-hydroxy-N-desmethyltamoxifen as a metabolite of tamoxifen in human bile. Cancer Res. 1988;48(8):2304–2308. [PubMed] [Google Scholar]
- 11.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310(3):1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 12.Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55(5):471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 13.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19(1):56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97(1):30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 15.Desta Z, Flockhart DA. Germline pharmacogenetics of tamoxifen response: have we learned enough? J Clin Oncol. 2007;25(33):5147–5149. doi: 10.1200/JCO.2007.13.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan VC. New insights into the metabolism of tamoxifen and its role in the treatment and prevention of breast cancer. Steroids. 2007;72(13):829–842. doi: 10.1016/j.steroids.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.American Cancer Society. Tamoxifen: some women don’t get full benefit. American Cancer Society; 2006. www.cancer.org/docroot/NWS/content/NWS 1 1x Tamoxifen Some Women Dont Get Full Benefit.asp. Last accessed November, 2008. [Google Scholar]
- 18.Lash TL, Lien EA, Sorensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10(8):825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegman P, Vainikka L, Stal O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7(3):R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okishiro M, Taguchi T, Jin KS, Shimazu K, Tamaki Y, Noguchi S. Genetic polymorphisms of CYP2D6*10 and CYP2C19*2,*3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115(5):952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 21.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91(3):249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 22.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9(1):R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101(1):113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 24.Newman WG, Hadfield KD, Latif A, et al. Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin Cancer Res. 2008;14(18):5913–5918. doi: 10.1158/1078-0432.CCR-07-5235. [DOI] [PubMed] [Google Scholar]
- 25.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25(33):5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19(8):1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 27.Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99(5):995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gor PP, Su HI, Gray RJ, et al. Cyclophosphamide-metabolizing enzyme polymorphisms and survival outcomes after adjuvant chemotherapy for node-positive breast cancer: a retrospective cohort study. Breast Cancer Res. 2010;12(3):R26. doi: 10.1186/bcr2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramon y Cajal T, Altes A, Pare L, et al. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat. 2010;119(1):33–38. doi: 10.1007/s10549-009-0328-y. [DOI] [PubMed] [Google Scholar]
- 30.Thompson AM, Johnson A, Quinlan P, et al. Comprehensive. CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res Treat. 2011;125(1):279–287. doi: 10.1007/s10549-010-1139-x. [DOI] [PubMed] [Google Scholar]
- 31.Abraham JE, Maranian MJ, Driver KE, et al. CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 2010;12(4):R64. doi: 10.1186/bcr2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302(13):1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28(8):1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehmann D, Nelsen J, Ramanath V, Newman N, Duggan D, Smith A. Lack of attenuation in the antitumor effect of tamoxifen by chronic CYP isoform inhibition. J Clin Pharmacol. 2004;44(8):861–865. doi: 10.1177/0091270004266618. [DOI] [PubMed] [Google Scholar]
- 35.Lash TL, Pedersen L, Cronin-Fenton D, et al. Tamoxifen’s protection against breast cancer recurrence is not reduced by concurrent use of the SSRI citalopram. Br J Cancer. 2008;99(4):616–621. doi: 10.1038/sj.bjc.6604533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahern TP, Pedersen L, Cronin-Fenton DP, Sorensen HT, Lash TL. No increase in breast cancer recurrence with concurrent use of tamoxifen and some CYP2D6-inhibiting medications. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2562–2564. doi: 10.1158/1055-9965.EPI-09-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lash TL, Cronin-Fenton D, Ahern TP, et al. Breast cancer recurrence risk related to concurrent use of SSRI antidepressants and tamoxifen. Acta Oncol. 2010;49(3):305–312. doi: 10.3109/02841860903575273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aubert RE, Stanek EJ, Yao J, et al. Risk. of breast cancer recurrence in women initiating tamoxifen with CYP2D6 inhibitors. J Clin Oncol. 2009;27(supplement):18S. [Google Scholar]
- 39.Kelly CM, Juurlink DN, Gomes T, et al. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dezentje VO, van Blijderveen NJ, Gelderblom H, et al. Effect of concomitant CYP2D6 inhibitor use and tamoxifen adherence on breast cancer recurrence in early-stage breast cancer. J Clin Oncol. 2010;28(14):2423–2429. doi: 10.1200/JCO.2009.25.0894. [DOI] [PubMed] [Google Scholar]
- 41.Chubak J, Buist DS, Boudreau DM, Rossing MA, Lumley T, Weiss NS. Breast cancer recurrence risk in relation to antidepressant use after diagnosis. Breast Cancer Res Treat. 2008;112(1):123–132. doi: 10.1007/s10549-007-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blichert-Toft M, Christiansen P, Mouridsen HT. Danish Breast Cancer Cooperative Group–DBCG: history, organization, and status of scientific achievements at 30-year anniversary. Acta Oncol. 2008;47(4):497–505. doi: 10.1080/02841860802068615. [DOI] [PubMed] [Google Scholar]
- 43.Union for International Cancer Control. TNM Classification of Malignant Tumours. 5th ed. Geneva, Switzerland: Springer; 1997. [Google Scholar]
- 44.Moller S, Jensen MB, Ejlertsen B, et al. The clinical database and the treatment guidelines of the Danish Breast Cancer Cooperative Group (DBCG); its 30-years experience and future promise. Acta Oncol. 2008;47(4):506–524. doi: 10.1080/02841860802059259. [DOI] [PubMed] [Google Scholar]
- 45.Greenland S, Finkle WD. A critical look at methods for handling missing covariates in epidemiologic regression analyses. Am J Epidemiol. 1995;142(12):1255–1264. doi: 10.1093/oxfordjournals.aje.a117592. [DOI] [PubMed] [Google Scholar]
- 46.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 47.Talman ML, Rasmussen BB, Andersen J, Christensen IJ. Estrogen receptor analyses in the Danish Breast Cancer Cooperative Group. History, methods, prognosis and clinical implications. Acta Oncol. 2008;47(4):789–794. doi: 10.1080/02841860801982741. [DOI] [PubMed] [Google Scholar]
- 48.Lash TL, Fox MP, Thwin SS, et al. Using probabilistic corrections to account for abstractor agreement in medical record reviews. Am J Epidemiol. 2007;165(12):1454–1461. doi: 10.1093/aje/kwm034. [DOI] [PubMed] [Google Scholar]
- 49.Thwin SS, Clough-Gorr KM, McCarty MC, et al. Automated inter-rater reliability assessment and electronic data collection in a multi-center breast cancer study. BMC Med Res Methodol. 2007;7(June 18):23. doi: 10.1186/1471-2288-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silliman RA, Guadagnoli E, Rakowski W, et al. Adjuvant. tamoxifen prescription in women 65 years and older with primary breast cancer. J Clin Oncol. 2002;20(11):2680–2688. doi: 10.1200/JCO.2002.08.137. [DOI] [PubMed] [Google Scholar]
- 51.Andersen KW, Mouridsen HT. Danish Breast Cancer Cooperative Group (DBCG). A description of the register of the nation-wide programme for primary breast cancer. Acta Oncol. 1988;27(6A) doi: 10.3109/02841868809091763. [DOI] [PubMed] [Google Scholar]
- 52.Weir B. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- 53.Lash TL, Fox MP, Fink AK. Applying Quantitative Bias Analysis to Epidemiologic Data. New York, NY: Springer; 2009. [Google Scholar]
- 54.Schroth W, Hamann U, Fasching PA, et al. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin Cancer Res. 2010;16(17):4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- 55.Seruga B, Amir E. Cytochrome P450 2D6 and outcomes of adjuvant tamoxifen therapy: results of a meta-analysis. Breast Cancer Res Treat. 2010;122(3):609–617. doi: 10.1007/s10549-010-0902-3. [DOI] [PubMed] [Google Scholar]
- 56.Ahern TP, Larsson H, Garne JP, Cronin-Fenton DP, Sorensen HT, Lash TL. Trends in breast-conserving surgery in Denmark, 1982-2002. Eur J Epidemiol. 2008;23(2):109–114. doi: 10.1007/s10654-007-9207-1. [DOI] [PubMed] [Google Scholar]
- 57.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60(2):284–295. [PMC free article] [PubMed] [Google Scholar]
- 58.Myrand S, Sekiguchi K, Man M, et al. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin Pharmacol Ther. 2008;84(3):347–361. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- 59.Erichsen R, Lash TL, Hamilton-Dutoit S, Bjerregaard B, Vyberg M, Pedersen L. Existing data sources for clinical epidemiology: the Danish National Pathology Registry and Data Bank. Clin Epidemiol. 2010 (August 9);2:51–56. doi: 10.2147/clep.s9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins LC, Marotti JD, Baer HJ, Tamimi RM. Comparison of estrogen receptor results from pathology reports with results from central laboratory testing. J Natl Cancer Inst. 2008;100(3):218–221. doi: 10.1093/jnci/djm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Badve SS, Baehner FL, Gray RP, et al. Estrogen- and progesterone-receptor status in ECOG 2197: comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol. 2008;26(15):2473–2481. doi: 10.1200/JCO.2007.13.6424. [DOI] [PubMed] [Google Scholar]
- 62.Jensena AR, Ewertz M, Cold S, Storm HH, Overgaard J. Time trends and regional differences in registration, stage distribution, surgical management and survival of breast cancer in Denmark. Eur J Cancer. 2003;39(12):1783–1793. doi: 10.1016/s0959-8049(03)00377-0. [DOI] [PubMed] [Google Scholar]
- 63.Owusu C, Buist DS, Field TS, et al. Predictors of tamoxifen discontinuation among older women with estrogen receptor-positive breast cancer. J Clin Oncol. 2008;26(4):549–555. doi: 10.1200/JCO.2006.10.1022. [DOI] [PubMed] [Google Scholar]
- 64.Geiger AM, Thwin SS, Lash TL, et al. Recurrences and second primary breast cancers in older women with initial early-stage disease. Cancer. 2007;109(5):966–974. doi: 10.1002/cncr.22472. [DOI] [PubMed] [Google Scholar]
- 65.Rae JM, Sikora MJ, Henry NL, et al. Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J. 2009;9(4):258–264. doi: 10.1038/tpj.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cole SR, Hernan MA. Fallibility in estimating direct effects. Int J Epidemiol. 2002;31(1) doi: 10.1093/ije/31.1.163. [DOI] [PubMed] [Google Scholar]
- 67.Lash TL, Rosenberg CL. Evidence and practice regarding the role for CYP2D6 inhibition in decisions about tamoxifen therapy. J Clin Oncol. 2010;28(8):1273–1275. doi: 10.1200/JCO.2009.26.7906. [DOI] [PubMed] [Google Scholar]
- 68.Ratliff B, Dietze EC, Bean GR, Moore C, Wanko S, Seewaldt VL. Re: active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2004;96(11):883–885. doi: 10.1093/jnci/djh170. [DOI] [PubMed] [Google Scholar]
- 69.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 70.Kavvoura FK, McQueen MB, Khoury MJ, Tanzi RE, Bertram L, Ioannidis JP. Evaluation of the potential excess of statistically significant findings in published genetic association studies: application to Alzheimer's disease. Am J Epidemiol. 2008;168(8):855–865. doi: 10.1093/aje/kwn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colhoun HM, McKeigue PM, Davey SG. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361(9360):865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 72.Peters BJ, Rodin AS, de BA, Maitland-van der Zee AH. Methodological and statistical issues in pharmacogenomics. J Pharm Pharmacol. 2010;62(2):161–166. doi: 10.1211/jpp.62.02.0002. [DOI] [PubMed] [Google Scholar]