Abstract
Background
The availability of high throughput experimental methods has made possible to observe the relationships between proteome and transcirptome. The protein abundances show a positive but weak correlation with the concentrations of their cognate mRNAs. This weak correlation implies that there are other crucial effects involved in the regulation of protein translation, different from the sole availability of mRNA. It is well known that ribosome and tRNA concentrations are sources of variation in protein levels. Thus, by using integrated analysis of omics data, genomic information, transcriptome and proteome, we aim to unravel important variables affecting translation.
Results
We identified how much of the variability in the correlation between protein and mRNA concentrations can be attributed to the gene codon frequencies. We propose the hypothesis that the influence of codon frequency is due to the competition of cognate and near-cognate tRNA binding; which in turn is a function of the tRNA concentrations. Transcriptome and proteome data were combined in two analytical steps; first, we used Self-Organizing Maps (SOM) to identify similarities among genes, based on their codon frequencies, grouping them into different clusters; and second, we calculated the variance in the protein mRNA correlation in the sampled genes from each cluster. This procedure is justified within a mathematical framework.
Conclusions
With the proposed method we observed that in all the six studied cases most of the variability in the relation protein-transcript could be explained by the variation in codon composition.
Background
The integration of large scale transcriptome and proteome data along with genome-wide sequence information can give insights into the molecular mechanisms that control cellular functions. Moreover, formulation of mathematical models, either mechanistic or statistic, to express such molecular mechanisms remains a challenging task to understand system properties [1]. The correlation between mRNA transcripts and their corresponding cognate proteins has been found to be positive, but it is not sufficiently good to predict protein levels based on their cognate transcript [2,3]. If all the mRNAs were translated at a constant rate the correlation between mRNA and protein concentration would be high. The observed lack of correlation is therefore due to the particularities of the translation mechanism. For instance, in yeast 73% of the variance in protein abundance is explained by the translation mechanism and only 27% due to the variations of the mRNA concentration [4,5]. To explain the differences in the responses between protein and transcript levels recent studies attempted to include information of the translation mechanism by using mechanistic modeling [6] or by using DNA sequence variables and statistic modeling [7]. Several publications have focused on the kinetics of translation; consisting of initiation, elongation and termination phases. For instance, using a gene-sequence-specific mechanistic model, Mehra and Hatzimanikatis [8] studied the rates of initiation, elongation and termination and found that the different response to mRNA levels is mainly dependent on the initiation step. Following these results, Zouridis and Hatzimanikatis [9] suggested that maximization of translation rate can be achieved by an interplay between ribosomal occupancy and ribosome distribution along the translated mRNA fragment. Subsequently, in a following study by the same authors [10], it was found that not only initiation is a controlling step, but also the elongation phase, which is function of the of tRNA concentration. The mentioned authors reformulated their mathematical model to include the competition between the different aminoacyl-tRNA's.
Codon usage has been shown to be correlated with the abundance of transcripts and proteins [11]. Sharp and Li [12] observed that the variability in mRNA levels of different genes is related to their codon usage and the genome-wide codon usage is related to the number of copies of tRNA genes [13]. Recent studies in E. coli have demonstrated experimentally that perturbation in the codon usage of a set of 40 proteins affected both the translation of the proteins and the tRNA levels in the cell [14].
Based on the analysis of published experimental proteome and transcriptome data for the yeast Saccharomyces cerevisiae (Additional file 1) we tried to evaluate how much the variance in the protein-mRNA correlation is affected by differences in codon usage; which has been demonstrated to be a relevant factor that affects the translation efficiency, either, by increasing the proofreading efficiency of the codon or modifying the folding energy of the mRNA [15,16]. The protein datasets used in this analysis are the result of experimental setups to quantify the peptides associated to each protein, therefore these techniques account for the amount of translated protein and, as it was suggested by Greenbaum et al [17], the protein level can be defined as the "translatome".
Methods
Molecular mechanisms of translation
Translation in yeast starts by the formation of the PIC (pre-initiation complex) which is formed in three steps: first, binding of the specific initiation Met-tRNA to the small ribosomal subunit; second, the resulting complex binds to the mRNA molecules localizing the start codon; and third, the attachment of large ribosomal subunit to generate the polysome structure. All these events are assisted by cis-acting proteins called translation factors. For the elongation process the polysome structure generates three binding sites (E,P,A). In each step an AA-tRNA has to reach the position of site A to place the correct amino acid in the peptide sequence [18,19]. Nevertheless, the existing wobble interactions generate a competition between the cognate and near cognates of charged tRNA (AA-tRNA). Thus, the elongation rate is the result of the time needed to transport the cognate AA-tRNA molecule to the site A in the ribosome [20]. As this is not an efficiently selective step, near cognates can interact in place causing delay due to proof reading and rejection (Figure 1).
Figure 1.
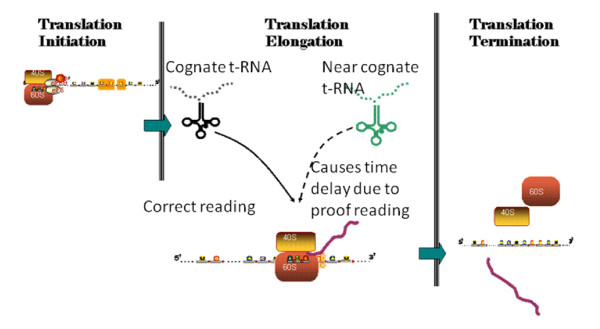
Translation of mRNA into proteins consists in three steps, initiation, elongation and termination. The elongation process consists in the attachment of the cognate tRNA in the right sequence position. Due to Wobble interactions near cognates compete for the position in the ribosome site A causing a delay in elongation time.
Mathematical framework
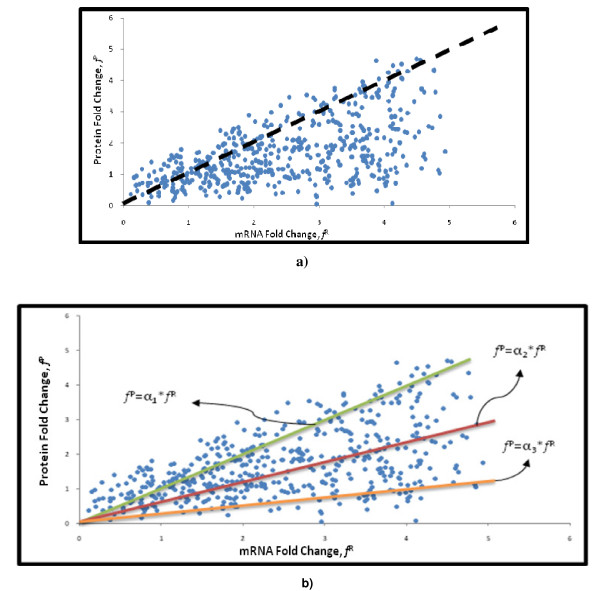
Conceptually there is a remarkable difference between correlating abundance expressed in molecules per cell units compared to fold change in abundance. For our analysis we have collected six datasets where fold changes were studied. For instance, in Figure 2a), the plot contains the values of protein and mRNA fold changes for different genes. If the protein concentration were proportional to mRNA concentration, the fold changes (fj) between conditions should be equal:
Figure 2.
Transcriptome and proteome correlations. a) the plot presents transcriptome and proteome experimental data where it is observed that there is a substantial deviation from the correlation one-to-one represented by the dashed line; b) the relationship between proteome and transcriptome is a function of the amplification factor α which accounts for different parameters such, tRNA availability, ribosome density, protein and transcript degradation rates, among others.
| (1) |
for j = 1...number of genes in the dataset. The superscript P and R correspond to Protein and mRNA quantities, respectively. If such relation were true, the experimental values should fall along the dashed line which is the one-to-one relationship, Figure 2a). If the proportionality constant between mRNA and protein concentrations changed between conditions, the expected graph would be a straight line with slope different from one. However what we found experimentally is a set of scattered points. This means that the proportionality constant not only changes between conditions but also does it differently for each protein.
| (2) |
where the constant α can take different positive values; plot b) in Figure 2. This constant can be seen as an amplification factor that implicitly contains the variation from different sources such as: posttranscriptional events, modification in the translation rates and protein half-lives.
The differential equation governing the concentration of a particular protein is the following one [21-23]:
| (3) |
Where [P] is the concentration of each protein, [mRNA] is the concentration of mRNA, ks,j and kdj are the protein synthesis and degradation rate constants; the dilution term is equal to the growth rate μ. In our approach we write the constant ks,j as the ratio of two characteristic parameters, the number of ribosomes united to each mRNA molecule ρRj and the elongation time of the protein tj. Note that this substitution is absolutely rigorous. The number of proteins synthesized per unit of time is equal to the number of ribosomes synthesizing the corresponding protein divided by the time that each ribosome takes to synthesize a protein.
| (4) |
The two negative terms in the equation correspond to the degradation rate and dilution of proteins as a result of the cellular growth. On the other hand, the elongation time depends on the gene codon composition in the following way
| (5) |
Where Sij is the number of codons i in the gene j and τi is the average time that will take to add the corresponding amino acid to the nascent peptide. This average time is specific for each codon and it depends on the concentration of the corresponding tRNA. The lower is the concentration of a particular tRNA, the longer the time that it takes to add it. The specific time also increases with the number of wrong proof readings that the ribosome performs before adding the right tRNA [20,24].
Assuming steady state for each protein and supposing that only the elongation time changes between proteins and all the other parameters can change in between conditions but not between proteins, we obtained the following relation between mRNA and protein fold changes.
| (7) |
Where the non-dimensional groups are,
| (8) |
The factor Tj depends on the protein composition and the tRNA concentrations in each of the two compared conditions, while the factor C groups all the effects that have been considered to vary only between conditions and do not depend on the protein. If this hypothesis were true, the genes with similar codon frequencies would show a similar behavior in their relation between protein and mRNA fold changes.
Clustering
In this paper we want to evaluate the effects of the codon frequency on protein translation. Proteins with similar codon contents (Sij) will have similar values for the coefficient Tj, if our hypothesis is correct, in a cluster of proteins with similar Tj the variability of the ratio fjP/fjR will be smaller than in the full proteome. We clustered genes using information about the codon composition which was extracted from the genome sequence downloaded from SGD (http://www.yeastgenome.org/). The codon usage has already been shown to be one of the sequence features most highly associated with protein expression [14,25]. The data were normalized using the total codon content of each gene (ΣiSij).
To cluster the proteins according to the codon usage data we used an unsupervised clustering method analysis, SOM, which is a clustering method based on neural networks, and it helps to visualize datasets by mapping a high dimensional data space into a two dimensional space [26]. SOM analysis provides a robust clustering method for outliers or data dispersion [27,28]. There is no theoretical background that dictates the number of map units (neurons) to build the grid; therefore we selected 20 units as it gave the best distribution of genes across the clusters (see Figure 3).
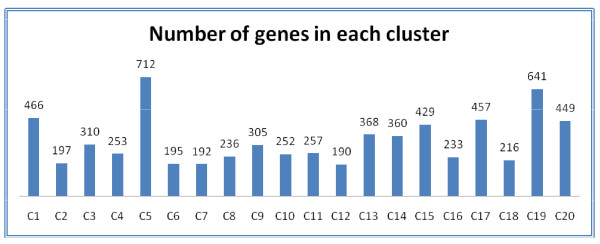
Figure 3.
Using the genome amino acid sequence content from yeast and applying SOM analysis, the result shows 20 different clusters with different numbers of ORFs.
GO enrichment analysis
To elucidate if the genes in each cluster shown functional enrichment we performed a Gene Ontology (GO) enrichment analysis. We performed hypergeometric tests using GO functional annotation from SGD to identify which GO biological process terms are enriched in each category. GO enrichment analysis was performed using BINGO tool [29]; a Cytoscape plug in. To identify which GO terms where significant we used a p-value less that 0.01 as a cutoff.
Analysis of variance
For each of the clusters obtained from the SOM analysis we calculated the ratio between the fold changes in transcriptome and proteome obtaining the value of α and applied the log2 transformation. Logarithmic transformation of data is commonly used as this transformation tends to provide values that are approximately normally distributed and for which ANOVA tests are appropriate [30]. Box plots and histograms showing the distribution of the data are in Additional File 2.
This was done for each protein within each cluster. The subsequent statistical tests will be performed on the following random variable:
| (9) |
ANOVA is a hypothesis test method suitable to compare the means across different groups; clusters in our case. Nevertheless, in this study we focus on quantifying the variance inside the clusters compared with the variance in the complete dataset. In this manner, the results will shed light on the amount of variance in expression levels due effects of the codon frequency and the associated tRNA competition in each of the different clusters. To calculate how much of the total variance for the whole data set was observed between clusters and within clusters the following mathematical formalism is needed. The total sum of squares is the sum of the squares within each cluster plus the sum of squares between the clusters.
| (10) |
Where:
| (11) |
and
| (12) |
The index j identifies each protein inside a given cluster and the index c identifies each cluster. The number of proteins in cluster c is noted as nc. The main question we are trying to answer is how much of the experimental variation in the fold changes can be explained by the variation in codon frequencies. The rest of the variation will be the result of changes in parameters such as degradation rate or number of ribosomes per mRNA molecule that we have grouped in the factor C in Eq.7.
Experimental data
We used six experimental datasets on transcriptome and proteome sampling of the yeast S. cerevisiae. All datasets were collected from the literature and each of them involves a different kind of cellular perturbation. To identify each of the datasets we used an ID which is composed using the last name of the first author: i.e, Griffin [31], Ideker [32], and Washburn [33]. For the dataset of Usaite [34,35] the ID is further specifying the type of deletion performed; e.g.Usaite.snf1 is the ID for deletion of the SNF1 gene in their study. The details for each dataset are presented in Additional File 1 (supplementary table S1). These data consist of fold change values, differently from other studies that have used abundance (molecules/cell) [36] to study the correlation between protein and mRNA and the co-variables that affect such correlation [15,37]. In a similar approach, Nie et al 2006 [38,39] used fold change ratios to demonstrate the correlation between mRNA and protein expression.
Results and Discussion
Correlation between proteome and transcriptome abundance in yeast has been widely studied and it has been observed to be weakly positive [2,3]. Fold changes have shown weak positive correlations as well [31]. In this analysis we used experimental transcriptome and proteome data from yeast (See table in Additional File 1 for more details) to investigate how much of the variance in the relationship between these two quantities is explained by the variance in codon usage [14,15,25,40,41]. More details of the experimental techniques of the datasets shown in Additional File 1 (supplementary table S2) can be seen elsewhere [31-35]. It has been demonstrated by Najafabadi et al. [14] that the codon usage content provides direct information about the translation elongation rate based on the demand of tRNA, which affects the fold change of the protein levels. Nevertheless, there are essential differences in the type of data and the method used for the analysis compared to our work. Najafabadi et al initially clustered the expression patterns using the "average" across several conditions in expression levels and expression "patterns" to perform the codon usage analysis and tRNA modulation. In our approach, we initially used the codon usage as a mean to identify sets of similar genes and performed the analysis using transcriptome and proteome levels independently for each of the considered conditions.
The initial analysis aimed to identify classes of genes with similar codon usage in their primary sequence using the whole annotated genome. From the SOM analysis we obtained a set of 20 different clusters in which the biggest cluster contained 712 ORFs, and the smallest 190 ORF's. The distribution of the clusters is shown in Figure 3.
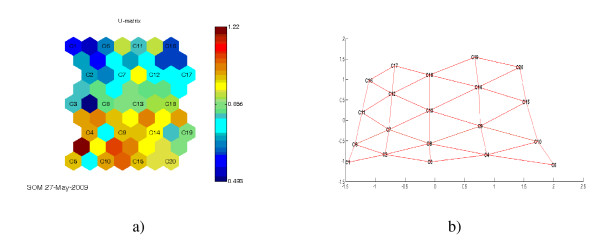
The results of applying SOM can be observed in Figure 4 which contains the unified distance matrix (U-matrix) showing the distances between clusters and also contains the PCA-like projection of the different clusters. Figure 4a) shows the distribution of the clusters and the distances between them. In the PCA-like projection, Figure 4b), it is shown that the separation of the clusters is uniform.
Figure 4.
a) U matrix with the 20 clusters (from C1-C20) and b) PCA-like projection. SOM clustering was based on the protein amino acid sequences. In the U-Matrix blue color separate neurons that are near to one another, and red to neurons that are distant from one another.
Each of the clusters contains a different number of genes (Figure 3) and to identify the functionality of these genes we applied a hypergeometric distribution test to assess the overrepresentation GO biological process. The BINGO tool [29], a Cytoscape plug in [42], was used to perform the analysis. In total the hypergeometric test reported 596 different GO biological process terms, out of which only 115 were repeatedly observed across the different clusters. The analysis shows enrichment of many terms, and by taking the 5 most significant GO terms (with a p-value < 0.01 and after multiple testing correction, FDR) we observed that there are few overlaps across clusters (see Table 1). The detailed GO analysis is contained in Additional file 3. This observation suggests that the primary structure of proteins can be naturally selected so that the proteins performing similar functions have similar codon frequencies [15,25,43]. The reason for that could be that proteins with similar codon frequencies respond in a similar way to changes in the transcription levels; as it was suggested also in Akashi H. (2003) and Tuller et al. (2007).
Table 1.
List of GO biological process terms in each cluster after overlap the results from all datasets.
| Cluster 1 | translation | biosynthetic process | cellular biosynthetic process | cellular protein metabolic process | protein metabolic process |
|---|---|---|---|---|---|
| Cluster 2 | Transport | establishment of localization | localization | transmembrane transport | glutamine family amino acid catabolic process |
| Cluster 3 | amine transport | establishment of localization | amino acid transport | transmembrane transport | carboxylic acid transport |
| Cluster 4 | GPI anchor biosynthetic process | GPI anchor metabolic process | phosphoinositide biosynthetic process | lipoprotein metabolic process | lipoprotein biosynthetic process |
| Cluster 6 | small molecule metabolic process | small molecule biosynthetic process | carboxylic acid metabolic process | oxoacid metabolic process | organic acid metabolic process |
| Cluster 7 | small molecule metabolic process | small molecule biosynthetic process | cellular nitrogen compound biosynthetic process | fatty acid catabolic process | organic acid catabolic process |
| Cluster 8 | telomere maintenance via recombination | ||||
| Cluster 10 | telomere maintenance via recombination | ||||
| Cluster 11 | small molecule metabolic process | small molecule biosynthetic process | heterocycle metabolic process | cellular nitrogen compound biosynthetic process | cellular ketone metabolic process |
| Cluster 12 | endocytosis | ||||
| Cluster 13 | transposition, RNA-mediated | transposition | cellular process | loss of chromatin silencing | cofactor biosynthetic process |
| Cluster 14 | transposition, RNA-mediated | transposition | regulation of biological process | regulation of cellular process | protein amino acid phosphorylation |
| Cluster 16 | ribosome biogenesis | ribonucleoprotein complex biogenesis | rRNA metabolic process | rRNA processing | ncRNA processing |
| Cluster 17 | cellular component biogenesis | nucleic acid metabolic process | macromolecular complex subunit organization | ribonucleoprotein complex biogenesis | RNA metabolic process |
| Cluster 18 | nucleic acid metabolic process | cellular response to stress | cellular component organization | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | response to DNA damage stimulus |
| Cluster 19 | cell cycle | cell cycle process | nucleic acid metabolic process | cellular component organization | cell cycle phase |
| Cluster 20 | regulation of biological process | biological regulation | M phase | regulation of cellular process | cell cycle phase |
*the genes in clusters 5, 9 and 15 were annotated to the GO term "biological process unknown"
Each cluster obtained from the SOM analysis contains genes that show similar codon frequencies. Thus, in order to investigate how much of the variance in the relationship between protein and mRNA fold change is the result of the differences in codon frequency, we estimated the amplification factor xj for each data point according to Eq. 9. The calculations were performed for each of the 6 considered datasets. Table 2 presents the sums of squares of the deviations from the average (Equations 9-13) between and within clusters. It can be seen that for all the datasets, the sum of squares between clusters is higher than the sum of squares within the clusters. For instance, for Usaite.snf1, the fraction of the variability within the clusters is 0.27 and the fraction of variability between the clusters is 0.73. This means that more similar proteins in terms of codon frequency, show similar responses in protein concentration to changes in mRNA, therefore most of the variability in the mRNA-protein relation can be explained by the codon frequency. The rest of the variability is attributed to factors such as protein degradation and seems to be lower compared to the effect of variability in the codon frequency. The F-test shows that except for one out of six datasets, the null hypothesis (e.g. all the clusters have the same average amplification factor) can be safely rejected.
Table 2.
The variance of the amplification factor in each cluster.
| Usaite.snf1 | Usaite.snf4 | Usaite.snf1.4 | Griffin | Ideker | Washburn | |
|---|---|---|---|---|---|---|
| Within/Total | 0.27 | 0.09 | 0.27 | 0.13 | 0.39 | 0.20 |
| Between/Total | 0.73 | 0.91 | 0.73 | 0.87 | 0.61 | 0.80 |
| F-test (B/W) | 2.70 | 10.06 | 2.75 | 6.63 | 1.54 | 4.09 |
| p-value | 0.001 | 1E-06 | 4.5E-5 | 0.015 | 0.55 | 2E-5 |
Alternatively to this analysis, we used exactly the same procedure but using amino acid content instead of codon frequency. In Additional File 1 the Table 2 presents the values of the variance comparing amino acid content and codon frequency. As it was expected, the same conclusions can be extracted both using codon frequency and amino acid content.
Conclusions
Experimentally, it has been observed that the correlation between transcriptome and proteome is positive but not high enough to predict protein levels based on their cognate mRNA transcript levels. In this work, by using experimental transcriptome and proteome data together with a statistical analysis, it was shown that most of the variability in the correlation between protein and mRNA concentration can be explained by the differences in codon usage. Thus, genes with similar codon frequencies show similar correlations between mRNA and protein levels. It was also observed that genes involved in the same cellular functions tend to have more similar codon frequencies. A possible explanation for this fact is the evolutionary advantage that would suppose that the concentrations of proteins involved in the same processes respond in similar ways to perturbations in the mRNA levels.
Authors' contributions
RO and SB developed the method and the mathematical framework. RO performed the data analysis. JN initiated, supervised and coordinated the project. All the authors wrote the manuscript and approved the final version.
Supplementary Material
Description and references for the experimental datasets and comparative table for variances in amino acid content. Supplementary Table S1. This is the list of the six datasets thet were used in this analysis containing expression values for protein and transcript. These datasets have been published on previous works and are considered as high quality data. Supplementary Table S2. It contains the variance in the amplification factor in clusters built using amino acid content and codon usage respectively.
Histograms and box plots of the experimental data. This file contains the histograms and boxplots showing the experimental distributions of the amplification factor, used in the analysis.
Cluster results and amplification factors data. This workbook contents the cluster number for each of the ORF annotated for Saccharomyces cerevisiae. The clusters were constructed using the codon sequence content which was normalized suing the total number of codons.
Contributor Information
Roberto Olivares-Hernández, Email: roberto.olivares@chalmers.se.
Sergio Bordel, Email: velasco@chalmers.se.
Jens Nielsen, Email: nielsenj@chalmers.se.
Acknowledgements
The authors are thankful to Chalmers Foundation and the EU-funded project SYSINBIO (KBBE-212766) for financial support. RO would like to thank to CONACYT-Mexico for the fellowship to support his studies during the first years.
References
- Nielsen J, Jewett MC. Impact of systems biology on metabolic engineering of Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8(1):122–131. doi: 10.1111/j.1567-1364.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19(11):7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25(1):117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924):255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Lee KH, Hatzimanikatis V. Insights into the relation between mRNA and protein expression patterns: I. Theoretical considerations. Biotechnol Bioeng. 2003;84(7):822–833. doi: 10.1002/bit.10860. [DOI] [PubMed] [Google Scholar]
- Nie L, Wu G, Culley DE, Scholten JCM, Zhang W. Integrative Analysis of Transcriptome and Proteomic Data: Challenges, Solutions and Applications. Critical Reviews in Biotechnology. 2007;27:63–75. doi: 10.1080/07388550701334212. [DOI] [PubMed] [Google Scholar]
- Mehra A, Hatzimanikatis V. An algorithmic framework for genome-wide modeling and analysis of translation networks. Biophys J. 2006;90(4):1136–1146. doi: 10.1529/biophysj.105.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouridis H, Hatzimanikatis V. A model for protein translation: polysome self-organization leads to maximum protein synthesis rates. Biophys J. 2007;92(3):717–730. doi: 10.1529/biophysj.106.087825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouridis H, Hatzimanikatis V. Effects of codon distributions and tRNA competition on protein translation. Biophys J. 2008;95(3):1018–1033. doi: 10.1529/biophysj.107.126128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32(17):5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Goodarzi H, Salavati R. Universal function-specificity of codon usage. Nucleic Acids Res. 2009;37(21):7014–7023. doi: 10.1093/nar/gkp792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Kupiec M, Ruppin E. Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput Biol. 2007;3(12):e248. doi: 10.1371/journal.pcbi.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA. 2010;107(8):3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13(1):56–63. doi: 10.1016/S0959-440X(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Fluitt A, Pienaar E, Viljoen H. Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput Biol Chem. 2007;31(5-6):335–346. doi: 10.1016/j.compbiolchem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Bailey JE. Analysis of growth rate effects on productivity of recombinant Escherichia coli populations using molecular mechanism models. Reprinted from Biotechnology and Bioengineering, Vol. 26, Issue 1, Pages 66-73 (1984) Biotechnol Bioeng. 2000;67(6):805–812. doi: 10.1002/(SICI)1097-0290(20000320)67:6<805::AID-BIT16>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Simulation of prokaryotic genetic circuits. Annu Rev Biophys Biomol Struct. 1998;27:199–224. doi: 10.1146/annurev.biophys.27.1.199. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94(3):814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyd A, Drew DA. A mathematical model for elongation of a peptide chain. Bull Math Biol. 2003;65(6):1095–1109. doi: 10.1016/S0092-8240(03)00076-4. [DOI] [PubMed] [Google Scholar]
- Lithwick G, Margalit H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 2003;13(12):2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesanto J, Himberg J, Alhoniemi E, Parhankangas J. SOM toolbox 2.0 for Matlab. 2005.
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96(6):2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiameli P, Chen SK, West D. A comparison of SOM neural network and hierarchical clustering methods. European Journal of Operational Research. 1996;93(2):402–417. doi: 10.1016/0377-2217(96)00038-0. [DOI] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mei-Ling TL. Analysis of Microarray Gene Expression Data. Springer US; 2004. [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1(4):323–333. doi: 10.1074/mcp.M200001-MCP200. [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100(6):3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaite R, Wohlschlegel J, Venable JD, Park SK, Nielsen J, Olsson L, Yates JR Iii. Characterization of global yeast quantitative proteome data generated from the wild-type and glucose repression saccharomyces cerevisiae strains: the comparison of two quantitative methods. J Proteome Res. 2008;7(1):266–275. doi: 10.1021/pr700580m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaite R, Jewett MC, Oliveira AP, Yates JR, Olsson L, Nielsen J. Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol Syst Biol. 2009;5:319. doi: 10.1038/msb.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Brockmann R, Beyer A, Heinisch JJ, Wilhelm T. Posttranscriptional expression regulation: what determines translation rates? PLoS Comput Biol. 2007;3(3):e57. doi: 10.1371/journal.pcbi.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W. Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations. Biochem Biophys Res Commun. 2006;339(2):603–610. doi: 10.1016/j.bbrc.2005.11.055. [DOI] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics. 2006;174(4):2229–2243. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithwick G, Margalit H. Relative predicted protein levels of functionally associated proteins are conserved across organisms. Nucleic Acids Res. 2005;33(3):1051–1057. doi: 10.1093/nar/gki261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4(9):e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164(4):1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description and references for the experimental datasets and comparative table for variances in amino acid content. Supplementary Table S1. This is the list of the six datasets thet were used in this analysis containing expression values for protein and transcript. These datasets have been published on previous works and are considered as high quality data. Supplementary Table S2. It contains the variance in the amplification factor in clusters built using amino acid content and codon usage respectively.
Histograms and box plots of the experimental data. This file contains the histograms and boxplots showing the experimental distributions of the amplification factor, used in the analysis.
Cluster results and amplification factors data. This workbook contents the cluster number for each of the ORF annotated for Saccharomyces cerevisiae. The clusters were constructed using the codon sequence content which was normalized suing the total number of codons.