Abstract
The solution structure of the second protein–protein complex of the Escherichia coli phosphoenolpyruvate: sugar phosphotransferase system, that between histidine-containing phosphocarrier protein (HPr) and glucose-specific enzyme IIAGlucose (IIAGlc), has been determined by NMR spectroscopy, including the use of dipolar couplings to provide long-range orientational information and newly developed rigid body minimization and constrained/restrained simulated annealing methods. A protruding convex surface on HPr interacts with a complementary concave depression on IIAGlc. Both binding surfaces comprise a central hydrophobic core region surrounded by a ring of polar and charged residues, positive for HPr and negative for IIAGlc. Formation of the unphosphorylated complex, as well as the phosphorylated transition state, involves little or no change in the protein backbones, but there are conformational rearrangements of the interfacial side chains. Both HPr and IIAGlc recognize a variety of structurally diverse proteins. Comparisons with the structures of the enzyme I–HPr and IIAGlc–glycerol kinase complexes reveal how similar binding surfaces can be formed with underlying backbone scaffolds that are structurally dissimilar and highlight the role of redundancy and side chain conformational plasticity.
Keywords: E.coli PEP:sugar PTS/glucose-specific enzyme IIAGlucose/histidine-containing phosphocarrier protein/NMR
Introduction
The phosphoenolpyruvate:sugar phosphotransferase system (PTS) of bacteria, first discovered in Escherichia coli over 35 years ago (Kundig et al., 1964), is a classical example of a signal transduction pathway involving phosphoryl transfer. Specifically, the transfer of a phosphoryl group originating on phosphoenolpyruvate (PEP) and ending on a sugar molecule occurs via a series of phosphoprotein intermediates involving an associative pathway in which successive protein–protein complexes between phosphoryl donor and acceptor molecules are formed (Herzberg and Klevit, 1994; Postma et al., 1996). The initial cascade involves a common pathway in which enzyme I (EI), autophosphorylated by PEP at His189 (in E.coli), transfers the phosphoryl group to His15 (in E.coli) of the histidine-containing phosphocarrier protein (HPr). The phosphoryl group associated with HPr is then available for transfer to a variety of sugar-specific IIA proteins. In addition to their role in sugar transport, proteins of the PTS pathway also function as regulatory factors (Postma et al., 1996). Thus, dephosphorylated HPr functions as a positive regulatory subunit of glycogen phosphorylase (Seok et al., 1997), while dephosphorylated IIAGlucose (IIAGlc) is a negative regulator of glycerol kinase (Novotny et al., 1985), as well as a variety of non-PTS permeases (Postma et al., 1996). In addition, phosphorylated IIAGlc is a positive regulator of adenyl cyclase activity (Peterkofsky et al., 1993). Crystal and solution NMR structures of individual components of the PTS pathway have been solved. These include: the N-terminal phosphoryl transfer domain of E.coli EI (EIN) (Liao et al., 1996; Garrett et al., 1997a), HPr from a variety of species (Herzberg et al., 1992; Wittekind et al., 1992; Jia et al., 1993; Kalbitzer and Henstenberg, 1993; van Nuland et al., 1994, 1995; Jones et al., 1997), IIAGlc (also referred to as IIIGlc) from a variety of species (Liao et al., 1991; Worthylake et al., 1991; Fairbrother et al., 1992; Hurley et al., 1993; Feese et al., 1997; Huang et al., 1998), IIAMan (Nunn et al., 1996), IIALac (Sliz et al., 1997) and IIAMtl (van Montfort et al., 1998), IIBGlc (Eberstadt et al., 1996), IIBCel (Ab et al., 1997; van Montfort et al., 1997) and IIBLev (Schauder et al., 1998). In contrast, only one protein–protein complex from the PTS pathway has been elucidated to date, namely that between E.coli EIN and HPr by NMR (Garrett et al., 1999). In this paper we present the NMR structure of the second complex in the glucose-specific arm of the PTS cascade, that between HPr and IIAGlc of E.coli.
Results and discussion
Structure determination
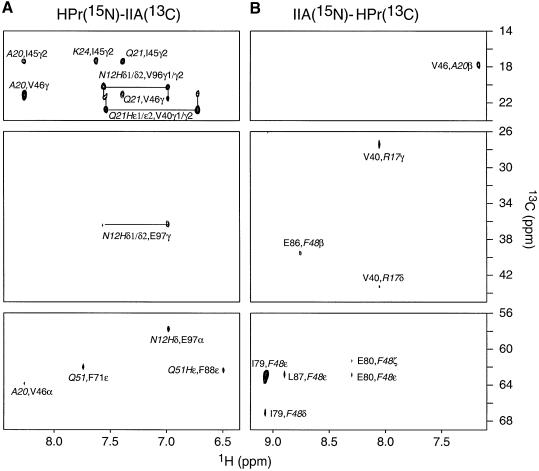
The HPr–IIAGlc complex is in fast exchange on the chemical shift time scale and the lower limit for the dissociation rate constant (as judged by the maximum observed 1HN chemical shift difference between the free and bound states) is ∼3000 s–1. The equilibrium association constant for the binding of HPr to IIAGlc, determined by following the 1H-15N cross-peaks of 15N-labeled IIAGlc upon titration with unlabeled HPr, is ∼105 M–1, consistent with previous biophysical measurements (Jablonski et al., 1983). The solution structure of the HPr–IIAGlc complex was solved by multi-dimensional heteronuclear NMR spectroscopy. A combination of isotopically (15N and/or 13C) labeled proteins was used to simplify the spectra for assignment purposes and to specifically observe intermolecular nuclear Overhauser enhancement (NOE) contacts between HPr and IIAGlc (Clore and Gronenborn, 1998a). Intermolecular NOEs were detected using four samples: HPr(15N/13C)–IIAGlc, HPr–IIAGlc(15N/13C), HPr(13C)–IIAGlc(15N) and HPr(15N)–IIAGlc(13C). Examples of the data quality are provided in Figure 1, which shows a complementary set of 2D 15N-filtered/13C-separated NOE experiments illustrating intermolecular contacts between protons attached to 15N on one protein and to 13C on the other. Other experiments used to identify intermolecular NOE contacts comprised 2D 13C-filtered/15N-separated and 13C-filtered/15N-filtered NOE spectra and 3D 13C-separated/12C-filtered NOE spectra.
Fig. 1. Intermolecular NOEs. 2D 15N-filtered/13C-separated NOE spectra (120 ms mixing time) recorded on (A) a 1:1 HPr(15N)–IIAGlc(13C) complex and (B) a 1:1 HPr(13C)–IIAGlc(15N) complex, specifically illustrating intermolecular NOE contacts from protons attached to 13C (F1 axis) to amide protons attached to 15N (F2 axis). Residues from HPr are denoted in italic. Intermolecular NOEs were assigned with the aid of complementary 2D 15N-separated/13C-filtered and 15N-filtered/13C-filtered NOE spectra. Note that the aromatic 13C resonances are folded.
The structure of the complex was calculated using the recently described procedure of rigid body minimization (Bewley and Clore, 2000; Clore, 2000), followed by constrained/restrained simulated annealing to refine the interfacial side chain positions and fine tune the relative orientation of the two proteins. In this approach, the intermolecular NOE distance restraints and dipolar couplings provide the necessary translational and orientational information to dock two proteins accurately, providing that no significant changes in the backbone of either protein occur upon complexation. In the case of both E.coli HPr and IIAGlc, high resolution crystal structures (1.5 and 2.0 Å resolution, respectively) are available for the free proteins (Jia et al., 1993; Feese et al., 1997). A comparison of the one-bond 15N-H backbone residual dipolar couplings (1DNH) measured on the complex (77 for HPr and 118 for IIAGlc) in a dilute liquid crystalline medium of tobacco mosaic virus (Clore et al., 1998a) with those calculated from the free X-ray structures by optimization of the magnitude and orientation of the alignment tensor yields dipolar coupling R factors (Rdip) (Clore and Garrett, 1999) of 16.7 and 15.0% for HPr and IIAGlc, respectively. [Note that the X-ray structure of IIAGlc comprises residues 19–168; residues 1–18 are disordered in the crystal structure of the free protein (Worthylake et al., 1991; Feese et al., 1997), in the free protein in solution (Pelton et al., 1991) and in the HPr–IIAGlc complex in solution as judged by the absence of any non-sequential NOEs.] Thus, one can safely conclude that no significant changes in backbone conformation occur upon complexation. This is further supported by the small 1HN and 15N chemical shift differences and nearly identical intramolecular NOE patterns between the free and bound states of the two proteins.
The structure calculation involves a two step procedure. (To facilitate discussion, residues of HPr are denoted in italic.) The starting coordinates come from the X-ray structures (with protons added) of E.coli HPr (Jia et al., 1993) and IIAGlc (Feese et al., 1997) in several different orientations with the Cα–Cα distance between the active site histidines (His15 of HPr and His90 of IIAGlc) ranging from 28 to 95 Å, including orientations where the two active site histidines are not opposed and where HPr is directed towards the face of IIAGlc opposite to the IIAGlc active site. The first step involves rigid body minimization on the basis of a target function comprising only three terms: the experimentally NOE-derived intermolecular interproton distance restraints, the dipolar coupling restraints and a simple quartic van der Waals repulsion potential. This system has only 9 degrees of freedom since one of the molecules (IIAGlc) is held fixed, the other molecule is free to rotate and translate (6 degrees of freedom) and the axis for the single dipolar coupling alignment tensor is free to rotate (3 degrees of freedom). In all cases, convergence to a single structure is obtained with an experimental data set comprising 74 intermolecular NOE-derived interproton distance restraints and 195 1DNH dipolar coupling restraints. The intermolecular van der Waals contacts in this structure, however, are very poor since the interfacial side chains have not been allowed to move in any way and, consequently, steric clash is inevitable. In the original paper (Clore, 2000), steric clash was relieved by subjecting the coordinates to conventional Cartesian coordinate constrained/restrained minimization in which all the coordinates were held fixed with the exception of the interfacial side chains (from the γ position onwards). In this case, however, some side chains alter their conformations significantly. For example, the χ1 rotamer of Phe48 of HPr changes from gauche– (g–) in the crystal structure of the free protein to trans (t) in the complex (determined by the measurement of three-bond heteronuclear 3JNCγ and 3JC′Cγ spin–spin coupling constants). In the second step we therefore made use of an alternative procedure with a much larger radius of convergence, which we term constrained/restrained simulated annealing. Only the interfacial side chains are allowed to alter their conformation; the backbone and non-interfacial side chains of one molecule (IIAGlc) are held completely fixed; the second molecule (HPr) can rotate and translate but the relative coordinates of its backbone and non-interfacial side chains are held fixed by the use of a non-crystallographic symmetry restraint to a duplicate molecule. This ensures that the atomic root mean square (r.m.s.) difference between the structure of HPr in the complex and that in the crystal structure of free HPr is maintained at <0.05 Å for the backbone and non-interfacial side chains and at <0.015 Å for the backbone N and HN atoms. The target function (Clore and Gronenborn, 1998b) for constrained/restrained simulated annealing comprises terms for covalent geometry, non-bonded contacts (in the form of a quartic van der Waals repulsion term and a conformational database potential of side chain torsion angles; Kuszewski and Clore, 2000), a non-crystallographic symmetry constraint, NOE, dipolar coupling (Clore et al., 1998c) and interfacial side chain torsion angle restraints and a radius of gyration restraint (Kuszewski et al., 1999). In addition to the intermolecular NOEs, 12 intramolecular NOEs relating to His15 and Arg17 of HPr were also included. The radius of gyration restraint is employed to ensure optimal packing of the interface and its target value is calculated using the formula 2.2N0.38, where N is the number of ordered residues [i.e. Rgyr(target) = 17.5 Å for 235 residues, 85 from HPr and 150 from IIAGlc] (Kuszewski et al., 1999). The force constant for the Rgyr term is rather weak and the value of Rgyr actually attained is 17.7 Å. After cooling, the structures were subject to a few cycles of constrained/restrained Cartesian coordinate minimization, followed by a few cycles of rigid body minimization.
To assess the accuracy of the determination of the relative orientations of HPr and IIAGlc, two sets of calculations were carried out using cross-validation (Clore and Garrett, 1999) in which the dipolar couplings were partitioned into two groups (A and B) of equal size, evenly distributed throughout the two proteins. In the first series of calculations, group A was employed as the work set and group B as the test set, while in the second series of calculations the allocation was reversed, with group B functioning as the work set and group A as the test set. In this manner, a working dipolar coupling R factor, Rdip(work), and a cross-validated one, Rdip(free), can be obtained. In both sets of calculations the values of Rdip(work) and Rdip(free) are comparable (Table I) and the atomic r.m.s. difference between the mean structures for the two sets of calculations (comprising 15 structures each) is 0.09 Å for the backbone atoms and 0.11 Å for all atoms, which is well within the precision of the coordinates (0.13 Å for backbone atoms and 0.25 Å for all atoms). In addition, the values of Rdip for the complex (calculated for a single alignment tensor) are comparable to those of the free X-ray structures (calculated from individual alignment tensors). One can therefore conclude that the quality of the fit to the experimental dipolar coupling data is good, that the dipolar coupling data have not been overfitted and that the accuracy with which the orientation of HPr and IIAGlc in the complex has been determined is high.
Table I. Structural statisticsa.
| <SA> | (SA―)r | |
|---|---|---|
| R.m.s. deviation from distance restraints (Å)b | ||
| intermolecular interproton distances (74) | 0.073 ± 0.011 | 0.061 |
| intramolecular interproton distances (12)c | 0.033 ± 0.027 | 0.000 |
| intermolecular salt bridge restraints (8)d | 0.020 ± 0.023 | 0.023 |
| R.m.s. deviation from torsion angle restraints (°) (61)b | 0.19 ± 0.07 | 0.16 |
| R factors for residual dipolar couplings (%)e | ||
| set 1 Rdip(work)/Rdip(free) | ||
| HPr (39/38) | 15.8 ± 0.1/18.1 ± 0.1 | |
| IIA (59/59) | 14.5 ± 0.1/15.8 ± 0.1 | |
| set 2 Rdip(work)/Rdip(free) | ||
| HPr (38/39) | 17.6 ± 0.2/16.2 ± 0.1 | |
| IIA (59/59) | 14.5 ± 0.1/15.8 ± 0.1 | |
| overall Rdip | ||
| HPr (77) | 16.9 | |
| IIA (118) | 15.2 | |
| Measures of structure qualityf | ||
| intermolecular repulsion energy (kcal/mol) | 0.09 ± 0.16 | 0 |
| intermolecular Lennard–Jones energy (kcal/mol) | –34 ± 2 | –33 |
| Coordinate precision (Å)g | ||
| backbone (N, Cα, C, O) | 0.13 | |
| interfacial side chains | 0.77 |
aThe notation of the NMR structures is as follows: <SA> is the final 30 simulated annealing structures; (SA―)r is the restrained regularized mean structure generated as described in the text. The number of terms for the various restraints is given in parentheses.
bNone of the structures exhibited interproton distance violations >0.5 Å or torsion angle violations >5°.
cThe intramolecular interproton distance restraints relate specifically to intramolecular NOEs involving His15 and Arg17 of HPr.
dAmbiguous distance restraints, represented by a (Σr–6)–1/6 sum with an upper bound of 5.5 Å, were included in the final stages of refinement for potential salt bridges identified using the criteria described (Omichinski et al., 1997).
eThe dipolar coupling R factor is defined as the ratio of the r.m.s. deviation between observed and calculated values to the expected r.m.s. deviation if the vectors were randomly oriented. The latter is given by {2Da2[4 + 3R2]/5}1/2, where Da is the magnitude of the axial component of the alignment tensor and R the rhombicity (Clore and Garrett, 1999). The values of Da and R for the 1DNH dipolar couplings, obtained directly from the powder pattern distribution of the measured dipolar couplings (Clore et al., 1998b), are –14.9 Hz and 0.2, respectively. For reference, the dipolar coupling R factors for the free X-ray structures of HPr and IIAGlc are 16.7 and 15.0%, respectively, the R factors for group A are 16.6 (39 dipolar couplings for HPr) and 14.2% (59 dipolar couplings for IIAGlc), respectively, and for group B 16.9 (38 dipolar couplings for HPr) and 15.8% (59 dipolar couplings for IIAGlc), respectively.
fThe intermolecular repulsion energy is given by the value of the intermolecular quartic van der Waals repulsion term calculated with a force constant of 4 kcal/mol/Å2 and a van der Waals radius scale factor of 0.8. The intermolecular Lennard–Jones–van der Waals energy is calculated using the CHARMM19/20 parameters and is not included in the target function employed in the structure calculations. The percentages of residues present in the most favorable region of the Ramachadran plot (Laskowski et al., 1993) are 93 and 89% for the X-ray structures of HPr (1POH) and IIAGlc (molecule 2 of 2F3G), respectively.
gDefined as the average r.m.s. difference between the final 30 simulated annealing structures and the mean coordinates. The value quoted for the backbone atoms provides only a measure of the precision with which the relative orientation of the two proteins have been determined and does not take into account the errors in the X-ray coordinates of HPr and IIAGlc.
An even more stringent test of quality is provided by calculations carried out in the absence of any dipolar coupling restraints. The backbone r.m.s. difference between the restrained minimized mean structures from the ensembles calculated with and without dipolar couplings is only 0.06 Å and the values of Rdip for the mean structure obtained without dipolar couplings (17.8% for HPr and 15.2% for IIAGlc) are only marginally higher than for the mean structure calculated with dipolar couplings (16.9% for HPr and 15.2% for IIAGlc). Thus, in this case the intermolecular NOE data alone is sufficient to determine the orientation of the two proteins correctly. The availability of dipolar couplings, however, provides considerable increased confidence in the resulting structure because the nature of the NOE and dipolar coupling data are so different (Clore, 2000).
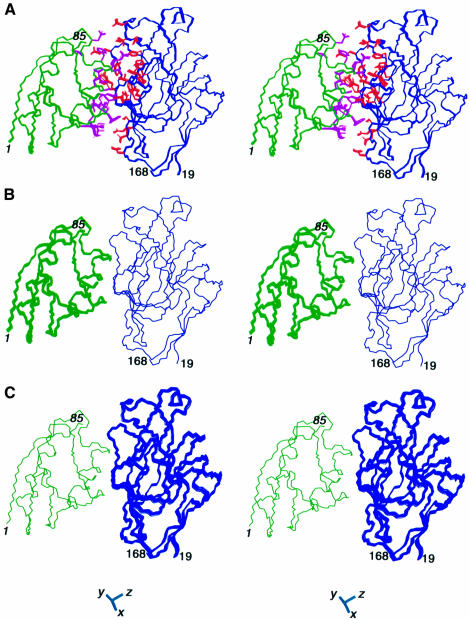
A summary of the structural statistics is provided in Table I and stereoviews of superpositions of the structures are shown in Figure 2.
Fig. 2. The structure of the E.coli HPr–IIAGlc complex. Three sets of superpositions are shown. (A) The structures are best fitted to all the backbone atoms; the backbone and side chains of HPr are shown in green and magenta, respectively, and the backbone and side chains of IIAGlc are shown in blue and red, respectively. (B) The structures are best fitted to the backbone atoms of IIAGlc only. (C) The structures are best fitted to the backbone atoms of HPr only. The latter two superpositions illustrate the precision with which the orientation of one molecule is determined relative to the other. Residues from HPr are denoted in italic. The axis of the alignment tensor is shown at the bottom of the figure.
Overall description of the HPr–IIAGlc interface
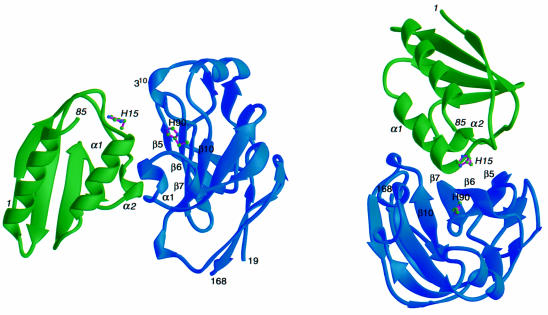
A ribbon diagram representation of the E.coli HPr–IIAGlc complex is shown in Figure 3 and a summary of the quantitative structural characteristics (Jones and Thornton, 1996) of the interface is provided in Table II. HPr is an α/β open faced sandwich protein consisting of three helices on one side of the molecule and a four-stranded antiparallel β-sheet on the other. Escherichia coli IIAGlc is a predominantly β-sheet sandwich protein comprising six antiparallel β-strands on either side of the molecule. The binding sites are close to circular and complementary in shape. The IIAGlc binding surface on HPr is convex and comprises helices α1 and α2, together with residues immediately N- and C-terminal to these two helices, consistent with previous chemical shift mapping (Wang et al., 2000). The HPr binding surface on IIAGlc, on the other hand, is concave and formed by four strands (β5, β6, β7 and β10) of the six-stranded antiparallel β-sheet (β5, β6, β7 and β10, β3, β2), bounded on three sides by a short α-helix and two short, distorted 310 helices. There are a total of 41 residues at the interface, 18 from HPr and 23 from IIAGlc. The total accessible surface area (ASA) buried at the interface of the two proteins is ∼1365 Å2, of which ∼735 Å2 originates from HPr and ∼630 Å2 from IIAGlc.
Fig. 3. Ribbon diagram showing two views of the E.coli HPr–IIAGlc complex. HPr is in green, IIAGlc in blue and the location of the active site histidines, His15 of HPr and His90 of IIAGlc, is indicated. Residues from HPr are denoted in italic. The secondary structure elements in the vicinity of the interface are indicated.
Table II. Comparison of overall features of the interfaces of the EIN–HPr, HPr–IIAGlc and IIAGlc–glycerol kinase complexes.
| Interface parameter | EIN–HPr | HPr-IIAGlc | IIAGlc–GK |
|---|---|---|---|
| ASA buried at interface (Å2)a,b | 950/995 | 735/630 | 615/690 |
| No. of residues at interfaceb | 21/23 | 18/23 | 17/16 |
| Polar atoms in interface (%)a | 35 | 42 | 40 |
| Non-polar atoms in interface (%)a | 65 | 58 | 60 |
| Electrostatic interactions | 11 | 8 | 6 |
| Gap index (Å)a | 2.1 | 2.5 | 2.9 |
| Planarity (Å)a | 2.7 | 2.5 | 1.9 |
| Circularitya,b | 0.82/0.90 | 0.77/0.82 | 0.76/0.72 |
| Secondary structureb | α/α | α/α,β | α,β/α |
aThese parameters were calculated using the program suite provided by the University College Protein–Protein Interaction Server (http://www.biochem.ucl.ac.uk/bsm/PP/server) as described (Jones and Thornton, 1996). The gap index, which is a measure of complementarity of the two partners, is given by the ratio of the gap volume between the molecules to the ASA buried at the interface; the circularity of the interfaces is calculated from the ratio of the length of the principal axes of the least squares plane through the atoms in the interface.
bThe first and second numbers refer to the first and second proteins, respectively, listed for each complex.
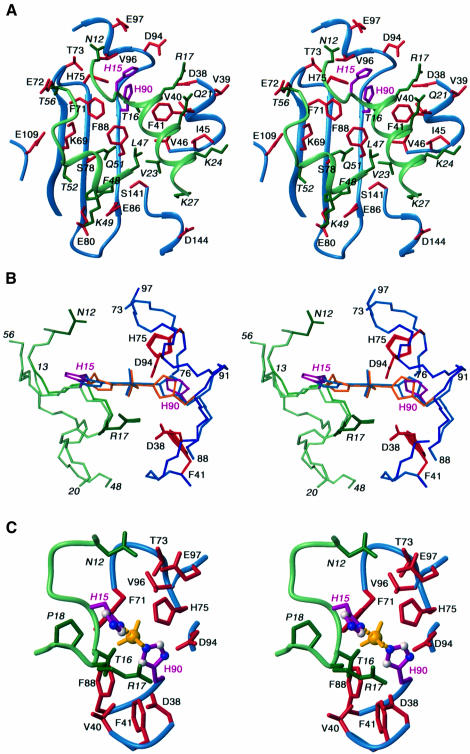
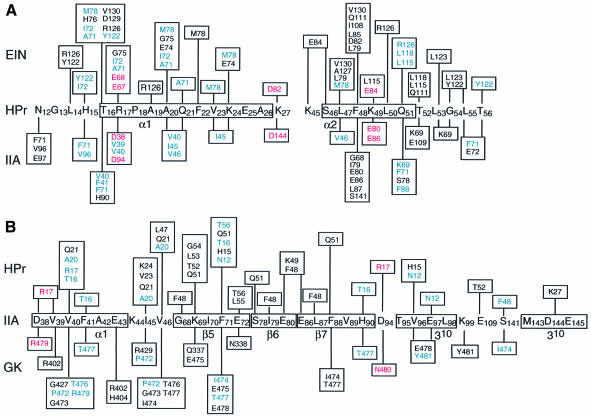
A detailed view of the interface between HPr and IIAGlc is shown in Figure 4A, a summary of the contacts is provided in Figure 5 and surface representations of the interface are displayed in Figure 6C and D (HPr residues in italic). The active site histidines, His15 and His90, are located at the top edge of the binding site in the views shown in Figures 4A and 6C and D. The central portion of each binding surface is predominantly hydrophobic, consisting in the case of HPr of the methyl groups of Thr16, Ala20, Val23 and Leu47 and the aromatic ring of Phe48, and in the case of IIAGlc of a ring of three phenylalanine residues, Phe41, Phe71 and Phe88, interspersed by three valines, Val40, Val46 and Val96. Phe48 of HPr is in direct contact with the backbone of strands β6 and β7 of IIAGlc and is located between the hydrophobic portions of the side chains of Ser78, Glu80, Glu86 and Phe88. The central hydrophobic patch is surrounded in both cases by polar and charged residues. The latter are entirely positively charged in the case of HPr and negatively charged in the case of IIAGlc. There are several electrostatic interactions at the interface (Figures 4A and 5) that were identified using criteria described previously (Omichinski et al., 1997). These include hydrogen bonding and ion pair interactions between the guanidino group of Arg17 and the carboxylates of Asp38/94 (which were also predicted by molecular modeling of the related Bacillus subtilis complex; Herzberg, 1992), the side chains of Asn12 and Glu97, the side chains of Gln51 and Ser78, and the backbone carbonyl of Leu53 and the side chain of Lys69. In addition, there are potential salt bridges between Lys27 and Asp144 and between Lys49 and Glu80/86.
Fig. 4. HPr–IIAGlc interactions in the unphosphorylated complex and in the transition state. (A) Stereoview of the unphosphorylated HPr–IIAGlc interface. The backbones of HPr and IIAGlc, depicted as ribbon diagrams, are shown in blue and light green, respectively, the side chains of HPr and IIAGlc are shown in dark green and red, respectively, and the active site histidines (His15 of HPr and His90 of IIAGlc) are depicted in purple. (B) Detailed view around the active site histidines, illustrating the backbone and side chain positions in the unphosphorylated complex, the dissociative transition state and the associative transition state. The color coding is the same as in (A) except that the active site histidines and pentacoordinate phosphoryl group (in the case of the transition states) are shown in purple for the unphosphorylated complex, in light blue for the dissociative transition state (Nδ1–Nε2 distance between His15 and His90 of ∼6 Å) and in orange for the associative transition state (Nδ1–Nε2 distance between His15 and His90 of ∼4 Å). The backbones for the unphosphorylated and dissociative transition state complexes are identical and only the positions of the active site histidines are different; in the associative transition state complex there are small alterations in the backbone of residues 13–17 of HPr and 89–91 of IIAGlc (see text for details). (C) Detailed view of the active site during the putative associative transition state illustrating the interactions that stabilize the phosphoryl group. The color coding is the same as in (A) and the phosphoryl group is shown in yellow. Residues from HPr are denoted in italic.
Fig. 5. Summary of interactions between (A) HPr and its partner proteins EIN and IIAGlc and (B) between IIAGlc and its partner proteins HPr and glycerol kinase (GK). Side chains that participate in analogous hydrophobic and electrostatic interactions in both partners are depicted in blue and red, respectively.
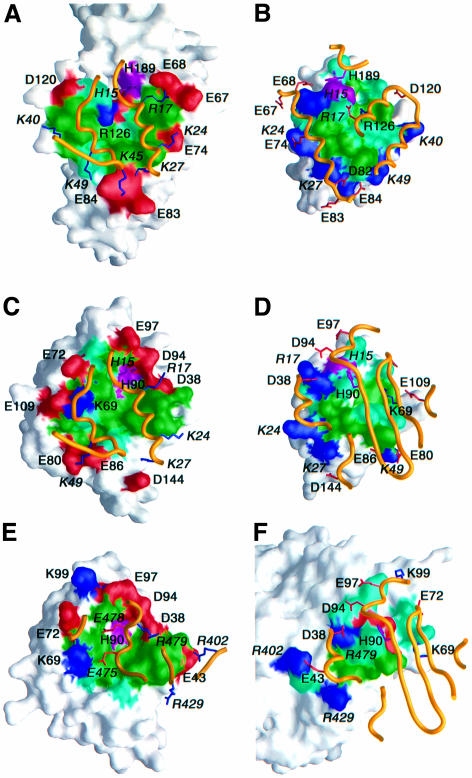
Fig. 6. Surface representations illustrating the binding surfaces involved in the (A and B) EIN–HPr, (C and D) HPr–IIAGlc and (E and F) IIAGlc–GK complexes. The HPr binding surface on EIN and IIAGlc are shown in (A) and (C), respectively; the EIN and IIAGlc binding surfaces on HPr are shown in (B) and (D), respectively; the GK binding surface on IIAGlc is shown in (E); the IIAGlc binding surface on GK is shown in (F). The binding surfaces are color coded with hydrophobic residues in green, polar residues in light blue, the active site histidines in purple, positively charged residues in dark blue and negatively charged residues in red. The relevant portion of the backbone of the partner protein is shown as a gold ribbon with positively charged side chains in dark blue and negatively charged ones in red. Only charged residues and the active site histidines are labeled, with residues from HPr and GK denoted in italic. Note that although the active site histidines of EIN (His189) and IIAGlc (His90) are in close contact with His15 of HPr, their direction of approach is different: His189 (EIN) approaches His15 from above (B), while His90 (IIAGlc) approaches His15 from below (D). The coordinates for the EIN–HPr and IIAGlc–GK complexes are taken from Garrett et al. (1999) (RCSB accession code 3EZA) and Hurley et al. (1993) (RCSB accession code 1GLA).
The transition state and phosphoryl transfer
HPr and IIAGlc are phosphorylated at the Nδ1 atom of His15 and Nε2 atom of His90, respectively (Meadow and Roseman, 1982; Weigel et al., 1982; Pelton et al., 1992, 1993; van Nuland et al., 1995; Jones et al., 1997). In the unphosphorylated HPr–IIAGlc complex the Cα–Cα and Nδ1–Nε2 distances between His15 and His90 are 11.5 and 8.0 Å, respectively. Long-range 1H-15N correlation experiments confirm that His15 is in the Nε2-H tautomeric state while His90 is in the Nδ1-H tautomeric state, as observed for the free proteins (van Dijk et al., 1990; Pelton et al., 1993). Since the phosphorylated state of the complex is short lived, it is not amenable to NMR structure determination. However, the transition state can readily be modeled from the structure of the unphosphorylated complex since it is known from isotope labeling experiments that odd and even numbers of phosphoryl transfer steps result in inversion and retention, respectively, of the configuration of the phosphorus (Begley et al., 1982). These data are indicative of a transition state involving a pentacoordinate phosphoryl group in a trigonal bipyramidal geometry, with the donor and acceptor atoms in apical positions and the oxygen atoms lying in the equatorial plane. The covalent P–N bond length is 1.78 Å for a phosphoamide (MacGillavry and Rieck, 1962) and the sum of the van der Waals radii of P (1.9 Å) and N (1.5 Å) is 3.4 Å. Thus, any P–N distance between 1.78 and 3.4 Å would indicate partial bonding. The actual distance between Nδ1(His15) and Nε2(His90) in the transition state depends on the exact mechanism of phosphoryl transfer, and specifically how associative it is. If fully dissociative, the distance would be 2 × 3.4 Å = 6.8 Å; if fully associative, the distance would be 2 × 1.78 Å = 3.56 Å. For an SN2 mechanism (i.e. 50% associative with a bond order of 0.5), the distance would be 2 × 1.96 = 3.92 Å, where the value of 1.96 comes from Pauling’s equation D(n) = D(1) – 0.6 logn, where n is the bond order and D(1) is a single bond length. Current data comparing linear free energy relationships for non-enzymatic and enzymatic phosphoryl transfer reactions suggest that, in the case of alkaline phosphatase at least, the transition state has substantial dissociative character (Hollfelder and Herschlag, 1995).
To model the transition state, we therefore proceeded along similar but not identical lines to those described in our paper on the EIN–HPr complex (Garrett et al., 1999). Specifically, we introduced a phosphoryl group to the coordinates generated subsequent to the first rigid body minimization step, with covalent geometry restraints relating to trigonal bipyramidal geometry at the phosphorus. The latter included: a symmetry restraint to maintain equal bond lengths for the Nδ1(His15)–P and Nε2(His90)–P bonds (without imposing a restraint for the actual bond length); the bond angle restraints Nδ1(His15)– P–Nε2(His90) = 180° and Cγ(His15)–Nδ1(His15)–P = Cε1(His15)–Nδ1(His15)–P = Cδ2(His90)–Nε2(His90)– P = Cε1(His90)–Nε2(His90)–P = 120°; two planarity restraints to ensure that the phosphorus atom lies in the plane of each imidazole ring. (Note that the imidazole rings of His15 and His90 are not required to be co-planar.) The phosphorus was initially placed approximately half way between His15 and His90 and the structure was subjected to constrained/restrained simulated annealing and a final round of rigid body minimization using exactly the same NOE-derived interproton distance restraints (with one exception), torsion angle restraints and dipolar coupling (including cross-validation) employed for the unphosphorylated HPr–IIAGlc complex. Only the distance restraint corresponding to the strong intramolecular NOE between the Hβ protons of Asn12 and the Hδ2 proton of His15 was removed, since this effectively limits the accessible χ1 torsion angle of His15 to a very narrow range. Thus, in these calculations the backbone is maintained at those of the X-ray coordinates of the free proteins and the side chains of His15 and His90 can move appropriately to accommodate the restraints imposed by inclusion of the phosphoryl group. An ensemble of 30 simulated annealing structures was calculated and, with the exception of the two active site histidines and the phosphoryl group, these structures are essentially identical to those of the unphosphorylated complex. Thus, the r.m.s. difference between the mean structures of the unphosphorylated complex and the transition state complex is 0.03 Å for the backbone atoms and 0.2 Å for the interfacial side chains (excluding His15 and His90). The Cα–Cα distance between His15 and His90 remains unchanged at 11.5 Å, but the Nδ1–Nε2 distance between His15 and His90 is reduced to 6 Å, with essentially idealized geometry of the phosphoryl group transition state (Figure 4B). The planes of the imidazole rings of His15 and His90 are oriented at ∼90° to each other. Thus, this model of the transition state corresponds to a largely dissociative mechanism in which the P–N bond of the donor histidine breaks before the P–N bond to the acceptor starts to form, producing a metaphosphate-like transition state. This is accompanied by relatively small changes in the side chain torsion angles of His15 and His90: in the unphosphorylated complex the χ1/χ2 angles of His15 and His90 are 51°/106° and 176°/–67°, respectively, while in the transition state complex the corresponding values are 89°/100° and 189°/–33°, respectively.
The above result does not favor a dissociative over an associative transition state. This is because small changes in backbone conformation in the immediate vicinity of the active site histidines could easily accommodate a shorter Nδ1(His15)–Nε2(His90) distance while preserving idealized geometry of the phosphoryl group in the transition state. To test this possibility, we subjected the dissociative model of the transition state to constrained/restrained minimization in which the coordinates of all the backbone atoms, with the exception of residues 13–17 of HPr and residues 89–91 of IIAGlc, and all the non-interfacial side chains were held completely fixed and in which the Nδ1(His15)–P and Nε2(His90)–P distances were restrained to 1.96 Å, corresponding to an SN2 associative transition state. The experimental NMR restraints and other covalent geometry restraints were the same as those used to compute the model of the dissociative transition state complex. As is evident from Figure 4B, only very minor changes in the backbone conformation of these eight residues, five for HPr and three for IIAGlc, are required to accommodate the shorter N–P distances. Indeed, the angular r.m.s. difference in the φ/ψ angles between the two transition state models is only 16° for the eight residues. Moreover, the χ1/χ2 torsion angles of His15 and His90, which have values of 85°/103° and –159°/–40°, respectively, are only minimally altered relative to those in the dissociative transition state complex.
A detailed view of the environment surrounding the phosphoryl group in the associative transition state complex is shown in Figure 4C. The phosphoryl group is surrounded by a cluster of hydrophobic residues from IIAGlc comprising Val40, Phe41, Phe71 and Val96. The phosphoryl group is stabilized by hydrogen bonds from the backbone amide and side chain hydroxyl protons of Thr16 and from the Hε2 proton of His75, as well as by potentially water-bridged hydrogen bonds from the backbone amides of Arg17, Asp94 and Val96. The Nε2-H tautomeric state of His75 is stabilized by a hydrogen bond in which the Nδ1 atom of His75 accepts a hydrogen bond from the hydroxyl group of Thr73. It is interesting to note that mutation of His75 to Gln (H75Q) decreases the rate of phosphoryl transfer between HPr and IIAGlc by a factor of ∼200 (Meadow and Roseman, 1996). While a Gln at position 75 can still donate a hydrogen bond to the phosphoryl group, it can no longer accept one from Thr73 (Pelton et al., 1996), thereby destabilizing the transition state.
Two other key features of the model of the transition state complex, which are also present in the structure of the unphosphorylated complex, stand out: the orientation of the imidazole ring of His15 is in part stabilized by hydrophobic contacts with Pro18 (Figure 4C); the negatively charged carboxylates on IIAGlc that are in the vicinity of the phosphoryl group are neutralized by additional hydrogen bonding interactions, specifically between the side chain amide of Asn12 and the carboxylate of Glu97 and between the guanidino group of Arg17 and the carboxylates of Asp38 and Asp94 (Figure 4B and C). The correct orientation of Arg17 is further stabilized by hydrophobic interactions between the aliphatic portion of the Arg17 side chain and the methyl groups of Val39 and Val40 (Figure 4A).
Since the N–P distances in the associative model of the transition state are close to the N–P bond distance, this structure also provides insight into the factors stabilizing the phosphorylated forms of uncomplexed IIAGlc and HPr. The interactions described above that stabilize the phosphoryl group in the transition state are consistent with previous NMR studies on the phosphorylated forms of free HPr (van Nuland et al., 1995; Jones et al., 1997) and IIAGlc (Pelton et al., 1992, 1993, 1996). Since the number of hydrogen bonding interactions to the phosphoryl group originating from HPr and IIAGlc is the same, one would predict that the equilibrium constant for the phosphoryl transfer reaction between HPr and IIAGlc would be close to 1. This is exactly what has been observed experimentally (Meadow and Roseman, 1996).
Relationship to the EIN–HPr and IIAGlc–glycerol kinase complexes
NMR chemical shift mapping has shown that HPr interacts with several apparently structurally unrelated proteins, namely EI, IIAGlc and glycogen phosphorylase, using a common binding surface (Chen et al., 1993; van Nuland et al., 1995; Wang et al., 2000). In addition, HPr interacts with a variety of IIA proteins that bear no structural resemblance to one another. Thus, while IIAGlc is a monomeric β-sandwich protein (Liao et al., 1991; Worthylake et al., 1991), IIAMtl is a monomeric α/β protein (van Montfort et al., 1998), IIAMan is a dimeric α/β protein (Nunn et al., 1996) and IIALac (Sliz et al., 1997) is a trimeric helical protein. Since His15 of HPr must be in close contact with the active site histidine of all IIA proteins, it follows that there must be extensive overlap of all the IIA binding surfaces on HPr. Likewise, IIAGlc also interacts with a variety of structurally unrelated proteins using a similar binding surface, namely its upstream partner (HPr; present study) and downstream partner (IIBGlc, chemical shift mapping; Gemmecker et al., 1997) in the PTS pathway, as well as glycerol kinase (GK) (crystallography; Hurley et al., 1993). Since structures of the EIN–HPr (Garrett et al., 1999), HPr–IIAGlc (this work) and IIAGlc–GK (Hurley et al., 1993) complexes are now available, a comparison of the three interfaces provides unique insights into the mode of protein–protein recognition, both within the PTS and in related regulatory pathways, and provides a molecular explanation for the ability of both HPr and IIAGlc to recognize and interact specifically with structurally diverse proteins. A comparison of all three interfaces is provided in Figures 5 and 6 and a summary of the structural characteristics of the interfaces is given in Table II.
There is a high species selectivity in the phosphoryl transfer reaction from EI to HPr; EI from E.coli only poorly transfers phosphorus to HPr from B.subtilis or Mycoplasma capricolum, and B.subtilis EI poorly transfers to E.coli HPr (Reizer et al., 1992; Zhu et al., 1994). In contrast, phosphoryl transfer from HPr to IIAGlc shows little species selectivity among the enzymes from E.coli, B.subtilis and M.capricolum. It is noteworthy that the three-dimensional structures of the IIAGlc proteins from these three species are quite similar [E.coli (Worthylake et al., 1991; Feese et al., 1997); B.subtilis (Liao et al., 1991); M.capricolum (Huang et al., 1998)]. Since it has now been established that other IIA proteins (IIAMan, IIALac and IIAMtl) have different three-dimensional structures, it might be predicted that E.coli HPr might show some species selectivity in phosphoryl transfer to those proteins. Appropriate kinetic studies with systems reconstituted from purified proteins have not yet been carried out.
The total ASA buried at the interface is somewhat larger for the EIN–HPr complex (1950 Å2) than for the other two complexes (1300–1350 Å2). The degree of complementarity, as measured by the gap index, decreases in the order EIN–HPr > HPr–IIAGlc > IIAGlc–GK. The binding sites on EIN and IIAGlc are located in shallow depressions, while those on HPr and GK are located on surface protrusions. The deviation from planarity of the EIN–HPr and HPr–IIAGlc interfaces is approximately the same, while that of the IIAGlc–GK interface is significantly more planar. All these structural parameters, however, fall within the expected range for heterocomplexes (Jones and Thornton, 1996). It would be tempting to postulate that the stability of the complexes could be directly correlated with these structural statistics, in which case one would predict that the equilibrium dissociation constant (Kdiss) would increase in the order EI–HPr < HPr–IIAGlc < IIAGlc–GK. However, it appears that the values of Kdiss for the three complexes are roughly comparable, lying in the 1–15 µM range (this study; Beneski et al., 1982; Novotny et al., 1985; Chauvin et al., 1996; Garrett et al., 1997b). This is not surprising since a decrease in contact surface or general complementarity, which would reflect a decrease in intermolecular van der Waals interactions, can readily be offset by a few intermolecular electrostatic interactions. Moreover, the association rate constant can be specifically modulated by long-range electrostatic effects involving charged residues located in the vicinity of the binding interface but not directly participating in intermolecular contacts (Selzer et al., 2000).
Comparison of the EIN–HPr and HPr–IIAGlc interfaces
The binding surfaces for EIN and IIAGlc on HPr are very similar, sharing 17 residues in common, out of a total of 18 that interact with IIAGlc and 23 with EIN (Figure 5A). The chief characteristic of this common convex binding surface is a central hydrophobic core surrounded by a ring of polar and positively charged residues (Figure 6B and D). The scaffolds used for the HPr binding surface on EIN and IIAGlc, however, are entirely different. The HPr binding surface on EIN is made up entirely of α-helices while that on IIAGlc is predominantly β-sheet with a short α-helix and two 310 helices located peripherally (Figure 6B and D). When surface representations of these two binding surfaces are compared, they appear to be very similar both in shape and residue type distribution (cf. Figure 6A and C). Thus, both surfaces are concave, approximately circular in shape, with a central, predominantly hydrophobic core surrounded by a ring of polar and negatively charged residues that are complementary to the binding surface on HPr. The majority of side chain contacts in the two complexes are similar in nature. For example, considering the hydrogen bonding and ion pair interactions, we find that Arg17 interacts with Glu67/Glu68 of EIN and with Asp38/Asp94 of IIAGlc, Lys27 with Asp82 of EIN and Asp144 of IIAGlc, Lys49 with Glu84 of EIN and Glu80/Glu86 of IIAGlc, and Gln51 with the Nε of Arg126 of EIN and the hydroxyl of Ser78 of IIAGlc (Figures 5 and 6A–D). Presumably, all these interactions play a role in assuring the correct orientation of the proteins within the two complexes. Even the single positively charged residues present on the HPr binding surfaces of IIAGlc (Lys69) and EIN (Arg126) are located in approximately the same relative position, just off-center of the binding surface, buried at the interface and hydrogen bonded to a backbone carbonyl (Leu53 and Leu14 of HPr for the IIAGlc and EIN complexes, respectively).
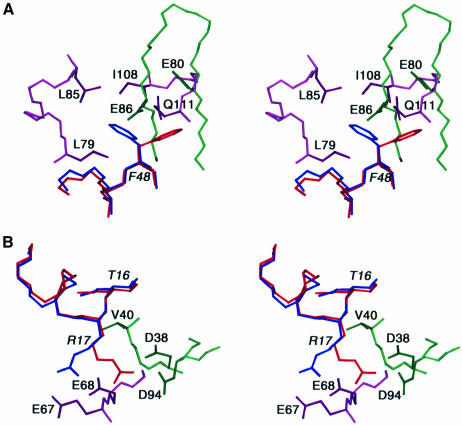
To achieve optimal intermolecular contacts some degree of side chain conformational plasticity is required. This is illustrated in Figure 7. For example, Phe48 of HPr is involved in key hydrophobic contacts. In the EIN–HPr complex the χ1 torsion angle of Phe48 is in the g– conformation, thereby permitting Phe48 to interact with Leu79, Leu85, Ile108 and Gln111 of EIN (Figure 7A). In the HPr–IIAGlc complex, on the other hand, χ1 of Phe48 is in the t conformation, thereby permitting the aromatic ring of Phe48 to interact with the backbone of strands β6 and β7 of the underlying β-sheet of IIAGlc (Figure 7A). Note that the χ1 rotamer of Phe48 in the two complexes is established unambiguously not only from analysis of 3JNCγ and 3JC′Cγ couplings but also from extensive intermolecular NOEs. Thus, the NOEs from Phe48 of HPr to Leu79, Leu85, Ile108, Gln111, Leu115 and Leu118 of EIN can only be accounted for by the g– χ1 rotamer, while those to Gly68, Ile79, Glu80, Glu86 and Leu87 of IIAGlc can only be accounted for by the t χ1 rotamer. Although the χ1 angle of Phe48 in the crystal structure of free HPr is fixed in the g– rotamer as a result of a crystal contact (Jia et al., 1993), solution NMR measurements on free HPr indicate that Phe48 undergoes rotamer averaging (van Nuland et al., 1995). Hence, Phe48 is primed to adopt different conformations for different partners. Another example is provided by Arg17 of HPr in Figure 7B. The χ1, χ2, χ3 and χ4 angles of Arg17 are in the t, g+, t and g– rotamers, respectively, in the EIN–HPr complex, but in the t, t, g– and t rotamers, respectively, in the HPr–IIAGlc complex. By this means, the guanidino group of Arg17 can form ion pairs with the carboxylates of Glu67 and Glu68 of EIN (Garrett et al., 1999) and with the carboxylates of Asp38 and Asp94 of IIAGlc. The importance of these ion pair interactions involving Arg17 is attested by the observation that mutation of Arg17 to a variety of residues has a large impact on phosphoryl transfer from EI to HPr and from HPr to IIAGlc (Sharma et al., 1991; Anderson et al., 1993; Kruse et al., 1993).
Fig. 7. Conformational side chain plasticity. Stereoviews of superpositions of selected regions of the interfaces of the HPr–IIAGlc and EIN–HPr complexes illustrating alternative conformations of (A) Phe48 and (B) Arg17 of HPr in the two complexes. HPr in the HPr–IIAGlc and EIN–HPr complexes is shown in red and blue, respectively; EIN is shown in purple and IIAGlc in green.
Some residues on HPr are not shared by the EIN and IIAGlc binding surfaces. In particular, Asn12 is involved in interactions with Phe71, Val96 and Glu97 of IIAGlc but has no counterpart in the EIN–HPr complex. Likewise, the interaction of Lys45 and Ser46 of HPr with Glu84 of EIN has no counterpart in the HPr–IIAGlc complex. These structural findings are completely consistent with the very different effects that mutation of Ser46 to Asp (S46D) has on the EI–HPr and HPr–IIAGlc systems. In particular, the S46D mutation results in an ∼2000-fold reduction in phosphoryl transfer activity (kcat/Km) between EI and HPr, mainly attributable to an ∼650-fold increase in the value of Km (Napper et al., 1996). In contrast, the same mutation has a minimal effect on the phosphoryl transfer activity between HPr and IIAGlc, reducing kcat/Km and increasing Km by only a factor of ∼4 (Reizer et al., 1992). Thus, the introduction of a negative charge at position 46 of HPr, either by mutation or by phosphorylation in the case of Gram-positive bacteria, will result in a significant reduction in the binding affinity of HPr for EI, as a result of unfavorable electrostatic interactions with Glu84, but will have only a small effect on formation of the HPr–IIAGlc complex. The latter can presumably be attributed to long-range electrostatic effects modulating the association rate constant (Selzer et al., 2000).
Comparison of the HPr–IIAGlc and IIAGlc–GK interfaces
Just as in the case of HPr binding, the interaction of GK with IIAGlc does not induce any change in the backbone conformation of IIAGlc. Thus, the backbone r.m.s. difference between the crystal structures of IIAGlc in the free state (Feese et al., 1997) and complexed to GK (Hurley et al., 1993) is only 0.4 Å, which is within the errors of the coordinates. There is considerable overlap of the HPr and GK binding surfaces on IIAGlc, with 16 residues in common out of 23 that interact with HPr and 17 with GK (Figures 5B and 6C and E). Once again, the scaffolds comprising the IIAGlc binding surfaces on HPr and GK are structurally distinct (Figure 6C and E). The binding surface on HPr involves two helices, while that on GK consists of one short helix and portions of three loops. The orientation of the GK helix on IIAGlc does not coincide with that of either of the two helices of HPr. However, just as in the case of HPr, the IIAGlc binding surface presented by GK is convex in shape. The location of the hydrophobic residues on the IIAGlc binding surfaces of HPr and GK approximately coincide, as do a significant portion of the polar and positively charged residues (Figure 6D and F) (HPr and GK residues in italic). A number of hydrogen bonding/ion pair interactions are also preserved. Thus, Tyr481 of GK fulfills an analogous role to Asn12 of HPr, making contacts with Val96 and Glu97 (as well as additional contacts with Lys99). Arg479 of GK occupies a similar position to Arg17 of HPr and forms an ion pair with Asp38. The side chains of Pro472, Ile474 and Thr476/477 of GK approximately coincide with those of Ala20, Phe48 and Thr16 of HPr and participate in a similar set of hydrophobic contacts with IIAGlc.
One key difference between the IIAGlc binding sites on HPr and GK is the presence of a single negatively charged residue, Glu478, on the surface of GK. Glu478 is located in close proximity to His90 of IIAGlc and explains why GK is only inhibited by unphosphorylated IIAGlc, since the presence of a negatively charged phosphoryl group bonded to His90 would result in unfavorable charge repulsion (Hurley et al., 1993).
Concluding remarks
The structure of the HPr–IIAGlc complex delineates the specific hydrophobic and electrostatic interactions at the interfaces and permits a critical evaluation of the various interactions. The key feature of the complex is the complementarity of the shape and amino acid composition of the binding surfaces. The binding surface on IIAGlc is concave and located in a shallow depression, while that on HPr is convex and located on a protrusion. The central region of each binding surface is hydrophobic and is surrounded by polar and charged residues. The latter are entirely positive in the case of HPr and, with only one exception, are negative in the case of IIAGlc. Important features of both HPr and IIAGlc are their ability to recognize structurally diverse proteins. Indeed, one might expect this to be generally the case for many proteins involved in signal transduction pathways where complex formation between the relevant protein partners is likely to be transient in nature. Comparison of the EIN–HPr, HPr–IIAGlc and IIAGlc–GK complexes demonstrates that similar binding surfaces can be obtained with structurally very different underlying scaffolds. For example, the HPr binding surfaces on EIN (Figure 6A) and IIAGlc (Figure 6C) are remarkably similar, although the underlying structural elements at the backbone level bear no similarity to one another. At the design level it is apparent that the ability to recognize multiple targets using the same binding surface is associated with a reasonably high degree of both redundancy and side chain conformational plasticity. Although the correct orientation of the proteins in the complexes is dictated both by surface complementarity and the presence of specific electrostatic interactions, one finds that only a subset are involved in specific ion pair and hydrogen bonding interactions. While many of the electrostatic and hydrophobic interactions are preserved between the various complexes, others are present in one but not the other. For example, Lys24 and Lys45 of HPr form ion pairs with Glu74 and Glu84 of EIN (Figure 6A and B) but have no counterparts in the HPr–IIAGlc complex (Figure 6C and D). Likewise, Glu43 of IIAGlc forms an ion pair with Arg402 of GK (Figure 6E and F) but has no counterpart in the HPr–IIAGlc complex (Figure 6C and D), while the ion pairs formed between Glu80 and Glu86 of IIAGlc and Lys49 of HPr (Figure 6C and D) have no counterpart in the IIAGlc–GK complex (Figure 6E and F). Side chain plasticity permits optimal interactions to be formed and relates not only to long linear side chains, such as those of Arg and Lys, but to others as well. For example, Phe48 of HPr, which participates in multiple intermolecular hydrophobic interactions, has distinct χ1 rotamer conformations in the EIN–HPr and HPr–IIAGlc complexes, namely g– and t, respectively. These features obviate the need for absolute preservation of the various binding surfaces and are likely to be a general characteristic of protein–protein interactions involving proteins that utilize approximately the same surface to interact with numerous disparate targets.
The completion of these studies (Garrett et al., 1999; this study) constitutes the elucidation of the structures of two of the protein–protein complexes in the glucose–PTS cascade. The determination of the structure of the final element in this pathway, that of the IIAGlc–IIBGlc complex, is currently underway.
Materials and methods
Expression and purification of proteins
Escherichia coli IIAGlc and HPr were expressed, purified and uniformly isotopically labeled with 15N (>95%) and 13C (>95%) as described previously (Reddy et al., 1991; Garrett et al., 1997b). Samples for NMR contained ∼1 mM 1:1 HPr–IIAGlc complex in 10 mM phosphate buffer, pH 7.1. The following samples were employed (only the presence of 15N and 13C isotopes are indicated; if no C or N isotope is mentioned, then the sample contained 12C or 14N at natural isotopic abundance): HPr(15N)–IIAGlc, IIAGlc(15N)–HPr, HPr(15N/13C)–IIAGlc, HPr–IIAGlc (15N/13C), HPr(15N)–IIAGlc(13C) and HPr(13C)–IIAGlc(15N).
NMR spectroscopy
All spectra were recorded at 35°C on Bruker DMX500, DMX600, DMX750 and DRX800 spectrometers equipped with x,y,z-shielded gradient triple resonance probes. Spectra were processed with the NMRPipe package (Delaglio et al., 1995) and analyzed using the programs PIPP, CAPP and STAPP (Garrett et al., 1991). 1H, 15N and 13C sequential assignments were obtained using 3D double and triple resonance through-bond correlation experiments (Clore and Gronenborn, 1991, 1998a; Bax and Grzesiek, 1993). 3J N–Cγ, C′–Cγ and Cα–Cδ couplings were measured using quantitative J correlation spectroscopy (Bax et al., 1994). Interproton distance restraints were derived from multi-dimensional NOE spectra with mixing times ranging from 75 to 120 ms. Three-dimensional experiments used for sequential assignments included HNCO, HNCA, HNCACB, CBCA(CO)NH, C(CCO)NH, HBHA(CBCACO)NH, H(CCO)NH, HCCH-COSY, CCH-COSY and HCCH-TOCSY experiments. NOE experiments included 3D 15N-separated, 13C-separated and 13C-separated/12C-filtered NOE spectra, 4D 15N/13C-separated and 13C/13C-separated NOE spectra and 2D 15N-separated/13C-filtered, 15N-filtered/13C-separated and 13C-filtered/15N-filtered NOE spectra. Residual 1DNH dipolar couplings were obtained by taking the difference in the corresponding J splittings measured in oriented (in an ∼60 mg/ml colloidal suspension of tobacco mosaic virus; Clore et al., 1998a) and isotropic (in water) HPr(15N)–IIAGlc and HPr–IIAGlc(15N) complexes using 2D IPAP {15N,1H}-HSQC experiments (Ottiger et al., 1998). In the latter samples the unlabeled protein was in 4-fold excess to ensure that the 15N-labeled protein (at a concentration of ∼0.3 mM) was entirely in the bound state. The magnitudes of the axial (–14.9 Hz) and rhombic (0.2) components of the alignment tensor were determined from the powder pattern distribution of the 1DNH dipolar couplings (Clore et al., 1998b), which range from –31 to +20 Hz. Attempts to measure dipolar couplings in other liquid crystalline media such as bacteriophages fd (Clore et al., 1998a) and pf1 (Hansen et al., 1998) or lipid bicelles (Ottiger and Bax, 1999) failed in this particular case owing to interactions between the complex and these liquid crystalline media. Long-range 1H-15N correlation spectra to correlate the Nδ1 and Nε2 15N shifts with the Hδ2 and Hε1 1H shifts of the imidazole ring (Pelton et al., 1993) were used to confirm the tautomeric states of the histidine residues in the complex.
Structure calculations
Intermolecular NOE-derived interproton distance restraints were classified into two ranges: 1.8–5.0 and 1.8–6.0 Å. An additional 0.5 Å was added to the upper bound for NOEs involving methyl groups and distances involving non-stereospecifically assigned protons were represented by a (Σr–6)–1/6 sum (Nilges, 1993). χ1 and χ2 torsion angle restraints were derived from analysis of heteronuclear 3J couplings and NOE/ROE experiments (Clore and Gronenborn, 1998a). Structures were calculated by a combination of rigid body minimization and constrained/restrained simulated annealing (Clore, 2000) using the NIH version (code written at NIH is available by anonymous ftp at portal.niddk.nih.gov in the directory /pub/clore/xplor_nih) of the program XPLOR (Brünger, 1993). The starting coordinates comprise the X-ray structures (with protons added) of E.coli HPr (RCSB accession code 1POH, 1.5 Å resolution) and IIAGlc (RCSB accession code 2F3G, molecule 2, 2.13 Å resolution). At all stages the relative coordinates of the backbone and non-interfacial side chains are maintained to those of the X-ray coordinates. Rigid body minimization, using exactly the protocol described (Clore, 2000), was used to dock the two proteins, followed by constrained/restrained simulated annealing with slow cooling from 3000 to 25 K to refine the interfacial side chain positions and fine tune the relative orientation of the two proteins. The simulated annealing schedule (Nilges et al., 1988) follows that described previously (Omichinski et al., 1997) with some minor modifications (see Results).
While there is no solution NMR structure for E.coli IIAGlc, there are NMR structures for E.coli HPr in both the free state (van Nuland et al., 1994) and complexed to EIN (Garrett et al., 1999). The rationale for choosing the X-ray over the NMR structure of E.coli HPr as a basis for docking is that one invariably finds that high resolution X-ray structures agree better with NMR observables, such as scalar coupling constants, chemical shifts and dipolar couplings, than the corresponding solution NMR structures (Clore and Gronenborn, 1998b). This is indeed the case here. Thus, Rdip (for the 1DNH dipolar couplings measured on the HPr–IIAGlc complex) is 38% for the NMR structure of free HPr (van Nuland et al., 1994) and 26% for the NMR structure of HPr in the EIN–HPr complex (Garrett et al., 1999), compared with 16.7% for the X-ray structure of free HPr (Jia et al., 1993). This is not a reflection of structural differences between solution and crystal structures or between the structures in the free and bound states, but is simply due to the lower degree of accuracy of the NMR coordinates relative to the X-ray ones. Indeed, the value of Rdip for the NMR structures is directly correlated with the backbone r.m.s. difference to the X-ray structure (1.1 Å for free HPr versus 0.6 Å for HPr in the EIN–HPr complex).
There are three sets of coordinates for E.coli IIAGlc in the RCSB protein data bank, representing either different crystal forms (2F3G versus 1F3Z) or different molecules in the unit cell (the two molecules in 2F3G) (Feese et al., 1997). The second molecule of 2F3G has the lowest value of the dipolar coupling R factor, Rdip (14.7%), and hence was chosen as the docking molecule in this study. The values of Rdip for the first molecule of 2F3G and for 1F3Z are 19.7 and 19.8%, respectively. These higher values are due to the presence of a few outliers: removal of the dipolar couplings for residues 134 and 145 for the first molecule of 2F3G reduces Rdip to 15.9%; similarly, removal of the dipolar couplings for residues 70, 134 and 136 of 1F3Z reduces Rdip to 16.4%. The pairwise backbone r.m.s. difference between these three sets of coordinates ranges from 0.29 to 0.33 Å and the precision of the backbone coordinates (given by the average backbone r.m.s. of the three individual structures to the mean coordinate positions) is 0.18 Å.
The coordinates of the mean NMR structure were obtained by averaging the coordinates of the 30 individual simulated annealing structures best fitted to each other (residues 1–85 of HPr and residues 19–168 of IIAGlc). The average coordinates were then used to create a template for constrained/restrained regularization to generate the so-called restrained minimized mean structure. The template is obtained by best fitting the X-ray coordinates of HPr and IIAGlc to the average coordinates and then substituting the backbone and non-interfacial side chains of the best fitted X-ray coordinates for those of the average structure. The resulting template is subjected to constrained/restrained regularization in which only the coordinates of the interfacial side chains (i.e. those obtained by averaging the coordinates of the individual simulated annealing structures) are allowed to move, all the remaining coordinates being held completely fixed. The target function includes the experimental NOE and torsion angle restraints, but not the dipolar coupling restraints, since the backbone positions are held fixed.
Structures were visualized and analyzed with the program VMD-XPLOR (C.Schwieters and G.M.Clore, unpublished). Figures were generated using VMD-XPLOR (C.Schwieters and G.M.Clore, unpublished), MOLMOL (Koradi et al., 1996), RIBBONS (Carson, 1991) and GRASP (Nicholls et al., 1991).
The coordinates have been deposited in the RCSB Protein Data Bank (RCSB accession code 1GGR).
Acknowledgments
Acknowledgements
We thank Al Mildvan and Greg Petsko for extensive discussions concerning the nature of the transition state, Carole Bewley, Dan Garrett, Jim Hurley and Attila Szabo for useful discussions, Osnat Herzberg for access to the unpublished coordinates of the modeled HPr–IIAGlc complex from Bacillus subtilis, Dan Garrett, Frank Delaglio, John Kuszewski and Charles Schwieters for software support, and Don Caspar and Ad Bax for the gift of tobacco mosaic virus and lipid bicelles, respectively. This work was supported in part by the AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C.)
References
- Ab E., Schuurmann-Wolters,G., Reizer,J., Saier,M.H.,Jr, Dijkstra,K., Scheek,R.M. and Robillard,G.T. (1997) The NMR side-chain assignments and solution structure of enyzme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of E.coli. Protein Sci., 6, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.W., Pullen,K., Georges,F., Klevitt,R.E. and Waygood,E.B. (1993) The involvement of the arginine 17 residue in the active site of the histidine-containing protein, HPr, of the phosphoenol pyruvate:sugar phosphotransferase system of Escherichia coli. J. Biol. Chem., 268, 12325–12333. [PubMed] [Google Scholar]
- Bax A. and Grzesiek,S. (1993) Methodological advances in protein NMR. Acc. Chem. Res., 26, 131–138. [Google Scholar]
- Bax A., Vuister,G.W., Grzesiek,S., Delaglio,F., Wang,A.C., Tschudin,R. and Zhu,G. (1994) Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol., 239, 79–106. [DOI] [PubMed] [Google Scholar]
- Begley G.S., Hanse,D.E., Jacobson,G.R. and Knowles,J.R. (1982) Stereochemical course of the reactions catalysed by the bacterial phosphoenolpyruvate:glucose phosphotransferase system. Biochemistry, 21, 5552–5556. [DOI] [PubMed] [Google Scholar]
- Beneski D.A., Nakazawa,A., Weigel,N., Hartman,P.E. and Roseman,S. (1982) Sugar transport by the bacterial phosphotransferase system: isolation and characterization of a phosphocarrier protein HPr from wild type and mutants of Salmonella typhimurium. J. Biol. Chem., 257, 14492–14498. [PubMed] [Google Scholar]
- Bewley C.A. and Clore,G.M. (2000) Determination of the relative orientation of the two halves of the domain-swapped dimer of cyanovirin-N in solution using dipolar couplings and rigid body minimization. J. Am. Chem. Soc., 122, 6009–6016. [Google Scholar]
- Brünger A.T. (1993) XPLOR: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Carson M. (1991) Ribbons 4.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Chauvin F., Fomenko,A., Johnson,C.R. and Roseman,S. (1996) The N-terminal domain of Escherichia coli enzyme I of the phosphoenolpyruvate/glucose phosphotransferase system: molecular cloning and characterization. Proc. Natl Acad. Sci. USA, 93, 7028–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Reizer,J., Saier,M.H.,Jr, Fairbrother,W.J. and Wright,P.E. (1993) Mapping of the binding interfaces of the proteins of the bacterial phosphotransferase system, HPr and IIAGlc. Biochemistry, 32, 32–37. [DOI] [PubMed] [Google Scholar]
- Clore G.M. (2000) Accurate and rapid docking of protein–protein complexes on the basis of intermolecular nuclear Overhauser enhancement data and dipolar couplings by rigid body minimization. Proc. Natl Acad. Sci. USA, 97, 9021–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M. and Garrett,D.S. (1999) R-factor, free R and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc., 121, 9008–9012. [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1991) Structures of larger proteins in solution: three- and four-dimensional heteronuclear NMR spectroscopy. Science, 252, 1390–1399. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1998a) Determining structures of larger proteins and protein complexes by NMR. Trends Biotechnol., 16, 22–34. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1998b) New methods of structure refinement for macromolecular structure determination by NMR. Proc. Natl Acad. Sci. USA, 95, 5891–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M., Starich,M.R. and Gronenborn,A.M. (1998a) Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc., 120, 10571–10572. [Google Scholar]
- Clore G.M., Gronenborn,A.M. and Bax,A. (1998b) A robust method for determining the magnitude of the fully asymmteric alignment tensor of oriented macromolecules in the absence of structural information. J. Magn. Reson., 133, 216–222. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Gronenborn,A.M. and Tjandra,N. (1998c) Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson., 131, 159–162. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Eberstadt M., Grdadolnik,S.G., Gemmecker,G., Kessler,H., Buhr,A. and Erni,B. (1996) Solution structure of the IIB domain of the glucose transporter of Escherichia coli. Biochemistry, 35, 11286–11292. [DOI] [PubMed] [Google Scholar]
- Fairbrother W.J., Gippert,G.P., Reizer,J., Saier,M.H.,Jr and Wright,P.E. (1992) Low resolution structure of Bacillus subtilis glucose permease IIA derived from heteronuclear three-dimensional NMR spectroscopy. FEBS Lett., 296, 673–677. [DOI] [PubMed] [Google Scholar]
- Feese M.D., Comolli,L., Meadow,N.D., Roseman,S. and Remington,S.J. (1997) Structural studies of the Escherichia coli signal transducing protein IIAGlc: implications for target recognition. Biochemistry, 36, 16087–16096. [DOI] [PubMed] [Google Scholar]
- Garrett D.S., Powers,R., Gronenborn,A.M. and Clore,G.M. (1991) A common sense approach to peak picking in two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson., 95, 214–220. [DOI] [PubMed] [Google Scholar]
- Garrett D.S., Seok,Y.-J., Liao,D.-I., Peterkofsky,A., Gronenborn,A.M. and Clore,G.M. (1997a) Solution structure of the 30 kDa N-terminal domain of enzyme I of the Escherichia coli phosphoenol pyruvate:sugar phosphotransferase system by multidimensional NMR. Biochemistry, 36, 2517–2530. [DOI] [PubMed] [Google Scholar]
- Garrett D.S., Seok,Y.-J., Peterkofsky,A., Clore,G.M. and Gronenborn,A.M. (1997b) Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry, 36, 4393–4398. [DOI] [PubMed] [Google Scholar]
- Garrett D.S., Seok,Y.-J., Peterkofsky,A., Gronenborn,A.M. and Clore,G.M. (1999) Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nature Struct. Biol., 6, 166–173. [DOI] [PubMed] [Google Scholar]
- Gemmecker G., Eberstadt,M., Buhr,A., Lanz,R., Grdadolnik,S.G., Kessler,H. and Erni,B. (1997) Glucose transporter of Escherichia coli: NMR characterization of the phosphocysteine form of the IIBGlc domain and its binding interface with the IIAGlc subunit. Biochemistry, 36, 7408–7417. [DOI] [PubMed] [Google Scholar]
- Hansen M.R., Rance,M. and Pardi,A. (1998) Observation of long 1H-1H distances in solution by dipolar coupling interactions. J. Am. Chem. Soc., 120, 11210–11211. [Google Scholar]
- Herzberg O. (1992) An atomic model for protein–protein phosphoryl group transfer. J. Biol. Chem., 267, 24819–24823. [PubMed] [Google Scholar]
- Herzberg O. and Klevit,R. (1994) Unravelling a bacterial hexose transport pathway. Curr. Opin. Struct. Biol., 4, 814–822. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Reddy,P., Sutrina,S., Saier,M.H.,Jr, Reizer,J. and Kapadia,G. (1992) Structure of the histidine-containing phospho carrier protein HPr from Bacillus subtilis at 2.0 Å resolution. Proc. Natl Acad. Sci. USA, 89, 2499–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollfelder F. and Herschlag,D. (1995) The nature of the transition state for enyzme-catalyzed phosphoryl transfer: hydrolysis of O-aryl phosphorothioates by alkaline phosphatase. Biochemistry, 34, 12255–12264. [DOI] [PubMed] [Google Scholar]
- Huang K., Kapadia,G., Zhu,P.-P., Peterkofsky,A. and Herzberg,O. (1998) A promiscuous binding surface: crystal structure of the IIA domain of the glucose-specific permease from Mycoplasma capricolum. Structure, 6, 697–710. [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Faber,H.R., Worthylake,D., Meadow,N.D., Roseman,S., Pettigrew,D.W. and Remington,S.J. (1993) Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science, 259, 673–677. [PubMed] [Google Scholar]
- Jablonski E.G., Brand,L. and Roseman,S. (1983) Sugar transport by the bacterial phosphotransferase system: preparation of a fluorescein derivative of the glucose-specific phophocarrier protein IIIGlc and its binding to the phosphocarrier protein HPr. J. Biol. Chem., 258, 9690–9699. [PubMed] [Google Scholar]
- Jia Z., Quail,J.W., Waygood,E.B. and Delbaere,L.T. (1993) The 2.0 Å resolution structure of the Escherichia coli histidine-containing phosphocarrier protein HPr: a redetermination. J. Biol. Chem., 268, 22940–22501. [DOI] [PubMed] [Google Scholar]
- Jones B.E., Rajgopal,P. and Klevit,R.E. (1997) Phosphorylation on histidine is accompanied by localized structural changes in phosphocarrier protein HPr from Bacillus subtilis. Protein Sci., 6, 2107–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. and Thornton,J.M. (1996) Principles of protein–protein interactions. Proc. Natl Acad. Sci. USA, 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbitzer H.R. and Hengstenberg,W. (1993) The solution structure of the histidine-containing protein (HPr) from Staphylococcus aureus as determined by two-dimensional 1H-NMR spectroscopy. Eur. J. Biochem., 216, 205–214. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich, K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kruse R., Hengstenberg,W., Beneicke,W. and Kalbitzer,H.R. (1993) Involvement of various amino- and carboxy-terminal residues in the active site of the histidine-containing protein HPr of the phosphoenolpyruvate-dependent phosphotransferase system of Staphylococcus carnosus: site directed mutagenesis with the ptsH gene, biochemical characterization and NMR studies of mutant proteins. Protein Eng., 6, 417–423. [DOI] [PubMed] [Google Scholar]
- Kundig W., Ghosh,S. and Roseman,S. (1964) Phosphate bound to histidine in a protein as an intermediate in a novel phospho transferase system. Proc. Natl Acad. Sci. USA, 52, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszewski J. and Clore,G.M. (2000) Source of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J. Magn. Reson., 146, 249–254. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn,A.M. and Clore,G.M. (1999) Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc., 121, 2337–2338. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Liao D.-I., Kapadia,G., Reddy,P., Saier,M.H.,Jr, Reizer,J. and Herzberg,O. (1991) Structure of the IIA domain of the glucose permease of Bacillus subtilis at 2.2 Å resolution. Biochemistry, 30, 9583–9594. [DOI] [PubMed] [Google Scholar]
- Liao D.-I., Silverton,E., Seok,Y.-J., Lee,B.R., Peterkofsky,A. and Davies,D.R. (1996) The first step in sugar transport: crystal structure of the amino terminal domain of enzyme I of the E.coli PEP:sugar phosphotransferase system and a model of the phosphotransfer complex with HPr. Structure, 4, 861–872. [DOI] [PubMed] [Google Scholar]
- MacGillavry C.H. and Rieck,G.D. (1962) International Tables for Crystallography, Vol. III. Kynoch Press, Birmingham, UK, p. 270. [Google Scholar]
- Meadow N.D. and Roseman,S. (1982) Sugar transport by the bacterial phosphotransferase system: isolation and characterization of a glucose specific phosphocarrier protein (IIIGlc) from Salmonella typhimurium. J. Biol. Chem., 257, 14526–14537. [PubMed] [Google Scholar]
- Meadow N.D. and Roseman,S. (1996) Rate and equilibrium constants for phosphoryl transfer between active site histidines of Escherichia coli HPr and the signal transducing protein IIIGlc. J. Biol. Chem., 271, 33440–33445. [DOI] [PubMed] [Google Scholar]
- Napper S., Anderson,J.W., Georges,F., Quail,J.W., Delbaere,L.T.J. and Waygood,E.B. (1996) Mutation of serine-46 to aspartate in the histidine-containing protein of Escherichia coli mimics the inactivation by phosphorylation of serine-46 in HPrs from gram-positive bacteria. Biochemistry, 35, 11260–11267. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1993) A calculational strategy for the structure determination of symmetric dimers by 1H NMR. Proteins, 17, 297–309. [DOI] [PubMed] [Google Scholar]
- Nilges M., Gronenborn,A.M., Brünger,A.T. and Clore,G.M. (1988) Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints: application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng., 2, 27–38. [DOI] [PubMed] [Google Scholar]
- Novotny M.J., Frederickson,W.L., Waygood,E.B. and Saier,M.H.,Jr (1985) Allosteric regulation of glycerol kinase by enzyme IIGlc of the phosphotransferase system in Escherichia coli and Salmonella typhimurium. J. Bacteriol., 162, 810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn R.S., Housley-Markovic,Z., Genovesio-Taverne,G., Flükiger,K., Rizkallah,P.J., Jansonius,J.N., Schirmer,T. and Erni,B. (1996) Structure of the IIA domain of the mannose transporter of Escherichia coli at 1.7 Å resolution. J. Mol. Biol., 259, 502–511. [DOI] [PubMed] [Google Scholar]
- Omichinski J.G., Pedone,P.V., Felsenfeld,G., Gronenborn,A.M. and Clore,G.M. (1997) The solution structure of a specific GAGA factor–DNA complex reveals a modular binding mode. Nature Struct. Biol., 4, 122–132. [DOI] [PubMed] [Google Scholar]
- Ottiger G. and Bax,A. (1999) Bicelle-based liquid crystals for NMR measurement of dipolar couplings at acidic and basic pH values. J. Biomol. NMR, 13, 187–191. [DOI] [PubMed] [Google Scholar]
- Ottiger M., Delaglio,F. and Bax,A. (1998) Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson., 131, 373–378. [DOI] [PubMed] [Google Scholar]
- Pelton J.G., Torchia,D.A., Meadow,N.D., Wong,C.Y. and Roseman,S. (1991) Secondary structure of the phosphocarrier protein IIIGlc, a signal-transducing protein from Escherichia coli determined by heteronuclear three-dimensional NMR spectroscopy. Proc. Natl Acad. Sci. USA, 88, 3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton J.G., Torchia,D.A., Meadow,N.D. and Roseman,S. (1992) Structural comparison of phosphorylated and unphosphorylated forms of IIIGlc, a signal-transducing protein from Escherichia coli using three-dimensional NMR technqiues. Biochemistry, 31, 5215–5224. [DOI] [PubMed] [Google Scholar]
- Pelton J.G., Torchia,D.A., Meadow,N.D. and Roseman,S. (1993) Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal transducing protein from Escherichia coli using two-dimensional heteronuclear NMR techniques. Protein Sci., 2, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton J.G., Torchia,D.A., Remington,S.J., Murphy,K.P., Meadow,N.D. and Roseman,S. (1996) Structures of the active site histidine mutants of IIIGlc, a major signal-transducing protein in Escherichia coli. J. Biol. Chem., 271, 33446–33456. [DOI] [PubMed] [Google Scholar]
- Peterkofsky A., Reizer,A., Reizer,J., Gollop,N., Zhu,P.-P. and Amin,N. (1993) Bacterial adenyl cyclases. Prog. Nucleic Acid Res. Mol. Biol., 44, 31–65. [DOI] [PubMed] [Google Scholar]
- Postma P.W., Lengeler,J.W. and Jacobson,G.R. (1996) Phosphoenol pyruvate:carbohydrate phosphotransfer systems. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1149–1174. [Google Scholar]
- Reddy P., Fredd-Kuldell,N., Liberman,E. and Peterkofsky,A. (1991) Overproduction and rapid purification of the phosphoenol pyruvate:sugar phosphotransferase system proteins enzyme I, HPr and protein IIIGlc of Escherichia coli. Protein Expr. Purif., 2, 179–187. [DOI] [PubMed] [Google Scholar]
- Reizer J., Sutrina,S.L., Wu,L.-F., Deutscher,J., Reddy,P. and Saier,M.H.,Jr (1992) Functional interactions between proteins of the phosphoenolpyruvate:sugar phosphotransferase systems of Bacillus subtilis and Escherichia coli. J. Biol. Chem., 267, 9158–9169. [PubMed] [Google Scholar]
- Schauder S., Nunn,R.S., Lanz,R., Erni,B. and Schirmer,T. (1998) Crystal structure of the IIB subunit of a fructose permease (IIBLev) from Bacillus subtilis. J. Mol. Biol., 276, 591–602. [DOI] [PubMed] [Google Scholar]
- Selzer T., Albeck,S. and Schreiber,G. (2000) Rational design of faster associating and tighter binding complexes. Nature Struct. Biol., 7, 537–541. [DOI] [PubMed] [Google Scholar]
- Seok Y.-J., Sondej,M., Badawi,P., Lewis,M.S., Briggs,M.C., Jaffe,H. and Peterkofsky,A. (1997) High affinity binding and allosteric regulation of Escherichia coli glycogen phosphorylase by the histidine phosphocarrier protein HPr. J. Biol. Chem., 272, 26511–26522. [DOI] [PubMed] [Google Scholar]
- Sharma S., Georges,F., Delbaerre,L.T.J., Lee,J.S., Klevitt,R.E. and Waygood,E.B. (1991) Epitope mapping by mutagenesis distinguishes between the two tertiary structures of the histidine-containing protein HPr. Proc. Natl Acad. Sci. USA, 88, 4877–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliz P., Engelmann,R., Hengstenberg,W. and Pai,E.F. (1997) The structure of enzyme IIAlactose from Lactobacillus casei reveals a new fold and points to possible interactions of a multicomponent system. Structure, 5, 775–788. [DOI] [PubMed] [Google Scholar]
- van Dijk A.A., de Lange,L.C.M., Bachovchin,W.W. and Robillard,G.T. (1990) Effect of phosphorylation on hydrogen-bonding interactions of the active site histidine of HPr determined by 15N NMR spectroscopy. Biochemistry, 29, 8164–8171. [DOI] [PubMed] [Google Scholar]
- van Montfort R.L., Pijning,T., Kalk,K.H., Reizer,J., Saier,M.H.,Jr, Thunissen,M.M., Robillard,G.T. and Dijkstra,B.W. (1997) The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein tyrosine phosphatases. Structure, 5, 217–225. [DOI] [PubMed] [Google Scholar]
- van Montfort R.L., Pijning,T., Kalk,K.H., Hangyi,I., Kouwijzer,M.L.C.E., Robillard,G.T. and Dijkstra,B.W. (1998) The structure of the Escherichia coli phosphotransferase IIAmannitol reveals a novel fold with two conformations of the active site. Structure, 6, 377–388. [DOI] [PubMed] [Google Scholar]
- van Nuland N.A.J., Hangyi,I.W., van Schaik,R.C., Berendsen,H.J.C., van Gunsteren,W.F., Scheek,R.M. and Robillard,G.T. (1994) The high-resolution structure of the histidine-containing phosphocarrier protein HPr from Escherichia coli determined by restrained molecular dynamics from nuclear magnetic resonance nuclear Overhauser effect data. J. Mol. Biol., 237, 544–559. [DOI] [PubMed] [Google Scholar]
- van Nuland N.A.J., Boelens,R., Scheek,R.M. and Robillard,G.T. (1995) High-resolution structure of the phosphorylated form of the histidine-containing phosphocarrier protein HPr from Escherichia coli determined by restrained molecular dynamics from NMR-NOE data. J. Mol. Biol., 246, 180–193. [DOI] [PubMed] [Google Scholar]
- Wang G., Sondej,M., Garrett,D.S., Peterkofsky,A. and Clore,G.M. (2000) A common interface on histidine-containing phosphocarrier protein for interaction with its partner proteins. J. Biol. Chem., 275, 16401–16403. [DOI] [PubMed] [Google Scholar]
- Weigel N., Kukuruzinska,M.A., Nakazawa,A., Waygood,E.B. and Roseman,S. (1982) Sugar transport by the bacterial phospho transferase system: phosphoryl transfer reactions catalyzed by enzyme I of Salmonella typhimurium. J. Biol. Chem., 257, 14499–14509. [PubMed] [Google Scholar]
- Wittekind M., Rajgopal,P., Cranchini,B.R., Reizer,J., Saier,M.H.,Jr and Klevit,R.E. (1992) Solution structure of the phosphocarrier protein HPr from Bacillus subtilis by two-dimensional NMR spectroscopy. Protein Sci., 1, 1363–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake D., Meadow,N.D., Roseman,S., Liao,D.-I., Herzberg,O. and Remington,S.J. (1991) Three-dimensional structure of the Escherichia coli phosphocarrier protein IIIGlc. Proc. Natl Acad. Sci. USA, 88, 10382–10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P.P., Reizer,J. and Peterkofsky,A. (1994) Unique dicistronic operon (ptsI-crr) in Mycoplasma capricolum encoding enzyme I and the glucose-specific enzyme IIA of the phosphoenolpyruvate:sugar phosphotransferase system: cloning, sequencing, promoter analysis and protein characterization. Protein Sci., 3, 2115–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]