Abstract
Human TAP is implicated in mRNA nuclear export and is used by simian type D retroviruses to export their unspliced genomic RNA to the cytoplasm of the host cell. We have determined the crystal structure of the minimal TAP fragment that binds the constitutive transport element (CTE) of retroviral RNAs. Unexpectedly, we find the fragment consists of a ribonucleoprotein (RNP) domain, which is not identifiable by its sequence, and a leucine-rich repeat (LRR) domain. The non-canonical RNP domain functions as the general RNA-binding portion of the fragment. The LRR domain is required in cis to the RNP domain for CTE RNA binding. The structural and biochemical properties of the domains point to a remarkable similarity with the U2B′′(RNP)–U2A′(LRR) spliceosomal heterodimer. Our in vitro and in vivo functional studies using structure-based mutants suggest that a phylogenetically conserved surface of the LRR domain of TAP may have different roles in the export of viral and cellular RNAs.
Keywords: crystal structure/CTE/nucleocytoplasmic transport/RNA export/RNP
Introduction
Eukaryotic RNAs are transported from their site of transcription in the nucleus to their site of function in the cytoplasm via interaction with factors that recognize and translocate individual RNA cargoes (for recent reviews see Mattaj and Englmeier, 1998; Nakielny and Dreyfuss, 1999; Görlich and Kutay, 1999). Specific factors mediate the nuclear export of different classes of RNA. As originally shown by microinjection experiments in Xenopus oocytes, uracil-rich small nuclear RNA (U snRNA), transfer RNA (tRNA), messenger RNA (mRNA) and ribosomal RNA (rRNA) do not compete for export, suggesting that they access distinct export pathways (Jarmolowski et al., 1994). The quest for the identification of the molecular mechanisms underlying cellular mRNA nuclear export is still in progress, but specific factors have recently been identified by genetic screens in yeast (Nakielny and Dreyfuss, 1999).
TAP is the metazoan homolog of the yeast mRNA export factor Mex67p and is implicated in the nuclear export of the unspliced genomic RNA of simian type D retroviruses (Grüter et al., 1998). A distinct requirement for retroviral replication is the delivery to the host cytoplasm of unspliced and partially spliced viral transcripts (Cullen, 1998). In contrast, cellular intron-containing pre-mRNAs are generally retained in the nucleus. To overcome nuclear retention, retroviruses encode cis-acting RNA elements that function as export signals for unspliced RNAs and interact, either directly or indirectly, with the cellular transport machinery. The cis-acting constitutive transport element (CTE) of simian type D retroviruses interacts directly with the cellular factor TAP/NXF1 (Grüter et al., 1998).
TAP is also implicated in cellular mRNA export. Titration of TAP by excess of CTE prevents cellular mRNA from exiting the nucleus (Pasquinelli et al., 1997; Saavedra et al., 1997; Grüter et al., 1998). The cellular function of TAP in mRNA export is probably evolutionarily conserved, as coexpression of human TAP and its partner p15 in yeast partially restores growth of the otherwise lethal mex67/mtr2 double knockout strain (Katahira et al., 1999). The viral function of human TAP/NXF1 is instead not shared by Mex67p or other TAP homologs, which do not directly stimulate viral CTE-dependent export (Herold et al., 2000; Stutz et al., 2000). Several lines of evidence suggest that the modes of interaction of TAP with viral and cellular RNAs are likely to be different. First, most cellular RNAs do not contain sequences similar to the CTE stem–loop structure (Tabernero et al., 1996; Ernst et al., 1997). Secondly, the in vitro affinity of TAP for the CTE RNA is three orders of magnitude higher than for mRNA (Braun et al., 1999). Finally, TAP stimulates the nuclear export of CTE-containing RNAs in Xenopus oocytes but not of cellular mRNAs, indicating that TAP is not limiting for the latter (Grüter et al., 1998; Braun et al., 1999). These observations suggest that mRNA may recruit TAP via protein–protein interactions while the CTE may access the mRNA export pathway by interacting directly with TAP (Grüter et al., 1998; Braun et al., 1999).
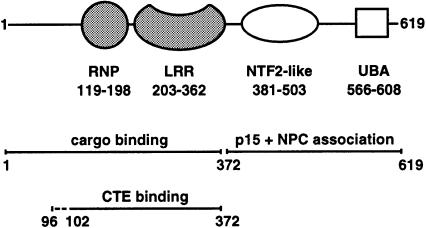
Human TAP/NXF1 is a multidomain protein of 70 kDa molecular weight (Figure 1) (Bear et al., 1999; Braun et al., 1999) that belongs to the NXF protein family (Herold et al., 2000). The C-terminal portion binds to p15 and to a subset of nucleoporins that line the nuclear pore complex (Katahira et al., 1999; Bachi et al., 2000; Suyama et al., 2000). The N-terminal half of TAP (residues 1–372) interacts with the CTE RNA and several RNA-binding proteins (Bachi et al., 2000; Strässer and Hurt, 2000; Stutz et al., 2000). The binding sites are overlapping but not identical, with the first 60 residues being dispensable for CTE binding but important for binding to hnRNP-like proteins (Braun et al., 1999; Bachi et al., 2000). The leucine-rich-repeat (LRR) region of TAP is essential for binding the CTE viral RNA and also affects cellular mRNA export when mutated (Braun et al., 1999).
Fig. 1. Domain structure of human TAP. The N-terminal region of the molecule includes the minimal CTE RNA-binding fragment (residues 102–372) (Braun et al., 1999). The domain boundaries of the RNP and the LRR domains (shaded in gray) are as identified from this work. The domain mapping of the C-terminal region is as previously described (Bachi et al., 2000; Suyama et al., 2000).
Here we report the three-dimensional structure of the minimal CTE-binding fragment of human TAP determined in two crystal forms at 2.9 and 3.15 Å resolution. The molecule comprises two tandem globular domains and an N-terminal disordered flexible region. The N-terminal domain has a ribonucleoprotein (RNP) fold (also known as RNA recognition motif, RRM, or RNA-binding domain, RBD), which is one of the most common RNA-binding modules. Its identifying features are two characteristic sequence motifs (RNP1 and RNP2), which are usually apparent from the amino acid sequence (Birney et al., 1993) but are divergent and as such cryptic in human TAP. The C-terminal domain folds into an LRR-containing protein homologous to the spliceosomal component U2A′. We show that the RNP and LRR domains of TAP have different RNA-binding capabilities, and are required in cis to mediate specific CTE RNA binding. The similarity to the U2B′′(RNP)–U2A′(LRR) spliceosomal heterodimer suggests that TAP might share a similar mechanism of RNA recognition and has been tested with site-directed mutants. More generally, our data indicate that functional RNP domains may be present in additional RNA-binding proteins but remain undetected due to the lack of primary sequence conservation.
Results and discussion
Structural overview
The three-dimensional structure of the minimal CTE-binding fragment of human TAP/NXF1 (102–372) was determined at 3.15 and 2.90 Å resolution in two different crystal forms, which contain four and two independent protein molecules per asymmetric unit, respectively (see Materials and methods). The structure was initially solved in the large tetragonal crystal form by multiple wavelength anomalous dispersion (MAD) methods with selenomethionine-substituted protein (Table I). The model was built and refined with the aid of multi-domain non-crystallographic symmetry (NCS) averaging and used to determine the structure in the small tetragonal crystal form by molecular replacement techniques (Table I). The structures have been refined to an Rfree of 27.8 and 30.3% in the 2.90 and 3.15 Å resolution crystal forms, respectively. The models have good stereochemistry with >98% of the residues lying in either the most favored or the additionally allowed regions of the Ramachandran plot (see Table I) and with one residue (Asn156), which is present in an ordered loop structure, displaying an unusual peptide conformation.
Table I. Data collection, phasing and refinement statistics.
| Data collection statistics | ||||
| crystal form | small tetragonal needle | large tetragonal bipyramid–SeMet | ||
| space group | P43212 | P41212 | ||
| cell dimensions (Å) | a = b = 96.4, c = 152.2 | a = b = 139.9, c = 206.7 | ||
| molecules per asymmetric unit | 2 | 4 | ||
| solvent content (%) | 57 | 70 | ||
| X-ray source | Elettra (Trieste) | ESRF ID14-4 (Grenoble) | ||
| wavelength (Å) | 0.98 | 0.9393 | 0.9789 | 0.9793 |
| (λ3, remote) | (λ2, peak) | (λ1, inflection) | ||
| resolution (Å) | 30–2.9 | 30–3.15 | 30–3.5 | 30–3.5 |
| total measurements | 67 191 | 227 961 | 171 633 | 157 557 |
| unique reflections | 16 300 | 35 832 | 26 644 | 26 331 |
| redundancy | 4.1 | 6.4 | 6.4 | 6.0 |
| completeness (%)a | 98.5 (93.0) | 99.6 (99.9) | 99.8 (98.9) | 99.8 (99.9) |
| I/σa | 21.0 (3.7) | 11.6 (7.0) | 14.3 (5.8) | 12.4 (3.9) |
| Rsym (%)a | 5.7 (23.5) | 9.9 (28.0) | 10.7 (31.2) | 10.7 (34.9) |
| Rano (%) | 3.7 | 5.9 | 5.1 | |
| MAD phasing statistics | ||||
| RCullis iso (centric) | 0.64 | 0.61 | – | |
| RCullis ano (acentric) | 0.96 | 0.81 | 0.77 | |
| phasing power iso (acentric) | 1.94 | 2.11 | – | |
| phasing power ano (acentric) | 0.83 | 1.70 | 1.81 | |
| FOM (acentric) | 0.46 | |||
| Refinement statistics | ||||
| resolution limits | 30–2.90 | 30–3.15 | ||
| Rfree(%) | 27.8 | 30.3 | ||
| Rworking(%) | 25.0 | 30.3 | ||
| φψ (%) most favored | 80.7 | 79.4 | ||
| φψ (%) additionally allowed | 19.0 | 19.4 | ||
| φψ (%) generously allowed | – | 0.9 | ||
| r.m.s.d. bond(Å) | 0.007 | 0.015 | ||
| r.m.s.d. angle (°) | 1.26 | 1.75 | ||
| protein residues | 378 | 864 | ||
| water molecules | 17 | – |
aValues for the outermost resolution shell are given in parentheses.
The CTE-binding domain of TAP features an RNP domain and a C-terminal LRR domain. The two crystal structures provide us with models of six independent LRR domains and of four independent RNP domains. The LRR domains are the most well-ordered part of the molecule, while the RNP domains are in general involved in fewer lattice contacts and either display higher B-factors or are partially disordered (see Materials and methods). The independent structures of each domain are very similar, with pairwise α-carbon root-mean-square deviations (r.m.s.d.) of <0.6 Å when comparing the LRR or RNP domains in the two different crystal forms, effectively ruling out a substantial contribution of crystal packing effects.
The discussion is based on the 3.15 Å resolution RNP domain and on the 2.9 Å resolution LRR domain that display the lowest average B-factors (59.0 and 53.1 Å2, respectively). The N-terminal glycine and arginine-rich portion of the molecule (residues 102–118) is disordered and may therefore be flexible in the absence of RNA. Similarly, no electron density is present for the 10 C-terminal residues and for the linker connecting the two domains. Due to the disorder of the interdomain linker and the particular lattice packing, it is not possible to assign unambiguously the multiple copies of each domain present in the asymmetric units to a single molecule and therefore to determine their spatial relationship. However, the paucity of interdomain contacts suggests that in the absence of RNA their spatial arrangement is determined by crystal contacts.
The N-terminal domain: an RNP fold with non-canonical sequence motifs
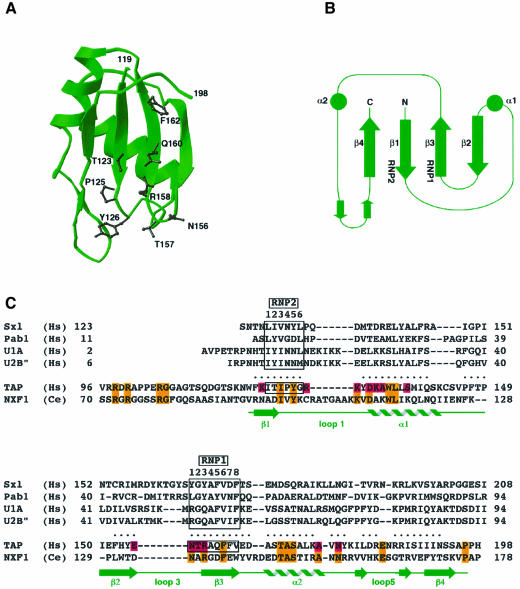
The region of human TAP encompassing residues 119–198 folds into a four-stranded antiparallel β-sheet with a pronounced right-handed twist (Figure 2A). Two perpendicularly oriented α-helices pack against one side of the β-sheet, whereas the other side is exposed to solvent. The C-terminal helix (α2) of the domain is connected to the C-terminal strand (β4) by an extended loop containing two small antiparallel β-strands (Figure 2B).
Fig. 2. The ribonucleoprotein (RNP) domain of human TAP. (A) A ribbon diagram showing in black the side chains of residues that are solvent exposed on the β-sheet platform and are part of the RNP1 and RNP2 consensus sequences of the domain. This and other ribbon diagrams were generated with the program Ribbons (Carson, 1991). (B) Topological arrangement of the secondary structural elements. The circles represent α-helices and the arrows β-strands. The secondary structural elements have been numbered sequentially as for the structurally homologous RNP domains. The positions of the RNP1 and RNP2 motifs are indicated. (C) Structure-based amino acid sequence alignment of the N-terminal domain of human TAP with the RNP domains of Sxl, Pab1 and the spliceosomal proteins U1A and U2B′′. The dots indicate residues in TAP that lie within 3.0 Å of corresponding RNP Cα atoms after optimal superposition of the domains. In green are the secondary structure elements of TAP (nomenclature as in B). The two boxes include the residues forming the octameric RNP1 and hexameric RNP2 consensus sequences, characteristic of RNP domains. A sequence alignment with TAP/NXF1 from C.elegans is included. Residues that have been mutated and analyzed for function are highlighted in pink, while conserved residues are orange.
The βαββαβ topology of this 80-residue domain is similar to the secondary structure elements of a prototype RNP domain (Burd and Dreyfuss, 1994; Varani and Nagai, 1998). Indeed, structural similarity searches of the Protein Data Bank using the program DALI (Holm and Sander, 1993) indicated statistically significant similarities with several RNP domains including the spliceosomal components U1A and U2B′′, and the single-stranded RNA-binding proteins Sex-lethal (Sxl) and poly(A)-binding protein Pab1 (Oubridge et al., 1994; Price et al., 1998; Deo et al., 1999; Handa et al., 1999). A structure-based sequence alignment between human TAP and the RNP domains of Sxl, Pab1, U1A and U2B′′ is shown in Figure 2C and reveals only one identity (Ala159 in β3) between equivalent residues in the RNP domains. In light of the extremely limited sequence homology, the structural similarity is remarkable. In each case, at least 70% of the RNP residues superpose with an r.m.s.d. of <3 Å in their α-carbon positions. The alignment shows that the RNP domain of human TAP lacks canonical RNP1 and RNP2 consensus sequence motifs, but maintains a loose sequence homology for the structurally important hydrophobic residues that are interspersed at particular positions in the sequence. Similarly, a cryptic RNA-binding domain has recently been revealed in the crystal structure of poly(A) polymerase (Martin et al., 2000).
The RNP1 sequence is defined by eight amino acid residues with consensus (K,R)OUT-G-(F,Y)OUT-(G,A)IN-FOUT-VIN-xOUT-(F,Y)IN (where x is any amino acid, and IN and OUT refer to the amino acid side chain pointing either into the core or out to the solvent). The canonical RNP2 sequence consists of the six residues (L,I)IN-(Y,F)OUT-(V,I)IN-(G,N)OUT-(G,N)OUT-(L,M)IN. As originally shown in the structure of U1A (Nagai et al., 1990), the two sequence motifs are juxtaposed on the two central β-strands β3 and β1 and serve both a structural and a functional role. The residues at positions 4, 6 and 8 of RNP1 and at positions 1, 3 and 6 of RNP2 point to the interior of the domain and are crucial for the formation of the hydrophobic core by packing with residues of the two α-helices. The charged and aromatic side chains typically present at the other positions are exposed to solvent and are usually involved in contacting the RNA (Oubridge et al., 1994; Price et al., 1998; Deo et al., 1999; Handa et al., 1999).
At the equivalent positions in the structure of TAP, the octameric RNP1 sequence NOUT-TOUT-ROUT-AIN-QOUT-FIN-FOUT-VIN (residues 156–163) and the hexameric RNP2 sequence IIN-TOUT-IIN-POUT-YOUT-GIN (residues 122–127) deviate significantly from the consensus. In RNP2, a glycine is found in place of the hydrophobic amino acid trademark of position 6, while the solvent-exposed residue at position 4 is an unusual proline, whose occurrence prematurely ends the β-strand conformation of the RNP2 motif. Furthermore, in RNP1 the conserved glycine at position 2 is replaced by a threonine. Not surprisingly, this is at the loop region displaying dihedral angles that are unfavorable for non-glycine amino acids.
The C-terminal domain: an LRR fold structurally homologous to U2A′
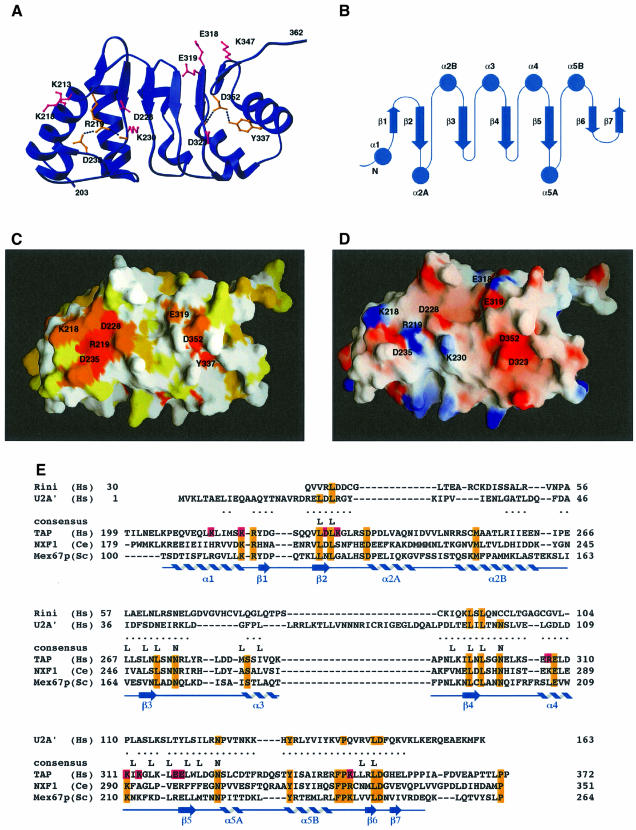
The C-terminal domain of the CTE-binding fragment spans residues 203–362. It includes not only the four tandem LRRs that had been detected previously by sequence analysis (Segref et al., 1997), but also the regions that flank the repeats forming a single structural unit (Figure 3A–D). The domain features a concave surface lined by β-strands and an outer surface formed by α-helices running roughly antiparallel to the strands.
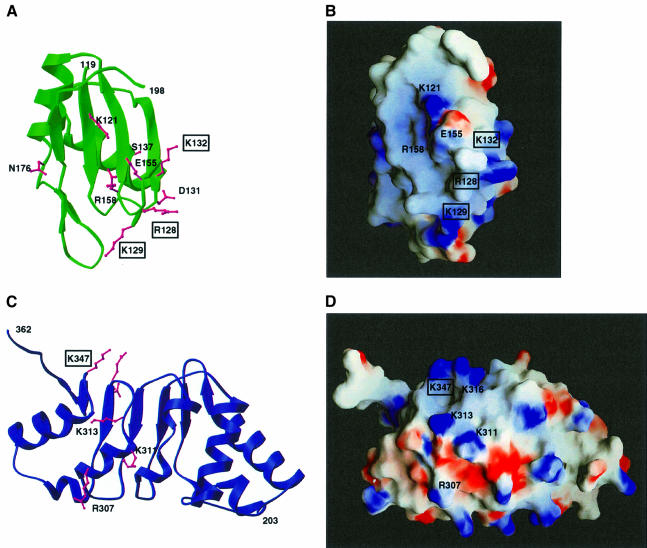
Fig. 3. The LRR domain of human TAP. (A) Ribbon diagram showing the concave β-sheet surface of the domain. The four central LRRs are flanked on either side by regions whose structure is maintained by conserved residues (in orange). In pink are the side chains of residues at the concave surface that have been mutated and tested for function (see also E). (B) Topology diagram of the secondary structural elements, with their sequential numbering indicated. (C) Surface representation of the concave β-sheet face of the LRR domain shown in a similar orientation to (A). The surface is colored according to sequence conservation within the NXF protein family, with a color gradient ranging from white (no sequence identity) to dark orange (100% sequence identity). Residues at the concave surface whose function is discussed in the text are labeled. This and other molecular surfaces were generated with the program GRASP (Nicholls et al., 1991). (D) Electrostatic surface representation of the concave β-sheet face of the LRR domain. Positive potential is colored blue and negative potential red. Residue numbers are indicated. (E) Structure-based amino acid sequence alignment of the LRR domain of human TAP with the ribonuclease inhibitor (Rini) and the spliceosomal protein U2A′. The dots indicate residues in TAP that lie within 3.0 Å of corresponding LRR Cα atoms after optimal superposition of the domains. The alignment with U2A′ takes into account the presence of an additional LRR motif, which has been chosen arbitrarily as the central one in the alignment. In blue are the secondary structure elements of TAP (as in Figure 2B). The sequence alignment with orthologs of human TAP from C.elegans and S.cerevisiae is included. Highlighted in orange are conserved residues in the NXF family, some of which are on the surface while others are in the core and are also conserved in the other two LRR-containing proteins. Pink represents residues that have been mutated and analyzed for function.
The individual repeats of the LRR correspond to β–α units analogous to those originally reported for the ribonuclease inhibitor structure (Kobe and Deisenhofer, 1993). Each repeat consists of ∼26 amino acids with the consensus sequence LIN-xOUT-LIN-xOUT-LIN-xOUT-xOUT-NIN (where x, IN and OUT are as defined above). As in other LRR structures (Kobe and Deisenhofer, 1995), the consensus sequence of the repeats maps onto the β-strands, which are relatively short and followed by a characteristic turn. The leucine side chains associated with the β-strands are involved in stacking interactions that are crucial for maintaining the hydrophobic core of the domain. The asparagine residues in the following turn hydrogen-bond to the backbone of the β-strands and contribute to the flatness of the β-sheet, which lacks the typical twist. The variable residues of the consensus sequence are instead exposed to solvent.
A database search with the program DALI (Holm and Sander, 1993) showed that among the known LRR structures, the TAP 203–362 domain shares remarkable similarity with the spliceosomal protein U2A′, which contains five tandem LRRs (Price et al., 1998). The similarity extends beyond the expected structural homology at the LRR motifs. In particular, the C-terminal region flanking the hydrophobic residues of the last LRR motif of TAP is anchored by the same set of hydrogen-bonding interactions found in U2A′. An invariant aspartic acid (Asp352 in TAP, 146 in U2A′) engages its side-chain carbonyl oxygens in two hydrogen bonds with a main-chain nitrogen and with a tyrosine side chain (Tyr337 in TAP, 131 in U2A′) (Figure 3A and E). The N-terminal flanking region similarly forms a hydrophilic shield on the buried hydrophobic residues of the first repeat and is structured by a salt bridge between two conserved and buried charged residues (Arg219 and Asp235) that also hydrogen-bond to a main-chain carbonyl oxygen (Figure 3A). Helix α2A in the N-terminal flanking region (Figure 3B) is the most variable region of the molecule, displaying some conformational flexibility when comparing the six independent molecules in the crystals.
General and specific RNA-binding activity of TAP domains
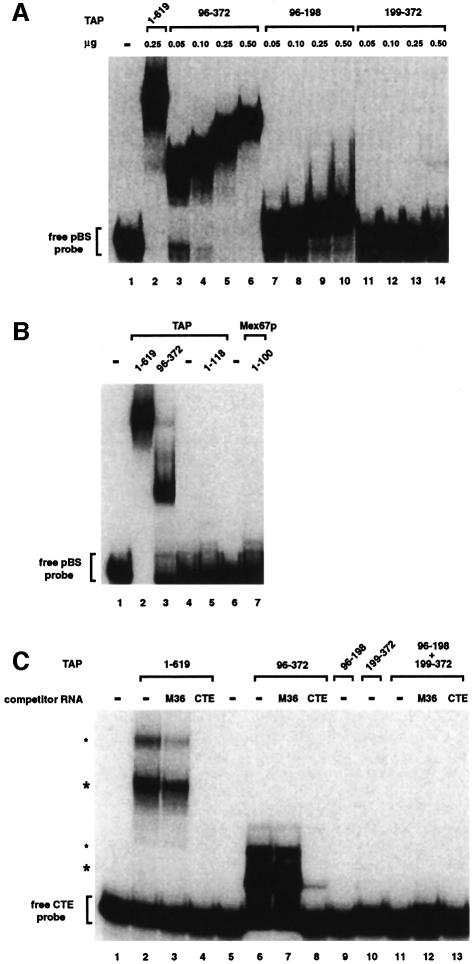
The general RNA-binding ability of TAP fragments was tested by an electrophoretic gel-mobility retardation assay using purified recombinant protein fragments and a non-specific single-stranded RNA probe (Figure 4A). Binding of the RNA probe to the TAP fragment 96–372 is as efficient as to the full-length protein (Figure 4A, lanes 2–6 and data not shown). The 96–372 fragment includes the RNP domain (119–198), the LRR domain (203–362) and an N-terminal region whose sequence (Figure 2C) and lack of electron density suggest flexibility in the absence of CTE RNA. Addition of seven residues N-terminal to the minimal CTE-binding fragment 102–372 (Braun et al., 1999) allows binding to the CTE RNA independently of the presence of the glutathione S-transferase (GST) fusion protein used for expression and purification (data not shown), and was therefore used for biochemical experiments. Upon testing the two domains separately, binding to the RNA probe is observed only with the N-terminal RNP-containing fragment (96–198) (Figure 4A, lanes 7–10), and not with the C-terminal LRR-containing fragment (199–372) (Figure 4A, lanes 11–14). The N-terminal region of human TAP upstream of the RNP domain (residues 1–118) does not exhibit detectable RNA-binding capabilities (Figure 4B, lane 5). The non-canonical RNP of TAP is therefore a bona fide RNA-binding domain despite having divergent sequence motifs. Its basal level of RNA-binding activity is not dependent simply on the presence of positively charged surface residues, as the LRR domain also features an electropositive surface area (Figure 6D) but no general RNA binding can be detected in this case.
Fig. 4. RNA-binding properties of TAP fragments. (A) The RNP domain exhibits general RNA binding affinity, while the LRR domain does not. An electrophoretic mobility retardation assay was performed with a labeled RNA probe derived from pBluescribe and the purified recombinant proteins indicated above the lanes with the quantities used. Binding reactions were performed in the presence of competitor tRNA (5 ng/µl) and herring sperm single-stranded DNA (0.5 ng/µl, mean size 600 nucleotides). The position of the free RNA probe (lane 1) is shown on the left. (B) A gel-mobility retardation assay was performed as described in (A). The purified recombinant proteins are indicated above the lanes and symbols are as in (A). (C) The RNP domain requires the LRR domain in cis for specific binding to the CTE RNA. A gel-mobility assay was performed with a labeled CTE RNA probe and the purified recombinant proteins are indicated above the lanes. For each protein, binding reactions were performed in the absence and presence of either CTE or M36 competitor RNAs (200 ng/µl) as indicated. All binding reactions contain competitor tRNA (200 ng/µl) and herring sperm single-stranded DNA (20 ng/µl). The concentration of the recombinant proteins in the binding reactions was 5 ng/µl, with the exception of fragment 96–372, which was tested at 2.5 ng/µl in order to have equimolar amounts relative to TAP. The positions of the free CTE RNA probe (lane 1) and of the TAP–CTE RNA complexes (asterisks) are indicated on the left. The upper complexes (small asterisks) may represent two molecules of TAP bound to the CTE RNA. Note that full-length TAP (1–619), TAP fragments 1–118 and Mex67p fragment 1–100 were fused to GST. Fragment 199–372 was expressed with a C-terminal His6 tag. Fragments 96–372 had no tag. Fragment 96–198 had no tag in (A), but was fused to GST in (C).
Fig. 6. Identification of putative macromolecular interaction surfaces. (A) Ribbon diagram of the RNP domain in green with the side chains of mutated residues in pink (see Figure 2C). Residues that are essential for CTE binding are boxed. (B) Surface representation of the RNP domain in a similar orientation to (A). The surface is colored according to electrostatic potential, with blue indicating positively charged areas and red indicating negatively charged areas. (C) Structure of the LRR domain viewed towards the convex α-helical surface. Residues at this surface that have been mutated are shown in pink (see Figure 3E). The molecule has been rotated 180° with respect to the view in Figure 3A. (D) Electrostatic surface of the convex outer surface of the LRR domain viewed as in (A), and colored as in (B). Residues whose function is discussed in the text are labeled, and boxed when essential.
Sequence comparison of members of the NXF family indicates that the RNP is one of the least conserved domains within the protein. It is present in C.elegans and Drosophila melanogaster NXF1, as well as in all human NXF proteins (Figure 2C; Herold et al., 2000), but it might not be present in Saccharomyces cerevisiae Mex67p. The region upstream of the LRR domain in S.cerevisiae Mex67p is half the size of that in human TAP and this N-terminal 100-residue fragment of Mex67p is unable to bind to the general RNA probe (Figure 4B, lane 7). Additional protein regions are likely to be required for the in vitro RNA binding reported for the S.cerevisiae Mex67p–Mtr2p complex (Santos-Rosa et al., 1998).
The specific CTE RNA-binding ability of TAP fragments was tested using a CTE probe in an in vitro binding assay similar to that described above. The full-length protein and the 96–372 fragment show high affinity in CTE recognition, both forming a complex even in the presence of high concentrations of competitor RNA (Figure 4C, lanes 2–8). Formation of these complexes is specifically competed by the presence of unlabeled CTE RNA but not by M36 RNA, a CTE derivative that does not bind TAP (Grüter et al., 1998) (Figure 4C, lanes 3, 4, 7 and 8). In the same conditions, the isolated RNP or LRR domains do not specifically interact with the CTE RNA (Figure 4C, lanes 9 and 10). Analogously, no specific binding is observed when the two domains are added in trans (Figure 4C, lanes 11–13). It is unclear at present whether the requirement of the two domains in cis might be due to an effective increase in their local concentration when covalently attached to each other or to sequence-specific RNA contacts of the amino acids in the linker that connects them.
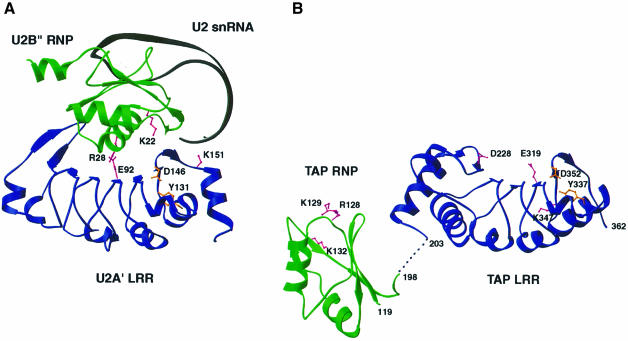
The requirement of both the RNP and LRR domains of TAP for cooperative binding to CTE RNA, their RNA-binding abilities and their structural homology to U2B′′ and U2A′, all point to a remarkable similarity with the spliceosomal U2B′′–U2A′ complex (Figure 5). The spliceosomal U2B′′ RNP protein is not able to recognize specifically the cognate U2 snRNA on its own, as instead is the case for the recognition of U1 snRNA by the homologous U1A RNP protein (Scherly et al., 1990a,b). Although the U2A′ LRR protein does not detectably bind to RNA in vitro (Scherly et al., 1990a), its association with U2B′′ is essential in increasing the protein–RNA contacts (Price et al., 1998). The concave β-sheet surface of the LRR domain associates with the α1 helix of the RNP domain, allowing the U2 snRNA to bind both to the β-sheet platform of the RNP and to a positively charged patch at the C-terminal edge of the U2A′ LRR. A few intersubunit salt bridges are critical for the interaction between U2B′′ and U2A′, which is impaired even by conservative mutations of the charged residues involved in these ionic interactions (such as Lys to Arg or Asp to Glu) (Scherly et al., 1990a). To determine whether the RNP and LRR domains of TAP are involved in a similar mode of macromolecular recognition with the cognate CTE RNA, selected site-directed mutants were tested in in vitro and in vivo functional assays as described below.
Fig. 5. Structural homology between TAP and the spliceosomal U2B′′–U2A′ complex. (A) The U2B′′ RNP domain (green) and the U2A′ LRR domain (blue) of the spliceosomal complex with the cognate RNA (in black) (Price et al., 1998). A few residues critical for the macromolecular interactions are highlighted in pink. In orange are two structurally important residues also present in TAP LRR. (B) The RNP domain (green) and LRR domain (in blue) of TAP in one copy in the asymmetric unit of the crystals. In the absence of RNA, the relative orientation of the two domains is likely to be due to crystal packing. Two residues that are structurally important are shown in orange, and some key residues discussed in the text are in pink.
Identification of macromolecular interaction surfaces
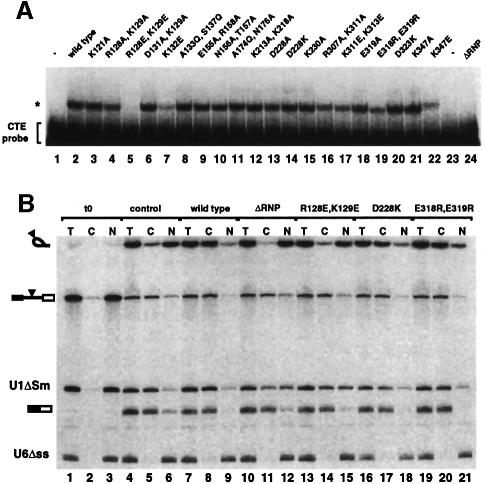
The surfaces of the RNP and LRR domains of TAP established from the structure were probed at multiple positions to identify potential sites of macromolecular interactions. Site-directed mutants were selected according to their amino acid type, phylogenetic conservation and location on the surface of the domains (Figures 3 and 6). The mutant proteins were synthesized in vitro and their ability to bind CTE RNA was assayed in the context of the full-length protein by an electrophoretic gel mobility retardation assay (Figure 7A). Interestingly, the CTE–TAP interaction is disrupted only by a few reverse-charged mutations in the RNP and LRR domains. More conservative substitutions of these residues to alanines does not detectably affect CTE binding (Figure 7A, compare lanes 4 and 5, or 21 and 22), suggesting that recognition is more likely mediated by multiple rather than a few crucial interactions. Although drastic, the effects of reverse-charged substitutions are specific, since at other positions they do not affect binding (Figure 7A, lanes 14 and 20).
Fig. 7. In vitro and in vivo functional studies with structure-based mutants. (A) A gel-mobility retardation assay was performed with a labeled CTE RNA probe and in vitro translated mutant proteins. The concentration of the mutants was similar, as judged by SDS–PAGE analysis and fluorography of the proteins translated in vitro in the presence of [35S]methionine (data not shown). Binding reactions were performed in the presence of competitor tRNA (300 ng/µl), herring sperm single-stranded DNA (30 ng/µl) and M36 RNA (0.5 µg/µl). Complex formation was resolved in a native polyacrylamide gel and visualized by autoradiography. Symbols are as in Figure 4A. (B) Xenopus oocyte nuclei were injected with a mixture of in vitro transcribed 32P-labeled U1ΔSm RNA, U6Δss RNA and a precursor RNA containing the SRV-1 CTE inserted at the intron (Ad-CTE). Purified recombinant GST–TAP and various TAP mutants (8 µM) were included in the injection mixtures as indicated. RNA samples from total oocytes (T), cytoplasmic (C) and nuclear (N) fractions were collected immediately after injection (lanes 1–3) or 2.5 h after injection (lanes 4–12). Products of the splicing reaction were resolved on 10% acrylamide–7 M urea denaturing gels. The mature products and intermediates of the splicing reaction are indicated diagrammatically on the left of the panels. The filled triangle represents the CTE.
The positively charged edge of the RNP β-sheet platform is essential for CTE binding. A double mutation of Arg128 and Lys129 (Figure 6A and B) to negatively charged residues abolishes CTE recognition (Figure 7A, lane 5). In the structure of U2B′′, a basic residue (Lys22) is similarly located in loop 1 (Figures 2C and 5) and interacts with RNA phosphate groups (Price et al., 1998). Deleterious effects in CTE binding are also produced by a reverse-charge mutation of Lys132 (Figure 7A, lane 7) in the RNP α1 helix pointing towards the side of the β-sheet platform (Figure 6A and B). No effect is observed when substituting Ala133 and Ser137 (Figure 7A, lane 8), which from helix α1 point towards the back of the β-sheet platform. Ala133 is in the equivalent structural position to Arg28 in U2B′′ (Figure 2C), a crucial residue for the interaction with U2A′ (Figure 5). It is unclear in this context whether Lys132 in the TAP RNP domain is involved in protein–RNA interactions or protein–protein interactions with the LRR domain.
Unexpectedly, substitution of several amino acids at the concave β-sheet surface of the LRR domain does not prevent CTE RNA recognition. In particular, a reverse-charge mutation of Asp228 results in equally strong CTE binding (Figure 7A, lane 14). The solvent-exposed Asp228 is conserved within the NXF family and is located in the N-terminal region flanking the first repeat (Figure 3A, C and D). The two neighboring conserved residues Arg219 and Asp235 (Figure 3C) play a structural role and their mutation was therefore avoided not to compromise the structural integrity of the protein. No effect in CTE recognition is observed upon mutating other surface residues (Figure 7A, lanes 12, 15 and 20), while a slight decrease in affinity is produced by the reverse-charge of Glu318 and Glu319 (Figure 7, lane 19) at the edge of the concave surface (Figure 3). The most dramatic effect within the LRR domain is caused by mutating the well-conserved Lys347 to an acidic residue (Figure 7, lane 22). Lys347 is located in the C-terminal region flanking the last repeat (Figure 3A or 6C) and lies within an electropositive patch at the edge of the outer convex surface (Figure 6D). The importance of this region is reinforced by the conservation of the interactions that play a structural role in TAP (Asp352, Tyr337) and remarkably also in U2A′ (Asp146, Tyr131). In the C-terminal region of U2A′, although not in an identical position, a positively charged residue (Lys151) is crucial for the recognition of the U2 snRNA stem–loop (Figure 5).
Selected mutants that either do or do not show in vitro CTE-binding abilities were tested for stimulation of CTE-dependent nuclear export in vivo. Purified recombinant proteins were injected into Xenopus oocyte nuclei together with a mixture of three labeled RNAs (Saavedra et al., 1997). This mixture consisted of an adenovirus-derived precursor mRNA bearing the CTE in the intron, U6Δss RNA and U1ΔSm RNA. U6Δss RNA is neither imported nor exported from the nucleus and serves as a control for accurate nuclear injection (Vankan et al., 1992). U1ΔSm is exported by a different pathway than the CTE RNA (Pasquinelli et al., 1997; Saavedra et al., 1997), and serves as a control for the specificity of the recombinant proteins. Immediately after injection all RNAs were found in the nuclear fraction (Figure 7B, lanes 1–3). Following 2.5 h of incubation, splicing of the precursor RNA was complete and 30% of the resulting CTE-bearing intron lariat was found in the cytoplasmic fraction (Figure 7B, lanes 4–6). Coinjection in oocytes of wild-type TAP stimulated the export of the intron lariat, ∼95% of which reached the cytoplasm (Figure 7B, lanes 7–9). The mutant proteins ΔRNP and R128E,K129E, which are unable to bind to CTE in vitro, did not stimulate the export of the intron lariat (Figure 7B, lanes 11, 12, 14 and 15). In contrast, efficient export to the cytoplasm was observed when injecting the mutant proteins D228K and E318R,E319R (Figure 7B, lanes 17, 18, 20 and 21), which retain CTE-binding activity in vitro. Export of the spliced Ad mRNA was not affected (Figure 7B, compare lanes 7–12 with 4–6) while we observed some non-specific effect on U1ΔSm RNA export. Thus, the in vivo behavior of the mutants tested is in agreement with the in vitro results.
Conclusions
The export of CTE-bearing retroviral RNAs to the host cytoplasm is achieved by their direct interaction with a fragment of the cellular protein TAP, which includes two independent globular domains. The N-terminal domain folds and functions as a bona fide RNP domain, despite lacking the canonical conserved sequence motifs. The C-terminal domain is an LRR-containing protein that does not show general RNA-binding activity but is required for specific binding to CTE RNA. The two independent domains have similar structural and biochemical properties to the U2B′′ and U2A′ components of the spliceosomal complex. Functional studies with structure-based mutants indicate that positively charged residues at the β-sheet platform of the RNP are likely to be involved in RNA binding, similarly to U2B′′ and to canonical RNP proteins in general. A residue identified on a helix at the back of the RNP platform plays an important role, either in RNP–LRR or RNP–RNA interactions. A positively charged patch at the outer convex surface of the LRR domain might also be involved in RNA binding, conferring specificity to CTE RNA recognition similarly to the positively charged patch on the U2A′ surface in U2 snRNA stem recognition. Despite these similarities, it is conceivable that the roles of the TAP and U2A′ LRRs in specific RNA binding are at least partly divergent. The most obvious difference is that recognition of CTE RNA requires the RNP and LRR domains to be present in cis in the same polypeptide together with an N-terminal flexible region, while U2B′′ and U2A′ function as separate proteins. Furthermore, substitution of residues along the conserved concave surface of the LRR domain of TAP does not disrupt CTE binding in vitro or its export in vivo, as would be expected for a U2B′′–U2A′ mode of protein–protein interaction.
How might this domain arrangement participate in cellular mRNA export? The RNP domain of TAP is not well conserved among the NXF family members, and in fact is not strictly required for their general RNA export activities (Herold et al., 2000). The LRR domain is conserved and mutations within it have been reported to affect cellular mRNA export (Braun et al., 1999; Strässer and Hurt, 2000). However, these mutations involve residues that have important structural roles and their substitution probably results in non-specific structural aberrations. Conserved residues cluster at the concave surface of the LRR domain, suggesting a plausible role in the cellular function of TAP if not in the viral CTE recognition and export. The positively charged patch at the outer convex surface is also conserved and might therefore be involved in interactions additional to viral RNA recognition. The degree of phylogenetic conservation and the compelling structural homology with the U2A′ component of the spliceosomal complex suggest that the LRR domain of TAP might function in the nuclear export of cellular mRNAs as a protein–protein interaction module for adapters, possibly RNP-containing proteins. We propose that the LRR domain, which acts in cis to promote CTE recognition, may more generally function in trans to interact with other RNP domain proteins associated with cellular RNAs in a manner analogous to the interaction of U2A′ with U2B′′.
Materials and methods
Plasmids and in vitro translation of recombinant proteins
Plasmids pGEXCS-TAP and pBSSK-TAP encoding full-length TAP have been described previously (Braun et al., 1999). Constructs encoding GST fusions of TAP fragments 1–118, 96–372, 96–198 and 102–372 were subcloned into pGEXCS. Fragment 199–372 was cloned in pQE-60 with an uncleavable C-terminal His tag, while the GST fusion Mex67p 1–100 construct is in pGEX-5X-2. Site-directed mutagenesis was performed using the Quick-change mutagenesis system (Stratagene), and confirmed by restriction mapping and sequencing. Plasmid pBSSK encoding wild-type TAP or various TAP mutants was used as a template in the in vitro transcription–translation coupled system (TnT) from Promega. Synthesis of 35S-labeled proteins was checked by SDS–PAGE and subsequent autoradiography.
In vitro RNA-binding assays
Binding to the CTE RNA probe was performed as described (Braun et al., 1999). For the RNA-binding assay shown in Figure 4A and B, a 64 nt RNA probe was synthesized using T3 RNA polymerase and pBluescribe vector linearized with EcoRI. Yeast tRNA or herring sperm single-stranded DNA was used as unlabeled competitor. Reactions were carried out in binding buffer [15 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol (DTT), 0.05 mg/ml bovine serum albumin (BSA) and 0.05% NP-40]. Final sample volumes were 10 µl. After 30 min at 4°C, 1 µl of a solution containing 0.05% bromophenol blue was added to the reaction mixtures. Samples were applied to a 5% non-denaturing polyacrylamide gel (19:1, acryl:bisacryl ratio). Electrophoresis was carried out at a constant voltage of 12 V/cm at 4°C in 0.5× TBE buffer. Complexes were visualized by autoradiography. The amount of recombinant proteins and of competitor RNA used is indicated in the figure legends.
Protein expression and purification
TAP fragments were expressed in Escherichia coli BL21(DE3) as GST fusion proteins linked by a Tev protease cleavage site and purified as described previously (Braun et al., 1999), with the addition of a cation-exchange last step of purification (Macroprep HiS, Bio-Rad). The protein was stored at 40 mg/ml in 20 mM HEPES pH 7.0, 100 mM NaCl, 10% glycerol. The selenomethionine variant of TAP 102–372 was produced by expression in B834(DE3)pLysS in minimal medium in the presence of 1 mM MgSO4, 0.02 mM CaCl2, 0.2% glucose, 1 mg/l thiamine, and supplemented with the 19 amino acids (excluding methionine) at 40 mg/l and l-selenomethionine (Acros) at 50 mg/l. The selenomethionine protein was purified with a similar protocol to that used for the native TAP fragment, the major difference being a decrease in NaCl content (from 100 to 20 mM) in the cation-exchange wash buffer to allow efficient binding to the resin. Analysis by mass spectrometry confirmed the full incorporation of the four selenomethionines in the TAP preparation (data not shown).
Crystallization and data collection
TAP 102–372 was crystallized by vapor diffusion at 4°C, after mixing the protein solution at 20 mg/ml with an equal volume of the well solution containing 100 mM cacodylate pH 6.8, 18% (w/v) polyethylene glycol (PEG) 8000 and 20 mM EDTA. Needle-shaped crystals appeared in 1 week and typically grew to a size of 50 × 50 × 400 µm. Crystals were cryo-protected in 100 mM cacodylate pH 6.8, 12% (w/v) PEG 8000, 10 mM EDTA, 20% glycerol and flash-frozen in liquid nitrogen-cooled propane. The crystals are in space group P43212 (a = b = 96.4 Å, c = 152.2 Å) with two molecules per asymmetric unit and 57% solvent content (Matthews, 1968). The data were collected using synchrotron radiation and processed with the Denzo/HKL package (Otwinowski and Minor, 1997). The statistics are summarized in Table I.
Selenomethionine-substituted TAP 102–372 was crystallized under similar conditions, occasionally yielding bipyramid-shaped tetragonal crystals with a larger unit cell. They are in space group P41212 (a = b = 139.9 Å, c = 206.7 Å) with four molecules in the asymmetric unit and 70% of solvent (Matthews, 1968). After exchange of the arsenate buffer with bis-tris propane in the stabilizing and harvesting solution, a three-wavelength MAD experiment was recorded around the absorption edge of selenium at the ESRF ID14-4 beamline (see Table I).
Structure determination and refinement: the large tetragonal crystal form
The structure was determined by selenomethionine MAD phasing using the large tetragonal bipyramid crystals (see Table I). Despite the relatively low resolution, the poor accuracy of the data and the small signal, the program SOLVE (Terwilliger and Berendzen, 1997) located 12 of the 16 sites present in the 120 kDa tetramer in the asymmetric unit. MAD phasing was treated as a special case of MIR, where the data set at the high-energy remote (λ3) was used as native and the other wavelength data were used as individual derivatives. The statistics of the phases computed with the program SHARP (de la Fortelle and Bricogne, 1997) are shown in Table I. The resulting electron density map allowed the identification of the asymmetric unit content, and the phases were improved by multidomain NCS averaging (Kleywegt and Jones, 1994).
The model was built with the program O (Jones et al., 1991) and refined using the maximum likelihood target in the program CNS (Brünger et al., 1998) against the high-energy remote data set extending to 3.15 Å resolution. No σ cut-off was applied to the data and a random sample containing 4% of the data was excluded. The progress of the refinement was judged by monitoring the agreement between calculated and observed structure factors for the excluded reflections (Rfree %). Due to the limited number of observations at this resolution, very tight NCS restraints were applied during refinement, both to the four large LRR domains and to the two small RNP domains. An initial NCS restraint weight of 300 effectively enforced the protomers to be close to identical, and was later released for residues at crystal contacts and at helix α2A, which assume apparently different conformations in the electron density.
The final model consists of residues 205–362 of the LRR domains and of residues 119–198 of the RNP domains. The Rfree is 30.3% at 3.15 Å resolution with statistics summarized in Table I. The average temperature factors of the four LRR domains are 75, 70, 67 and 64 Å2. The average temperature factors of the small RNP domain A is 59 Å2, while for the small RNP domain B and the incomplete RNP domain E the values are 100 and 93 Å2. Despite the very high B-factors of the RNP domains B and E, features in the electron density clearly indicated their scattering contribution, which was further confirmed by monitoring the Rfree value. Coordinates have been deposited in the RSCB Protein Data Bank (accession No. 1FT8).
Structure determination and refinement: the small tetragonal crystal form
Phasing of the needle-shaped small tetragonal crystal form was achieved using the molecular replacement method with the program AMoRe (CCP4, 1994) and the LRR domain as search model. The model has been refined to 2.9 Å resolution to an Rfree of 27.8% and includes two LRR domains (residues 203–362) with average temperature factors of 57.9 and 53.1 Å2, and one partially disordered RNP domain (residues 123–191) with average temperature factor 96.2 Å2. After superposition, the LRR domains superpose with each other with an r.m.s. separation of corresponding Cα atoms of 0.28 Å, and an r.m.s. separation of <0.6 Å when comparing with the four LRR domains in the large tetragonal crystal form. Coordinates have been deposited in the RSCB Protein Data Bank (accession No. 1FO1).
Acknowledgments
Acknowledgements
The technical support of Michaela Rode is gratefully acknowledged. We are very grateful to Raymond Ravelli and Gordon Leonard for advice and assistance on MAD data collection at beamline ID14-4 (ESRF, Grenoble) and to Luca Olivi at Elettra (Trieste). We wish to thank Mathias Wilm for performing mass spectrometry analysis, and Iain Mattaj and Peter Brick for critical reading of the manuscript. This study was supported by the European Molecular Biology Organization.
References
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J., Tan,W., Zolotukhin,A.S., Tabernero,C., Hudson,E.A. and Felber,B.K. (1999) Identification of novel import and export signals of human TAP, the protein that binds to the CTE element of the type D retrovirus mRNAs. Mol. Cell. Biol., 19, 6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach,E., Schmitt,C. and Izaurralde,E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallographic and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions in RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. (1998) Retroviruses as model systems for the study of nuclear RNA export pathways. Virology, 249, 203–210. [DOI] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy atom parameter refinement for multiple isomorphous replacement and multiwavelength diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- Ernst R.K., Bray,M., Rekosh,D. and Hammarskjold,M.-L. (1997) Secondary structure and mutational analysis of the Mason–Pfizer monkey virus RNA constitutive transport element. RNA, 3, 210–222. [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Grüter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homologue of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Herold A., Suyama,M., Rodrigues,J.P., Braun,I., Kutay,U., Carmo-Fonseca,M., Bork,P. and Izaurralde,E. (2000) TAP/NXF1 belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens,W.C., Izaurralde,E. and Mattaj,I.W. (1994) Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol., 124, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Katahira J., Strässer,K., Podtelejnikov,A., Mann,M., Jung,J.U. and Hurt,E. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Halloween. .. Masks and Bones. SERC Daresbury Laboratory, Warrington, UK. [Google Scholar]
- Kobe B. and Deisenhofer,J. (1993) Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature, 366, 751–756. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1995) Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol., 5, 409–416. [DOI] [PubMed] [Google Scholar]
- Martin G., Keller,W. and Doublie,S. (2000) Crystal structure of a mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J., 19, 4193–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. (1968) Solvent content of protein crystals. J. Mol. Biol., 33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nagai K., Oubridge,C., Jessen,T.H., Li,J. and Evans,P.R. (1990) Crystal structure of the RNA binding domain of the U1 small nuclear ribonucleoprotein A. Nature, 348, 515–520. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Oubridge C., Ito,N., Evans,P.R., Teo,C.H. and Nagai,K. (1994) Crystal structure at 1.92Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Ernst,R.K., Lund,E., Grimm,C., Zapp,M.L., Rekosh,D., Hammarskjold,M.-L. and Dahlberg,J.E. (1997) The constitutive transport element (CTE) of Mason–Pfizer monkey virus (MPMV) accesses an RNA export pathway utilized by cellular messenger RNAs. EMBO J., 16, 7500–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price S.R., Evans,P.R. and Nagai,K. (1998) Crystal structure of the spliceosomal U2B′′–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature, 394, 645–650. [DOI] [PubMed] [Google Scholar]
- Saavedra C., Felber,B.K. and Izaurralde,E. (1997) The simian retrovirus-1 constitutive transport element (CTE), unlike HIV-1 RRE, utilizes factors required for cellular RNA export. Curr. Biol., 7, 619–628. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Panté,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherly D., Boelens,W., Datham,N.A., van Venrooij,W.J. and Mattaj,I.W. (1990a) Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B′′ and their cognate RNAs. Nature, 345, 502–506. [DOI] [PubMed] [Google Scholar]
- Scherly D., Dathan,N.A., Boelens,W., van Venrooij,W.J. and Mattaj,I.W. (1990b) The U2B′′ RNP motif as a site of protein–protein interaction. EMBO J., 9, 3675–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma,K., Doye,V., Hellwig,A., Huber,J., Luhrmann,R. and Hurt,E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K. and Hurt,E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Séraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Doerks,T., Braun,I.C., Sattler,M., Izaurralde,E. and Bork,P. (2000) Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep., 1, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero C., Zolotukhin,A.S., Valentin,A., Pavlakis,G.N. and Felber,B.K. (1996) The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J. Virol., 70, 5998–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1997) Bayesian MAD phasing. Acta Crystallogr. D, 53, 571–579. [DOI] [PubMed] [Google Scholar]
- Vankan P., McGuigan,C. and Mattaj,I.W. (1992) Roles of U4 and U6 snRNAs in the assembly of splicing complexes. EMBO J., 11, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]