Abstract
Background
Segmentation is a hallmark of the arthropods; most knowledge about the molecular basis of arthropod segmentation comes from work on the fly Drosophila melanogaster. In this species a hierarchic cascade of segmentation genes subdivides the blastoderm stepwise into single segment wide regions. However, segmentation in the fly is a derived feature since all segments form virtually simultaneously. Conversely, in the vast majority of arthropods the posterior segments form one at a time from a posterior pre-segmental zone. The pair rule genes (PRGs) comprise an important level of the Drosophila segmentation gene cascade and are indeed the first genes that are expressed in typical transverse stripes in the early embryo. Information on expression and function of PRGs outside the insects, however, is scarce.
Results
Here we present the expression of the pair rule gene orthologs in the pill millipede Glomeris marginata (Myriapoda: Diplopoda). We find evidence that these genes are involved in segmentation and that components of the hierarchic interaction of the gene network as found in insects may be conserved. We further provide evidence that segments are formed in a single-segment periodicity rather than in pairs of two like in another myriapod, the centipede Strigamia maritima. Finally we show that decoupling of dorsal and ventral segmentation in Glomeris appears already at the level of the PRGs.
Conclusions
Although the pair rule gene network is partially conserved among insects and myriapods, some aspects of PRG interaction are, as suggested by expression pattern analysis, convergent, even within the Myriapoda. Conserved expression patterns of PRGs in insects and myriapods, however, may represent ancestral features involved in segmenting the arthropod ancestor.
Background
The expression of the pair rule genes (PRGs) in seven transversal stripes is the first sign of metamerization in Drosophila. Primary PRGs are regulated in a double segmental periodicity by upstream-acting maternal-effect genes and zygotically-expressed gap genes at the blastoderm stage. The primary PRGs then regulate the expression of the secondary PRGs in a similar double segmental pattern (reviewed in [1,2]). This expression pattern in alternating segments is also reflected in the phenotypes of null mutants of these genes: the loss of alternating segmental structures (hence, the name "pair rule") [3]. Later in the extended germ band stage secondary stripes of many PRGs intercalate between the primary stripes and the genes function as segment polarity genes at this time of development [4,5].
However, this mode of segmentation where all segments are produced simultaneously (the so-called long-germ developmental mode) is derived within the insects and is apparently correlated with the high speed of Drosophila embryonic development [6,7]. In the majority of arthropods (including many holometabolous insects) only the anterior segments form simultaneously, and all posterior segments are formed sequentially from a posterior segment addition zone (SAZ) [8]. This mode of segment formation is called the short-germ developmental mode. Theoretically, this mode of sequential segment formation does not require the pair rule gene mechanisms operating during Drosophila segmentation. However, studies in the beetle Tribolium castaneum have shown that although the developmental mode is that of a typical short-germ arthropod, some of the PRG orthologs function as "true" pair rule genes [9,10]. Functional studies in more basal hemimetabolous insects, such as Oncopeltus fasciatus or Gryllus bimaculatus, support the idea that PRGs are involved in segmentation, but question the existence of a pair rule mechanism [11,12]. Studies on the expression of PRGs in chelicerates [13], crustaceans [14,15] and myriapods [16] support this thesis. An exception is seen in the centipede Strigamia maritima, where posterior segments are initially determined in a two-segmental periodicity, revitalizing the question of whether an ancestral pair rule mechanism might exist in arthropods [17,18].
Part of the hierarchic interactions of the PRGs as known from Drosophila is also conserved in the beetle Tribolium [9,10] and may also be conserved in a spider [13]. However, some levels of PRG interaction are obviously divergent and it is, thus, unclear to what extent the interactions of PRGs are generally conserved in arthropods.
In this paper we present the expression profiles of orthologs of most of the known Drosophila PRG orthologs in the pill millipede Glomeris marginata during trunk segmentation. The data support the idea that PRG orthologs are generally involved in segmentation, but do not function as classical pair rule genes like in Drosophila and Tribolium. Our data show that posterior segments are formed one at a time and not in a double segmental periodicity. The interaction of PRG orthologs in Glomeris, as inferred from gene expression data, displays more similarities to Tribolium than to Drosophila. Also some of the PRG orthologs appear to have either ventral or dorsal specific functions, suggesting that the earlier reported decoupling of ventral and dorsal segmentation [19,20] is already obvious with the function of the PRGs. Finally, we discuss our data in the context of the assumed arthropod segmentation clock acting in the addition of posterior segments under the control of Delta-Notch signaling.
Materials and methods
Gene cloning
RNA isolation and cDNA synthesis were carried out as described previously [19]. Initial fragments of the Glomeris even-skipped, runt and hairy-1 gene orthologs were amplified using degenerate primer sets directed against the homeodomain (for even-skipped), the runt domain (for runt) and the helix-loop-helix domain (for hairy), respectively, as described by Damen et al. 2000 [21]. The degenerate primers for the isolation of Glomeris sloppy-paired, odd-paired, odd-skipped and pairberry-1 orthologs have been described in Damen et al. 2005 [13] and Schoppmeier and Damen 2005 [22]. To obtain larger fragments of the genes we subsequently performed 3'-RACE PCR using the MARATHON RACE Kit (Clontech, Heidelberg, Germany).
Sequences of all fragments were determined from both strands of several clones on a 3100 automated sequencer, using Big Dye dye-terminators version 3.1 (Big Dye Terminator Cycle Sequencing Kit; Perkin-Elmer Applied Biosystems, Foster City, CA, USA). The sequences of Glomeris pair rule gene orthologs are available under the accession numbers AM279687 (Gm-eve), AM279688 (Gm-h1), FR715033 (Gm-h2), FR715033 (Gm-h3), AM279692 (Gm-run), FR715039 (Gm-slp), FR715035 (Gm-opa), FR715036 (Gm-odd), FR715037 (Gm-pby1) and FR715038 (Gm-pby2).
Sequence analysis
For the similarity analysis, we searched GenBank [23] using the pairwise alignment program Gapped BLAST [24]. Sequences were aligned using the multiple alignment program Clustal X [25]. The alignments were calculated from the blocks substitution matrix BLOSUM 62 using gap costs at 20 for existence and 0.2 for extension. Maximum likelihood analysis was performed using the Quartet Puzzling method [26] as implemented in PAUP 4.0b10 [27].
In situ hybridization and nuclei staining
Whole mount in situ hybridizations were performed as described in Prpic and Tautz (2003) [28]. Double in situ staining with digoxigenin (DIG) and fluorescein (FL) labeled RNA probes in parallel is described in Janssen et al. (2008) [29]. For reasons of enhanced signal clearness (yolk stains yellow when using INT/BCIP or FastRed) embryos were flat mounted prior to photography. A detailed in situ hybridization protocol is available from the authors upon request.
Cell nuclei distribution was visualized by using the fluorescent dye 4-6-Diamidin-2-phenylindol (DAPI). Incubation in 1 μg/ml DAPI in phosphate buffered saline with 0.1% Tween-20 (PBST) for 30 minutes was followed by extensive washes in PBST. Embryos were analyzed under a Leica dissection microscope (Leica, Heerbrugg, Switzerland) equipped with either an Axiocam (Zeiss, Jena, Germany) or a D70 digital camera (Nikon, Tokyo, Japan). Brightness, contrast, and color values were adjusted in all images using the image processing software Adobe Photoshop CS2 (Version 9.0.1 for Apple Macintosh (Adobe Systems Inc. San Jose, CA, USA).
Results
Transcripts and phylogenetic analysis
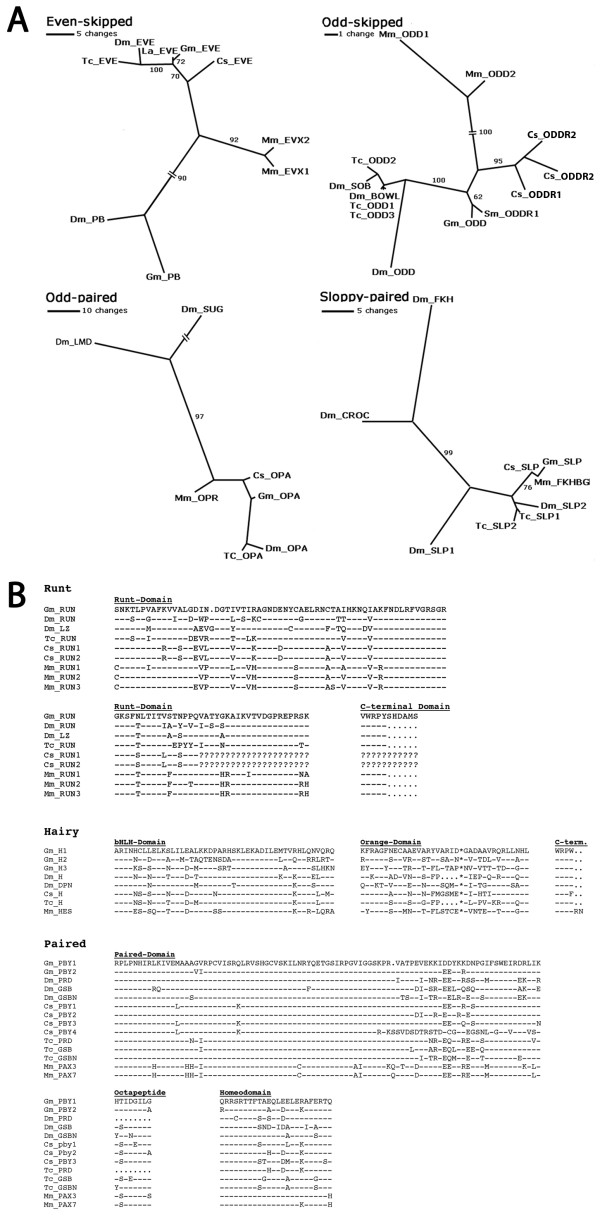
We recovered gene fragments with significant similarity to the even-skipped, runt, sloppy-paired, odd-skipped and odd-paired genes from Drosophila melanogaster and other arthropods that we designated as Glomeris even-skipped (Gm-eve), Glomeris runt (Gm-run), Glomeris sloppy-paired (Gm-slp), Glomeris odd-skipped (Gm-odd) and Glomeris odd-paired (Gm-opa), respectively (Figure 1). The Gm-eve fragment covers almost the complete homeodomain (except its very N-terminal end), the complete C-terminal part of the open reading frame (ORF) and a subsequent 3'untranslated region (UTR) ending in a poly-A tail. Within a reasonable distance from the poly-A sequence is a putative polyadenylation signal (AATAAA). The Gm-run fragment covers most of the runt-domain (except its N-terminal end), the complete C-terminal part of the ORF including the characteristic C-terminal WRPY motif (Figure 1B) [30]. Remarkably, this C-terminal WRPY motif is not at the very end of the ORF in Glomeris as it is in other arthropod and vertebrate runt orthologs, but is followed by a sequence of an additional six amino acids (Figure 1B). The subsequent 3'UTR ends in a poly-A stretch, but an obvious polyadenylation signal is not present.
Figure 1.
Phylogenetic analysis of Glomeris pair rule gene orthologs. (A) Phylograms of orthologs of the Drosophila pair rule genes Even-skipped, Odd-skipped, Odd-paired and Sloppy-paired from various arthropods and the mouse. Closely-related genes from the Drosophila genome serve as outgroups. Un-rooted majority rule consensus trees are shown, computed from 1000 intermediate trees produced with the Quartet Puzzling Method [25]. Reliability values are indicated at the edges. Abbreviations for animals: Cs, Cupiennius salei (spider); Dm, Drosophila melanogaster (fly); Gm, Glomeris marginata (millipede); La, Lithobius atkinsoni (centipede); Mm, Mus musculus (mouse); Sm; Strigamia maritima (centipede); Tribolium castaneum (beetle). Abbreviations for gene names: LMD, Lame duck; SUG, Sugarbabe; BOWL, Brother of odd with entrails limited; CROC, Crocodile; EVE, Even-skipped; EVX, vertebrate Even-skipped ortholog; FKH, Forkhead; FKHBG, Forkhead box G1; ODD, Odd-skipped; ODDR, Odd-skipped-related; OPA, Odd-paired; OPR, Odd-paired-related; PB, Proboscipedia; SLP, Sloppy-paired; SOB, Sister of odd and bowl. (B) Alignment of Runt, Hairy and Paired sequences. Orthologs from Drosophila melanogaster, Tribolium castaneum, Cupiennius salei and mouse are aligned with the available Glomeris marginata gene fragments. Aligned is part of the Runt domain and the C-terminal consensus (for Runt); part of the bHLH domain and the orange domain (for Hairy); part of the Paired domain, the octapeptide and part of the homeodomain (for Paired/Pairberry). Dashes denote identical amino acids, dots indicate gaps introduced to improve the alignment. Question marks stand for missing sequence information. Species abbreviations as in (A). Gene name abbreviations as in (A). Additional abbreviations: DPN, Deadpan; GSB, Gooseberry; GSBN, Gooseberry-neuro; Pax3/7, Pax group III genes; PRD, Paired; H, Hairy; HES, Hairy/Enhancer of split; LZ, Lozenge; PBY, Pairberry; RUN, Runt.
The Gm-slp fragment encodes the C-terminal part of the protein including part of the forkhead-domain and the complete 3'-UTR as indicated by the presence of a putative polyadenylation signal (AATAAA). Gm-odd and Gm-opa encode part of the zinc-finger domains. The 5'and 3'ends of these two genes have not been recovered.
We isolated three gene fragments that show significant similarities to Drosophila hairy (Figure 1B). Drosophila possesses several bHLH transcription factors related to hairy with deadpan and side showing most similarities to hairy (Figure 1B). Since the order of branches in our phylogenetic analysis is statistically unresolved (trees not shown; cf. [21]) we designated the Glomeris fragments simply as Gm-h1, Gm-h2, and Gm-h3. All fragments encode the bHLH-domain (except its basic N-terminal part), the orange-domain, and the complete C-terminal part of the ORF each ending in a WRPW motif which, like the orange-domain, is characteristic for the hairy-class of bHLH-domain genes. All Glomeris hairy fragments contain a 3'UTR ending in a poly-A tail with a putative polyadenylation signal (that is, AATAAA for Gm-h1 and ATTAAA for Gm-h2 and Gm-h3) just upstream.
We obtained partial fragments for two Glomeris PaxIII group genes, which we named pairberry-1 (Gm-pby1) and pairberry-2 (Gm-pby-2) because of their sequence similarity to the Drosophila PaxIII group genes (that is, paired, gooseberry-neuro and gooseberry). The conceptually translated protein fragment contains the paired-domain (except its very N-terminal part), the octapeptide domain and the homeodomain (except its very C-terminal part) (Figure 1B).
Expression of primary PRG orthologs during Glomeris trunk segmentation
Three of the known pair rule genes (PRGs) from Drosophila, that is, even-skipped, runt and hairy, are described as primary since they act upstream of the so-called secondary PRGs and control the expression of the latter [1]. The following section focuses on the expression of the orthologs of the primary PRGs during trunk segmentation in Glomeris.
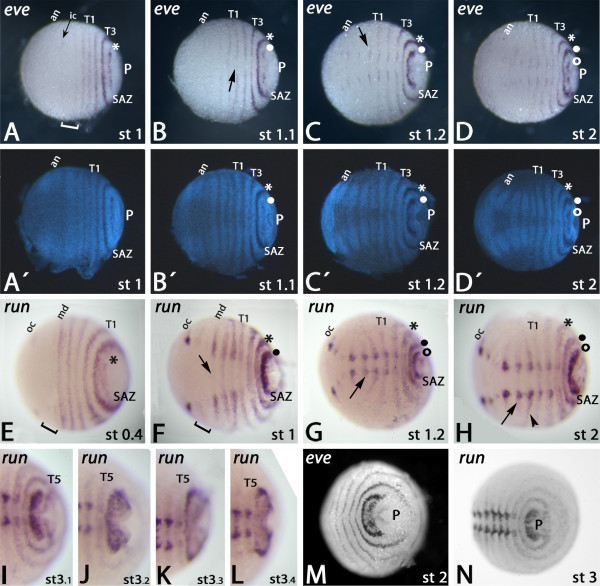
Gm-eve is expressed in segmental stripes. During the process of segment formation it is dynamically expressed in the SAZ (Figures 2 A-D, M and 3A, B). This dynamic expression starts with two small patch-like expression domains on either side of the proctodaeum in the posterior part of the SAZ (Figure 2B, D) that soon after broadens into a single patch-like expression domain (Figure 2A) and finally refines into a single stripe of expression in the very anterior of the nascent segment and posterior adjacent to the segmental expression of engrailed (en) (Figures 2A-D, M and 3A, B). This segmental eve expression covers the ventral and dorsal tissue in nascent segments and the SAZ (Figure 2C, D). The dorsal segmental expression persists slightly longer than the ventral (Figure 2C, D). eve is expressed in circles surrounding the proctodaeum in the most posterior segments (Figures 2M and 3A). At later stages this is less obvious since the SAZ and its surrounding tissue narrows. Apart from its expression during segment formation, Gm-eve is also expressed in the developing ventral nervous system (CNS) (Figure 2C, D).
Figure 2.
Expression of Glomeris even-skipped and runt. Expression of even-skipped (A-D, M) and runt (E-L, N). (A'-D') show DAPI counter-staining of the embryos shown in (A-D). (A) Stage 1 embryo. Segmental stripes in every segment. Bracket marks premandibular and mandibular segments. Asterisk marks stripe of expression in the SAZ. (B) Stage 1.1 embryo. Arrow points to fading ventral expression. Note that the expression in the SAZ in (A) (asterisk) is now in the nascent segment anterior to the SAZ (asterisk in (B)) and a new expression domain is forming in the SAZ (filled circle). (C) Stage 1.2 embryo. Arrow points to expression in the CNS. Note that the expression domain seen in the SAZ in (B) has developed into a single broad domain (filled circle). (D) Stage 2 embryo. Former expression in the SAZ has transformed into a stripe in the nascent segment (filled circle). A new expression domain forms in the SAZ (open circle). (A'-D') DAPI counter-staining of the embryos shown in (A-D). (E) Stage 0.4 embryo. Bracket marks premandibular and antennal segments. Expression in the SAZ (asterisk). (F) Stage 1 embryo. Arrow points to disappearing ventral expression. Expression seen in the SAZ in A now lies in the nascent segment anterior to the SAZ (asterisk). A new domain of expression has formed in the SAZ (filled circle). Bracket as in (E). (G) Stage 1.2 embryo. Arrow points to expression in the CNS. Expression in the SAZ seen in (F) is now in the nascent segment anterior to the SAZ (filled circle). Expression seen in the SAZ in (E) (asterisk) is now one segment anterior to it. A new domain appears in the SAZ (open circle). (H) Stage 2 embryo. Arrow as in (G). Expression in the SAZ (open circle) expands. Arrowhead points to strengthened expression in the postmaxillary segment. (I-L) Dynamic expression of runt in the SAZ in four consecutive developmental stages (that is, stage 3.1 to 3.4). (M, N) Concentric expression of even-skipped (M) and runt (N) around the proctodaeum. In all pictures anterior is to the left. Abbreviations: an, antennae; ic, intercalary (premandibular) segment; md, mandibular segment; oc, ocular field; P, proctodaeum; SAZ, segment addition zone; T, trunk segment.
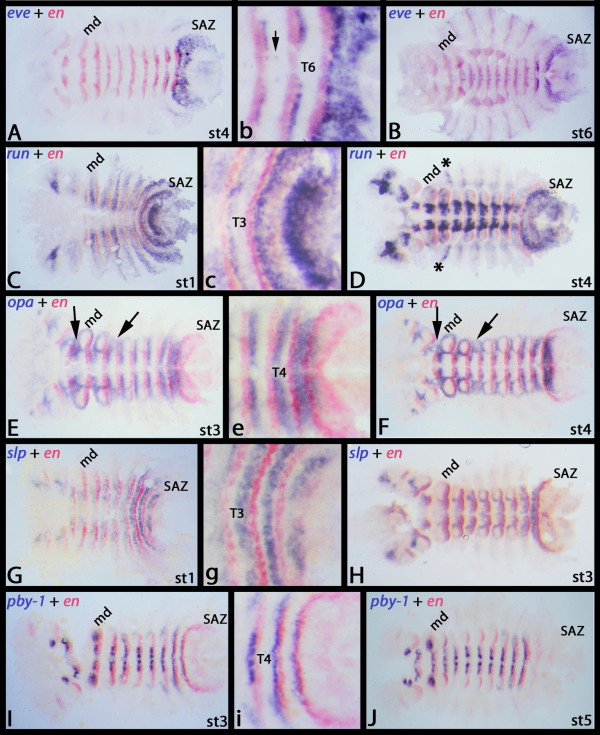
Figure 3.
Double staining of Glomeris PRGs (purple) with engrailed (red). Pictures denoted in capital letters are whole mounts; pictures denoted in small letters show close-ups. All embryos are flat-mounted; yolk removed. (A, B) engrailed (en) is expressed anterior adjacent to even-skipped (eve). Arrow in (B) points to dot-like expression in the CNS. (C, D) en is expressed anterior adjacent to runt (run). Asterisks in (D) denote enhanced dorsal expression in the postmaxillary segment. (E, F) en is expressed posteriorly adjacent to opa. The arrows mark opa expression at the position where the premandibular and postmaxillary appendages should form, but are repressed by unknown mechanisms. (G, H) en is expressed posterior to sloppy-paired (slp) at the anterior edge of the SAZ. (I, J) en is expressed posterior to and partially overlapping with pairberry-1 (pby-1) in nascent segments. Abbreviations: md, mandibular segment; SAZ, segment addition zone; st, stage; T, trunk segment.
Gm-run is expressed in segmental stripes at early segmentation stages (Figure 2E). Like Gm-eve, Gm-run is also dynamically expressed in the SAZ (Figures 2E-L, N and 3C, D). In contrast to the dynamic expression of eve, run initially appears in a small domain in the SAZ anterior adjacent to the proctodaeum (Figures 2E, G and 3C), that later broadens and finally fills the complete SAZ (Figure 2F, H). Then this domain disappears from the SAZ and only leaves a distinct stripe of expression in the very anterior of the nascent segment (Figures. 2K and 3C, D). The ventral portion of this stripe disappears soon after while the dorsal part persists (Figure 2L). As described for eve, the expression of run in the most posterior segments is also in circles (Figure 2N). The segmental expression persists longer in younger stages (Figures 2E-H and 3C) than in older stages (Figure 3D). Apart from its expression during segment formation, Gm-run is also expressed in broad, but well-defined, domains in the CNS (Figures 2G-L, N and 3D) including the ocular region (Figures 2F-H and 3C, D). This expression is initially weaker in the antennal and premandibular segment (Figures 2F-H and 3C). The dorsal expression in the postmaxillary segment is enhanced (Figures 2H and 3D).
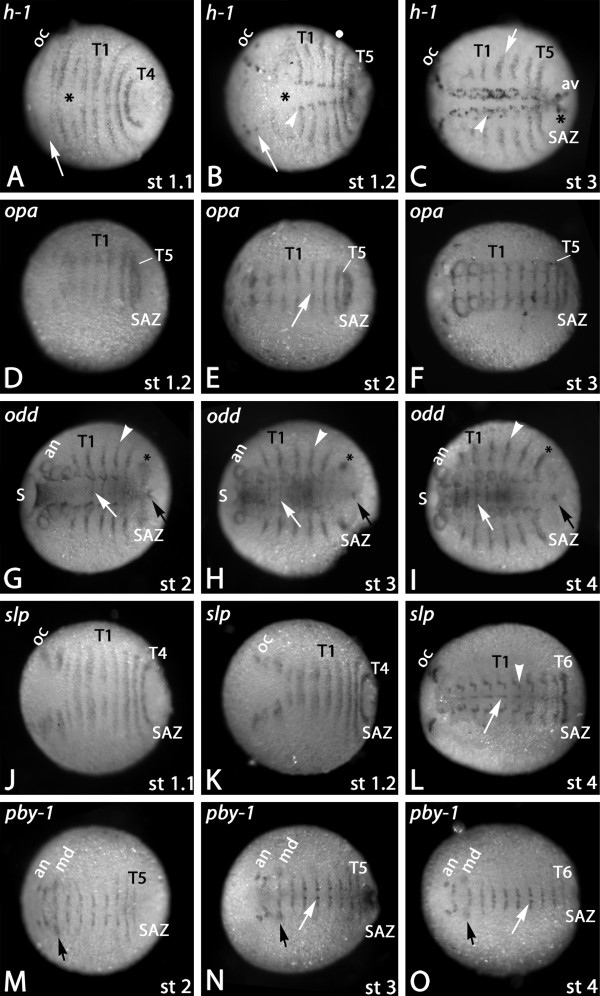
Gm-h1 is expressed in transversal segmental stripes at early segmentation stages (Figure 4A). Unlike eve and run, h1 is not expressed dynamically in the SAZ, but only appears at the very anterior of it, where it covers ventral and dorsal tissue (Figure 4A-C). Again the segmental expression is more obvious at younger stages. In older embryos the ventral segmental expression disappears soon after its appearance and only the dorsal segmental expression persists (Figures 4A-C and Additional file 1: Figure S1A, B). The dorsal stripes of h1 expression lie anterior to en (Additional file 1: Figure S1A, B). The dorsal expression reaches into the dorsal extraembryonic tissue (Additional file 1: Figure S1K). Apart from the expression during segment formation, h1 is also expressed in the anal valves (Figure 4C) and in part of the CNS including the ocular region (Figures 4B, C and Additional file 1: Figure S1C). The expression profile of Gm-h2 gene is similar to that of Gm-h1 (Additional file 1: Figure S1D/E). However, at younger developmental stages the expression is very weak compared to that of h1 and no dynamic expression in the SAZ is detectable. Like h1, also h2 is expressed in the dorsal embryonic tissue (Additional file 1: Figure S1L). Gm-h3 is expressed in the neuroectoderm, in segmental spots in the dorsal tissue, in the anal valves and at later stages in the tips of the appendages (Additional file 1: Figure S1F-H).
Figure 4.
Expression of Glomeris hairy-1, odd-paired, odd-skipped, sloppy-paired and pairberry-1. Expression of hairy-1 (h1) (A-C), odd-paired (opa) (D-F), odd-skipped (odd) (G-I), sloppy-paired (slp) (J-L) and pairberry-1 (pby-1) (M-O). (A) Stage 1.1 embryo. Arrow points to antennal segment that does not express h1 at this stage. Asterisk marks weaker expression near the ventral midline. (B) Stage 1.2 embryo. Asterisk as in (A). Note that the segmental expression proceeds into the extra-embryonic tissue (filled circle). Enhanced patch-like expression in the central nervous system (CNS) (arrowhead), the ocular region (oc) and the antennal segment (arrow). (C) Stage 3 embryo. Segmental stripes are restricted to dorsal tissue (arrow). Dotted expression in the CNS (arrowhead) and in the anal valves (asterisk). (D) Stage 1.2 embryo. (E) Stage 2 embryo. Expression is dis-continuous in the anterior segments (arrow). (F) Stage 3 embryo. Note that expression is restricted to ventral tissue only. (G) Stage 2 embryo. Asterisk marks dynamic expression in the SAZ. Arrowhead points to dorsal segmental stripes. White arrow points to ventral segmental stripes. Black arrow points to expression in the proctodaeum. (H) Stage 3 embryo. Note the transformation of the SAZ-expression (asterisk in (G)) into dorsal patches (asterisk in H) in parallel with a clearance of expression from the SAZ. Arrows and arrowhead as in (G). (I) Stage 4 embryo. Transformation of the dorsal patches (asterisk in (H)) into a dorsal stripe (asterisk in (I)). Arrows and arrowhead as in (G) and (H). (J) Stage 1.1 embryo. (K) Stage 1.2 embryo. (L) Stage 4 embryo. Segmental expression is reduced to the ventral midline (arrow), the ventral part of the appendages and small segmental domains in the CNS (arrowhead). (M) Stage 2 embryo. Note that expression does not extend into the appendage buds, except for the mandibles (black arrow). (N) Stage 3 embryo. Enhanced expression in the CNS (white arrow). Black arrow as in (M). (O) Stage 4 embryo. Arrows as in (N). Anterior is to the left in all pictures. Abbreviations as in Figure 2. Additional abbreviations: av, anal valves; S, stomodaeum
Expression of secondary PRG orthologs during Glomeris trunk segmentation
The Glomeris odd-paired ortholog (Gm-opa) is expressed in segmental stripes. Its expression, however, is strictly restricted to the ventral part of the embryo (Figures 3E, F and 4D-F). opa is expressed posterior adjacent to the expression of en (Figure 3E, F). opa is also expressed in the SAZ (Figures 3E, F and 4D-F), but in a way that is different from the dynamic expression of eve and run. Initially the anterior SAZ expresses opa in a broad domain, that later transforms into a single small stripe in the newly formed segment (Figure 4D-F). Gm-opa is expressed in rings around the outgrowing appendages, with the exception of the labrum. The pre-mandibular and postmaxillary segments do not form appendages and here opa is expressed in the tissue corresponding to where the appendages form in the other segments (Figure 3E, F).
Gm-odd is expressed dynamically in the SAZ (Figure 4G-I). This dynamic expression, however, never results in ventral segmental stripes, but only in dorsal stripes. Initially a faint propeller-shaped expression domain appears in the SAZ (Figure 4G, I) that later becomes more clearly visible (Figure 4G). This domain then disappears from its ventral part and continues to strengthen in its dorsal part, leaving a strong patch of expression in the dorsal tissue of the nascent segment (Figure 4H). This then transforms into dorsal stripes (Figure 4I). The dorsal segmental expression of odd lies posterior to that of the segmental marker engrailed (en) (Additional file 1: Figure S1I, J). As with h1 and h2, odd expression also extends dorsally into the dorsal extraembryonic tissue (Additional file 1: Figure S1M). Gm-odd is also strongly expressed in the region of the forming stomodaeum (Figure 4G), the ventral portion of the developing appendages (Figure 4G), a dot-like domain anterior to the proctodaeum (Figure 4G-I) and in the CNS (Figure 4G-I).
Gm-slp is expressed in segmental stripes anteriorly abutting the expression of en (Figures 3G, H and 4J-L). The segmental expression initially extends into dorsal tissue, but soon after this expression pattern disappears (Figures 3G, H and 4L). There is no dynamic expression in the SAZ. The most posterior segmental stripe appears anterior to en in the anterior part of the SAZ or the nascent segment (Figures 3G, H and 4J-L). At later stages the segmental expression ceases in the anterior segments and remains only in the ocular region, the antennae, the walking legs, a restricted ventral region in the CNS, and segmentally reiterated stripes along the ventral midline (Figure 4L). The posterior part of the SAZ remains free from slp transcripts at all stages (Figures 3G, H and 4J-L).
Gm-pby1 is neither expressed dynamically in the SAZ nor in stripes in the posterior part of the SAZ, but appears first in a segmental stripe in the anterior of the SAZ or the nascent segment (Figures 3I, J and 4M-O). However, its expression is restricted to the ventral tissue only. The segmental expression disappears soon after its appearance in the ventral midline and the legs (Figures 3I, J and 4M-O). Its intra-segmental position overlaps with the posterior part of wingless (wg) (not shown) and the anterior part of en (Figure 3I, J). Gm-pby2 expression is restricted to the tips of the legs at late embryonic stages (not shown).
Discussion
Pair rule gene orthologs are involved in myriapod segmentation
So far only expression data of a few PRG orthologs in centipede myriapods (Chilopoda) have been published, but no data have been available for PRG orthologs of millipedes (Diplopoda). Since functional studies have not yet been established for any myriapod species our knowledge on PRGs in myriapods is restricted to sequence and expression data [16-18]. All canonical PRGs are transcription factors with a spatially and temporally regulated expression pattern and it is reasonable to suspect that they should, therefore, be functional more or less exclusively in those cells where they are expressed. Thus, in the absence of functional data the PRGs expression data can provide us with clues as to their likely role in segmentation. Segmentation genes on the level of the segment polarity genes (SPGs) and PRGs are in most cases expressed in transversal stripes within the segments or at least in the posterior SAZ where the segments are formed, clearly suggesting a role in segment specification and formation (Figure 5). Some functional studies in insects, however, report possible exceptions from this rule. In Tribolium, for example, RNAi did not uncover a function of the primary PRG hairy in posterior segmentation [31], although its expression pattern suggests this [32,33]. Furthermore, in the same species fushi-tarazu (ftz) and odd-paired (opa) did not show segmentation defects in RNAi studies, although they are expressed in the typical transversal stripes [9,34]. In most cases, however, gene expression data in comparable developmental studies give a valuable clue on what a gene's function may be and if this function could be conserved in different organisms.
Figure 5.
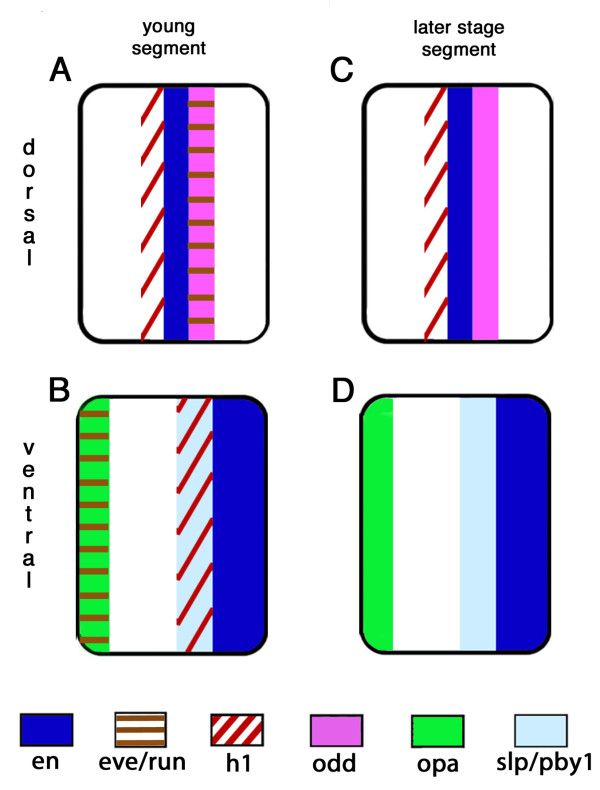
Schematic summary of Glomeris pair rule gene expression patterns in dorsal and ventral segments. Segmental expression of PRG orthologs in young (A, B) and older (C, D) segments. (B) and (D) represent ventral segments, (A) and (C) represent dorsal segments. The genes are denoted in different colors and patterns and are explained in the legend below the drawings. Note that expression of the primary PRGs eve and run is present in the nascent segments, but absent from older segments, both in ventral and dorsal tissue. The patterning of dorsal and ventral segments is very different at the level of the PRGs. Some genes are exclusively expressed in dorsal or ventral segments, respectively (for example, odd, pby1, opa). Other genes are expressed in both ventral and dorsal segments, but show differences in their temporal expression profile or their intrasegmental position (for example, eve, run, h1). These results support earlier results that suggested a decoupling between dorsal and ventral segments, and indicate that this decoupling is already present at the level of the PRGs. Abbreviations: en, engrailed; eve, even-skipped; h1, hairy1; odd, odd-skipped; pby1, pairberry1; run, runt; slp, sloppy-paired; wg, wingless.
The expression profiles of the PRGs examined here suggest a general role in segment formation, or at least the development of segmentally-iterated structures. Even if some of the reported PRGs in Glomeris - despite their expression patterns that are typical for segmentation genes - might not be involved in segmentation, the data from a wide range of arthropods including spiders and myriapods support the idea that PRGs are generally important factors in arthropod segmentation [13,21].
Posterior segments form in a single segmental periodicity
It is still an open question whether PRGs act in a pair rule like manner in arthropods other than Drosophila and Tribolium [1,9,10,35].
In the hemimetabolous insect Schistocerca americana and the spider-mite Tetranychus urticae the prd/pby orthologs Sg-pby1 and Tu-Pax3/7 appear with delay in every other segment in the anterior embryo which may hint at an ancestral double segmental mechanism [36,37]; discussed in [2]. However, outside the higher insects such a pair rule mechanism may be restricted to the anterior part of the embryo, which is patterned similarly as the Drosophila embryo. Therefore, mechanisms found in anterior patterning are likely to be more conserved between short germ and long germ arthropods than it is the case for posterior patterning (discussed in [38-40]). Since expression data on PRGs in anterior patterning are scarce outside the insects, this theory remains open for further debate and study.
There is, however, one example that supports a posterior pair-rule like mechanism outside higher insects. In the centipede Strigamia maritima posterior segments are added with a double segmental periodicity [17,18]. This pair-rule like mechanism appears, however, to be the result of parallel evolution rather than a conserved feature of arthropod posterior segmentation, because some of the genes involved, like, for example, caudal, appear to be co-opted for this special purpose [17], discussed in [41,42]. The expression of another PRG, eve, in the centipede Lithobius atkinsoni, however, does not show any kind of a double segmental pattern [16] further supporting the thesis that the double segmental mechanism found in Strigamia may be a peculiarity of geophilomorphs rather than a general feature of myriapods or even short germ arthropods. This is also supported by data from the spider Cupiennius salei where PRGs show no double segmental expression [13,21,22].
We find that none of the investigated PRGs are expressed in alternating segments in the trunk. In fact, the dynamic expression patterns of Gm-eve and Gm-run demonstrate clearly that the posterior trunk segments appear with a single segmental period (Figure 2). The dynamic expression in the SAZ of either of these genes can be followed into each nascent segment demonstrating that every single stripe of eve or run is correlated with the formation of a single new posterior segment.
Conserved aspects of early PRG interaction
In Drosophila the PRGs are subdivided into two classes; the primary PRGs, which are under control of maternal factors and the gap genes; and the secondary PRGs, which receive their input from the primary PRGs [1]. Recent studies in the beetle Tribolium have examined the possibility of this PRG hierarchy being conserved [9]. Although a number of differences exist in the regulation of the PRGs, the principal hierarchy of PRGs between Drosophila and Tribolium is conserved. eve and run act high in the regulatory network in both species [9,43-45] and prd and slp are under the control of these primary PRGs [9,45,46]. Data from the spider Cupiennius support the idea that a hierarchic order of PRGs may even be conserved in all arthropods, since Cs-eve and Cs-run are expressed more posterior (that is, earlier) in the SAZ than Cs-slp and Cs-pby-3 that are restricted to the anterior of the SAZ and the anterior rim of the SAZ, respectively [13,21,22].
In Glomeris eve and run both display the most prominent expression pattern in the SAZ passing through it from its very posterior to its anterior and finally into the nascent segments (Figure 2). Furthermore these two PRG orthologs are the only ones in Glomeris being additionally expressed in concentric circles around the proctodaeum, as is the case for odr-1 in Strigamia [17] or eve in Lithobius [16] (Figure 2M, N). The dominant expression profiles of eve and run in the SAZ thus make them good candidates for segmentation genes acting at high level in a possible network, as it is the case for their orthologs in Drosophila [45], Tribolium [9] and possibly also Cupiennius [13,21].
The Glomeris orthologs of the secondary PRGs (that is, Gm-slp and Gm-pby1) are never expressed dynamically in the SAZ, but appear relatively late in the SAZ shortly before the inter-segmental borders form (Figure 4J-O). Notably these two genes also display highly conserved intra-segmental expression patterns (discussed below). In addition, both orthologs are never expressed in the posterior part of the SAZ in Cupiennius, but only in the anterior SAZ (that is, Cs-pby-3) or at the anterior rim of the SAZ (that is, Cs-slp) [13]. This led Damen and colleagues [13] to suggest that these genes may be under the control of genes like eve and run, which are active earlier (that is, already in the posterior SAZ). Since the relative tempo-spatial expression of some of the putative primary and secondary PRGs in the SAZ is conserved between Tribolium, Cupiennius and Glomeris, we postulate that at least eve and run are acting at a high level in a possible regulatory network of PRG orthologs in all arthropods.
In Glomeris the odd gene, which is a primary PRG in Tribolium and a secondary PRG in Drosophila, is also expressed dynamically in the entire SAZ (Figure 4G-I), suggesting that odd may play an important role in segment formation in Glomeris. Moreover, expression of the odd ortholog odr-1 in the centipede Strigamia also suggests a crucial role of this gene in posterior segment formation [17]. Taken together these data imply that odd may already have gained a dominant function in segmentation in lower arthropods. Later, it may then have been recruited as a primary PRG in Tribolium. Indeed, in a bioinformatics study on transcriptional regulation of segmentation gene interaction in Drosophila odd rather behaves like a primary PRG although functionally it is a secondary one [47]. This result strengthens the idea that odd orthologs fulfill important tasks in arthropod segmentation, even in Drosophila, where its function in segmentation may be understated by the fact that it acts like a secondary PRG.
Conserved segmental expression of secondary PRGs
As already discussed above, there appears to be a hierarchy of PRGs in Glomeris where putative primary PRGs are expressed early and dynamically in the Glomeris SAZ; whereas the expression of putative target genes such as the secondary PRGs (and SPGs) is restricted to anterior regions of the SAZ and the nascent segment. However, in Drosophila some of the PRGs are also expressed at later stages, where they function to stabilize the parasegment boundary (PSB) [4]. Several studies have shown that the parasegment boundary is a conserved entity among arthropods and characterized by the expression of SPGs, like engrailed and wingless, at either side of the parasegment boundary [8,19,29,48-51].
We, therefore, also analyzed the intra-segmental expression of the Glomeris PRGs and discuss here to what extend this late function/interaction of the Glomeris PRGs may be conserved or have diverged. Expression and function of most PRGs is known only from Drosophila and Tribolium, which are both rather derived holometabolous insects. Apart from that, the only almost-complete set of PRG orthologs is known from the spider Cupiennius. But the focus of this study is on the early expression in the SAZ and does not include the examination of later segmental expression patterns [13]. The data on "late" PRG expression are thus very limited except for data on Pax3/7 group genes (that is, the Gm-pby1 orthologs) that are very abundant due to the availability of cross-reacting antibodies detecting all genes of this family in various arthropods (that is, orthologs of Drosophila paired, gooseberry and gooseberry-neuro) [15,36].
The segmental expression patterns of the primary PRGs in Glomeris are different from that in Tribolium and/or Drosophila. For example, Gm-eve and Gm-run are expressed posterior to Gm-en in the ventral segments with no or only very little overlap (Figure 3A-D), whereas, in Drosophila and Tribolium eve and run expression is overlapping with en [9].
The segmental expression of the secondary PRG opa is only reported from the fly Drosophila and the spider Cupiennius. In the fly an initial ubiquitous expression later refines into faint segmental stripes, but the intra-segmental position of these stripes is unclear [52]. In the spider the gene is expressed in stripes in the SAZ and the newly formed segments, but again the intra-segmental position of its expression is unclear [13]. In Tribolium however opa is expressed posterior abutting to en (with little overlap) (CP Choe and SJ Brown personal communication). This expression is thus conserved at least between Tribolium and Glomeris, and may therefore represent the ancestral expression profile of opa (Figure 6).
Figure 6.
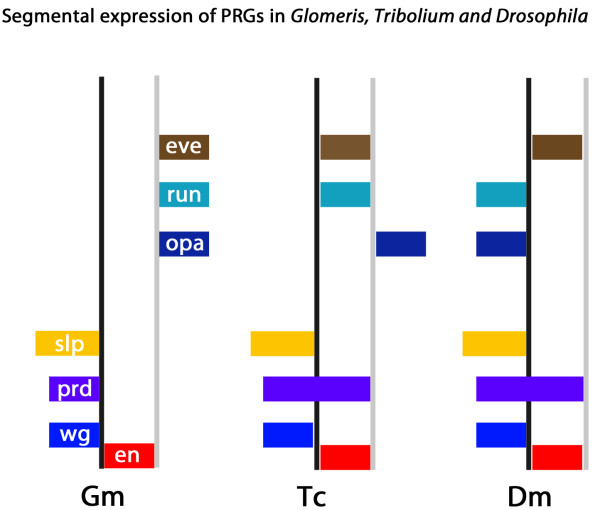
Segmental expression of PRGs in Glomeris (ventral segments only), Tribolium and Drosophila. Schematic drawing and colour coding is based on [70]. Expression of en (red), wg (blue), prd (purple), slp (yellow), opa (dark blue), run (petrol) and eve (brown). Black lines represent parasegment boundaries; grey lines represent segment boundaries. The expression of the segment polarity genes en and wg is conserved, as well as that of the PRG slp. Expression of prd is conserved with modification in every other segment in Tribolium and Drosophila. Intra-segmental position of opa is conserved between Tribolium and Glomeris, which may reflect the ancestral pattern. Co-expression of eve and run is also conserved Tribolium and Glomeris, but at a different intra-segmental position.
The intra-segmental expression of Gm-slp and Gm-pby1 is conserved between Glomeris and various arthropods (for pby1) [15,36] and Glomeris, Drosophila and Tribolium (for slp) [5,10]. In all cases slp is expressed anteriorly adjacent to en, and the Pax3/7 ortholog pby-1 is expressed overlying the PSG boundary and, thus, partially overlapping the expression of wg and of en (Figure 6).
We, therefore, propose that the late function of slp and Pax3/7 orthologs is conserved in the formation of the PSB in that slp represses en, and in that pby1 might activate wg and en. Whether the segmental expression profiles of the other Glomeris PRG orthologs is conserved among basal arthropods and what their late function in segmentation may be has to be examined in the future. Data on expression and function of all PRG orthologs from various basal arthropods would possibly help in answering this question.
Decoupled dorsal and ventral segmentation
In Glomeris the germ band consists of ventral tissue from the beginning of germ band extension on, but dorsal tissue does not start developing before stage 3 (staging after [19]). We reported earlier that Glomeris dorso-ventral segmentation is decoupled at the level of the segment polarity (SPGs) and Hox genes suggesting that the process of decoupling must have its origin at earlier regulatory levels [19,20]. Since in Drosophila, the SPGs are under control of the pair rule genes (PRGs) it appeared likely that the decoupling of segmentation in Glomeris is attributed to the level of the PRGs. Our screen on Glomeris PRGs now revealed that some genes display a profile that suggests restricted function in dorsal segmentation, while others are predominantly expressed in ventral tissue (Figure 5). By contrast, the genes opa and pby-1 are expressed in stripes only in the ventral segments. Thus, they are likely to be involved in ventral segmentation, but they are not required for the formation of the dorsal segments. The remaining genes are initially expressed in ventral and dorsal segmental tissue, but show dorso-ventral differences in their persistence of expression. The expression of eve, runt and h1 stripes persist longer in dorsal tissue, whereas the slp stripes persist longer on the ventral side (Figures 4 and 5). Our data thus suggest that decoupling of dorsal and ventral segmentation already starts with the action of the PRG orthologs. Especially those genes that are initially expressed on both sides, but then "decide" for either ventral or dorsal persistence, provide further insight into the possible mechanisms of dorso-ventral decoupling. These genes are likely members of a core segmentation network that is common to both ventral and dorsal segments, but then are regulated differently on both sides to achieve the specific metamerisms of the ventral and dorsal sides, respectively.
The finding that the ventral segmentation gene expressions in Glomeris show similarities to those in other arthropods not only on the level of the SPGs but also of the PRGs, while the dorsal patterns deviate on this level as well, strengthens the notion that the ventral metamerism in Glomeris is homologous to the segmentation in other arthropods, while the dorsal metamerism mechanisms have diversified independently in different arthropod lineages [19].
The idea of an arthropod "segmentation clock"
Segmentation in short-germ arthropods and vertebrates displays fundamental morphological similarities. Separate body units (that is, segments and somites, respectively) are added sequentially from a posterior region in the embryo: the SAZ in arthropods and the presomitic mesoderm (PSM) in vertebrates [53,54]. The cyclic expression of genes that is involved in the formation of new somites in vertebrates very closely resembles the dynamic expression of the Glomeris PRGs eve, run and also odd. The cyclic gene expression in vertebrates is under control of a so-called "segmentation clock" or "oscillator", of which Notch/Delta signaling is a key component [54]. Recent work on the spider Cupiennius salei and the cockroach Periplaneta americana [55,56] has shown that Notch/Delta signaling is a main component of arthropod segmentation as well, and implies that the lack of Notch/Delta signaling in Drosophila segmentation is the derived state. The morphological similarities of segment formation in arthropods and vertebrates and the common usage of Notch/Delta expression suggest that segmentation in both phyla traces from a common ancestor [55]. However, the functional similarities between segmentation in arthropods and vertebrates are more extensive than just using Notch/Delta signaling. Recent work in the spider Achaearanea tepidariorum has shown that a posterior expression of the Wnt8 gene is involved in the maintenance of SAZ activity [57-59] and the ortholog of this gene, Wnt8 is required for the proper function of the PSM in vertebrates [60,61].
It is currently unclear how these newly discovered vertebrate-like mechanisms feed into the more canonical segmentation mechanisms in arthropods, that is, the level of the pair rule and segment polarity genes. It has been suggested that these levels are under control of a posterior segmentation clock involving Notch/Delta signaling [39]. So far there is only circumstantial evidence that supports this idea. First, the vertebrate Her/Hes genes, that are orthologs of the arthropod PRG hairy, are controlled by the Notch/Delta oscillator [62,63], thus providing a link between these two components in vertebrates. It is possible that this link has been conserved and extended during arthropod evolution. Indeed, in the spider Cupiennius hairy expression is changed upon Notch and Delta RNAi [55,64]. Second, the PRG odd is regulated downstream of Notch signalling in Drosophila leg development [65]. This fact caused Chipman et al. [17] to suggest that odd might be under the control of Notch/Delta signaling in other processes as well, including segment formation. In agreement with this idea, in Glomeris and two other myriapods, Strigamia and Lithobius, Notch and Delta are expressed dynamically in the SAZ [18,66,67], strikingly similar to the dynamic patterns that we find in the present work for the segmentation genes. In Glomeris this dynamic Notch pattern results in dorsal segmental Notch stripes [66] and we have shown here that the odd stripes in Glomeris also persist only in the dorsal segments, thus pointing to a possible link between Notch signaling and odd activation. Another argument for the involvement of Notch/Delta signalling in dorsal segment formation comes from the expression of Gm-h1 (Figure 4B, C). Like Gm-Notch and Gm-odd also Gm-h1 becomes restricted to dorsal segments soon after its expression in the SAZ (Figure 4B, C). The expression profiles of Notch, Delta, odd, and h1 would be in agreement with the suggested ancestral interaction of a posterior "segmentation clock" mechanism and PRG orthologs [39,68].
Expression of PRGs in dorsal extraembryonic tissue
In most arthropods a thin layer of dorsal ectodermal tissue connects the separated dorsal edges of the developing embryo [69]. The nature and function of this tissue is not very well understood and the lack of expression of the segmentation genes in this tissue in most species suggests that it is not metamerized or subdivided into separate developmental units. However, in Glomeris the primary PRGs, eve and run are expressed in circles around (or within) the SAZ, which is especially clear at early developmental stages (Figure 2); the segmental expression of these genes thus includes extraembryonic tissue. A similar expression profile has also been reported for pair rule genes, for example, odr1 (odd ortholog) and eve1/2, in Strigamia [18] and it was, thus, not unexpected to find similar patterns in Glomeris as well. In Glomeris, however, h1 and odd are not only expressed in the dorsal segments (discussed above), but their segmental expression in the extraembryonic tissue persists even in older stages (Additional file 1: Figure S1). Since neither the segment polarity genes nor the Hox genes are expressed in the extraembryonic tissue it appears unlikely that this tissue is patterned or metamerized in the classical sense. But what then is the function of h1, h2 and odd expression in the extraembryonic tissue?
We postulate that the expression of the hairy genes (h1 and h2) and odd may be involved in "guiding" the two dorsal sides of the embryo in order to assure that "matching" tissue meets during the process of dorsal closure. This would be especially important, because the matching cannot be guided via the connection across the ventral side, as the dorsal segments are functionally decoupled from the ventral side.
Conclusions
The activation of the pair rule genes (PRGs) appears to be a crucial step in arthropod segmentation as it marks the transition from a non-periodic pattern to a periodic pattern. It is, therefore, surprising to some degree that this step of the segmentation mechanisms appears to be the most variable and diverse level, especially when it is compared to the evolutionarily conserved steps of the SPGs or the Hox genes [40,70,71]. Expression of all investigated PRGs in the millipede Glomeris is consistent with a role in segmentation. The PRGs in Glomeris seem to act in a hierarchic manner, and can be subdivided into primary and secondary PRG, similar as has been described for insects and a spider [9,13]. However, the exact interactions between the Glomeris PRGs remain unclear due to the lack of functional methods. The expression of the primary PRGs strongly supports the idea that the initial patterning of the segments is in a single segmental period. A similar single segmental period has also been observed in other arthropods implying that it represents the ancestral mode for segment patterning and formation [22].
Our work is the first analysis in a non-insect arthropod in which the late expression of the PRGs has been studied. The intrasegmental positions of the expression patterns show similarities but also differences among the species analysed (Figure 6A). The comparison of insect data (that is, Tribolium and Drosophila) with data from a myriapod may help to distinguish between conserved and derived features of segmentation and thus provide clues about the origin and evolution of segmentation mechanisms in the arthropods.
In both Tribolium and Glomeris the intra-segmental expression of opa, slp and prd/pby1 is conserved, and although its function is unclear, it may be part of the ancestral patterning system (Figure 6).
An interesting case of "partial" conservation is provided by the genes run and eve. These genes are co-expressed in Tribolium and Glomeris (but not in Drosophila) (Figure 6). However, the co-expression stripes of run and eve do not have the same intrasegmental position in the two species. Thus, co-expression of these two genes appears to be a conserved feature already present in the common ancestor of insects and myriapods, but the mechanisms that position the run/eve stripe within a segment must differ in the two species.
Our results also add to the previous finding that the segmentation mechanisms in the dorsal and ventral segments of Glomeris are decoupled [19]. Previous results have shown that dorsal and ventral segmentation mechanisms differ mainly in their usage of the segmentation genes of the segment polarity group, but our present data show that strong dorso-ventral differences already exist in the expression of the orthologs of the PRGs.
The ancestral role of the PRGs in arthropod segmentation remains unclear until comparative data on expression and function are available for representatives of all groups of arthropods, as well as their assumed sister-groups, the onychophorans and the tardigrades [72]. Shared conserved or diverged aspects of the segmentation process may then also contribute to the unraveling of arthropod phylogeny [73], which appears still not fully resolved [74,75].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RJ carried out the experiments, wrote the first draft of the manuscript and was mainly responsible for the experimental outline. GEB, NMP and WGMD were involved in drafting the final version of the manuscript. NMP and WGMD discussed the experimental outline. RJ and WGMD initiated work on Glomeris segmentation.
Supplementary Material
Figure S1. Additional aspects of h1 and odd expression, expression of h2 and h3 and single-colour expression detection of h1+en and odd+en. (A, B) Double single-colour staining of h1+en. The black arrows point to expression of en; the white arrows point to expression of h1 anteriorly abutting the expression of en. (C) Expression of h1 in a stage 5 embryo (cf. similar expression of h2 in (E)). White arrow points to expression in the ventral nervous system. Asterisk marks expression in the brain. (D, E) Expression of h2. White arrow and asterisk in (E) as in (C). (F-H) Expression of h3. Arrows in (F) point to expression in the brain and the ventral nervous system. Arrowhead points to expression in the SAZ. Asterisk marks expression in the labrum. (G) Same embryo as in (F) but slightly rotated to show a rear aspect and the posterior expression in the SAZ (arrowhead). (H) De novo expression in lateral tissue (arrow) and in the appendages (arrowhead). (H') Same embryo as in (H); DAPI stained; arrow and arrowhead point to same position as in (H). (I) Double single-colour staining of odd+en. (I') Same embryo as in (I); DAPI stained. (J) Close-up on dorsal tissue of the embryo shown in (I/I'). Black arrow points to expression of en. White arrow points to expression of odd. (K) Dorsolateral view of a stage 5 embryo stained for h1. Dorsal segmental expression is connected by weak expression (asterisks) in the dorsal extraembryonic tissue (e). (L) Dorsal view. Expression of h2 in the (e). Asterisks as in (K). (M) Dorsal view. Expression of odd in the (e). Asterisks as in (K). Abbreviations: (e), dorsal extraembryonic tissue; en, engrailed, h, hairy; odd, odd-skipped; st, stage.
Contributor Information
Ralf Janssen, Email: ralf.janssen@geo.uu.se.
Graham E Budd, Email: graham.budd@pal.uu.se.
Nikola-Michael Prpic, Email: nprpic@uni-goettingen.de.
Wim GM Damen, Email: wim.damen@uni-jena.de.
Acknowledgements
We would like to thank Hilary Pioro ( neé Dove) for Notch and Delta clones and two anonymous reviewers for their helpful comments and suggestions. This work has been supported by the Swedish Research Council (VR: grant to GEB), the European Union via the Marie Curie Training network "ZOONET" (MRTN-CT-2004-005624 (to GEB, RJ and WGMD)). The work of NMP is funded by the University of Göttingen, the Graduate School for Neurosciences and Molecular Biosciences (GGNB), and the Deutsche Forschungsgemeinschaft.
References
- Pankratz MJ, Jäckle H. In: The development of Drosophila melanogaster. Bate M, Martinez Arias A, editor. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. Blastoderm segmentation; pp. 467–516. [Google Scholar]
- Davis GK, Patel NH. Playing by pair rules? BioEssays. 2003;25:425–429. doi: 10.1002/bies.10278. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Baker NE, Martinez-Arias A. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even skipped. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994;8:899–913. doi: 10.1101/gad.8.8.899. [DOI] [PubMed] [Google Scholar]
- Sanson B. Generating patterns from fields of cells. EMBO reports. 2001;2:1083–1088. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GK, Patel NH. Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu Rev Entomol. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- Janssen R, Le Gouar M, Pechmann M, Poulin F, Bolognesi R, Schwager EE, Hopfen C, Colbourne JK, Budd GE, Brown SJ, Prpic NM, Kosiol C, Vervoort M, Damen WGM, Balavoine G, McGregor AP. Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol Biol. p. 374. [DOI] [PMC free article] [PubMed]
- Choe CP, Miller SC, Brown SJ. A pair rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci USA. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. Evolutionary flexibility of pair rule patterning revealed by functional analysis of secondary pair rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol. 2007;302:281–94. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PZ, Kaufman TC. even-skipped is not a pair rule gene but has segmental and gap-like functions in Oncopeltus fasciatus, an intermediate germband insect. Development. 2005;132:2081–2092. doi: 10.1242/dev.01807. [DOI] [PubMed] [Google Scholar]
- Mito T, Kobayashi C, Sarashima I, Zhang H, Shinahara W, Miyawaki K, Shinmyo Y, Ohuchi, Noji S. even-skipped has gap-like, pair rule-like, and segmental functions in the cricket Gryllus bimaculatus, a basal, intermediate germ insect (Orthoptera) Dev Biol. 2007;303:202–213. doi: 10.1016/j.ydbio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Damen WGM, Janssen R, Prpic NM. Pair rule gene orthologs in spider segmentation. Evol Dev. 2005;7:618–628. doi: 10.1111/j.1525-142X.2005.05065.x. [DOI] [PubMed] [Google Scholar]
- Copf T, Rabet N, Celniker SE, Averof M. Posterior patterning genes and the identification of a unique body region in the brine shrimp Artemia franciscana. Development. 2003;130:5915–5927. doi: 10.1242/dev.00835. [DOI] [PubMed] [Google Scholar]
- Davis GK, D'Alessio JA, Patel NH. Pax3/7 genes reveal conservation and divergence in the arthropod segmentation hierarchy. Dev Biol. 2005;285:169–184. doi: 10.1016/j.ydbio.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Exploring myriapod segmentation: the expression patterns of even-skipped, engrailed, and wingless in a centipede. Dev Biol. 2002;247:47–61. doi: 10.1006/dbio.2002.0683. [DOI] [PubMed] [Google Scholar]
- Chipman AD, Arthur W, Akam M. A double segment periodicity underlies segment generation in centipede development. Curr Biol. 2004;14:1250–1255. doi: 10.1016/j.cub.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Chipman AD, Akam M. The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair rule gene homologues. Dev Biol. 2008;319:160–169. doi: 10.1016/j.ydbio.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Janssen R, Prpic NM, Damen WGM. Gene expression suggests decoupled dorsal and ventral segmentation in the millipede Glomeris marginata (Myriapoda: Diplopoda) Dev Biol. 2004;268:89–104. doi: 10.1016/j.ydbio.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Janssen R, Damen WGM. The ten Hox genes of the millipede Glomeris marginata. Dev Genes Evol. 2006;216:451–465. doi: 10.1007/s00427-006-0092-5. [DOI] [PubMed] [Google Scholar]
- Damen WGM, Weller M, Tautz D. The expression patterns of hairy, even-skipped, and runt in the spider Cupiennius salei imply that these genes were segmentation genes in a basal arthropod. Proc Natl Acad Sci USA. 2000;97:4515–4519. doi: 10.1073/pnas.97.9.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppmeier M, Damen WGM. Expression of Pax group III genes suggests a single-segmental periodicity for opisthosomal segment patterning in the spider Cupiennius salei. Evol Dev. 2005;7:160–167. doi: 10.1111/j.1525-142X.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2003;31:23–27. doi: 10.1093/nar/gkg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PsiBLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates, Sunderland, Massachusetts; 2002. Version 4. [Google Scholar]
- Prpic NM, Tautz D. The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. Dev Biol. 2003;260:97–112. doi: 10.1016/s0012-1606(03)00217-3. [DOI] [PubMed] [Google Scholar]
- Janssen R, Budd GE, Damen WG, Prpic NM. Evidence for Wg-independent tergite boundary formation in the millipede Glomeris marginata. Dev Genes Evol. 2008;218:361–370. doi: 10.1007/s00427-008-0231-2. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Sakate M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda M, Marques-Souza H, Bayer T, Tautz D. The role of the segmentation gene hairy in Tribolium. Dev Genes Evol. 2008;218:465–477. doi: 10.1007/s00427-008-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, Tautz D. Involvement of an orthologue of the Drosophila pair rule gene hairy in segment formation of the short germ-band embryo of Tribolium (Coleoptera) Nature. 1993;361:448–450. doi: 10.1038/361448a0. [DOI] [PubMed] [Google Scholar]
- Eckert C, Aranda M, Wolff C, Tautz D. Separable stripe enhancer elements for the pair rule gene hairy in the beetle Tribolium. EMBO Rep. 2004;5:638–642. doi: 10.1038/sj.embor.7400148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Hilgenfeld RD, Denell RE. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc Natl Acad Sci USA. 1994;91:12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderspacher F, Bucher G, Klingler M. Pair rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev Genes Evol. 1998;208:558–568. doi: 10.1007/s004270050215. [DOI] [PubMed] [Google Scholar]
- Davis GK, Jaramillo CA, Patel NH. Pax Group III genes and the evolution of insect pair rule patterning. Development. 2001;128:3445–3458. doi: 10.1242/dev.128.18.3445. [DOI] [PubMed] [Google Scholar]
- Dearden PK, Donly C, Grbic M. Expression of pair rule gene homologues: early patterning of the two-spotted spider mite Tetranychus urticae. Development. 2002;129:5461–5472. doi: 10.1242/dev.00099. [DOI] [PubMed] [Google Scholar]
- Minelli A. A three-phase model of arthropod segmentation. Dev Genes Evol. 2001;211:509–521. doi: 10.1007/s004270100180. [DOI] [PubMed] [Google Scholar]
- Peel A, Akam M. Evolution of segmentation: Rolling back the clock. Curr Biol. 2003;13:R708–R710. doi: 10.1016/j.cub.2003.08.045. [DOI] [PubMed] [Google Scholar]
- Peel A. The evolution of arthropod segmentation mechanisms. BioEssays. 2004;26:1108–111. doi: 10.1002/bies.20097. [DOI] [PubMed] [Google Scholar]
- Damen WGM. Arthropod segmentation: Why centipedes are odd. Curr Biol. 2004;14:R557–R559. doi: 10.1016/j.cub.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Chipman AD. Parallel evolution of segmentation by co-option of ancestral gene regulatory networks. BioEssays. 2010;32:60–70. doi: 10.1002/bies.200900130. [DOI] [PubMed] [Google Scholar]
- Harding K, Rushlow C, Doyle HJ, Hoey T, Levine M. Cross-regulatory interactions among pair rule genes in Drosophila. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Frei E, Noll M. Complex regulation of early paired expression: initial activation by gap genes and pattern modulation by pair rule genes. Development. 1993;117:609–623. doi: 10.1242/dev.117.2.609. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A. Interactions between segment polarity genes and the generation of the segmental pattern in Drosophila. Mech Dev. 1991;35:77–87. doi: 10.1016/0925-4773(91)90059-f. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DI, MacNicol AM, Patel NH. Functional conservation of the wingless-engrailed interaction as shown by a widely applicable baculovirus misexpression system. Curr Biol. 1999;9:1288–1296. doi: 10.1016/s0960-9822(00)80050-0. [DOI] [PubMed] [Google Scholar]
- Damen WG. Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. Development. 2002;129:1239–1250. doi: 10.1242/dev.129.5.1239. [DOI] [PubMed] [Google Scholar]
- Prpic NM. Parasegmental appendage allocation in annelids and arthropods and the homology of parapodia and arthropodia. Front Zool. 2008;5:17. doi: 10.1186/1742-9994-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 1994;8:105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- Pourquie O. Notch around the clock. Curr Opin Genet Dev. 1999;9:599–565. doi: 10.1016/s0959-437x(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Pourquie O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Schoppmeier M, Damen WGM. Involvement of Notch and Delta genes in spider segmentation. Nature. 2003;423:863–865. doi: 10.1038/nature01682. [DOI] [PubMed] [Google Scholar]
- Pueyo JI, Lanfear R, Couso JP. Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. PNAS. 2008;105:16614–16619. doi: 10.1073/pnas.0804093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Feitosa NM, Kruck S, Aranda M, Damen WG. Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Curr Biol. 2008;18:1619–1623. doi: 10.1016/j.cub.2008.08.045. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Damen WG. An ancestral regulatory network for posterior development in arthropods. Commun Integr Biol. 2009;2:174–176. doi: 10.4161/cib.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat S, Hopfen C, McGregor AP. The function and evolution of Wnt genes in arthropods. Arthropod Struct Dev. 2010;39:446–452. doi: 10.1016/j.asd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neuroectoderm patterning. Dev Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Li HY, Bourdelas A, Carron C, Gomez C, Boucaut JC, Shi DL. FGF8, Wnt8 and Myf5 are target genes of Tbx6 during anteroposterior specification in Xenopus embryo. Dev Biol. 2006;290:470–481. doi: 10.1016/j.ydbio.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Takke C, Campos-Ortega JA. her1, a zebrafish pair rule like gene, acts downstream of notch signalling to control somite development. Development. 1999;126:3005–3014. doi: 10.1242/dev.126.13.3005. [DOI] [PubMed] [Google Scholar]
- Sieger D, Tautz D, Gajewski M. her11 is involved in the somitogenesis clock in zebrafish. Dev Genes Evol. 2004;214:393–406. doi: 10.1007/s00427-004-0427-z. [DOI] [PubMed] [Google Scholar]
- Schoppmeier M, Damen WGM. Suppressor of Hairless and Presenilin phenotypes imply involvement of canonical Notch-signalling in segmentation of the spider Cupiennius salei. Dev Biol. 2005;280:211–224. doi: 10.1016/j.ydbio.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Irvine KD. Notch-mediated segmentation and growth control of the Drosophila leg. Dev Biol. 1999;210:339–350. doi: 10.1006/dbio.1999.9273. [DOI] [PubMed] [Google Scholar]
- Dove H, Stollewerk A. Comparative analysis of neurogenesis in the myriapod Glomeris marginata (Diplopoda) suggests more similarities to chelicerates than to insects. Development. 2003;130:2161–2171. doi: 10.1242/dev.00442. [DOI] [PubMed] [Google Scholar]
- Kadner D, Stollewerk A. Neurogenesis in the chilopod Lithobius forficatus suggests more similarities to chelicerates than to insects. Dev Genes Evol. 2004;214:367–379. doi: 10.1007/s00427-004-0419-z. [DOI] [PubMed] [Google Scholar]
- Damen WGM. Evolutionary conservation and divergence of the segmentation process in arthropods. Dev Dyn. 2007;236:1379–1391. doi: 10.1002/dvdy.21157. [DOI] [PubMed] [Google Scholar]
- Anderson D. Embryology and Phylogeny in Annelids and Arthropods. Oxford: Pergamon Press; 1973. [Google Scholar]
- Choe CP, Brown SJ. Genetic regulation of engrailed and wingless in Tribolium segmentation and the evolution of pair-rule segmentation. Dev Biol. 2009;325:482–491. doi: 10.1016/j.ydbio.2008.10.037. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DO, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, Sörensen MV, Haddock SHD, Schmidt-Rhaesa A, Okusu A, Kristensen RM, Wheeler WC, Martindale MQ, Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Budd GE. The place of phylogeny and cladistics in Evo-Devo research. Int J Dev Biol. 2003;47:479–490. [PubMed] [Google Scholar]
- Budd GE, Telford MJ. The origin and evolution of arthropods. Nature. 2009;457:812–817. doi: 10.1038/nature07890. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Campbell L, Brinkmann H, Edgecombe GD, Longhorn SJ, Peterson KJ, Pisani D, Phillipe H, Telford MJ. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. Proc Biol Sci. 2011;278:298–306. doi: 10.1098/rspb.2010.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Additional aspects of h1 and odd expression, expression of h2 and h3 and single-colour expression detection of h1+en and odd+en. (A, B) Double single-colour staining of h1+en. The black arrows point to expression of en; the white arrows point to expression of h1 anteriorly abutting the expression of en. (C) Expression of h1 in a stage 5 embryo (cf. similar expression of h2 in (E)). White arrow points to expression in the ventral nervous system. Asterisk marks expression in the brain. (D, E) Expression of h2. White arrow and asterisk in (E) as in (C). (F-H) Expression of h3. Arrows in (F) point to expression in the brain and the ventral nervous system. Arrowhead points to expression in the SAZ. Asterisk marks expression in the labrum. (G) Same embryo as in (F) but slightly rotated to show a rear aspect and the posterior expression in the SAZ (arrowhead). (H) De novo expression in lateral tissue (arrow) and in the appendages (arrowhead). (H') Same embryo as in (H); DAPI stained; arrow and arrowhead point to same position as in (H). (I) Double single-colour staining of odd+en. (I') Same embryo as in (I); DAPI stained. (J) Close-up on dorsal tissue of the embryo shown in (I/I'). Black arrow points to expression of en. White arrow points to expression of odd. (K) Dorsolateral view of a stage 5 embryo stained for h1. Dorsal segmental expression is connected by weak expression (asterisks) in the dorsal extraembryonic tissue (e). (L) Dorsal view. Expression of h2 in the (e). Asterisks as in (K). (M) Dorsal view. Expression of odd in the (e). Asterisks as in (K). Abbreviations: (e), dorsal extraembryonic tissue; en, engrailed, h, hairy; odd, odd-skipped; st, stage.