Abstract
Iron–sulfur (Fe–S) clusters are cofactors found in many proteins that have important redox, catalytic or regulatory functions. In mammalian cells, almost all known Fe–S proteins are found in the mitochondria, but at least one is found in the cytosol. Here we report cloning of the human homologs to IscU and NifU, iron-binding proteins that play a critical role in Fe–S cluster assembly in bacteria. In human cells, alternative splicing of a common pre-mRNA results in synthesis of two proteins that differ at the N-terminus and localize either to the cytosol (IscU1) or to the mitochondria (IscU2). Biochemical analyses demonstrate that IscU proteins specifically associate with IscS, a cysteine desulfurase that is proposed to sequester inorganic sulfur for Fe–S cluster assembly. Protein complexes containing IscU and IscS can be found in the mitochondria as well as in the cytosol, implying that Fe–S cluster assembly takes place in multiple subcellular compartments in mammalian cells. The possible roles of the IscU proteins in mammalian cells and the potential implications of compartmentalization of Fe–S cluster assembly are discussed.
Keywords: human/iron/IRP1/iscU/localization
Introduction
Iron–sulfur (Fe–S) clusters are abundant and pervasive cofactors that are essential for a wide variety of processes, including facilitation of electron transfer processes in oxidative phosphorylation, catalysis of enzymatic reactions in aconitase and dehydratases, and maintenance of structural integrity in the DNA repair enzyme endonuclease III (Beinert and Holm, 1997). In addition, Fe–S clusters are critical in enabling cells to sense intracellular iron and/or oxidant levels, as in the case of iron regulatory protein 1 (IRP1), a sensor of intracellular iron levels, and the bacterial proteins SoxR and FNR, sensors of intracellular oxygen species (Rouault and Klausner, 1996; Beinert and Kiley, 1999). Over the last few decades, much has been learned about the structure and function of Fe–S clusters, but, until recently, little was known about the in vivo cluster assembly process.
Enzymes involved in Fe–S cluster assembly were first identified within the nif (nitrogen fixation) operon, which encodes proteins critical in the biosynthesis of functional nitrogenase in the nitrogen-fixing bacterium Azotobacter vinelandii. Genetic studies in these bacteria showed that disruption of two of the open reading frames (ORFs) in the nif operon, nifS or nifU, resulted in loss of nitrogenase activity (Jacobson et al., 1989). Although nitrogenase polypeptides were synthesized, the Fe–S clusters essential for activity were absent, implicating nifS and nifU in the process of Fe–S cluster assembly. More recently, homologs of nifS and nifU have been identified in non-nitrogen-fixing organisms, including Escherichia coli, yeast and higher eukaryotes, leading to the proposal that these genes are involved in the general process of Fe–S cluster assembly (Zheng et al., 1998). Biochemical studies have demonstrated that NifS and its homologs mobilize inorganic sulfide (Zheng et al., 1993), and NifU homologs bind [2Fe–2S] clusters (Fu et al., 1994). Hence, they have been designated as isc genes to indicate their proposed roles in iron--sulfur cluster assembly (Zheng et al., 1998).
Within the last 2 years, several yeast proteins with high homology to bacterial nifU have been identified and revealed to have unexpected importance in mitochondrial iron homeostasis and function. Isu1 and Isu2 are two distinct genes in Saccharomyces cerevisiae that encode protein homologs of the N-terminal domain of bacterial NifU (Garland et al., 1999), whereas Nfu1 is an S.cerevisiae homolog of the C-terminal domain of bacterial NifU (Schilke et al., 1999). Genetic studies have demonstrated that these gene products are essential in S.cerevisiae, as yeast strains harboring deletions in both isu1 and isu2 loci (Δisu1Δisu2) are inviable. Activities of several mitochondrial Fe–S proteins decrease in Δisu1, Δisu2 or Δisu1Δnfu1 mutant strains, consistent with the proposal that these NifU homologs function in Fe–S cluster assembly in yeast mitochondria (Garland et al., 1999; Schilke et al., 1999). In strains that express an activated allele of the iron-sensing transcription factor, Aft1, expression of isu1 and isu2 increases, indicating that these genes are regulated by the iron status of the cell (Garland et al., 1999). Significant mitochondrial iron accumulation occurs in Δisu1, Δisu2 or Δisu1Δnfu1 strains (Garland et al., 1999; Schilke et al., 1999), implying that NifU homologs play an important role not only in Fe–S cluster synthesis, but also in maintenance of normal mitochondrial iron homeostasis. While extensive studies have revealed much about iron uptake, storage and regulation in the yeast cytosol (Askwith and Kaplan, 1998), much remains to be learned about mitochondrial iron homeostasis. Elucidation of the function of isc genes in mitochondria should lead to better understanding of iron homeostasis in mitochondria.
The complexity and importance of mitochondrial iron metabolism are underscored by recent studies that indicate a link between mitochondrial iron homeostasis and the neurodegenerative disease Friedreich’s ataxia. Frataxin, the disease gene in Friedreich’s ataxia (Campuzano et al., 1996), is a mitochondrial protein required for maintenance of normal mitochondrial iron levels (Babcock et al., 1997; Radisky et al., 1999). In the absence of functional frataxin, iron accumulates in mitochondria, causing oxidative damage to mitochondrial DNA and various mitochondrial Fe–S proteins.
In addition to understanding Fe–S protein biosynthesis in the mitochondria, another important aim in pursuing studies of Fe–S cluster assembly is to understand the molecular basis for regulation of the mammalian cytosolic iron-sensing protein IRP1. IRP1 and IRP2 are proteins that regulate expression of numerous iron metabolism proteins, including ferritin and transferrin receptor, to ensure that uptake and sequestration of iron are appropriate to cellular needs. In iron-replete cells, IRP1 contains a [4Fe–4S] cluster and is a functional aconitase, interconverting citrate and isocitrate in the cytosol. In cells that are depleted of iron, IRP1 lacks the Fe–S cluster, and the apoprotein binds RNA stem–loop sequences known as iron-responsive elements (IREs) that are found in transcripts of several iron metabolism proteins. The position of the IRE in the transcript determines whether IRP binding will inhibit translation of transcripts such as ferritin, or mRNA turnover in transcripts such as transferrin receptor (reviewed in Hentze and Kuhn, 1996; Rouault and Klausner, 1997). Since the Fe–S cluster is the key to IRP1 function, it is of great interest to understand how Fe–S clusters are assembled.
To understand fully the regulation of the cytosolic protein IRP1, and to understand iron metabolism in mitochondria, we have cloned and characterized the human homolog of NifU. Our results indicate that alternative splice products encode distinct isoforms that are targeted to the mitochondria, the cytosol and the nucleus. Furthermore, human IscU proteins are specifically associated with distinct forms of human IscS proteins in different subcellular compartments, indicating that Fe–S cluster assembly is compartmentalized in mammalian cells.
Results
Cloning of human iscU homolog
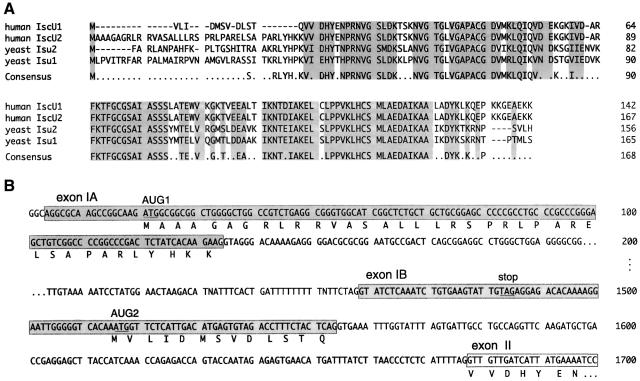
The human expressed sequence tag (EST) database was searched, and analysis of multiple ESTs revealed that there are two ORFs that show high homology to yeast Isu1 and Isu2 (Figure 1A). The complete cDNA sequences were derived from sequencing of EST clones and 5′ RACE (rapid amplification of cDNA ends) products. Sequencing analysis revealed that these two cDNAs are identical in most of the coding region and the 3′ UTR, differing only in the 5′ UTRs and the beginning of the coding sequences. IscU1 mRNA was predicted to encode a protein with a 13 amino acid N-terminal sequence unique to IscU1. Conversely, iscU2 was predicted to encode a protein with a 39 amino acid N-terminal sequence unique to IscU2.
Fig. 1. Molecular cloning and sequence analysis of human iscU. (A) Predicted amino acid sequences of two human IscU isoforms compared with yeast Isu1 and Isu2. The cDNA sequences of human IscU were obtained by EST database analysis and 5′ RACE experiments. Sequence analysis of 5′ RACE products, EST and genome databases has revealed polymorphisms at codon 7 (G or F) and codon 12 (V or A) in the N-terminus of IscU2. Only one variant is shown here. (B) Genomic sequence of the 5′ end of human iscU obtained by PCR and EST database analysis.
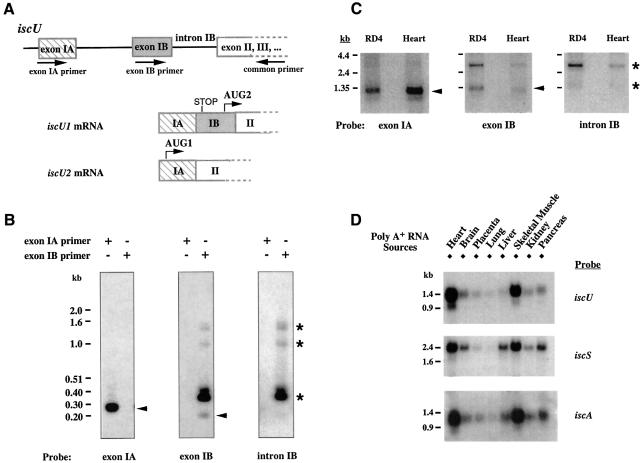
A genomic fragment that contained the 5′ end of iscU was cloned, and mapping of the intron–exon boundaries suggested that alternative splicing could result in generation of the two different isoforms. As shown in Figure 1B, exon IA contains the N-terminal sequence information included in the IscU2 mRNA sequence, whereas exon IB contains the N-terminal sequence information included in the IscU1 mRNA sequence. To assess whether alternative transcription initiation sites were used, the 5′ cDNA ends of iscU were synthesized from human heart (or liver) poly(A)+ RNA and amplified by nested PCR. Sequencing analysis indicated that both the iscU1 and iscU2 mRNAs shared the same transcription initiation site. They differed in that iscU2 mRNA contained exon IA but not exon IB (Figure 2A), whereas iscU1 mRNA contained both exon IA and exon IB. However, in the iscU1 transcript, initiation at AUG1 would result in synthesis of a short upstream ORF due to termination at an in-frame stop codon present in exon IB. Synthesis of functional IscU1 protein would instead initiate at AUG2. Taken together, these 5′ RACE experiments indicated that IscU1 and IscU2 share a common pre-mRNA and are encoded by alternatively spliced transcripts.
Fig. 2. Alternative splicing of iscU pre-mRNA results in two isoforms with different predicted N-terminal sequences. (A) Schematic representation of iscU1 and iscU2 5′ cDNA ends. Nested PCR of a 5′ RACE fragment was carried out using a cap-binding primer, in combination with iscU gene-specific primers. Sequence analysis indicated that iscU1 and iscU2 are alternative splice products. (B) Nested PCR of a 5′ RACE fragment was carried out using a 3′ primer within exon III in combination with 5′ primers specific to either exon IA or exon IB. Southern blot analysis was carried out using exon IA, exon IB or intron IB probe. Arrowheads denote bands of the appropriate sizes as expected from iscU1 and iscU2 mRNA sequences. Asterisks denote bands containing intron sequence and are derived from splicing intermediates. (C) Northern blot analysis of poly(A)+ RNA from RD4 cells and human heart (Clontech) was carried out using exon IA, exon IB or intron IB probe. Arrowheads denote bands of the appropriate sizes as expected from iscU1 and iscU2 mRNA sequences. Asterisks denote splicing intermediates. (D) Human multiple tissue northern blotting (Clontech) revealed that iscU, iscS and iscA are all expressed predominantly in heart and skeletal muscle.
A separate set of nested PCR experiments also confirmed alternative splicing of human iscU. As shown in Figure 2B, nested PCR of the 5′ RACE products using a primer within exon IA and a primer within exon III generated a 240 bp product that contained exon IA but not exon IB, as expected from the IscU2 mRNA sequence. PCR using a primer within exon IB and a primer within exon III generated the 200 bp product as predicted from the mRNA sequence for IscU1 (Figure 2B). In addition, this reaction also amplified several fragments of higher molecular weight (Figure 2B, middle panel). Southern hybridization using intron IB as a probe indicated that these bands represented partially spliced intermediates (Figure 2B, right panel), the sizes of which are consistent with predicted splicing intermediates based on genomic DNA sequence. Northern blot analyses performed with probes specific to each 5′ end indicated that endogenous iscU2 mRNA is ∼1.3 kb and iscU1 mRNA is ∼1.4 kb (Figure 2C), sizes that are consistent with the cDNA data. Although the sizes of these mRNAs are similar, the size difference is discernible in northern blots (Figure 2C). Furthermore, two lines of evidence indicate that these two bands represent two different IscU transcripts. First, experiments shown in Figure 2B indicate that the exon IA and exon IB probes are very specific and that there are no discernible cross-reactions. Secondly, the 1.3 kb band observed using exon IA as a probe is clearly stronger in human heart mRNA than in RD4 mRNA, whereas the 1.4 kb band observed using exon IB as a probe is weaker in human heart mRNA than in RD4 mRNA. These results provide additional evidence that the two probes are hybridizing with different transcripts. In a human multiple tissue northern blot (Clontech) probed with sequences common to both cDNAs, the ∼1.35 kb band detected predominantly in heart and skeletal muscle (Figure 2D) is likely to arise from overlap of signals from the two cDNAs.
In summary, the iscU1 cDNA comprises 1090 bp and contains an ORF encoding a protein of 142 amino acids with a predicted molecular mass of 15.4 kDa. The iscU2 cDNA comprises 990 bp and contains an ORF encoding a protein of 168 amino acids with a predicted molecular mass of 17.9 kDa.
Transiently expressed IscU1 and IscU2 are targeted to different subcellular compartments
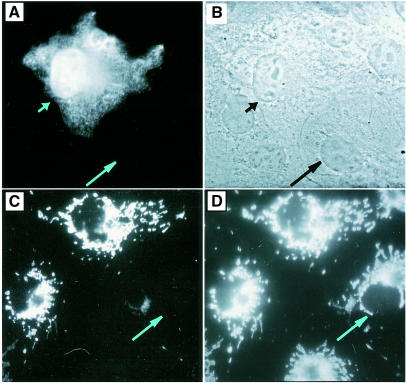
Analysis of the two different N-terminal sequences of human IscU1 and IscU2 suggested that these two proteins might be targeted to different subcellular locations. Sorting programs identified the N-terminal sequence of IscU2 as a putative mitochondrial targeting sequence, whereas IscU1 was predicted to be a cytosolic protein (Nakai and Kanehisa, 1992). The expression and localization of IscU1 and IscU2 were examined by immunofluorescence microscopy in COS cells that were transfected with expression constructs containing the two different ORFs (Figure 3). In cells transfected with the IscU1 construct, staining was detected in the cytosol and the nucleus. In contrast, the IscU2 proteins were detected in the mitochondria, as judged by co-localization with the mitochondrial marker rhodamine 123.
Fig. 3. IscU1 is localized to cytosol and nucleus, whereas IscU2 is localized to mitochondria. COS cells transfected with HA-tagged IscU1 were stained with monoclonal anti-HA antibodies and analyzed by immunofluorescence microscopy (A) and phase contrast microscopy (B). (C) COS cells transfected with myc-tagged IscU2 were stained with monoclonal anti-myc antibodies and analyzed by immunofluorescence microscopy. (D) Mitochondrial staining by the mitochondrial marker rhodamine 123 of transfected COS cells shown in (C). Short arrows denote transfected cells, whereas long arrows denote untransfected cells.
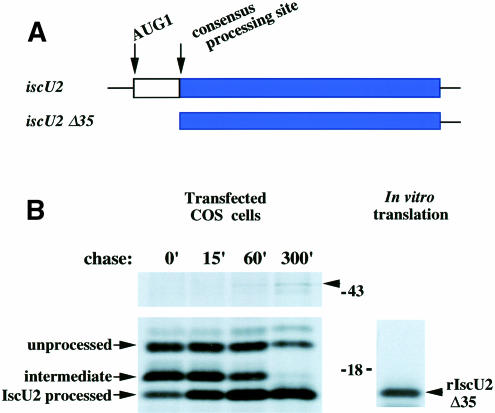
Transient expression of IscU2 in COS cells also revealed that the IscU2 translation product underwent a rapid, two-step processing into a shorter cleavage product of ∼15 kDa (Figure 4B), consistent with proteolytic removal of the N-terminal targeting sequence upon import into the mitochondrial matrix (Pfanner et al., 1997). Comparison with the consensus sequence for mitochondrial matrix processing sites suggested that proteolytic cleavages would occur initially at E27 and subsequently at Y35. To verify this hypothesis, we analyzed the in vitro translation product from a construct in which the N-terminal sequence up to Y35 was deleted. The IscU2Δ35 co-migrated with the fully processed IscU2 in transfected COS cells, thus supporting the conclusion that IscU2 is cleaved at or near Y35 upon import into mitochondria (Figure 4B). In addition, the in vitro translation product of IscU2Δ27, a construct from which the first 27 amino acids of IscU2 were deleted, co-migrated with the processing intermediate identified in transfected COS cells in Figure 4 (data not shown).
Fig. 4. IscU2 protein is rapidly processed and becomes associated with an ∼47 kDa protein upon maturation. (A) Schematic representation of the full-length IscU2 construct and IscU2Δ35, a construct that lacks sequence 5′ of the putative processing site. (B) COS cells transfected with a construct encoding a full-length myc-tagged IscU2 were pulse labeled with [35S]methionine for 3 min, and chased for 0, 15, 60 and 300 min, respectively. Immunoprecipitation with a monoclonal anti-myc antibody indicated a two-step processing of the IscU2 protein.In vitro translation of myc-tagged IscU2Δ35 generated a protein that co-migrated with the processed IscU2 in transfected COS cells. At the 60 and 300 min time points, a 47 kDa band (arrowhead) was co-immunoprecipitated with IscU.
Subcellular localization of endogenous IscU proteins
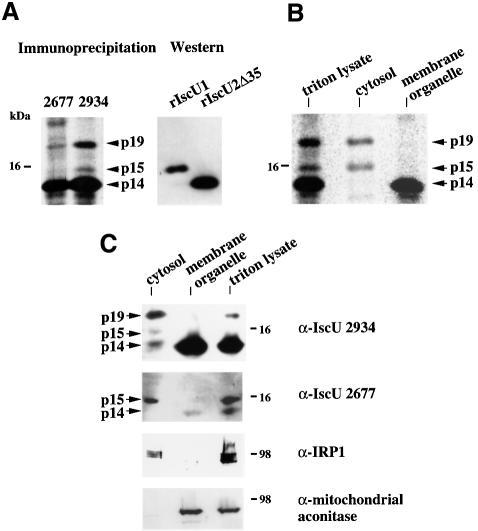
To analyze the endogenous IscU proteins, antibodies were raised against two different peptides within the region common to both isoforms. Metabolic labeling and immunoprecipitation experiments indicated that both antibodies 2677 and 2934 immunoprecipitated a 14 kDa protein (p14) from RD4 cell lysates, strongly suggesting that this is an endogenous IscU protein (Figure 5A). In addition, a weaker p15 protein was also identified in immunoprecipitations using one of the antibodies (α-IscU 2934). Comparison with recombinant IscU proteins expressed in E.coli indicated that p14 co-migrated with IscU2Δ35, whereas p15 co-migrated with IscU1 (Figure 5A). These results suggested that p14 is the processed form of mitochondrial IscU2, whereas p15 is a form of IscU1.
Fig. 5. Endogenous IscU proteins are present in the cytosol and in the mitochondria of RD4 cells. (A) Immunoprecipitation experiments revealed multiple forms of endogenous IscU proteins in RD4 cells. Immunoprecipitations were carried out using affinity-purified anti-IscU peptide antibodies 2934 and 2677. Western blot analyses of recombinant IscU1 (rIscU1) and recombinant IscU2Δ35 (rIscU2Δ35) are shown for comparison. (B) Immunoprecipitation of endogenous IscU proteins in different subcellular fractions of RD4 cell lysates using anti-IscU 2934. (C) Western blot analyses of endogenous IscU proteins in different subcellular fractions of RD4 cell lysates. Western blot analyses of mitochondrial aconitase and cytosolic IRP1 are shown for comparison.
The subcellular localizations of endogenous IscU proteins were further assessed by fractionation experiments. Immunoprecipitation experiments (Figure 5B) and western blot analyses (Figure 5C) using two different antibodies showed that p14 is the sole form of IscU in the membrane organelle fraction, whereas p15 is detected in the cytosolic fraction. Taken together, these results support the hypothesis that IscU2 is a mitochondrial form of IscU, whereas IscU1 is a cytosolic isoform.
In addition to p14 and p15, experiments shown in Figure 5 also identify a 19 kDa protein (p19) as another form of cytosolic IscU. As shown in Figure 5A, p19 was immunoprecipitated with two α-IscU antibodies that bind to different regions of the antigen, consistent with the possibility that p19 is also an endogenous IscU. Subcellular fractionation experiments (Figure 5B) and western blot analysis (Figure 5C) indicated that p19 is present in the cytosolic fraction, suggesting that p19 represents either a cytosolic precursor of mitochondrial IscU2 or a modified form of cytosolic IscU1. It is not yet clear why a form of cytosolic IscU1 migrates as a 19 kDa band. Unusual behavior on SDS–PAGE has previously been reported for another putative Fe–S cluster assembly protein in A.vinelandii: ferredoxin IV (FdIV) (Jung et al., 1999a). Although mass spectrometry data indicated that native FdIV apoprotein had a mass of 12.43 kDa and recombinant FdIV apoprotein had a mass of 12.48 kDa, SDS–PAGE indicated that native FdIV apoprotein migrated as a 25 kDa band while recombinant FdIV apoprotein migrated as a 14 kDa band (Jung et al., 1999a). Likewise, A.vinelandii ferredoxin I (FdI) has a mass of 12 kDa, but migrated as a 24 kDa band (Jung et al., 1999b). These results suggest that a small modification of low molecular weight Fe–S proteins like IscU, FdI or FdIV can result in anomalous behavior on SDS–PAGE.
Co-immunoprecipitation of IscU with IscS inRD4 cells
Since previous studies have shown that different forms of human IscS proteins localize to the mitochondria, the cytosol and the nucleus (Land and Rouault, 1998), we addressed the possibility that there are distinct Fe–S cluster assembly complexes in different subcellular compartments. In these ‘immunoprecipitation–recapture’ experiments, IscU and its associated proteins were first immunoprecipitated with α-IscU from RD4 Triton lysate. Immunoprecipitates were denatured by treatment with SDS–dithiothreitol (DTT) and heat, and a second immunoprecipitation (recapture) was carried out with α-IscS. Only the 47 kDa mitochondrial form of IscS was detected in association with IscU in RD4 lysate (Figure 6A), consistent with the observation that there is much more precipitable mitochondrial IscU than cytosolic IscU (Figure 5).
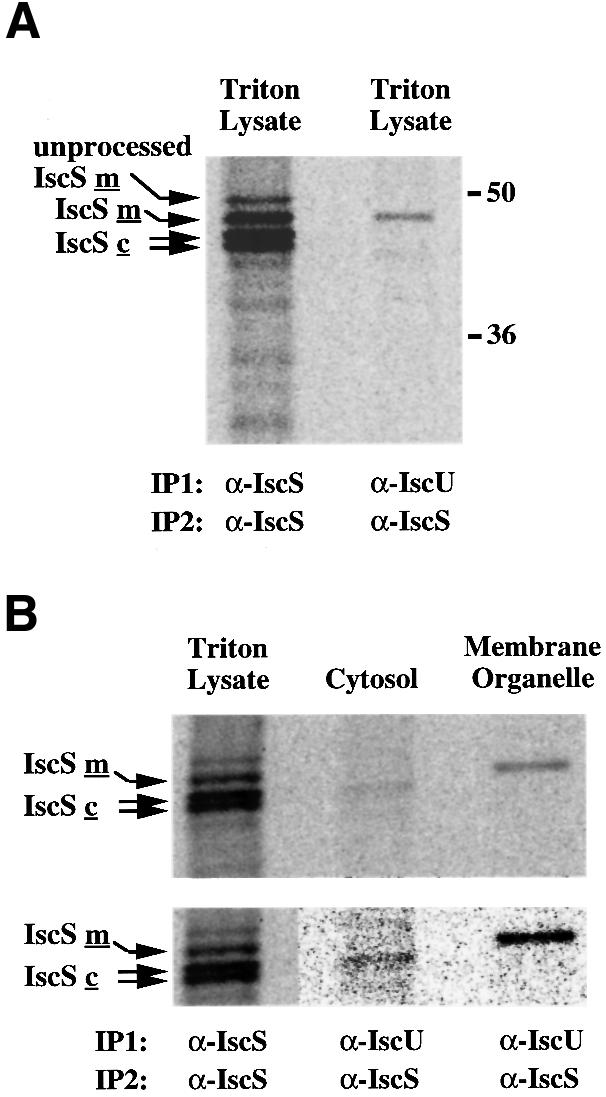
Fig. 6. Co-immunoprecipitation of IscU and IscS proteins in the cytosol and in the mitochondria of RD4 cells. (A) Coimmuno-precipitation of mitochondrial IscS with IscU in RD4 Triton lysates. The mitochondrial IscU–IscS complex was first immunoprecipitated from [35S]methionine-labeled RD4 cell lysate using either α-IscS or α-IscU 2934. Following dissociation of the immunoprecipitates from the antibodies with SDS–DTT and heating, a second immuno precipitation (recapture) using α-IscS was carried out. (B) Cytosolic IscU1 co-immunoprecipitates with one of the cytosolic IscS proteins (IscSc), and mitochondrial IscU2 co-immunoprecipitates with mitochondrial IscS (IscSm) in RD4 cell lysates (upper panel). The lower panel shows the lanes containing the cytosolic and membrane organelle fractions at high contrast to allow better visualization and alignment of the cytosolic IscS band.
To enhance the probability of detecting specific cytosolic complexes, co-immunoprecipitations were performed on fractionated lysates. IscU and its associated proteins were first immunoprecipitated with α-IscU from the cytosolic or the membrane organelle fraction of RD4 cells. A 45 kDa protein was recaptured by IscS antibodies from the cytosolic fraction, whereas a 47 kDa band was recaptured in the membrane organelle fraction (Figure 6B). These IscS bands co-migrated with bands that have been previously shown to represent cytosolic and mitochondrial forms of IscS, respectively (Land and Rouault, 1998), clearly indicating that separate Fe–S cluster assembly complexes are present in the cytosol and in the mitochondria.
Discussion
Iron–sulfur cluster assembly in mammalian cells
In the last few years, elegant work in bacteria has revealed that the assembly of Fe–S clusters is mediated by a number of proteins, including enzymes that generate inorganic sulfur, enzymes that incorporate iron into rudimentary clusters and chaperone molecules that direct the unfolding of the recipient proteins (Zheng et al., 1993, 1998; Agar et al., 2000; Yuvaniyama et al., 2000). The tasks of assembly and insertion of clusters into appropriate recipient proteins are complex in mammalian cells, in part because proteins that require Fe–S clusters for function are found not only in the mitochondria, but also in the cytosol. In addition, mammalian cells may have mechanisms that allow discrimination amongst potential recipients of Fe–S clusters. Mammalian zinc finger proteins are known to acquire Fe–S clusters sometimes when overexpressed in bacteria (Li et al., 1991; Archer et al., 1994), but Fe–S clusters are not inappropriately inserted into zinc finger proteins in mammalian cells.
To gain understanding of the process of Fe–S cluster assembly in mammalian cells, we have undertaken systematic cloning of mammalian homologs of bacterial isc proteins. Previously, we have cloned and characterized the human homolog of bacterial NifS and demonstrated that an unusual type of translational regulation involving alternative utilization of in-frame AUGs allows synthesis of IscS proteins that are targeted either to mitochondria or to cytosol and nucleus. These results suggested that separate Fe–S cluster assembly complexes could exist in different compartments of the cell (Land and Rouault, 1998).
Here, we report cloning of a second human Fe–S cluster assembly enzyme, IscU. Preliminary biochemical studies showed that human IscU protein binds iron and can facilitate Fe–S cluster assembly in IRP1 (W.-H.Tong and T.Rouault, unpublished data), indicating that human IscU proteins are functional homologs of the bacterial and yeast IscU proteins. Analyses of genomic DNA, transcripts and translation products indicate that alternative splicing of iscU leads to expression of two iscU proteins that contain different subcellular targeting information. In accordance with the results on human IscS, the present study demonstrated that human IscU proteins are present in the cytosol as well as in the mitochondria. Furthermore, the co-immunoprecipitation experiments clearly indicate that mitochondrial IscU2 associates with the mitochondrial form of IscS, whereas IscU1 in the cytosol associates with the cytosolic form of IscS. These data therefore imply that separate Fe–S cluster assembly complexes are present in the mitochondria and in the cytosol in higher eukaryotes.
It is interesting to note that in the case of both human iscS and iscU, the mechanism of generating multiple isoforms from a single gene ensures that the proteins differ only at the N-terminus, while the functional portions of the proteins remain identical. In the case of the IscS protein, generation of multiple isoforms is mediated by differential utilization of in-frame AUGs of a single transcript (Land and Rouault, 1998), whereas in the case of IscU, generation of the multiple isoforms is achieved by alternative splicing. The use of a single gene to generate distinct isoforms for different subcellular compartments may enable cells to synchronize Fe–S cluster assembly in different compartments in the cell. Previous studies have shown that lowering the pH of the culture media results in an increase in synthesis of the mitochondrial IscS, accompanied by a proportional decrease in the cytosolic/nuclear forms (Land and Rouault, 1998). These results suggest that cells may vary the ratio of mitochondrial and cytosolic/nuclear IscS in accordance with changes in metabolic status. It will be interesting to see how expression of the two IscU isoforms is affected by variation in pH and iron availability.
Although distinct IscS and IscU complexes have been identified in the cytosol and in the mitochondria in human cells, IscU and IscS in yeast have previously been detected only in the mitochondria (Garland et al., 1999; Kispal et al., 1999; Li et al., 1999; Schilke et al., 1999). It has been proposed that mitochondria are the only site of Fe–S synthesis in yeast and that fully formed Fe–S clusters are exported from mitochondrial matrix for use in the cytosol (Kispal et al., 1999). However, recent studies of IscA, another member of the isc operon in S.cerevisiae, have shown that IscA1 is in the mitochondrial matrix, whereas IscA2 is in the intermembrane space (Jensen and Culotta, 2000), suggesting that yeast might also require isc gene products both inside and outside of the mitochondrial matrix.
Likewise, it is interesting to note that a significant amount of IscU1 localizes to the nucleus when transiently expressed in COS-7 cells (Figure 3A). Preliminary western blot analysis revealed that endogenous IscU1 (p15) is also present in the nuclear fraction of RD4 cells (data not shown), suggesting that Fe–S cluster assembly also takes place in the nucleus. Although the sequence of IscU1 does not contain a recognizable nuclear localization signal, previous immunofluoresence and western blot analyses have shown that human IscS, which contains a nuclear localization motif, is also present in the nucleus (Land and Rouault, 1998). It is therefore tempting to speculate that IscU1 is targeted to the nucleus by association with IscS. The potential requirement for Fe–S cluster assembly machinery in the nucleus is underscored by recent identification of nuclear proteins that contain Fe–S binding motifs. Ntg2p (also known as Scr2) in S.cerevisiae is a functional homolog of an E.coli Fe–S protein, endonuclease III (You et al., 1998). Spectroscopic studies of purified Ntg2p showed that Ntg2p also possesses an Fe–S cluster (You et al., 1998) and fluorescence microscopy revealed that an Ntg2p–green fluorescent protein (GFP) fusion protein localizes to the nucleus (You et al., 1999). Furthermore, a human homolog of Ntg2p, hNth1, has been shown to localize to both the mitochondria and the nucleus when transiently expressed in COS-7 or HeLa cells (Takao et al., 1998). An objective of future work will be to determine whether the Fe–S cluster assembly complex is also present in the nucleus and to address the possible biological significance of nuclear Fe–S cluster assembly.
Intracellular iron homeostasis
The fact that iron accumulates in mitochondria in yeast strains with IscU mutations (Garland et al., 1999; Schilke et al., 1999) raises the question of what additional roles these gene products may play in mitochondrial and cytosolic iron homeostasis. The mechanisms by which iron is transported to target apoproteins in different subcellular compartments are unknown. Based on the recent advances of intracellular copper transport, it is possible that similar mechanisms exist for the intracellular transport of iron. In yeast, intracellular copper trafficking involves the action of a family of low molecular weight, soluble, copper-binding proteins referred to as copper chaperones. These proteins bind copper ions and transport them to various cellular destinations, including cytochrome oxidase in the mitochondria (Glerum et al., 1996), superoxide dismutase SOD1 in the cytosol (Culotta et al., 1997) and copper transporter Ccc2p in the secretory pathway (Lin et al., 1997). It is conceivable that intracellular trafficking of other biologically active trace metals such as iron is mediated through the action of another family of metal chaperones. In this respect, IscU may be part of a network of iron-trafficking genes required for delivery of Fe to IRP1 in the cytosol and to various Fe–S proteins in the mitochondria. As copper chaperone proteins are thought to protect cells against copper toxicity by sequestering excess free copper (Pufahl et al., 1997), ‘iron chaperones’ could potentially protect cells against iron toxicity. In the cytosol of mammalian cells, protection from the harmful effects of free iron is in large measure accomplished by sequestration of excess iron by ferritin. In mitochondria, however, iron-sequestration proteins have not been identified thus far. Iron-binding proteins such as mitochondrial IscU may possibly serve to sequester free iron in the mitochondria and thus protect mitochondrial components against iron toxicity. Alternatively, mitochondrial IscU may be important in delivering excess free iron to unidentified iron-sequestration proteins in the mitochondrial matrix.
The biochemical complexity of Fe–S cluster assembly in vivo is reflected in the number and organization of genes involved, as demonstrated by studies in A.vinelandii and S.cerevisiae. Dean and co-workers identified an iscSUA–hscBA–fdx gene cluster in both A.vinelandii and E.coli (Zheng et al., 1998). The co-expression of iscSUA genes with chaperone genes hscBA suggested the presence of a multiprotein complex for assembly and insertion of Fe–S clusters. The co-immunoprecipitation of human IscU and IscS proteins reported here provides further support for this idea. The demonstration of a stable IscU–IscS protein complex also raises the possibility of identifying other associated proteins by similar methods. In particular, a human IscA homolog has also been identified based on sequence homology. Multiple tissue northern blot analysis indicated remarkably similar tissue distribution between human iscU, iscS and iscA (Figure 2C), suggesting a common role for these genes. Identification of other proteins in the complex will allow characterization of Fe–S cluster assembly in vivo and further dissection of the factors that regulate cellular and mitochondrial iron homeostasis.
Materials and methods
Cloning and sequence analysis of human iscU homolog
Clones containing sequences similar to bacterial nifU were obtained from the human EST database and analyzed by sequencing. To complete the cDNA sequences, 5′ RACE (Clontech SMART PCR cDNA synthesis) were carried out using human heart (or liver) poly(A)+ RNA. The 5′ RACE amplification products were cloned into Bluescript vector pSK+ (Stratagene) or pNoTA/T7 vector (Eppendorf-5 Prime) and sequenced. Mapping of the intron–exon boundaries was carried out on the genomic sequence obtained by PCR (Advantage cDNA PCR).
Southern and northern blot analysis
RACE amplification products were separated on 1.5% agarose– formaldehyde gels and transferred to GeneScreen (Dupont NEN). 32P-labeled DNA probes were prepared using the Megaprime DNA labeling system (Amersham) and hybridizations were carried out in Hybrisol (Oncor). For northern blot analysis, poly(A)+ RNA from human heart (Clontech) and RD4 cells was separated on 1.5% agarose– formaldehyde gels, transferred to GeneScreen and hybridized to human iscU probes.
Antibody production
Synthetic multiple antigenic peptide (MAP) corresponding to amino acids 74–91 and 110–124 of human IscU2 was synthesized by Bio-Synthesis, Inc. Rabbit polyclonal antibodies 2934 and 2677 were generated by Covance Laboratories Inc. (Vienna, VA). Antibody to human IscS was generated as described previously (Land and Rouault, 1998). Immunoaffinity purification of antisera was carried out using peptides immobilized on CNBr-activated Sepharose 4B (Pharmacia).
Cell culture, expression vectors and DNA transfection
RD4 and COS cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 9% fetal calf serum, 100 U/ml penicillin, 100 U/ml streptomycin and 2 mM l-glutamine (DMEM-complete medium). Human iscU1 and iscU2 cDNAs were subcloned into pSX vector for COS cell transfections. Constructs encoding IscU1 contain 150 bp 5′ of AUG2 and the complete iscU1 ORF sequence. Constructs encoding IscU2 contain 10 bp 5′ of AUG1 and the complete iscU2 ORF sequence. In some experiments, a myc epitope tag (MEQKLISEEDLN) or an HA tag (CYPYDVPDYASL) was added to the C-termini of the IscU constructs. Transfections were carried out by electroporation of COS cells with 10 µg of DNA (four pulses of 0.5 kV × 99 µs; BTX Electrosquare porator model T820) and incubation for 48 h before immunofluorescence or metabolic labeling experiments.
Immunofluorescence
Transfected COS cells were fixed in 2% paraformaldehyde at room temperature for 20 min, permeabilized with methanol for 2 min, and stained with a monoclonal anti-myc or a monoclonal anti-HA antibody (Babco), followed by fluorescein-conjugated donkey antibodies to mouse IgG. The samples were photographed by immunofluorescence and phase contrast microscopy as described.
Metabolic labeling experiments
Transfected COS cells were incubated at 37°C for 30 min in methionine-free DMEM-complete medium (depletion medium). Pulse labelings were carried out at 37°C in depletion medium supplemented with 2 mCi/ml [35S]methionine (ICN). At the end of the labeling interval, cells were washed once in chase medium (DMEM-complete medium supplemented with 5 mM methionine) at 4°C, then incubated in chase medium at 37°C for the appropriate intervals. At the end of the chase intervals, cells were washed four times with phosphate-buffered saline (PBS) before analysis.
RD4 cells were incubated at 37°C for 30 min in depletion medium, and then labeled at 37°C for 2 h in depletion medium supplemented with 2 mCi/ml [35S]methionine.
Cell lysis and fractionation
For RD4 Triton lysates, cells were washed with ice-cold PBS and lysed in Triton X-100 lysis buffer (Land and Rouault, 1998). For subcellular fractionation, cell pellets were suspended in digitonin lysis buffer [0.08% digitonin, 210 mM mannitol, 70 mM sucrose, 4 mM HEPES pH 7.2, 1 mM AEBSF (ICN), 0.5 µg/ml leupeptin]. The lysates were centrifuged at 600 g for 10 min at 4°C. The supernatants were centrifuged at 7000 g for 9 min at 4°C. The pellets were washed and lysed with Triton X-100 lysis buffer and considered as the membrane organelle fraction. The supernatants were centrifuged at 14 000 g for 10 min at 4°C, and the final supernatants were considered as the cytosolic fraction.
Immunoprecipitation and western blotting
Protein A–Sepharose beads with pre-absorbed antibodies (prepared by tumbling at 4°C for 1 h in lysis buffer) were added to cell lysates or fractions, and the mixture was tumbled at 4°C for 1–2 h. The beads were washed four times with cold lysis buffer, once with PBS and then incubated in elution buffer (1% SDS, 0.1 M Tris–HCl pH 7.4, 10 mM DTT) for 5 min at room temperature, 5 min at 65°C and 5 min at 95°C to elute bound proteins. The eluate was diluted 10-fold with lysis buffer supplemented with 0.1% (w/v) bovine serum albumin and 10 mM iodoacetic acid, and then used for a second immunoprecipitation. Lysates or immunoprecipitates were analyzed on SDS–polyacrylamide gels, followed by autoradiography or phosphoimaging analysis.
For western blot analysis, samples were separated by SDS– polyacrylamide gels and transferred to nitrocellulose. Blots were incubated in antibody diluted in blocking buffer (5% non-fat dry milk, 0.1% Triton X-100, PBS) for 1–2 h at room temperature. Horseradish peroxidase-conjugated secondary antibodies were detected by an ECL system (Pierce).
DDBJ/EMBL/GenBank accession numbers
The DDBJ/EMBL/GenBank accession numbers for IscU1 and IscU2 are AY009127 and AY009128, respectively.
Acknowledgments
Acknowledgements
The authors appreciate help from Tiit Land and Ken Takekoshi for cloning and sequencing work in the early stages of this project and thank their colleagues in the laboratory for helpful discussion and critical reading of the manuscript.
References
- Agar J.N., Zheng,L., Cash,V.L., Dean,D.R. and Johnson,M.K. (2000) Role of IscU protein in iron–sulfur cluster biosynthesis: IscS-mediated assembly of a [Fe2S2] cluster in IscU. J. Am. Chem. Soc., 122, 2136–2137. [Google Scholar]
- Archer V.E., Breton,J., Sanchez-Garcia,I., Osada,H., Forster,A., Thomson,A.J. and Rabbitts,T.H. (1994) Cysteine-rich LIM domains of LIM-homeodomain and LIM-only proteins contain zinc but not iron. Proc. Natl Acad. Sci. USA, 91, 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith C. and Kaplan,J. (1998) Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci., 23, 135–138. [DOI] [PubMed] [Google Scholar]
- Babcock M., de Silva,D., Oaks,R., Davis-Kaplan,S., Jiralerspong,S., Montermini,L., Pandolfo,M. and Kaplan,J. (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science, 276, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Beinert H. and Holm,R.H. (1997) Iron–sulfur clusters: nature’s modular, multipurpose structures. Science, 277, 653–659. [DOI] [PubMed] [Google Scholar]
- Beinert H. and Kiley,P.J. (1999) Fe–S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol., 3, 152–157. [DOI] [PubMed] [Google Scholar]
- Campuzano V. et al. (1996) Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- Culotta V.C., Klomp,L., Strain,J., Casareno,R., Krems,B. and Gitlin,J.D. (1997) The copper chaperone for superoxide dismutase. J. Biol. Chem., 272, 23469–23472. [DOI] [PubMed] [Google Scholar]
- Fu W., Jack,R.F., Morgan,T.V., Dean,D.R. and Johnson,M.K. (1994) nifU gene product from Azotobacter vinelandii is a homodimer that contains two identical [2Fe–2S] clusters. Biochemistry, 33, 13455–13463. [DOI] [PubMed] [Google Scholar]
- Garland S.A., Hoff,K., Vickery,L.E. and Culotta,V.C. (1999) Saccharomyces cerevisiae ISU1 and ISU2: Members of a well-conserved gene family for iron–sulfur cluster assembly. J. Mol. Biol., 294, 897–907. [DOI] [PubMed] [Google Scholar]
- Glerum D.M., Shtanko,A. and Tzagoloff,A. (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem., 271, 14504–14509. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. and Kuhn,L.C. (1996) Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric acid and oxidative stress. Proc. Natl Acad. Sci. USA, 93, 8175–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M.R., Cash,V.L., Weiss,M.C., Laird,N.F., Newton,W.E. and Dean,D.R. (1989) Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Gen. Genet., 219, 49–57. [DOI] [PubMed] [Google Scholar]
- Jensen L.T. and Culotta,V.C. (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol., 20, 3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.S., Gao-Sheridan,H.S., Christiansen,J., Dean,D.R. and Burgess,B.K. (1999a) Purification and biophysical characterization of a new [2Fe–2S] ferredoxin from Azotobacter vinelandii, a putative [Fe–S] cluster assembly/repair protein. J. Biol. Chem., 274, 32402–32410. [DOI] [PubMed] [Google Scholar]
- Jung Y.S., Roberts,V.A., Stout,C.D. and Burgess,B.K. (1999b) Complex formation between Azotobacter vinelandii ferredoxin I and its physiological electron donor NADPH-ferredoxin reductase. J. Biol. Chem., 274, 2978–2987. [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere,P., Prohl,C. and Lill,R. (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J., 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land T. and Rouault,T.A. (1998) Targeting of a human iron–sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell, 2, 807–815. [DOI] [PubMed] [Google Scholar]
- Li J., Kogan,M., Knight,S.A.B., Pain,D. and Dancis,A. (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron–sulfur cluster proteins, cellular iron uptake and iron distribution. J. Biol. Chem., 274, 33025–33034. [DOI] [PubMed] [Google Scholar]
- Li P.M., Reichert,J., Freyd,G., Horvitz,H.R. and Walsh,C.T. (1991) The LIM region of a presumptive Caenorhabditis elegans transcription factor is an iron–sulfur- and zinc-containing metallodomain. Proc. Natl Acad. Sci. USA, 88, 9210–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.J., Pufahl,R.A., Dancis,A., O’Halloran,T.V. and Culotta,V.C. (1997) A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem., 272, 9215–9220. [PubMed] [Google Scholar]
- Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Craig,E.A. and Honlinger,A. (1997) Mitochondrial preprotein translocase. Annu. Rev. Cell Dev. Biol., 13, 25–51. [DOI] [PubMed] [Google Scholar]
- Pufahl R.A., Singer,C.P., Peariso,K.L., Lin,S.-L., Schmidt,P.J., Fahrni,C.J., Culotta,V.C., Penner-Hahn,J.E. and O-Halloran,T.V. (1997) Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science, 278, 853–856. [DOI] [PubMed] [Google Scholar]
- Radisky D.C., Babcock,M.C. and Kaplan,J. (1999) The yeast frataxin homologue mediates mitochondrial iron efflux. Evidence for a mitochondrial iron cycle. J. Biol. Chem., 274, 4497–4499. [DOI] [PubMed] [Google Scholar]
- Rouault T.A. and Klausner,R.D. (1996) Post-translational regulation of genes of iron metabolism in mammalian cells. J. Biol. Inorg. Chem., 1, 494–498. [Google Scholar]
- Rouault T. and Klausner,R. (1997) Regulation of iron metabolism in eukaryotes. Curr. Top. Cell Regul., 35, 1–19. [DOI] [PubMed] [Google Scholar]
- Schilke B., Voisine,C., Beinert,H. and Craig,E. (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 10206–10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M., Aburatani,H., Kobayashi,K. and Yasui,A. (1998) Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res., 26, 2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H.J., Swanson,R.L. and Doetsch,P.W. (1998) Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry, 37, 6033–6040. [DOI] [PubMed] [Google Scholar]
- You H.J. et al. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: Broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama P., Agar,J.N., Cash,V.L., Johnson,M.K. and Dean,D.R. (2000) NifS-directed assembly of a transient [2Fe–2S] cluster within the NifU protein. Proc. Natl Acad. Sci. USA, 97, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., White,R.H., Cash,V.L., Jack,R.F. and Dean,D.R. (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl Acad. Sci. USA, 90, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Cash,V.L., Flint,D.H. and Dean,D.R. (1998) Assembly of iron–sulfur clusters: Identification of an iscSUA–hscBA–fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem., 273, 13264–13272. [DOI] [PubMed] [Google Scholar]