Abstract
Bacterial artificial chromosome (BAC) vectors enable stable cloning of large DNA fragments from single genomes or microbial assemblages. A novel shuttle BAC vector was constructed that permits replication of BAC clones in diverse Gram-negative species. The “Gram-negative shuttle BAC” vector (pGNS-BAC) uses the F replicon for stable single-copy replication in E. coli and the broad-host-range RK2 mini-replicon for high-copy replication in diverse Gram-negative bacteria. As with other BAC vectors containing the oriV origin, this vector is capable of an arabinose-inducible increase in plasmid copy number. Resistance to both gentamicin and chloramphenicol is encoded on pGNS-BAC, permitting selection for the plasmid in diverse bacterial species. The oriT from an IncP plasmid was cloned into pGNS-BAC to enable conjugal transfer, thereby allowing both electroporation and conjugation of pGNS-BAC DNA into bacterial hosts. A soil metagenomic library was constructed in pGNS-BAC-1 (the first version of the vector, lacking gentamicin resistance and oriT), and recombinant clones were demonstrated to replicate in diverse Gram-negative hosts, including Escherichia coli, Pseudomonas spp., Salmonella enterica, Serratia marcescens, Vibrio vulnificus and Enterobacter nimipressuralis. This shuttle BAC vector can be utilized to clone genomic DNA from diverse sources, and then transfer it into diverse Gram-negative bacterial species to facilitate heterologous expression of recombinant pathways.
Keywords: bacterial artificial chromosome, vector, metagenome, heterologous expression
1. Introduction
BAC vectors using the modified F plasmid are commonly used for construction and analysis of genomic libraries; greatly facilitating research that relies upon the stable maintenance of very large DNA inserts (Shizuya et al., 1992). Early versions of BAC vectors (e.g., pBELOBAC11) provided excellent stability of recombinant clones, but due to the single copy number of the BAC replicon the concentration of plasmid DNA necessary for library construction or clone analysis required large culture volumes to achieve sufficient vector DNA (Kim et al., 1996). The introduction of an additional origin of replication (oriV) from a broad host-range RK2 plasmid permitted a stable but inducible copy system, wherein the copy number was controlled by an arabinose-inducible replicator protein (TrfA) inserted into the E. coli chromosome (Wild et al., 2002, Wild and Szybalski, 2004a). With inducible copy number, BAC clones can be maintained at single copy under control of the F replicon and then induced to multiple copies (50- to 150-fold induction) by the addition of 0.01% arabinose to the culture medium. These vectors were further developed by Szybalski’s lab to a new class of pBAC/oriV “copy-control tightly regulated expression vectors” (Wild and Szybalski, 2004b).
While providing significant advantages, these commercially available inducible BAC vectors (e.g., CopyRight v2.0 BAC, Lucigen Corp., Middleton, WI) are limited to replication within an E. coli host. For sequence-based mapping and molecular analysis, maintenance in E. coli is sufficient. However, for construction and functional screening of metagenomic libraries it is advantageous to transfer recombinant clones into multiple bacterial expression hosts to improve heterologous expression of cloned metagenomic DNA (Craig et al., 2010; Handelsman et al., 1998; Rondon et al., 1998). Various shuttle vectors have been used to transfer recombinant clones into alternative heterologous hosts, such as Streptomyces and Pseudomonas spp. (Martinez et al., 2004; Wang et al., 2000). By incorporating into a BAC vector both oriV and trfA, which is the mini-replicon necessary for RK2 plasmid replication (Thomas et al., 1981), a much greater host range can be achieved, including most Gram-negative bacterial species.
A previous phylogenetic analysis of a soil metagenomic library indicated the very low prevalence of 16S rRNA genes from Gram-positive phyla (Liles et al., 2003), reflecting the poor lysis of Gram-positive cells when attempting to clone large DNA fragments. Therefore, a Gram-negative shuttle BAC vector would be particularly advantageous in cloning DNA derived from the diverse Gram-negative bacteria preferentially represented within large-insert metagenomic libraries. With any environmental sample, cloning into a shuttle vector would allow for conjugal transfer and heterologous expression of metagenomic cloned DNA in multiple bacterial hosts. One such example is pRS44, a RK2-based broad-host-range cloning vector (Aakvik et al., 2009). Unlike the pGNS-BAC vector, which has the complete RK2 mini-replicon contained within the vector (i.e., oriV and trfA), the pRS44 vector system requires transposon-mediated insertion of the trfA gene within the desired host species. Each heterologous bacterial host may have unique transcriptional regulatory factors that can affect expression of cloned genes; thus, the pGNS-BAC vector increases the probability of identifying clones with specific functions by expanding the range of genomic library hosts for recombinant gene expression.
2. Materials and Methods
2.1 Bacterial strains and media
E. coli strain DH10B was used as the primary host for transformations. Cultures were grown at 37°C in Luria-Bertani broth or agar plates supplemented with the appropriate antibiotics. Concentrations of antibiotics were 12.5 μg/ml chloramphenicol (Cm) and 30 μg/ml gentamicin sulfate (Gm). Pseudomonas putida, Pseudomonas aeruginosa, Pseudomonas stutzeri, Pseudomonas fluorescens, Salmonella enterica, Serratia marcescens, Vibrio vulnificus, and Enterobacter nimipressuralis were used as recipients to test the host range of pGNS-BAC-1 (Table 1).
Table 1.
Bacterial strains and plasmids.
| Bacteria | Plasmids of interest | Plasmid-encoded antibiotic resistance or other characteristic | Source |
|---|---|---|---|
| E. coli strain DH10B | pJW544 | BAC vector, CmR, oriV | Wild et al., 2004 |
| E. coli strain DH10B | pGNS-BAC1 | BAC vector, CmR, oriV, BamHI site-minus | This study |
| E. coli strain SM10 | pGNS-BAC | BAC vector, CmR, GmR, oriV, oriT, sacB | This study |
| Pseudomonas putida | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
| Pseudomonas aeruginosa | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
| Pseudomonas stutzeri | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
| Pseudomonas fluorescens | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
| Salmonella enterica | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
| Vibrio vulinificus | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
| Enterobacter nimipressuralis | pGNS-BAC1 or pGNS-BAC |
CmR, or CmR and GmR | This study |
2.2 Construction of pGNS-BAC-1
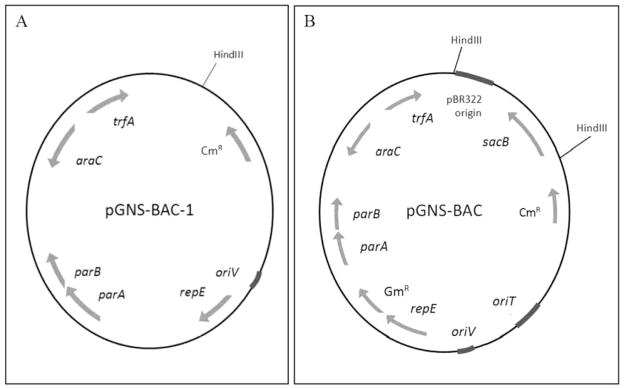
The ParaBAD promoter of plasmid pJW544 drives expression of the trfA gene, and the TrfA replication initiation protein then binds to oriV iterons (Perri et al., 1991). A BamHI restriction site within the promoter was destroyed by restriction digestion and subsequent fill-in with Klenow DNA polymerase and dNTPs. Plasmid DNAs were extracted from E. coli cultures using a Promega Wizard Plus SV Minipreps kit (Madison, WI). Restriction and DNA sequence analysis was conducted to confirm loss of the BamHI site, and induction of plasmid copy number with 0.01% arabinose was performed to confirm that the copy-inducible phenotype was still functional. The resulting plasmid was named pGNS-BAC-1 (Fig. 2A).
Figure 2.
Annotated plasmid map for pGNS-BAC-1 (Panel A) and pGNS-BAC (Panel B).
2.3 Soil metagenomic library construction
To determine if recombinant BAC clones in the vector pGNS-BAC-1 were capable of replication within Gram-negative bacterial hosts, a small-insert BAC library was constructed from bacterial cells that were first extracted from the soil prior to DNA isolation (Liles et al., 2008). Briefly, the extracted and washed bacterial cells were incorporated into agarose plugs, lysed, and then high molecular weight (HMW) metagenomic DNA was electrophoresed from the plug. Purification by a formamide denaturation step (70% final concentration) resulted in removal of associated nuclease activity from the HMW DNA and improved cloning efficiency (Liles et al., 2008). The formamide-treated metagenomic DNA was partially restriction digested with HindIII, electroeluted from an agarose gel, and ligated into a Hind-III digested and dephosphorylated pGNS-BAC-1 vector. The ligated vector and insert DNA was transformed into E. coli strain DH10B, and transformants were selected on LB containing 12.5 μg/ml Cm. Transformants were robotically picked into a 96-well format and stored in 10% glycerol at −80°C.
2.4 Electroporation of BAC DNA into bacterial strains
Random clones were selected from the soil metagenomic library in pGNS-BAC-1. DNA was extracted using a manual alkaline lysis protocol and characterized by restriction fragment length polymorphism (RFLP) analysis using HindIII (Promega). Clones with insert DNA were transformed into electrocompetent Serratia marcescens, V. vulnificus, and Pseudomonas putida (1 mm gap cuvette, 1.8 kV, 600 Ohms, 10 μF) Cells were grown in. SOC recovery medium for 1 hour at 37°C and plated on LB agar supplemented with Cm. id DNA was extracted and subjected to RFLP analysis as above to test for the presence of the recombinant BAC DNA in each bacterial host.
2.5 Construction of pGNS-BAC
A Gm resistance cassette was obtained from plasmid pBSL141 (Alexeyev et al., 1995) as a NheI restriction fragment and ligated into an Eco47III site of the vector pGNS-BAC-1. Transformants were selected on LB containing both Cm and Gm. Restriction digests with EcoRV established the presence of the Gm-resistance cassette, resulting in the plasmid pGNS-BAC-2.
A cloning region from the vector pSMART BAC v2.0 (Lucigen Corporation, Middleton, WI) containing the counter-selectable sacB gene and pUC19 origin of replication was cloned into pGNS-BAC-2 to reduce the background of transformants without inserts (i.e., by sacB-mediated counter-selection) and to provide very high copy number for preparation of empty vector DNA (pUC19 origin). The cloning region was PCR amplified using flanking primers, purified, blunt-ended, and ligated to a filled-in HindIII restriction site of the pGNS-BAC-2 vector. The ligation was transformed into electrocompetent E. coli strain DH10B and plated onto LB containing Cm and Gm. Transformants were screened for sucrose sensitivity. Plasmid DNA was extracted from sucrose-sensitive clones and restriction digested with HindIII to confirm the addition of the cloning region to pGNS-BAC-2. The resulting BAC vector was designated as pGNS-BAC-3.
To introduce the ability to conjugally transfer the BAC vector, the oriT (mob) gene from pLOF-Km was PCR amplified using the primers mobF (5′-GATCCTCGAGGGATCCTTTTTGTCCG) and mobR (5′-GATCCTCGAGCAGCCGACCAGGCT) (Herrero et al., 1990). The PCR primers include 5′-XhoI restriction sites (bold). After amplification and XhoI digestion, the amplicon was ligated into the XhoI site of pGNS-BAC-3, transformed into E. coli strain DH10B, and selected on LB containing Cm and Gm. Clones containing the oriT were identified via PCR using the mobF and mobR primers, and the resultant plasmid was verified by restriction digestion with Sau3AI. The final vector construct, pGNS-BAC-4, also referred to as the pGNS-BAC vector (Fig. 2B), was stored as a glycerol stock at −80°C. The pGNS-BAC vector was sequenced completely, and the sequence was deposited within the GenBank database (Accession number HQ245711).
2.6 Conjugal transfer of BAC vector DNA into bacterial strains
The pGNS-BAC vector was electroporated (1 mm gap cuvette, 1.8 kV, 600 Ohms, 10 μF) into E.coli strain SM10, which permits conjugal transfer of oriT-containing plasmids (Simon et al., 1983). Cells were grown in SOC recovery medium for 1 hour at 37°C and plated on LB agar supplemented with Cm (12.5 μg/ml) and Gm (30 μg/ml). E. coli strain SM10 having the pGNS-BAC vector was used as the donor for conjugation experiments. Confirmation of the ability of oriT to mediate conjugal transfer was performed using S. marcescens as the recipient. LB broth supplemented with Cm and Gm was used to grow the donor, and LB broth without antibiotics was used to grow the recipient. Cultures were grown overnight at 37°C with aeration. Donor and recipient were mixed in a ratio of 1:4 (50 μl and 200 μl respectively) and treated with 1 ml of 10 mM MgSO4. After mixing thoroughly, centrifugation was carried out at 15,000 × g for 10 minutes. One ml of supernatant was discarded, and the cells were resuspended in the remaining liquid and spread on a nitrocellulose membrane placed on the surface of an LB agar plate. Following incubation at 37°C for 4 hours the membrane was transferred to an LB agar plate containing 1 mM IPTG and incubated at 37°C for another 12 hours. Cells were then washed off the membrane with 3 ml of 10 mM MgSO4 and collected in a tube. Different dilutions of this cell mixture were then spread on LB agar containing Cm and Gm (to select against the recipient) and colistin (10 μg/ml, to select against the donor). This procedure allows exclusive selection of the S. marcescens (or other recipient host) transconjugants.
Transconjugants were selected and screened for the presence of pGNS-BAC vector DNA by isolating plasmid DNA from the recipient hosts after varying times of cultivation, in the presence and absence of Cm and/or Gm and/or 0.01% arabinose. The presence of plasmid DNAs was confirmed by restriction analysis. The cell counts of donor, recipient, and transconjugants were estimated by plating a range of serial dilutions on suitable media.
2.7 Amplification of the pGNS-BAC vector in E.coli strain DH10B and Serratia marcescens
Two sets of E. coli and S. marcescens cultures containing the pGNS-BAC vector were grown overnight in 10 ml of LB containing 12.5 μg/ml Cm. Cultures were grown with shaking for 30 minutes and then one set of cultures was induced with 50 μl of 2% L-Arabinose (Sigma) [0.01% final concentration]. After 4 hrs of induction the DNA was extracted using Wizard plus SV Minipreps DNA Purification System (Promega). DNA was prepared from 4.5 ml of induced and not induced cultures and eluted from the column with 50 μl of nuclease free water. To evaluate the integrity and relative amount of DNA purified from each culture, a restriction digestion with NcoI (cuts twice within pGNS-BAC) was performed at 37°C. After 1hr of incubation the restriction digest was electrophoresed through a 0.6% agarose gel and restriction fragments were visualized by staining with EtBr.
2.8 Increase in MIC of Cm and Gm conferred by pGNS-BAC
Minimum inhibitory concentration (MIC) testing using the macrodilution method was carried out to test the degree of resistance to gentamicin or chloramphenicol conferred by pGNS-BAC on E. coli or S. marcescens. Both bacterial species were tested in the presence and absence of the BAC vector and with or without addition of 0.01% arabinose to the cation-adjusted Mueller-Hinton broth (CAMHB) (Table 2). Antibiotic stock solutions of Gm (960 μg/ml), Cm (400 μg/ml), and arabinose (0.01% and 0.02%) were made using CAMHB. The final concentration range tested for Gm was from 7.5 μg/ml with a twofold consecutive increase up to 960 μg/ml and likewise for Cm from 3.125 μg/ml up to 400 μg/ml. The experiment was conducted in triplicate, with inclusion of the controls: 1) bacterial strains without vector DNA, 2) bacterial growth without any antibiotics added, and 3) media only. Tubes were incubated overnight at 37ºC and turbidity was measured the next day to determine the MIC of the antibiotic.
Table 2.
MIC for Cm and Gm conferred by pGNS-BAC.
| Bacterial strain | pGNS-BAC | Arabinose | MIC (μg/ml) | |
|---|---|---|---|---|
| Cm | Gm | |||
| E. coli | + | − | 25 | 60 |
| E. coli | + | + | 200 | 480 |
| E. coli | − | − | 6.25 | 15 |
| E. coli | − | + | 6.25 | 15 |
| S. marcescens | + | − | 12.5 | 30 |
| S. marcescens | + | + | 200 | 240 |
| S. marcescens | − | − | 6.25 | 15 |
| S. marcescens | − | + | 6.25 | 15 |
A gradient agar plate method was used to confirm increase in antibiotic resistance as a result of plasmid amplification (Bryson and Szybalski, 1952). A Cm gradient agar plate with antibiotic concentration ranging from no added Cm to 500 μg/ml Cm in the presence and absence of 0.01% arabinose was used to streak overnight cultures of E. coli and S. marcescens. Plates were incubated at 37ºC and observed and recorded after 24 hours.
3. Results
3.1 pGNS-BAC-1 construction and analysis
The pGNS-BAC-1 plasmid was tested as a shuttle vector under control of either of its two origins of replication (i.e., F and RK2) (Table 1). The pGNS-BAC-1 vector is maintained in E. coli as a single-copy plasmid by repressing the RK2 origin of replication with the addition of 0.1% glucose to the growth medium. Induction of plasmid copy number in E. coli was achieved by supplementation with 0.01% arabinose. The pGNS-BAC-1 vector was electroporated into P. putida, P. aeruginosa, P. stutzeri, P. fluorescens, S. enterica, S. marcescens, V. vulinificus, and E. nimipressuralis (Table 1). Isolated colonies from each transformation were used to inoculate LB broth cultures containing Cm, and the plasmid DNAs extracted from each host revealed a banding pattern identical to the pGNS-BAC-1 plasmid (data not shown). In some cases, the DNA isolated from non-E. coli hosts (e.g., S. marcescens) was retransformed into E. coli, yielding Cm-resistant clones with a pGNS-BAC-1 restriction profile (data not shown).
3.2 Construction of a soil metagenomic library and lateral transfer of recombinant clones
To determine if recombinant pGNS-BAC-1 clones can also stably replicate in different bacterial hosts, a metagenomic library was constructed within pGNS-BAC-1. Metagenomic DNA was extracted from soil at the Bonanza Creek Experimental Forest near Fairbanks, AK, and was partially restriction digested and ligated into pGNS-BAC-1. E. coli transformants were picked into 96-well plates, and random clones were analyzed by RFLP to identify large-insert containing clones. Random clones containing DNA inserts of approximately 75.0 kb, 79.8 kb, 83.9 kb, and 86.0 kb were electroporated into S. marcescens, V. cholerae, and E. nimipressuralis. Cm-resistant transformants were successfully isolated for each of the clones in each of the bacterial hosts. The range of transformation efficiencies for the clones containing inserts relative to the empty pGNS-BAC-1 vector was 94.9% to 259% for S. marcescens, 45.7% to 76.2% for V. cholerae, and 55.9% to 104.9% for E. nimipressuralis. Recovery of recombinant clones with intact inserts from the transformants in S. marcescens was confirmed by pulsed field gel electrophoresis.
3.3 pGNS-BAC construction and analysis
Although pGNS-BAC-1 was maintained in multiple Gram-negative bacteria, its utility as a shuttle vector was limited due to the presence of only a single antibiotic resistance gene and an inability to be conjugally transferred to recipient hosts. Therefore, a Gm resistance cassette and an oriT were added to pGNS-BAC-1.
An improved multiple cloning region with a removable counter-selectable marker was also added to pGNS-BAC-1 to provide much lower background during transformations, resulting in the final pGNS-BAC vector construct. Cells containing intact pGNS-BAC vector are sucrose-sensitive due to the presence of the sacB gene within the cloning region. This region is removed as a restriction fragment during preparation of the vector for cloning. The final vector size is 11.9 Kb, and recombinant clones are sucrose-resistant (data not shown). The complete sequence of pGNS-BAC was determined and annotated and submitted to GenBank (Accession number HQ245711).
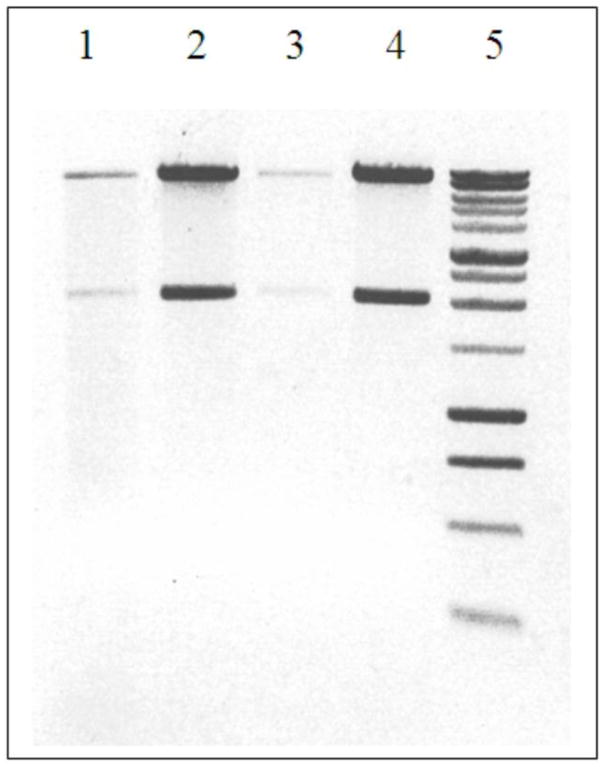
E. coli strain SM10 containing pGNS-BAC was mixed with S. marcescens to test its ability to be conjugally transferred and to replicate within a bacterial host other than E. coli. Transconjugants that were CmR and GmR were readily obtained (> 1 × 105 transconjugants ug−1 DNA). Representative transconjugants were inoculated into broth cultures with and without antibiotic selection, and after 12 to 16 hours of growth, plasmid DNAs were isolated and restriction digested to determine plasmid yield and stability. Plasmid DNAs corresponding to the pGNS-BAC restriction profile were observed by RFLP. Fig. 1 shows the comparison of pGNS-BAC DNA isolated from E. coli strain DH10B and Serratia marcescens with and without arabinose induction.
Figure 1.
Isolation of BAC vector DNA from E. coli and S. marcescens. Lanes 1–2, S. marcescens containing pGNS-BAC without (lane 1) or with arabinose induction (lane 2); Lanes 3–4, E. coli strain DH10B containing pGNS-BAC without (lane 3) or with arabinose induction (lane 4); Lane 5, molecular weight marker (Bench Top 1kb DNA ladder, Promega). By comparing the DNA band intensity in lane 1 relative to lane 2 and lane 3 relative to lane 4 the degree of arabinose-mediated plasmid copy-induction can be estimated.
In the absence of arabinose copy-induction, E. coli (pGNS-BAC) had an MIC for Cm of 25 μg/ml and an MIC for Gm of 60 μg/ml, whereas S. marcescens (pGNS-BAC) had an MIC for Cm of 12.5 μg/ml and an MIC for Gm of 30 μg/ml (Table 2). In the presence of arabinose, E. coli (pGNS-BAC) had an MIC for Cm of 200 μg/ml and 480 μg/ml for Gm, and S. marcescens (pGNS-BAC) had an MIC for Cm of 200 μg/ml and an MIC for Gm of 240 μg/ml (Table 2). Thus, both E. coli and S. marcescens harboring pGNS-BAC had a ca. 32-fold increase in resistance to Cm as a result of arabinose-mediated copy-induction, and a similar increase in resistance to Gm in the presence of arabinose (32-fold for E. coli, and 16-fold for S. marcescens). However, in the absence of the pGNS-BAC vector no arabinose-induced changes in MIC levels were observed (Table 2).
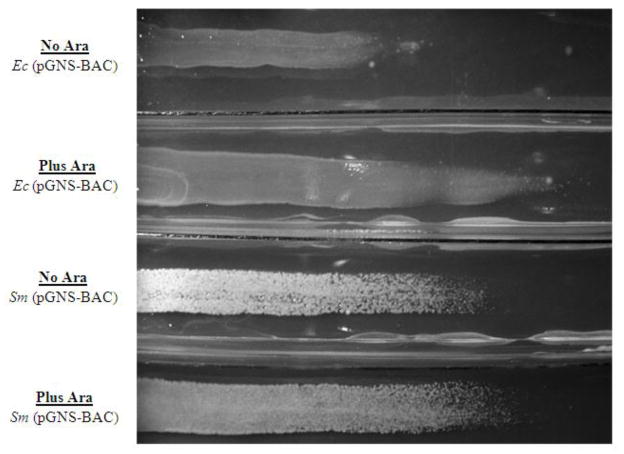
On a Cm gradient agar plate ranging from no added Cm to 500 μg/ml Cm, both E. coli and S. marcescens with the pGNS-BAC vector exhibited higher resistance to Cm in the presence of arabinose (Fig. 3). Comparatively less growth was observed in the absence of arabinose. This confirms that pGNS-BAC plasmid amplification leads to increase in the level of antibiotic resistance in multiple bacterial hosts.
Figure 3.
Growth of E. coli with the pGNS-BAC vector (Ec (pGNS-BAC)) and S. marcescens with the pGNS-BAC vector (Sm (pGNS-BAC)) in the presence (Plus Ara) and absence (No Ara) of arabinose, and on a Cm gradient agar plate ranging from no added Cm (left) to 500 μg/ml Cm (right).
4. DISCUSSION
The pGNS-BAC vector provides the ability to clone DNA inserts and maintain recombinant clones at single copy in E. coli, utilizing the well-described stability of the F plasmid. The addition of arabinose results in induction of pGNS-BAC copy number mediated by trfA located on the plasmid. Copy-induction greatly increases plasmid DNA yield and could improve heterologous expression of cloned DNA via a gene-dosage mechanism (Rine et al., 1983). The RK2 mini-replicon that affords the copy-inducible phenotype in E. coli also permits replication in a phylogenetically diverse range of Gram-negative bacteria. Large-insert clones within the first version of the shuttle vector pGNS-BAC-1 were demonstrated to be capable of transfer and replication within diverse Proteobacteria species. The final pGNS-BAC vector construct has a significantly expanded host range compared to pGNS-BAC-1 due to the addition of genes for Gm resistance and its ability to be conjugally transferred.
This inducible-copy and Gram-negative shuttle vector can be employed for metagenomic analysis of diverse environments, most of which contain abundant Gram-negative species, as well as to heterologously express specific genetic pathways. Construction of a soil metagenomic library in the pGNS-BAC vector provides the ability to transfer entire libraries, or specific recombinant clones, into bacterial hosts that may be more closely related to the bacterial taxa from which the cloned DNA was derived. Ideally, metagenomic libraries from a given source DNA could be constructed in both pGNS-BAC and a Gram-positive shuttle vector, potentially providing the widest possible range of heterologous expression hosts.
The rapidly advancing science of metagenomics requires molecular tools to enhance the heterologous expression of cloned DNAs. The metagenomic libraries constructed in pGNS-BAC will have all of the properties valued in previous libraries, such as stable maintenance of large inserts, with added features that have great potential to facilitate manipulation and expression of recombinant clones in a variety of different Gram negative hosts.
Acknowledgments
We thank the members of the Liles, Goodman, and Szybalski laboratories for their help in the execution of these experiments and their critical analysis of this publication. We acknowledge the contribution of Larissa Parsley in the MIC experiments. Strains used in this work were provided by the Bacteriology Department, University of Wisconsin. Madison, WI. Funding for this work was provided by the National Institutes of Health (grant # 1R21AI083852-01).
List of abbreviations in the order that they appear in the manuscript
- BAC
bacterial artificial chromosome
- DNA
deoxyribonucleic acid
- pGNS-BAC
Gram-negative shuttle BAC vector
- oriV
origin of replication
- oriT
origin of transfer
- IncP plasmid
plasmid of incompatibility group P
- TrfA
replication initiation protein
- trfA
gene encoding the replication initiation protein
- rRNA
ribosomal RNA
- Cm
chloramphenicol
- Gm
gentamicin sulfate
- dNTP
deoxyribonucleoside triphosphate
- HMW
high molecular weight
- RFLP
restriction-fragment length polymorphism
- LB
Luria-Bertani
- sacB
gene encoding Bacillus subtilis levansucrase
- PCR
polymerase chain reaction
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- EtdBr
ethidium bromide
- MIC
minimum inhibitory concentration
- CAMHB
cation-adjusted Mueller-Hinton broth
- kb
kilobase(s) or 1000 bp
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakvik T, Degnes KF, Dahlsrud R, Schmidt F, Dam R, Yu L, Volker U, Ellingsen TE, Valla S. A plasmid RK2-based broad-host-range cloning vector useful for transfer of metagenomic libraries to a variety of bacterial species. FEMS Microbiol Lett. 2009;296:149–58. doi: 10.1111/j.1574-6968.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- Alexeyev MF, Shokolenko IN, Croughan TP. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160(1):63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- Bryson V, Szybalski W. Microbial Selection. Science. 1952;116:45–51. [PubMed] [Google Scholar]
- Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol. 2010;76:1633–41. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in Gram-negative bacteria. J Bacteriol. 1985;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J, Rondon MR, Brady S, Clardy J, Goodman RM. Molecular biology provides access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistanceselection markers for cloning and stable chromosomalinsertion of foreign genes in Gram-negativebacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UJ, Birren BW, Slepak T, Mancino V, Boysen C, Kang HL, Simon MI, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Liles MR, Manske BF, Bintrim SB, Handelsman J, Goodman RM. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl Environ Microbiol. 2003;69 (5):2684–2691. doi: 10.1128/AEM.69.5.2684-2691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, Handelsman J. Recovery, purification, and cloning of high molecular weight genomic DNA from soil microorganisms. Appl Environ Microbiol. 2008;74:3302–3305. doi: 10.1128/AEM.02630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Kolvek SJ, Yip CLT, Hopke J, Brown KA, MacNeil IA, Osburne MS. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol. 2004;70:2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri S, Helinski DR, Toukdarian A. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host-range plasmid RK2. J Biol Chem. 1991;266:12536–12543. [PubMed] [Google Scholar]
- Rine J, Hansen W, Hardeman E, Davis RW. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci USA. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Osburne MS, Clardy J, Handelsman J, Goodman RM. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Thomas CM, Stalker DM, Helinski DR. Replication and incompatibility properties of segments of the origin region of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]
- Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J. Novel natural products from soil DNA libraries in a streptomycete host. Org Lett. 2000;2:2401–2404. doi: 10.1021/ol005860z. [DOI] [PubMed] [Google Scholar]
- Wild J, Hradecna Z, Szybalski W. Conditionally Amplifiable BACs: Switching From Single-Copy to High-Copy Vectors and Genomic Clones. Genome Res. 2002;2002–12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J, Szybalski W. Copy-control pBAC/oriV vector for genomic cloning. In: Balbas P, Lorence A, Walker JM, editors. Methods in Molecular Biology, Recombinant Gene Expression. Reviews and Protocols. 267 Chap 10. Humana Press Inc; Totowa NJ: 2004a. pp. 145–154. [Google Scholar]
- Wild J, Szybalski W. Copy-control tightly regulated expression vectors based on pBAC/oriV. In: Balbas P, Lorence A, Walker JM, editors. Methods in Molecular Biology, Recombinant Gene Expression. Reviews and Protocols. 267 Chap 11. Humana Press Inc; Totowa NJ: 2004b. pp. 155–167. [DOI] [PubMed] [Google Scholar]