Abstract
Holliday junctions are four-way branched DNA structures formed during recombination, replication and repair. They are processed in Escherichia coli by the RuvA, RuvB and RuvC proteins. RuvA targets the junction and facilitates loading of RuvB helicase and RuvC endonuclease to form complexes that catalyse junction branch migration (RuvAB) and resolution (RuvABC). We investigated the role of RuvA in these reactions and in particular the part played by the acidic pin located on its DNA-binding surface. By making appropriate substitutions of two key amino acids (Glu55 and Asp56), we altered the charge on the pin and investigated how this affected junction binding and processing. We show that two negative charges on each subunit of the pin are crucial. They facilitate junction targeting by preventing binding to duplex DNA and also constrain branch migration by RuvAB in a manner critical for junction processing. These findings provide the first direct evidence that RuvA has a mechanistic role in branch migration. They also provide insight into the coupling of branch migration and resolution by the RuvABC resolvasome.
Keywords: branch migration/Escherichia coli/helicase/recombination/resolution
Introduction
Homologous recombination is a vital cellular process that acts both to promote genetic diversity and to conserve genomic integrity. The key to the reaction is the formation and subsequent processing of Holliday junctions, a branched DNA intermediate in which the branchpoint is free to migrate through regions of homology. In Escherichia coli, Holliday junctions are formed during DNA pairing and strand exchange reactions catalysed by RecA protein (Lloyd and Low, 1996; West, 1997; Kowalczykowski, 2000). They also arise from stalled replication forks by regression of the forked DNA and annealing of the nascent strands, and provide means to clear the lesion and resume replication (Seigneur et al., 1998; McGlynn and Lloyd, 2000; Michel, 2000). Once formed, they are targeted and processed by proteins that recognize the four-way branched structure of the DNA. The RuvA, RuvB and RuvC proteins appear to have evolved specifically for this purpose (West, 1997). A tetramer of RuvA targets the junction and holds it in an open conformation (Figure 1A, i) that allows two RuvB hexameric rings to assemble on diametrically opposed arms (Figure 1B). RuvC, a junction-specific endonuclease, binds the exposed face of the junction between the two RuvB rings, forming a resolvasome complex in which ATP-dependent branch migration driven by the RuvAB helicase motor (Figure 1B) locates the junction at sequences that can be cleaved by RuvC (Davies and West, 1998; van Gool et al., 1998, 1999; Zerbib et al., 1998). These reactions catalysed by RuvABC are late steps in RecA-mediated recombination that remove junctions and enable chromosomes to segregate at cell division. However, when RuvABC targets a junction formed from a stalled replication fork, they may also act to initiate recombination (Seigneur et al., 1998; McGlynn and Lloyd, 2000). Both roles are consistent with the increased sensitivity of ruv mutant strains to UV light and other DNA-damaging agents, and their reduced ability to promote recombination (Lloyd et al., 1984).
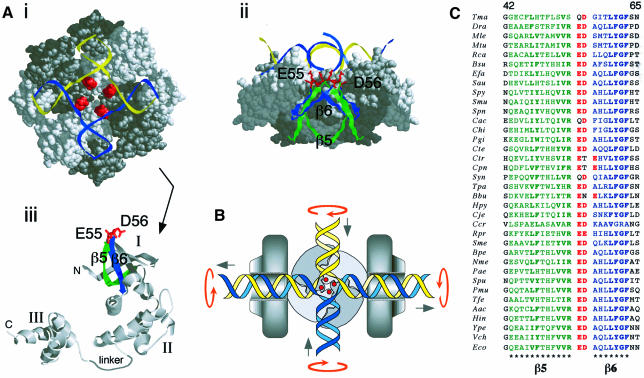
Fig. 1. Interactions between RuvA, RuvB and Holliday junction DNA and analysis of the acidic pin of RuvA. (A) Structure of RuvA showing: (i) the DNA-binding surface of the RuvA tetramer in a complex with junction DNA and the location of the acidic pin (red); (ii) a side view of the RuvA–junction complex projected from the end of a duplex DNA arm docked within its groove; and (iii) a single subunit of RuvA, its three domains and the β5 (green) and β6 (blue) strands supporting the acidic pin residues (E55 and D56). (B) Model of a RuvAB–junction complex illustrating how rotation of the duplex arms bound by the two RuvB hexamer rings draws the DNA through the complex to catalyse branch migration via symmetrical strand exchange at the crossover. (C) Alignment of RuvA sequences from diverse bacterial species showing conservation of the acidic pin residues (ED) and the flanking β5 and β6 strands. Sequences for the RuvA proteins from Neisseria gonorrhoeae and Salmonella typhimurium are identical in this region to those of Neisseria meningitidis (Nme) and Escherichia coli (Eco), respectively. The full species names, abbreviated on the left, are listed in Sharples et al. (1999) except for Rhodobacter capsulatus (Rca), Staphylococcus aureus (Sau), Chlamydia pneumoniae (Cpn), Sinorhizobium meliloti (Sme), Pasteurella multocida (Pmu), Thiobacillus ferrioxidans (Tfe) and Vibrio cholerae (Vch).
The specificity required for efficient targeting of Holliday junctions is provided by the RuvA subunit. Crystallization of RuvA, both alone (Rafferty et al., 1996) and in complexes with Holliday junctions (Hargreaves et al., 1998; Roe et al., 1998; Ariyoshi et al., 2000), has revealed a tetrameric architecture in which the identical subunits form a concave platform with a basic surface uniquely suited to bind the Holliday junction structure. Four grooves formed at the interface between the subunits accommodate the duplex arms of the open junction (Figure 1A). Two helix–hairpin–helix (HhH) motifs in domain II of each subunit contact the phosphodiester backbone of the duplex on the minor groove side and have been shown to be essential for DNA binding (Rafferty et al., 1998). The grooves in RuvA radiate from a central acidic pin formed by the four symmetry-related turns between strands β5 and β6 in domain I (Glu55 and Asp56, shown in red in Figure 1A). The open junction sits neatly over the central pin, with the DNA backbone repelled by the negative charges on Glu55 and Asp56 (Ariyoshi et al., 2000).
Two types of RuvA–junction complexes have been detected by gel-retardation assays: complex I in which the junction is bound to a single tetramer of RuvA, and complex II in which the junction is sandwiched between two tetramers (Whitby et al., 1996). Crystal structures providing examples of both types have been obtained (Hargreaves et al., 1998; Roe et al., 1998; Ariyoshi et al., 2000). In the two examples of complex I analysed, the junction deviates from a strict square-planar arrangement, with the point of crossover being pushed closer to the RuvA surface than was suggested by modelling (Rafferty et al., 1996). The complex resolved at 3.1 Å by Ariyoshi et al. (2000) also shows that two base pairs located symmetrically at the crossover are disrupted. Each unpaired nucleotide may form water-mediated hydrogen bonds with the acidic side chains of Glu55 and Asp56. In contrast, the junction sandwiched in complex II is withdrawn from the central pin and adopts a planar conformation. The duplex arms also make different contacts with the HhH motifs (Roe et al., 1998).
The significance of complex II is uncertain. Its formation would exclude RuvC and prevent the coupling of branch migration and resolution (Whitby et al., 1996). Furthermore, studies in vivo have shown that DNA repair becomes critically dependent on RecG helicase when RuvC alone is mutated (Mandal et al., 1993; Mahdi et al., 1996; McGlynn and Lloyd, 2000). Since RecG drives branch migration of Holliday junctions very efficiently, at least in vitro (Lloyd and Sharples, 1993; Whitby et al., 1993), these studies raise the possibility that a RuvAB complex may be unable to drive branch migration without RuvC or has no function. However, the binding of RuvAB may preserve junctions until RuvC becomes available to form an active resolvasome.
Here we describe an investigation of the RuvA pin structure and show that its negative charge provides the specificity needed for efficient targeting of Holliday junctions by preventing abortive loading on duplex DNA. More importantly, we provide evidence that the negative pin severely constrains branch migration by RuvAB and show that this is critical for efficient junction processing by RuvABC. The data presented provide the first direct evidence that RuvA has a positive role in the catalysis of branch migration.
Results
Conservation of the pin structure in eubacterial RuvA proteins and construction of mutant proteins
The alignment of RuvA sequences from 36 diverse eubacterial species shown in Figure 1C reveals that the critical residues forming the central acidic pin (Glu55 and Asp56 in the E.coli protein) are very well conserved. There are three instances in which the glutamate is replaced with glutamine, and three others in which the aspartate is replaced with either threonine or asparagine. However, in these latter three, there is an associated acidic substitution (Glu) at the next position, which may compensate. Residues in β strands 5 and 6 supporting the pin (Figure 1A, ii) are also highly conserved. Arg54 is present in all but two of the sequences, which may reflect the need to establish contact with the DNA backbone of the junction at the point of strand exchange (Hargreaves et al., 1998; Ariyoshi et al., 2000).
The high conservation of Glu55 and Asp56 indicates that an acidic pin structure providing eight negative charges within a tetramer may be important for activity. To test this directly, we altered the charge on the pin of E.coli RuvA and tested the mutant proteins both in vitro and in vivo. Removal of one of the negative charges on each subunit was achieved with conservative E55Q or D56N substitutions, but introducing positive charges on one or both residues required the more radical E55R and D56K substitutions. The E55D substitution maintains the wild-type negative charge on the pin and was generated as a control to assess the significance of the high conservation of a glutamate as an acidic residue at this position. Each of the mutant proteins (Table I) was purified readily using the procedure designed for wild-type RuvA, which indicates that the changes made have little effect on overall structure.
Table I. DNA repair activities of wild-type and mutant RuvA proteins.
| RuvA protein | Pin structure | Charge on residues 55 and 56a | Survival of UV-irradiated strainsb |
||
|---|---|---|---|---|---|
| SR2210 (ruvA) | AB1157 (ruv+) | TNM1208 (rus-1 ΔruvAC) | |||
| None | 0.00004 | 0.68 | 0.28 | ||
| Wild type | E55D56 | – – | 0.04 | 0.43 | 0.00007 |
| E55D | D55D56 | – – | 0.056 | 0.32 | 0.00048 |
| E55Q | Q55D56 | N – | 0.011 | 0.053 | 0.000019 |
| E55R | R55D56 | + – | 0.0012 | 0.082 | 0.00039 |
| D56N | E55N56 | – N | 0.0000039 | 0.008 | 0.000091 |
| D56K | E55K56 | – + | 0.0012 | 0.13 | 0.00016 |
| E55R,D56K | R55K56 | + + | 0.00021 | 0.16 | 0.087 |
aN, neutral; –, negative; +, positive.
bThe strains indicated carrying the pT7-7 vector (none) or derivatives encoding the RuvA proteins indicated were irradiated with a UV dose of 45 J/m2 and the number of cells surviving to form colonies was determined as a fraction of unirradiated controls. Values are the means of at least two independent experiments.
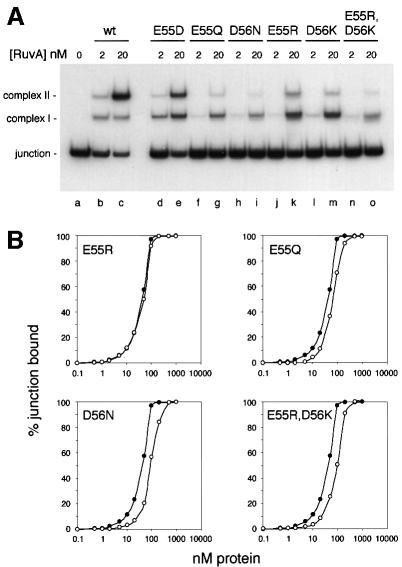
DNA binding activities of pin mutants
We investigated the ability of the mutant proteins to bind a small synthetic Holliday junction substrate, J11, using a standard band-shift assay. In each case, we observed complexes identical in mobility to complexes I and II formed by wild-type RuvA (Figure 2A). These data indicate that the mutants retain the ability to form tetramers and also that the changes made to the pin do not interfere with the ability of two tetramers to sandwich the junction. The affinity of the proteins for junction DNA also remained high, although some reduction relative to the wild-type protein was noticeable with the E55Q and D56N proteins, both of which have four negative charges eliminated from the pin, and also with the E55R,D56K double mutant in which all eight negative charges on the pin are replaced with positive charges (Figure 2B). Substituting four positive charges at either position (E55R or D56K) had little or no effect, and the E55D protein also bound junction DNA with the same affinity as wild-type RuvA (Figure 2B and data not shown).
Fig. 2. Holliday junction binding by wild-type and mutant RuvA proteins. (A) Gel-retardation assay showing formation of complex I and complex II with J11 DNA. Reactions contained 0.16 nM J11 DNA and proteins as indicated. (B) Binding curves showing the relative affinities of wild-type (filled circles) and mutant (open circles) RuvA proteins for J11 DNA. Reactions contained 0.16 nM J11. Wild-type and mutant proteins were analysed in parallel on the same gel, and the values shown (means of two independent experiments) are based on the fraction of the total DNA retarded.
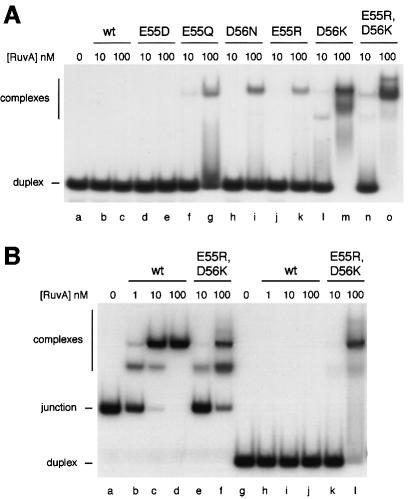
Negative charges on the pin are required to prevent binding to duplex DNA
When binding to J11 was monitored over a wide range of RuvA concentrations, we could not detect any effect of the mutations on the relative yields of complexes I and II (Figure 3A and B, and data not shown). In all cases, complex II replaced complex I as the concentration of protein increased. Thus, there is no indication of changes in the relative stability of these complexes. The only major variation on this pattern was seen with the positively charged E55R,D56K pin double mutant (Figure 3C). In this case, complexes I and II were formed in a concentration-dependent manner at the lower end of the range of protein used, but additional complexes migrating more slowly than complex II were formed as the concentration increased. These new complexes formed a distinct ladder of decreasing mobility. At very high levels of protein, some of the bound DNA failed to migrate out of the wells (Figure 3C, lanes k–m). A single novel complex was also observed with the E55R protein (Figure 3B, lane m) and with D56K, but not with E55Q, D56N or E55D (data not shown).
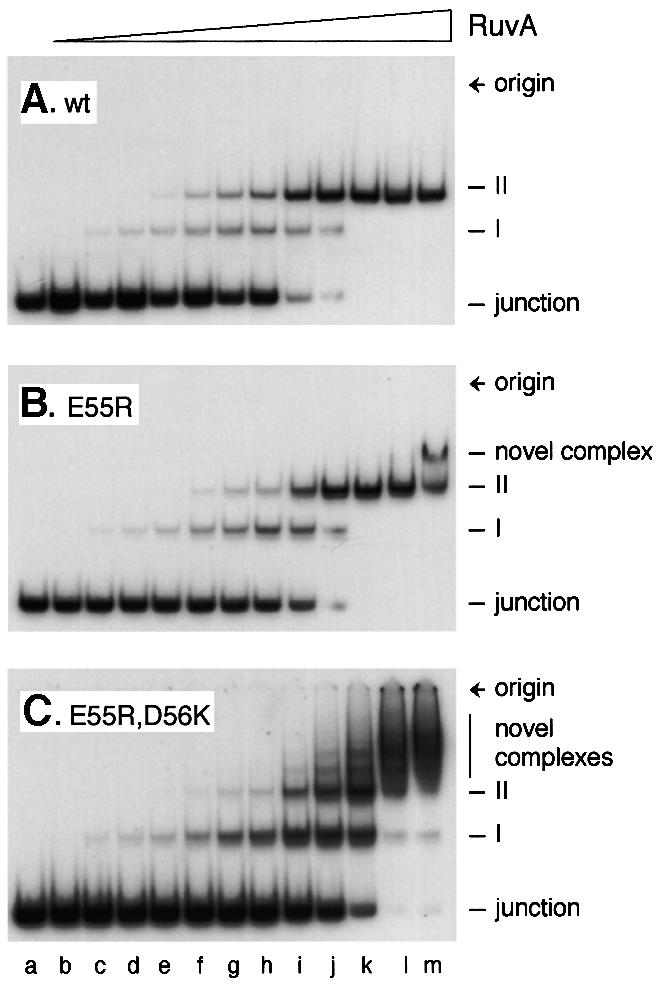
Fig. 3. Formation of novel complexes on junction DNA by mutant RuvA proteins. Reactions contained 0.16 nM J11 DNA and RuvA at 0.1, 0.5, 1, 2, 5, 10, 20, 50, 100, 200, 500 and 1000 nM in lanes b–m, respectively.
Previous studies with the RusA Holliday junction resolvase revealed that a ladder of complexes similar to that shown in Figure 3C, lanes i–m, may be formed when the protein binds to the duplex arms of a junction already bound at the crossover (Chan et al., 1998; Bolt et al., 1999). Wild-type RuvA does not bind duplex DNA with any significant affinity (Parsons et al., 1992) and does not form complexes migrating more slowly than complex II even at concentrations higher than those described in Figure 3 (data not shown). To test directly whether the alterations made to the pin allow RuvA to bind duplex DNA, band-shift assays were conducted using a 50mer linear duplex DNA substrate. No binding could be detected with wild-type RuvA or with the E55D mutant (Figure 4A). However, all the other mutants showed clear evidence of binding, forming either a single retarded complex (E55Q, D56N and E55R) or series of complexes (D56K and E55R,D56K). Duplex binding was particularly strong with the D56K substitution, either alone or in conjunction with E55R. All of the substrate was retarded when the D56K and E55R,D56K proteins were present in the binding reaction at 100 nM (Figure 4A, lanes m and o). To compare the relative affinities of wild-type RuvA and the E55R,D56K mutant for 50mer junction and linear duplex DNAs, binding reactions were conducted in parallel and analysed on the same gel. An example is shown in Figure 4B. The same amount of DNA was used for each substrate, which equates to twice as many linear duplexes as junctions per reaction. This experiment confirmed that the double mutant binds junctions with reduced affinity relative to the wild type (Figure 4B, lanes c–f), but binds linear duplex molecules with about the same affinity as it binds junctions (lanes f and l). Furthermore, the strongest retarded band with the duplex DNA (lane l) co-migrates with complex II formed with junction DNA (lane f). Minor species with the same mobility as junction complex I or migrating more slowly than complex II are also detectable. These results indicate that as with junctions, linear duplex molecules may be sandwiched between two tetramers of RuvA E55R,D56K. From these data, we cannot exclude the possibility that the E55R,D56K protein fails to target the branch point with 100% efficiency and sometimes binds instead to the duplex arms. This may explain the more diffuse nature of the retarded species identified as complexes I and II in Figure 3C.
Fig. 4. Binding of mutant RuvA proteins to linear duplex and junction DNA. (A) Gel assay showing binding of mutant RuvA proteins to linear duplex DNA. Reactions contained 0.32 nM 50 bp linear duplex DNA and protein as indicated. (B) Gel assay showing the relative affinities of wild-type and E55R,D56K RuvA proteins for junction and linear duplex DNA substrates. Reactions contained 0.08 nM J12 (lanes a–f) or 0.16 nM 50mer linear duplex DNA (lanes g–l) to give the same amount of DNA in each case, and proteins as indicated.
Further analysis of linear duplex DNA binding by the D56K and E55R,D56K proteins was conducted using molecules of 88, 129 and 205 bp. The retarded complexes became more diffuse as the length of the molecule increased (data not shown). This is consistent with binding of multiple RuvA tetramers at any point along the DNA and excludes the possibility of binding only at the DNA ends. Thus, the mutant proteins must be able to bind a linear duplex by docking the DNA into two diametrically related grooves on the protein surface such that it bridges the altered central pin. RuvA has a footprint on junction DNA extending 13 bp from the crossover along each duplex arm (Hiom and West, 1995). Therefore, a 50 bp linear duplex molecule should be able to accommodate two RuvA tetramers or double tetramers bound adjacent to one another. At least three discrete complexes are visible with the E55R,D56K protein (Figure 4A, lane o). The 50 bp junction, J11, has a core of homology of 11 bp within which the junction can branch migrate, which means it can extend two duplex arms of up to 31 bp. This is probably enough to allow simultaneous binding of RuvA at the crossover and on one or both of the extended arms. Such arrangements would explain the ladder of retarded complexes observed with this substrate (Figure 3C).
Taken together, these data lead to three important conclusions. First, two negative charges are needed on each subunit of the central pin to exclude binding of RuvA to linear duplex DNA. Secondly, placing a positive charge at position 56 (D56K) or at both 55 and 56 (E55R,D56K) generates RuvA proteins that can bind duplex DNA with particularly high affinity. Thirdly, the linear DNA bound by the mutant proteins appears to dock across the central pin.
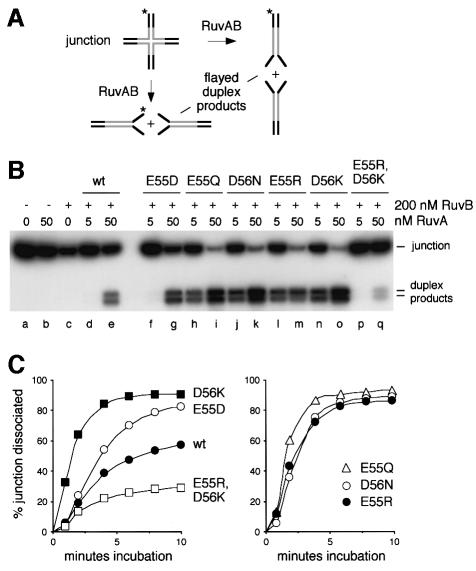
The acidic pin modulates the rate of branch migration by RuvAB
Wild-type RuvA interacts with RuvB ring helicase to form a complex at a Holliday junction that can drive the branchpoint along the DNA. With synthetic junctions such as J11, this ATP-dependent branch migration reaction also unwinds the heterologous arms and dissociates the structure into flayed duplex products (Parsons et al., 1992). Each molecule can be dissociated in either of two orientations so that a mixture of four flayed duplexes is generated during reactions in vitro, two of which are labelled if the junction carries 32P at the 5′ end of one oligonucleotide (Figure 5A). Dissociation depends on the presence of both RuvA and RuvB, and an efficient reaction requires an appropriate ratio of the two (Figure 5B, lanes b–e). Each of the six RuvA pin mutants retained the ability to interact with RuvB to promote this reaction (Figure 5B, lanes f–q). No products were detected with any of the mutants if either RuvB or ATP was omitted (data not shown), which confirms that dissociation remains dependent on the helicase activity of RuvB. The E55D mutant dissociated slightly more of the substrate than wild-type RuvA (lane g). However, the E55R, E55Q, D56N and D56K proteins were much more active (lanes i, k, m and o). The dissociation achieved using 5 nM of these four mutant proteins (lanes h, j, l and n) almost matched that with 50 nM wild-type RuvA. Time course assays revealed that the initial reaction rate with D56K in particular, but also with E55Q, E55R and D56N, was substantially higher than that catalysed by wild-type RuvA (Figure 5C). This increase in activity is even more surprising in the case of the E55Q and D56N proteins given that their affinity for junction DNA is, if anything, reduced slightly (Figure 2B). These remarkable findings indicate that two negative charges on each subunit of the RuvA pin structure act to limit the branch migration activity of RuvAB and that this limitation may be relaxed by removing either of these charges or by substituting a single positive charge. However, the dissociation activity of the E55R,D56K double mutant was reduced relative to wild-type RuvA (Figure 5C). We do not know whether this is because the protein binds to the duplex arms and blocks dissociation or whether eight positive charges on the pin interfere directly with the branch migration reaction when the protein targets the crossover.
Fig. 5. Branch migration of Holliday junctions by RuvAB. (A) Diagram illustrating a 32P-labelled (*) synthetic junction in which the branch point is located within a central core of homologous sequences (grey) flanked by four heterologous arms (black), and its dissociation to flayed duplex products by RuvAB in either of the two possible orientations. (B) Gel assay showing dissociation of J11 DNA to flayed duplex products. Reactions contained 0.16 nM J11 DNA and proteins as indicated, and were incubated for 30 min. (C) Rates of dissociation by wild-type and mutant RuvA proteins. Reactions contained 0.16 nM J11, 20 nM RuvA and 60 nM RuvB. Values are the means of 2–4 independent experiments using the same batch of junction DNA, and each mutant protein was analysed in parallel with wild-type RuvA. The results are separated into two panels for clarity of presentation.
Pin mutations reduce Holliday junction processing by RuvABC
Genetic studies revealed that Holliday junction resolution by RuvC protein in vivo depends on RuvAB (Mandal et al., 1993; Mahdi et al., 1996), and recent biochemical studies demonstrated the formation of a tripartite RuvABC complex on junction DNA that stimulates junction cleavage by RuvC by coupling the branch migration and resolution reactions (Whitby et al., 1996; Eggleston et al., 1997; Davies and West, 1998; van Gool et al., 1998, 1999; Zerbib et al., 1998). We therefore investigated whether the mutant RuvA proteins retained the ability to form a functional RuvABC resolvasome by testing their ability to stimulate RuvC in the presence of RuvB. In this experiment, we used a χ DNA as a substrate, a large Holliday junction structure in which the junction is located within a 300 bp homologous core containing several sequences matching the consensus target for cleavage by RuvC, flanked by duplex arms of 0.8–1.6 kb (Zerbib et al., 1997). Resolution of a χ molecule labelled at the 5′ end of all four arms generates four nicked duplex products that are readily separated on agarose gels. The level of RuvC used was such that in the absence of RuvAB, only a fraction (∼15%) of χ molecules were resolved (Figure 6A, i and ii, lane b). As shown previously (Zerbib et al., 1998; McGlynn and Lloyd, 2000), the addition of wild-type RuvA and RuvB under conditions allowing branch migration increased the amount of χ resolved ∼3-fold (lane d). Stimulation of resolution was also observed when wild-type RuvA was substituted with some of the mutant proteins. However, the level of stimulation was reduced as compared with wild-type RuvA in the order E55D (lane e), E55Q (lane f) and D56K (lane g). With E55R,D56K, no stimulation was observed (lane h). Indeed, cleavage by RuvC appeared to be somewhat inhibited by this mutant. The long duplex arms of the χ structure may titrate E55R,D56K. This may explain the lack of stimulation of cleavage. It may also explain why it has an inhibitory effect since such binding would interfere with spontaneous branch migration of the junction, especially if it occurred within the homologous core.
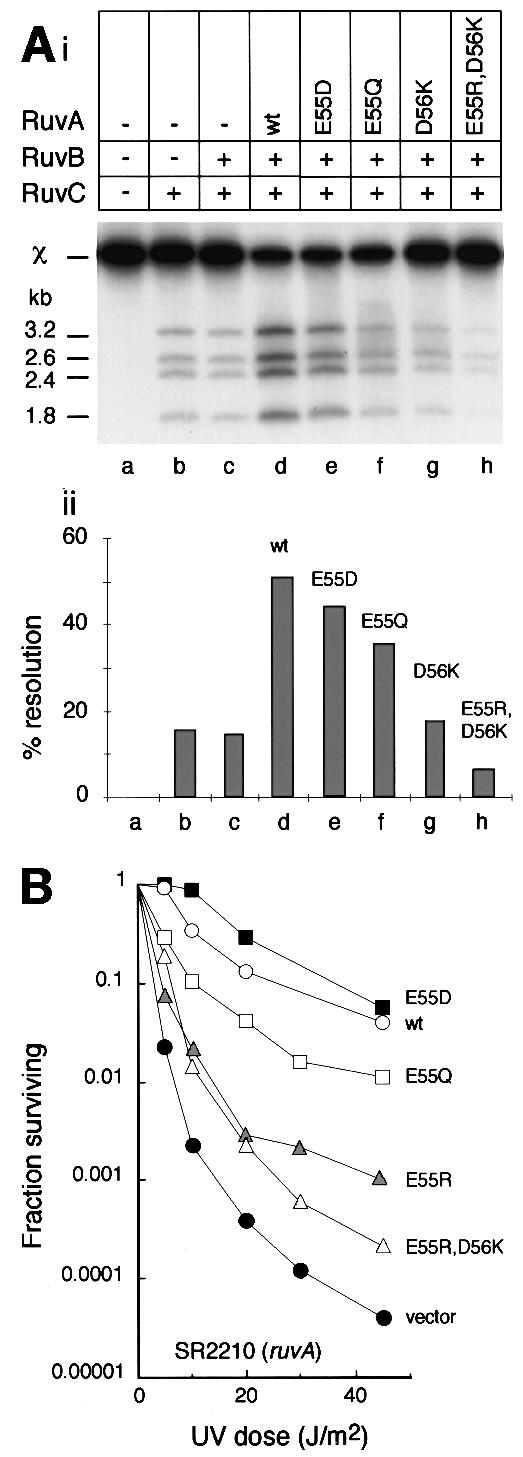
Fig. 6. Effect of RuvA pin mutations on the activity of RuvABC. (A) In vitro assay showing (i) products of cleavage of χ DNA by RuvC and (ii) quantification of this resolution activity (means of two independent experiments). (B) In vivo assay measuring survival of UV-irradiated ruvA mutant strain SR2210 expressing RuvA from a multicopy plasmid. Values are the means of at least two independent experiments.
Eight negative charges on the pin are required for RuvA to promote repair
Although χ DNA is a better mimic of in vivo Holliday junctions as compared with small substrates like J11, it is clear that RuvA has to target Holliday junctions in vivo in the presence of a vastly greater excess of duplex DNA than is represented even in χ DNA. We therefore tested the mutant proteins for their effects on DNA repair to gain biologically more significant measures of their ability to process junctions. Three different strains were used for these experiments: a ruvA mutant to assess the ability to interact with wild-type RuvB and RuvC; a ruv+ strain to reveal any interference with the activity of wild-type RuvAB and/or RuvABC; and a rus-1 ΔruvAC strain to assess junction targeting. The rus-1 mutation activates expression of the RusA Holliday junction resolvase, which promotes repair very efficiently in the absence of RuvABC (Mandal et al., 1993; Sharples et al., 1994; Mahdi et al., 1996). However, its activity is inhibited by RuvA, which competes more effectively for junction DNA (Chan et al., 1997). Thus, RuvA interferes with RusA’s ability to promote repair in vivo (Mandal et al., 1993), especially when RuvA is amplified from a multicopy plasmid (Rafferty et al., 1998).
The results obtained are summarized in Table I. All the mutant proteins reduced survival of the UV-irradiated rus-1 ΔruvAC strain TNM1208. Mutant proteins with four of the negative charges on the pin neutralized (E55Q and D56N) or substituted with positive charges (E55R and D56K) had quite a severe effect. These proteins clearly retain the ability to interfere with the activity of RusA, and by implication to target Holliday junctions in vivo. In contrast, the E55R,D56K protein with eight positive charges on the pin clearly has a much weaker inhibitory effect on DNA repair in strain TM1208 (Table I). This result implies that E55R,D56K targets junctions with much lower efficiency. This is consistent with its reduced affinity for junction DNA in vitro (Figures 2B and 4B) and its increased affinity for duplex DNA (Figure 4), which would also limit the amount available to target junctions and interfere with RusA.
Results with the ruvA strain SR2210 revealed a correlation between cell survival and the charge change on the mutant pin. The more negative the pin, the greater the ability of RuvA to act with wild-type RuvB and RuvC to promote repair. This correlation is clear in the case of changes made at position 55 (Figure 6B). The E55Q and E55R proteins also reduce survival of the ruv+ strain AB1157, which indicates that they interfere with the ability of wild-type RuvA to promote repair. Expression of ruvA and ruvB is induced following damage to DNA as part of the SOS response (Shurvinton and Lloyd, 1982; Benson et al., 1988; Shinagawa et al., 1988). However, in ruv+ strains carrying a mutant ruvA gene on a multicopy plasmid, the mutant RuvA protein may predominate in the formation of RuvAB and RuvABC complexes, at least immediately following irradiation. The D56K protein behaved very much like E55R, which may reflect the equal number of positive and negative charges on the pin in both cases. However, the D56N protein differed significantly from E55Q, despite the neutralization of four negative charges in both cases. It had a much stronger dominant-negative effect on survival of the irradiated ruv+ strain and also increased the sensitivity of the ruvA mutant substantially. The latter observation indicates that D56N blocks repair mediated by other factors such as RecG, which is known to be critical in the absence of RuvABC (Lloyd, 1991). The E55R,D56K protein had little ability to promote repair, which may reflect its weakened ability to target junctions and to support branch migration.
Taken together, these in vivo studies support the notion that the pin structure of RuvA is a critical feature of the protein required to limit binding to duplex DNA and, in conjunction with RuvB and RuvC, to promote efficient resolution of Holliday junctions.
Discussion
The RuvA protein targets Holliday junction intermediates in recombination and DNA repair to create a platform that facilitates processing of these intermediates by the RuvAB branch migration motor and the RuvABC resolvasome. By making appropriate substitutions of the highly conserved acidic amino acids forming the central pin structure on the DNA-binding surface of RuvA, we have shown that negative charges on the pin facilitate junction targeting by preventing binding to duplex DNA. They also severely constrain the rate of branch migration by RuvAB in a way that appears crucial for junction processing in vivo. These findings provide the first evidence that RuvA has a mechanistic role in branch migration that may relate to the efficiency of junction resolution.
Structure specificity of DNA binding
The grooves radiating from the central pin on the DNA-binding surface of RuvA provide a means to establish specific contacts with each of the four duplex arms of an open Holliday junction (Rafferty et al., 1996). Binding is consolidated in particular by two HhH motifs along one side of each groove (Hargreaves et al., 1998; Rafferty et al., 1998; Ariyoshi et al., 2000). In complex I, these interactions tilt the arms some 12° from the plane of the junction and bring the crossover into proximity with the acidic pin where water-mediated hydrogen bonds may establish further contacts between unpaired nucleotides at the crossover and the acidic side chains of Glu55 and Asp56 (Ariyoshi et al., 2000). We found that mutant proteins with altered charges on the pin retain the ability to bind junction DNA with high affinity and to form both complex I and complex II. However, they also bind duplex DNA. Reducing the charges may allow a duplex molecule to be bound in one groove, with its end approaching the altered pin. However, from the range of complexes detected, we suspect that a DNA duplex may dock across the altered pin into two diametrically related grooves. Wild-type RuvA cannot form such a complex. Presum ably, stable contacts within two diametrically related grooves would require the DNA to bend towards the protein surface, leading to charge clashes with the acidic pin. This is consistent with our finding that the most stable duplex DNA binding was observed with the E55R,D56K protein. The eight positive charges on the altered pin may help to keep the duplex DNA in the conformation required to stabilize interactions with the HhH motifs.
Although the charge mutants are able to bind duplex DNA, we discovered that all but the E55R,D56K protein were able to target junctions very effectively in vivo, as revealed by their ability to block the activity of RusA resolvase. Even so, these proteins had a reduced ability to promote repair in a ruv mutant strain and, moreover, their activity in this respect could be related to the net charge on the pin—the more positive the pin, the greater the failure to promote repair. A similar correlation was found with respect to the stimulation of junction resolution by RuvC induced by RuvAB, although we did not investigate all the proteins in detail. Taken together, these findings suggest that the acidic pin may have additional and more critical functions unrelated to junction targeting.
Branch migration by RuvAB and resolution by RuvABC
We made the remarkable discovery that reducing the negative charge on the pin increased the rate of the RuvAB-mediated branch migration reaction quite markedly. The assay used measured dissociation of a synthetic junction with heterologous arms. Thus, we cannot exclude the possibility that the mutant RuvAB complex simply has an improved ability to drive branch migration through such heterology. However, this would not explain why all the charge mutants except E55R,D56K target junctions in vivo as well or nearly as well as wild-type RuvA and yet promote DNA repair with significantly reduced efficiency.
In the crystal structure of RuvA–junction complex I studied by Ariyoshi et al. (2000), the unpaired nucleotides at the crossover are located ∼6 Å from the carboxyl side chains of Glu55 and Asp56, which is close enough to allow water-mediated hydrogen bonds to be formed between these acidic side chains and the unpaired nucleotides. However, the negative charges on the pin are also thought to repel the phosphate backbones of the four DNA strands at the point of strand exchange (Ariyoshi et al., 2000). Such interactions may be critical features of the branch migration reaction catalysed by RuvAB as the RuvB helicase rings step through cycles of ATP binding, hydrolysis and DNA translocation to pull the DNA through the complex (Ariyoshi et al., 2000; Singleton et al., 2000). Altering the charges on the pin, especially substituting positive ones, might disrupt these interactions and thus affect the rate of the reaction. The changes we made significantly reduced the ability of RuvAB to stimulate junction cleavage by RuvC and to promote DNA repair in vivo. Whether these negative effects are a direct consequence of the accelerated rate of branch migration manifested by the mutant proteins in vitro remains to be determined. In the RuvABC resolvasome, RuvC is assumed to scan the DNA for cleavable sequences at the crossover as the strands are drawn across the surface of RuvA (Whitby et al., 1996; Eggleston et al., 1997; Davies and West, 1998; van Gool et al., 1998, 1999; Zerbib et al., 1998). The rate at which RuvAB drives branch migration may critically affect the ability of RuvC to make the necessary base-specific contacts or to interact subsequently with the junction in a productive manner. An increase in the rate, together with any reduction in the ability of RuvA to target Holliday junctions, may explain the correlation observed between the charge on the mutant RuvA pin and the ability of RuvABC to promote DNA repair.
Exactly how RuvC targets strand cleavage remains to be determined, but from the analysis presented it may be that interactions at the acidic pin on the RuvA surface have a role in nucleotide recognition by RuvC. Thus, while RuvB provides the motor for driving branch migration and RuvC the nuclease activity for strand cleavage, RuvA may have a mechanistic role that influences and refines both activities, and links the two together. Such interactions illustrate the constraints on co-evolution of protein complexes and the refinements that lead to efficient molecular machines.
Materials and methods
Strains, plasmids and general methods
Escherichia coli K-12 strains AB1157 (ruv+), SR2210 (ruvA200), TNM1208 (rus-1 ΔruvAC65) and the SI171 (ΔruvAC65) derivative of E.coli BL21 (DE3) have been described, as has the vector plasmid pT7-7 and its ruvA+ derivative pAM159 (Rafferty et al., 1998). Strains were grown in LB medium with antibiotic selection as required, and sensitivity to UV light was measured as described (Mandal et al., 1993).
Construction of RuvA pin mutants
RuvA proteins with defined amino acid substitutions at Glu55 and/or Asp56 were made by introducing the appropriate codon changes into the ruvA gene in pAM159 using the Quikchange™ procedure (Stratagene), Pfu DNA polymerase and mutagenic oligonucleotides of 25–33 nucleotides in length as PCR primers. GAA165 (E55) was changed to CAG (Q), CGT (R) or GAT (D), and GAC168 (D56) was changed to AAC (N) or AAA (K). The desired mutation was verified by sequencing the ruvA insert in each of the plasmids generated.
Proteins
Wild-type and mutant RuvA proteins were purified following expression of the appropriate pT7-7-derived ruvA construct in strain SI171 as described (Rafferty et al., 1998). RuvB and RuvC were purified as described (Tsaneva et al., 1992). Protein concentrations were estimated by a modified Bradford assay using a Bio-Rad assay kit and bovine serum albumin (BSA) as standard. Amounts are expressed as moles of the monomeric protein.
DNA substrates
Synthetic X-junctions J11 and J12 are 50mer synthetic Holliday junctions with mobile cores of homology of 11 and 12 bp, respectively, and were prepared by annealing the appropriate oligonucleotides, as described (Whitby et al., 1996). One oligonucleotide was labelled with 32P at the 5′ end prior to annealing. The 50mer duplex DNA molecule used the same labelled oligonucleotide as used for J12, plus its complement. χ DNA, a large Holliday structure with a 300 bp homologous core flanked by heterologous arms of 0.8–1.6 kb, was made and labelled on all four arms as described (McGlynn and Lloyd, 2000).
DNA binding assays
Reaction mixtures (20 µl) contained 32P-labelled junction (J11) or linear duplex DNA in 50 mM Tris–HCl pH 8.0, 5 mM EDTA, 1 mM dithiothreitol (DTT), 100 µg/ml BSA and 6% (v/v) glycerol. Reactions containing RuvA as indicated were left on ice for 10 min prior to loading on a 4% polyacrylamide gel in low ionic strength buffer (6.7 mM Tris–HCl pH 8.0, 3.3 mM sodium acetate, 2 mM EDTA). Gels were run at 160 V for 90 min, dried and analysed by autoradiography and phosphoimaging.
Branch migration assays
32P-labelled J11 DNA was mixed with RuvA and/or RuvB protein as indicated in helicase buffer (20 mM Tris–HCl pH 7.5, 2 mM DTT, 100 µg/ml BSA) in a final volume of 20 µl. RuvA was added before RuvB and reactions were incubated at 37°C for 30 min before adding 5 µl of stop mix (2.5% SDS, 200 mM EDTA, 10 mg/ml proteinase K) and incubating for a further 10 min. DNA products were analysed by native PAGE, using 10% gels in TBE buffer. Gels were dried, and labelled products analysed as described. For time courses, 20 µl samples were removed at intervals from bulk reactions and processed as described.
RuvC-mediated resolution of χ DNA
Reactions contained 0.012 nM 32P-labelled χ DNA and proteins as indicated in 50 mM Tris–HCl pH 8.0, 15 mM Mg(OAc)2, 20 mM KOAc, 1 mM DTT, 2 mM ATP, 20 mM phosphocreatine and 100 µg/ml BSA, in a final volume of 20 µl. In reactions containing RuvA, RuvB and RuvC, RuvB was added to the reaction mixture containing the DNA and left for 5 min at 37°C before adding RuvC and RuvA. Incubation was continued for 60 min before addition of 5 µl of stop mix and incubating for a further 10 min. DNA products were resolved by agarose gel electrophoresis and analysed as described.
Acknowledgments
Acknowledgements
We are grateful to John Rafferty for advice on the structure of RuvA–DNA complexes, Peter McGlynn for χ DNA, and Tim Moore for helping with Figure 1A. We also thank Lynda Harris, Carol Brown and Lisa Corbett for excellent technical support. S.M.I. was in receipt of a studentship from the Biotechnology and Biological Sciences Research Council, and the work was supported by a Medical Research Council programme grant awarded to R.G.L. and G.J.S.
References
- Ariyoshi M., Nishino,T., Iwasaki,H., Shinagawa,H. and Morikawa,K. (2000) Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl Acad. Sci. USA, 97, 8257–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F.E., Illing,G.T., Sharples,G.J. and Lloyd,R.G. (1988) Nucleotide sequencing of the ruv region of Escherichia coli K-12 reveals a LexA regulated operon encoding two genes. Nucleic Acids Res., 16, 1541–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt E.L., Sharples,G.J. and Lloyd,R.G. (1999) Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J. Mol. Biol., 286, 403–415. [DOI] [PubMed] [Google Scholar]
- Chan S.N., Harris,L., Bolt,E.L., Whitby,M.C. and Lloyd,R.G. (1997) Sequence-specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem., 272, 14873–14882. [DOI] [PubMed] [Google Scholar]
- Chan S.N., Vincent,S.D. and Lloyd,R.G. (1998) Recognition and manipulation of branched DNA by the RusA Holliday junction resolvase of Escherichia coli. Nucleic Acids Res., 26, 1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A.A. and West,S.C. (1998) Formation of RuvABC–Holliday junction complexes in vitro. Curr. Biol., 8, 725–727. [DOI] [PubMed] [Google Scholar]
- Eggleston A.K., Mitchell,A.H. and West,S.C. (1997) In vitro reconstitution of the late steps of genetic recombination in E.coli. Cell, 89, 607–617. [DOI] [PubMed] [Google Scholar]
- Hargreaves D., Rice,D.W., Sedelnikova,S.E., Artymiuk,P.J., Lloyd,R.G. and Rafferty,J.B. (1998) Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nature Struct. Biol., 5, 441–446. [DOI] [PubMed] [Google Scholar]
- Hiom K. and West,S.C. (1995) Branch migration during homologous recombination: assembly of a RuvAB–Holliday junction complex in vitro. Cell, 80, 787–793. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- Lloyd R.G. (1991) Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG.J. Bacteriol., 173, 5414–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R.G. and Low,K.B. (1996) Homologous recombination. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella Cellular and Molecular Biology, 2nd edn. ASM Press, Washington, DC, pp. 2236–2255. [Google Scholar]
- Lloyd R.G. and Sharples,G.J. (1993) Dissociation of synthetic Holliday junctions by E.coli RecG protein. EMBO J., 12, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R.G., Benson,F.E. and Shurvinton,C.E. (1984) Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol. Gen. Genet., 194, 303–309. [DOI] [PubMed] [Google Scholar]
- Mahdi A.A., Sharples,G.J., Mandal,T.N. and Lloyd,R.G. (1996) Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J. Mol. Biol., 257, 561–573. [DOI] [PubMed] [Google Scholar]
- Mandal T.N., Mahdi,A.A., Sharples,G.J. and Lloyd,R.G. (1993) Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB and ruvC mutations. J. Bacteriol., 175, 4325–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P. and Lloyd,R.G. (2000) Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell, 101, 35–45. [DOI] [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Parsons C.A., Tsaneva,I., Lloyd,R.G. and West,S.C. (1992) Interaction of E.coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc. Natl Acad. Sci. USA, 89, 5452–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty J.B., Sedelnikova,S.E., Hargreaves,D., Artymiuk,P.J., Baker,P.J., Sharples,G.J., Mahdi,A.A., Lloyd,R.G. and Rice,D.W. (1996) Crystal structure of the DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science, 274, 415–421. [DOI] [PubMed] [Google Scholar]
- Rafferty J.B., Ingleston,S., Sharples,G.J., Lloyd,R.G. and Rice,D.W. (1998) Structural similarities between E.coli RuvA protein and other DNA binding proteins and a mutational analysis of its binding to the Holliday junction. J. Mol. Biol., 278, 105–116. [DOI] [PubMed] [Google Scholar]
- Roe S.M., Barlow,T., Brown,T., Oram,M., Keeley,A., Tsaneva,I.R. and Pearl,L.H. (1998) Crystal structure of an octameric RuvA–Holliday junction complex. Mol. Cell, 2, 361–372. [DOI] [PubMed] [Google Scholar]
- Seigneur M., Bidnenko,V., Ehrlich,S.D. and Michel,B. (1998) RuvAB acts at arrested replication forks. Cell, 95, 419–430. [DOI] [PubMed] [Google Scholar]
- Sharples G.J., Chan,S.C., Mahdi,A.A., Whitby,M.C. and Lloyd,R.G. (1994) Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J., 13, 6133–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples G.J., Ingleston,S.M. and Lloyd,R.G. (1999) Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG and RusA. J. Bacteriol., 181, 5543–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H., Makino,K., Amemura,M., Kimura,S., Iwasaki,H. and Nakata,A. (1988) Structure and regulation of the Escherichia coli ruv operon involved in DNA repair and recombination. J. Bacteriol., 170, 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C.E. and Lloyd,R.G. (1982) Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol. Gen. Genet., 185, 352–355. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Sawaya,M.R., Ellenberger,T. and Wigley,D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed] [Google Scholar]
- Tsaneva I.R., Illing,G.T., Lloyd,R.G. and West,S.C. (1992) Purification and physical properties of the RuvA and RuvB proteins of Escherichia coli. Mol. Gen. Genet., 235, 1–10. [DOI] [PubMed] [Google Scholar]
- van Gool A.J., Shah,R., Mezard,C. and West,S.C. (1998) Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J., 17, 1838–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool A.J., Hajibagheri,N.M., Stasiak,A. and West,S.C. (1999) Assembly of the Escherichia coli RuvABC resolvasome directs the orientation of Holliday junction resolution. Genes Dev., 13, 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. (1997) Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- Whitby M.C., Ryder,L. and Lloyd,R.G. (1993) Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell, 75, 341–350. [DOI] [PubMed] [Google Scholar]
- Whitby M.C., Bolt,E.L., Chan,S.N. and Lloyd,R.G. (1996) Interactions between RuvA and RuvC at Holliday junctions: inhibition of junction cleavage and formation of a RuvA–RuvC–DNA complex. J. Mol. Biol., 264, 878–890. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Colloms,S.D., Sherratt,D.J. and West,S.C. (1997) Effect of DNA topology on Holliday junction resolution by Escherichia coli RuvC and bacteriophage T7 endonuclease I. J. Mol. Biol., 270, 663–673. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Mézard,C., George,H. and West,S.C. (1998) Coordinated actions of RuvABC in Holliday junction processing. J. Mol. Biol., 281, 621–630. [DOI] [PubMed] [Google Scholar]