Abstract
An Arabidopsis thaliana line that is mutant for the R2R3 MYB gene, AtMYB4, shows enhanced levels of sinapate esters in its leaves. The mutant line is more tolerant of UV-B irradiation than wild type. The increase in sinapate ester accumulation in the mutant is associated with an enhanced expression of the gene encoding cinnamate 4-hydroxylase, which appears to be the principal target of AtMYB4 and an effective rate limiting step in the synthesis of sinapate ester sunscreens. AtMYB4 expression is downregulated by exposure to UV-B light, indicating that derepression is an important mechanism for acclimation to UV-B in A.thaliana. The response of target genes to AtMYB4 repression is dose dependent, a feature that operates under physiological conditions to reinforce the silencing effect of AtMYB4 at high activity. AtMYB4 works as a repressor of target gene expression and includes a repression domain. It belongs to a novel group of plant R2R3 MYB proteins involved in transcriptional silencing. The balance between MYB activators and repressors on common target promoters may provide extra flexibility in transcriptional control.
Keywords: dose dependence/plants/repressor/R2R3 MYB transcription factor/UV-B tolerance
Introduction
Because plants require sunlight for photosynthesis they are inevitably exposed to ultraviolet light, including that in the wavelength range of 280–320 nm (ultraviolet-B, UV-B). UV-B light is potentially very damaging to DNA and proteins, and increases free radical production. To provide a sunscreen, most plants synthesize secondary metabolites that can absorb UV-B and also act as free radical scavengers to counteract the damaging consequences of irradiation. Prominent amongst these sunscreens are the phenylpropanoids, and from these compounds some plants produce flavonoids, others produce hydroxycinnamic acid derivatives and yet others, such as Arabidopsis thaliana, produce sinapate esters (hydroxycinnamic acid derivatives) in their leaves, but augment this protection with flavonoid production (Li et al., 1993).
The control of production of UV-B-protecting metabolites has been intensively studied in several plant species and has been shown to be predominantly transcriptional (Chappell and Hahlbrock, 1984; Douglas et al., 1987; Schulze-Lefert et al., 1989; Li et al., 1993; Hartmann et al., 1998). The most detailed analysis of transcriptional regulation has been undertaken for the promoter of the chalcone synthase gene (CHS) where light responsive units (LRUs), consisting of an ACGT-containing element (ACE) and a MYB recognition element (MRE), confer responsiveness to UV-containing white light. Two bZIP transcription factors bind the ACE and are believed to be involved in transcriptional activation in response to UV-containing light (Feldbrugge et al., 1994; Sprenger-Haussels and Weisshaar, 2000). Two MYB-related proteins have also been shown to interact with the MRE (Sablowski et al., 1994; Feldbrugge et al., 1997). Other members of the R2R3 MYB gene family have been implicated in regulation of other branches of phenylpropanoid metabolism, including phlobaphene biosynthesis (P in maize; Grotewold et al., 1994), flavonol synthesis (AmMYB305 and AmMYB340) in Antirrhinum majus (Moyano et al., 1996) and hydroxycinnamic acid and monolignol biosynthesis (AmMYB308 and AmMYB330) in tobacco (Tamagnone et al., 1998a). Of these, the action of AmMYB308 in regulating flux to soluble hydroxycinnamic acid derivatives could represent an important control point in regulating the synthesis of UV-protecting compounds, and in the acclimation to UV-B stress of those plants that use these particular phenylpropanoids as sunscreens.
AmMYB308 was isolated from A.majus and overexpression in tobacco caused an inhibition of hydroxycinnamic acid and monolignol biosynthesis, implying that it could repress transcription of the biosynthetic genes. However, MYB-related proteins have generally been shown to operate as transcriptional activators (Weston and Bishop, 1989; Foos et al., 1994). c-MYB has been shown to contain an activation domain, although negative regulatory domains within the C-terminus of the protein may limit the activation by the protein through interaction with negative regulatory proteins (Dubendorff et al., 1992; Wang et al., 1999). B-MYB, although reported to act as a repressor (Foos et al., 1992; Watson et al., 1993), also contains an activation domain (Ansieau et al., 1997). In plants, the view that MYB-related transcription factors function predominantly as transcriptional activators follows from the demonstration of activation domains in the maize MYB-related proteins C1 and P (Sainz et al., 1997), and the presence of regions with a high probability of forming amphipathic α-helix within the predicted C-termini of many other R2R3 MYB-related proteins from plants (Martin and Paz-Ares, 1997; Kranz et al., 1998). Given the evidence for R2R3 MYB-related proteins activating transcription of structural genes in different branches of phenylpropanoid metabolism (Martin and Paz-Ares, 1997), the action of AmMYB308 in repressing expression of some hydroxycinnamic acid synthesizing genes is unusual. Its effect in tobacco might reflect that it can bind to the same target sites as endogenous MYB-related activators, but not activate from these sites in a heterologous host.
To determine the biological role of MYB-related transcription factors in production of hydroxycinnamic acid derivatives, we have characterized a gene from A.thaliana, AtMYB4, orthologous to AmMYB308, and we have identified a knockout mutant. The mutant demonstrates that AtMYB4 plays a significant role in controlling sinapate ester formation and in the development of sunscreens offering protection to UV-B light. AtMYB4 appears to work as a repressor, particularly of one key target gene—that encoding cinnamate 4-hydroxylase. Through regulation of the expression of this gene, AtMYB4 negatively modulates sinapate ester formation in the absence of UV-B light. Expression of AtMYB4 is downregulated in response to UV-B, resulting in increased sinapate ester production.
Results
Identification of AtMYB4 and a knockout mutant from A.thaliana
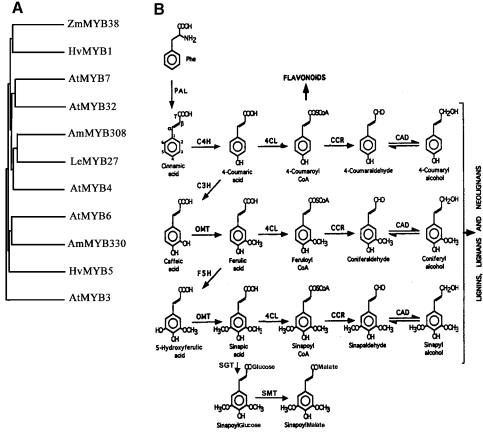
The R2R3-related transcription factor AmMYB308 had been shown to downregulate the C4H, 4CL and CAD genes (Table I; Figure 1B) of hydroxycinnamic acid/monolignol biosynthesis when overexpressed in transgenic tobacco (Tamagnone et al., 1998a). To determine the normal physiological role of AmMYB308 we needed to find a mutation of the gene; the most appropriate strategy was to identify the orthologous gene from A.thaliana and screen the numerous insertion-mutagenized populations for knockout mutations. This task was facilitated by the near-complete cloning of R2R3 MYB genes from A.thaliana (Kranz et al., 1998; Romero et al., 1998). Comparison of sequences encoded by these genes to that of AmMYB308 revealed AtMYB4 to encode the most similar protein (Figure 1A). Both proteins belong to R2R3 subgroup 4 proteins, which share both a conserved motif (pdLNLD/ELxiG/S) and a region potentially forming a zinc finger domain at their C-termini (Kranz et al., 1998). Given the near completion of the A.thaliana genome sequence and the identification of 111 R2R3 MYB genes from a total predicted number of ∼125, it is highly likely that AtMYB4 is the orthologous gene in A.thaliana to AmMYB308 in A.majus.
Table I. Biosynthetic enzymes of phenylpropanoid metabolism.
| Enzyme | Abbreviation | Isoform used | Accession No. |
|---|---|---|---|
| Phenylalanine ammonia lyase | PAL | PAL2 | L33678 |
| Cinnamate 4-hydroxylase | C4H | D78596 | |
| p-coumaroyl 4-CoA ligase | 4CL | 4CL1 | U18675 |
| 4CL3 | AF106088 | ||
| p-coumarate 3-hydroxylase | C3H | ||
| Caffeic acid o-methyl transferase | OMT | U70424 | |
| Caffeoyl CoA o-methyl transferase | CCoAOMT | L40031 | |
| Ferulate 5-hydroxylase | F5H | AF068574 | |
| Caffeoyl CoA reductase | CCR | ||
| Cinnamoyl alcohol dehydrogenase | CAD | CAD1 | L37883 |
| Sinapic acid UDPG:sinapoyl transferase | SGT | ||
| Sinapoylglucose malate:sinapoyl transferase | SMT | ||
| Chalcone synthase | CHS | AF012810 |
Fig. 1. (A) Dendrogram showing structural relationships between members of R2R3 MYB subgroup 4 from different plant species. Zm signifies Zea mays; Hv, Hordeum vulgare; Le, Lycopersicon esculentum; At, Arabidopsis thaliana; Am, Antirrhinum majus. For gene references see Kranz et al. (1998). (B) Pathway of hydroxycinnamic acid metabolism in A.thaliana leaves. Interconversions producing the precursors for flavonoid biosynthesis, monolignols and sinapate esters are indicated (for key to enzymes see Table I). CCoAOMT can use caffeoyl CoA and 5-hydroxyferuloyl CoA as substrates to produce feruloyl CoA and sinapoyl CoA in an alternative methylation pathway for sinapoyl ester production.
A mutant of AtMYB4 was sought by PCR-mediated reverse genetic screening of several insertion mutagenized populations, and a stable dSpm insertion was identified (ecotype Colombia; Meissner et al., 1999). The insertion was in the first exon, 92 bp after the initiating ATG. It disrupted the sequence encoding the first repeat (R2) of the DNA binding domain of AtMYB4. Heterozygous plants were identified from pooled seed, and plants segregating as homozygous wild type or homozygous mutants were selected for phenotypic analysis. Two individuals, homozygous for the dSpm insertion, were propagated by selfing (atmyb4– A and B) as examples of the mutant, and one homozygous wild-type sibling line (WT) was similarly propagated. The effect of the dSpm insertion on the expression of AtMYB4 was established by northern blots of RNA from leaves, where no transcript was visible in the mutant compared with a transcript of 1.2 kb in leaves from the wild-type plant. This was confirmed by RT–PCR (Figure 2A) of cDNA made from RNA isolated from leaves.
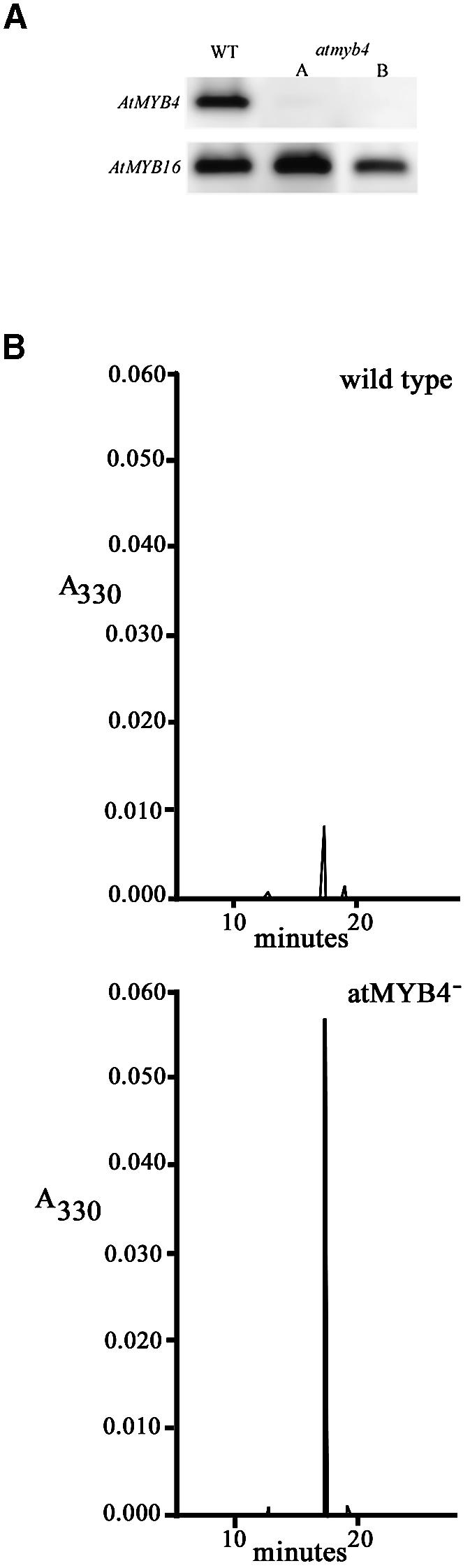
Fig. 2. (A) Expression of AtMYB4 in leaves of wild-type (WT) and atmyb4– mutant plants. Expression was monitored by RT–PCR amplification of first strand cDNA. The transcript of another MYB gene expressed in leaves, AtMYB16, was amplified from the same cDNA as a control. (B) HPLC traces of soluble phenolics in methanolic extracts from leaves of wild-type and atmyb4– mutant plants. A single major peak absorbing at A330 was confirmed to be sinapoyl malate by liquid chromatography mass spectrometry (LCMS). In the mutant the levels of sinapoyl malate were 6- to 7-fold higher than in wild type.
Phenotype of atmyb4– mutant plants
Visual inspection of atmyb4– plants showed no obvious differences in growth rate or appearance in comparison to wild-type plants. Wild-type and mutant lines were grown on soil in the greenhouse for 12 days and methanolic extracts of leaves were analysed by HPLC. A single major peak corresponding to sinapoyl malate was apparent from wild-type leaves (Figure 2B). This is the major soluble phenolic in A.thaliana leaves and is synthesized via the hydroxycinnamic acid pathway (Figure 1B; Chapple et al., 1992; Li et al., 1993; Landry et al., 1995; Lorenzen et al., 1996). In the mutant seedlings this peak was ∼6-fold greater, indicating a significant increase in the production of sinapoyl malate in atmyb4– mutant plants. These experiments were repeated with leaf material from seedlings grown on agar, and consistently showed significantly higher levels of sinapate esters in the mutant lines compared with wild type, although the differences were not as great as the difference observed in the first analysis (Table II).
Table II. Sinapoyl malate content (µg/g fresh weight leaves) of A.thaliana lines.
| Line | Plants grown under |
|
|---|---|---|
| Short days | Long days | |
| Wild type | 458.9 | 295.7 |
| atmyb4– A | 524.4 | 389.8 |
| atmyb4– B | 557.7 | 352.8 |
| 35S-AtMYB4A | 372.9 | 252.4 |
| 35S-AtMYB4B | 258.2 | 238.7 |
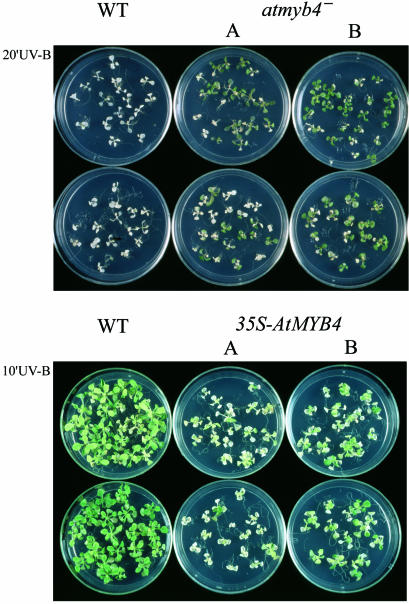
The enhanced levels of sinapoyl malate in the atmyb4– mutant lines suggested that these plants might be more tolerant of UV-B (Li et al., 1993; Landry et al., 1995). A dose–response curve showed that a 20 min exposure to UV-B light (3.2 mW/cm2) caused >80% death of wild-type seedlings. Treatment of seedlings homozygous for the atmyb4– mutation for the same length of time gave significantly reduced plant death (36%, 21%), establishing that loss of AtMYB4 activity increased sinapoyl malate levels, affording greater tolerance to UV-B (Figure 3A).
Fig. 3. Effect of mutation of AtMYB4 on the tolerance of A.thaliana seedlings to UV-B. The upper panel shows seedlings exposed to 20 min UV-B (3.2 mW/cm2). More seedlings survived in duplicate plates of atMYB4– mutants than did from the wild type. The lower panel shows the enhanced sensitivity of AtMYB4-overexpressing plants to UV-B. Plants were exposed to 10 min UV-B (3.2 mW/cm2). Two independent transgenic lines showed enhanced seedling death on duplicate plates compared with wild type.
Expression of AtMYB4 in response to light and wounding
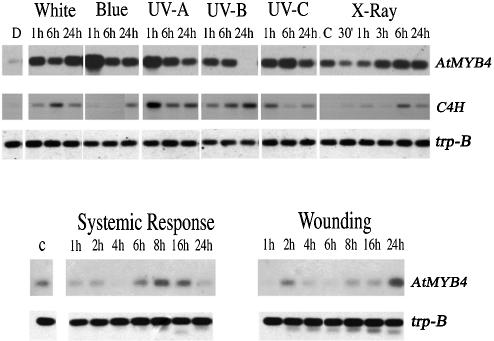
Given that AtMYB4 represses sinapoyl malate production, and that mutation of this gene will make A.thaliana seedlings more tolerant of UV-B, it was of interest to determine the normal response of AtMYB4 expression to varying light stimuli. Generally, AtMYB4 expression was induced by all types of light (Figure 4). However, while prolonged (24 h) exposure to white light left AtMYB4 expression more or less the same as after 1 h, prolonged exposure to blue or UV light gave reduced levels of AtMYB4 expression. This was true for UV-A, UV-B and UV-C light, but the most dramatic change occurred in response to UV-B. Twenty-four hour exposure to relatively low levels of UV-B (0.2 mW per cm2) gave complete loss of AtMYB4 expression. This response was reproducible in independent extracts from seedlings.
Fig. 4. Expression of AtMYB4 in seedlings grown under different light conditions. Transcript levels were determined from total RNA from leaves. The expression of the C4H gene was determined for the same samples. As a control, the β subunit of tryptophan synthase (trpB) was amplified. The expression of AtMYB4 was also determined for wounded leaves and for leaves from wounded plants (systemic response).
In addition, the response of AtMYB4 expression to wounding was analysed (Figure 4). Transcript levels of AtMYB4 decreased in leaves 4–6 h after wounding, both in wounded leaves and systemically in non-wounded leaves from the same rosette. AtMYB4 transcript levels recovered after ∼24 h.
Identification of genes regulated by AtMYB4
To analyse which genes AtMYB4 regulates, RNA was extracted from leaves of wild-type and mutant, soil-grown plants. First strand cDNA was synthesized and primer pairs were designed for each of the genes encoding enzymes in hydroxycinnamic acid and monolignol biosynthesis, where the gene sequence had been published. These were used initially to amplify the relevant transcripts from wild-type RNA (Table I). cDNAs were subcloned and their identity was confirmed by sequencing.
The same primers used to amplify the biosynthetic gene transcripts were used for quantitative RT–PCR of cDNA from wild type and mutant. Compared with wild-type RNA, only one gene showed higher transcript levels in the two mutant lines—that encoding C4H (Table I; Figure 5). Given that atmyb4– mutant plants routinely showed higher levels of sinapate esters, this suggested that the expression of the C4H gene can significantly affect flux along this branch of phenylpropanoid metabolism. In other words, C4H represents an important control point in the synthesis of sinapate esters with an implied high flux-control coefficient.
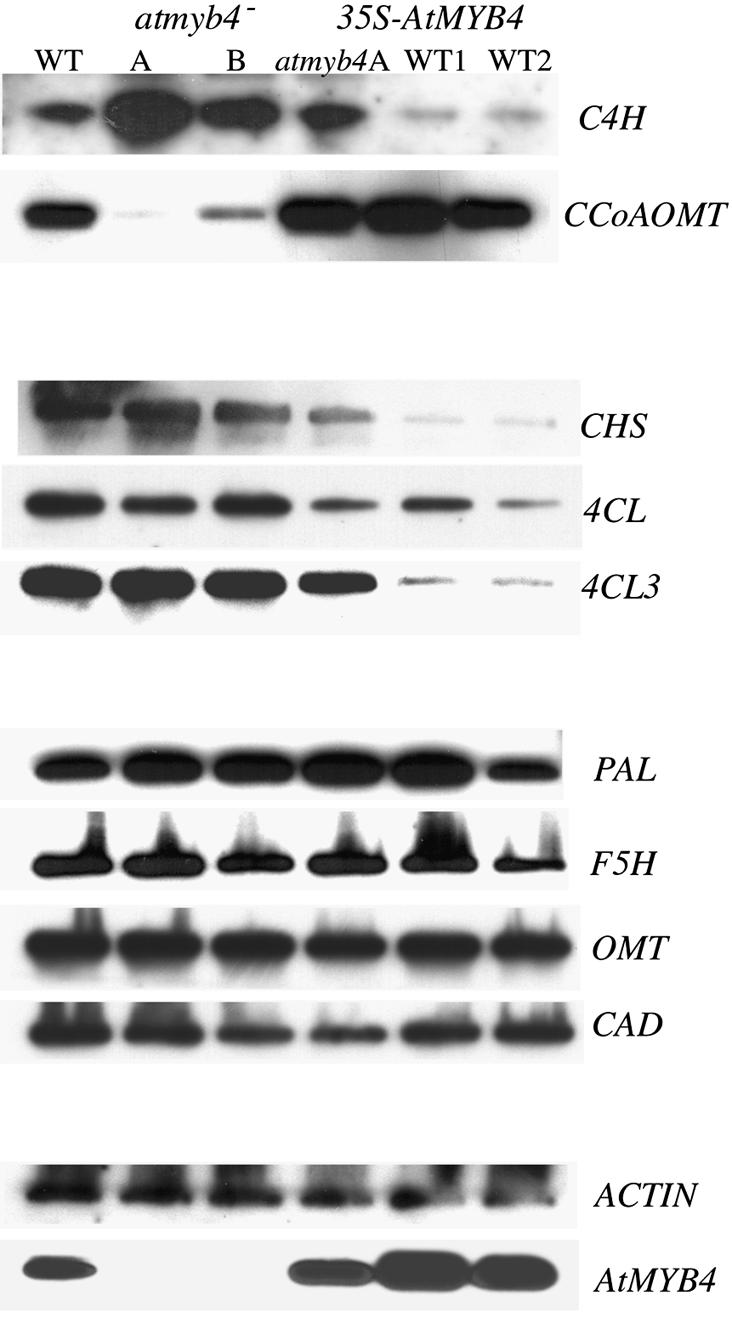
Fig. 5. Effect of mutation of AtMYB4 and overexpression of the gene on transcript levels of the genes of hydroxycinnamic acid and flavonoid metabolism. Transcript levels were determined by quantitative RT–PCR. Expression of C4H increased in the mutant and was decreased in overexpressing lines. The overexpression construct in the atmyb4– mutant background gave an intermediate transcript level. The CCoAOMT gene showed complementary responses; its transcript levels were decreased in the mutant and enhanced in the overexpressing lines. CHS, 4CL1 and 4CL3 genes were downregulated by overexpression of AtMYB4.
Only one other gene showed a difference in expression between the atmyb4– mutants and the wild type, that encoding CCoAOMT (Figure 5). Interestingly, this showed reduced transcript levels in the mutant compared with the wild type, implying that its expression was upregulated in response to AtMYB4 activity.
We examined expression of the gene encoding C4H in relation to expression of AtMYB4. Under blue or UV-B light there was a negative correlation between AtMYB4 expression and C4H expression (Figure 4). This suggested that under these conditions AtMYB4 activity is the principal factor influencing expression of the C4H gene, but under other light conditions other factors are probably more influential.
Overexpression of AtMYB4 in tobacco and A.thaliana
Overexpression of AtMYB4 gave rise to the same phenotype in transgenic tobacco that had been observed for AmMYB308 (Figure 6): slow-growing plants with pale leaves that showed premature white lesions on the mature leaves indicative of reduced levels of hydroxycinnamic acid derivatives (Tamagnone et al., 1998b). AtMYB4 overexpression reduced the steady state transcript levels of the C4H, 4CL1 and CAD genes in tobacco in the same way as AmMYB308 (data not shown; Tamagnone et al., 1998a).
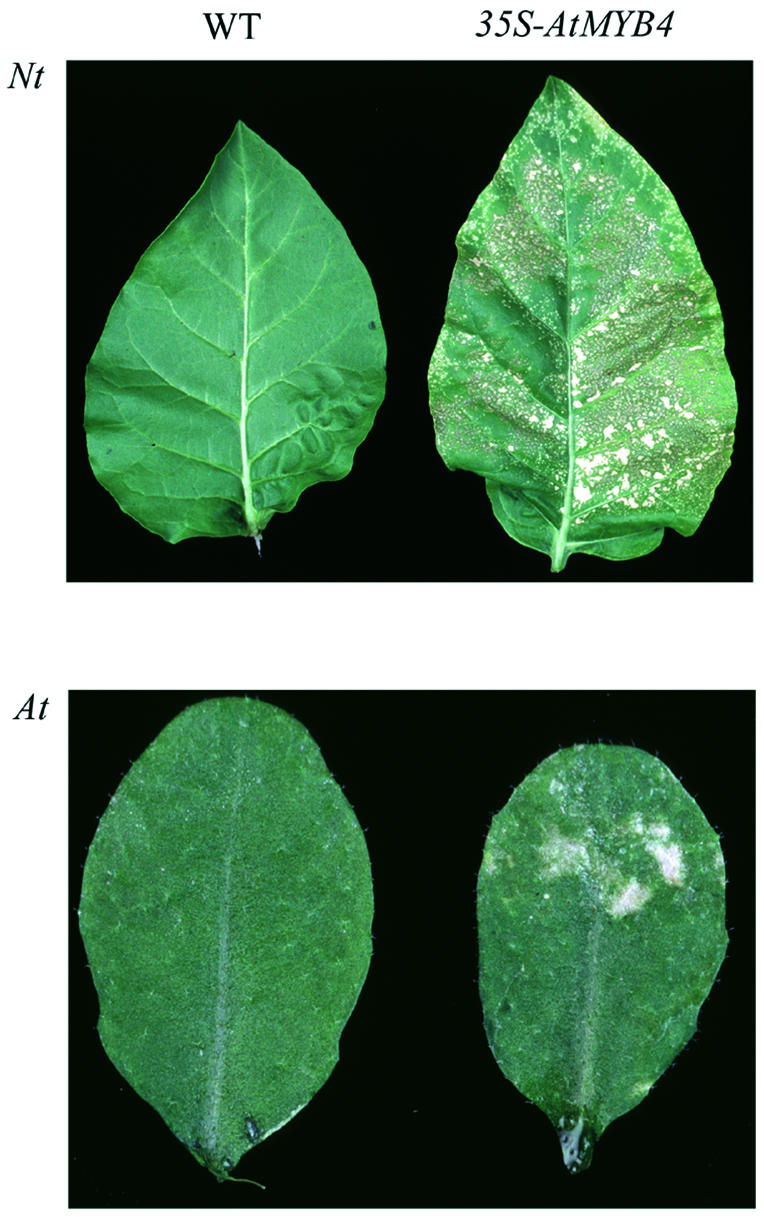
Fig. 6. Phenotypic effects of AtMYB4 overexpression in tobacco (Nt) and A.thaliana (At). In both species overexpression caused the appearance of white lesions on older leaves very similar to those observed for AmMYB308 overexpression in tobacco (Tamagnone et al., 1998b).
In A.thaliana plants overexpressing AtMYB4, white lesions were apparent in older leaves (Figure 6). The atMYB4– mutant line was also transformed with the construct giving high levels of AtMYB4 expression. This line showed no phenotype compared with wild type.
Plants overexpressing AtMYB4 were more sensitive to UV-B. Exposure to a transilluminator for 10 min, which caused no death in wild-type plants, resulted in 60 and 37% plant death in two independent lines overexpressing AtMYB4 (Figure 3B). Analysis of the phenylpropanoids in these overexpressing lines confirmed that they had reduced levels of sinapate esters (Table II), but their flavonoid composition was not affected (data not shown).
Identification of genes regulated by AtMYB4 in overexpressing A.thaliana lines
Comparison of transcript levels for the biosynthetic genes in two independent overexpressing lines with wild-type plants showed that expression of the C4H gene was considerably reduced by AtMYB4 overexpression (Figure 5). Interestingly, the CCoAOMT gene showed increased transcript levels in both overexpressing lines, and increased expression in the mutant line overexpressing AtMYB4. The transcript levels of PAL2, F5H, OMT and CAD1 were unaffected by overexpression of AtMYB4, in contrast to the effects of overexpression of the gene in tobacco where CAD was downregulated. This result suggests that regulation of the CAD genes from tobacco is distinct from regulation of the CAD1 gene in A.thaliana. Interestingly, overexpression of AtMYB4 also affected three genes that were unaffected in the atmyb4– mutant, CHS, 4CL1 and 4CL3 (Figure 5), which all showed reduced transcripts in overexpressing lines, while the atmyb4– mutant overexpressing AtMYB4 showed an intermediate effect.
Dose-dependent selection of target genes by AtMYB4
One of the most significant aspects of our data on AtMYB4 action was that it appeared to regulate new genes when present in high concentrations. Comparison of the transcript levels in wild-type and atmyb4– knockout plants suggested that only the C4H gene was negatively regulated by AtMYB4, and that the CCoAOMT gene was positively regulated by the transcription factor. However, in plants with higher activity of AtMYB4, as achieved by expression of the cDNA under the control of the double CaMV 35S promoter, the CHS, 4CL1 and 4CL3 genes were also negatively regulated by AtMYB4. Additional experiments were undertaken to determine whether dose-dependent selection of target genes by AtMYB4 occurred under physiological conditions.
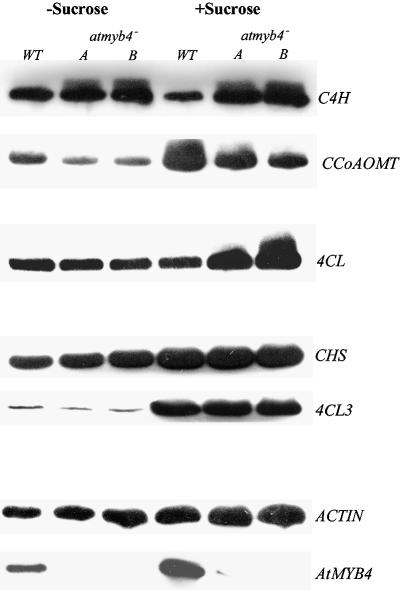
The steady state transcript levels of AtMYB4 increased in plants grown on agar containing sucrose compared with plants grown on agar alone (Kranz et al., 1998). Consequently, wild-type and atmyb4– mutant plants were grown on agar alone and on agar supplemented with 100 mM sucrose. This resulted in an induction of AtMYB4 transcript levels in the wild-type plants grown on agar plus sucrose, confirming the previous observations (Figure 7). Comparison of transcript levels for the C4H gene in wild-type and mutant plants showed reduced transcripts in wild-type plants grown on sucrose compared with those without sucrose, and higher levels of transcripts in the mutant lines compared with the wild type under both conditions, as expected. Also, as predicted, the CCoAOMT gene was upregulated in plants grown on sucrose, but atmyb4– mutant plants showed lower transcript levels than wild type when grown on agar with or without sucrose. Interestingly, the 4CL1 gene (Ehlting et al., 1999) showed no difference in transcript levels between wild type and mutants on agar without sucrose, but a higher level of transcript was observed in mutant lines compared with wild type where plants were grown on agar plus sucrose. These data suggested that the induction of AtMYB4 expression by sucrose results in elevation of AtMYB4 activity sufficient to bring the 4CL1 gene under control of the repressor, and that the dose-dependent changes in the target specificity of AtMYB4 can play a physiological role in regulating target gene expression in vivo.
Fig. 7. Dose dependence of target gene repression by AtMYB4. Transcript levels were analysed by quantitative RT–PCR. The effect of AtMYB4 was monitored by comparing transcript levels in wild type and mutants (atmyb4– A and B). The C4H gene was downregulated by AtMYB4 in plants grown with or without sucrose. The CCoAOMT gene was upregulated by AtMYB4 in plants grown with or without sucrose. On high sucrose, when AtMYB4 expression was elevated, the 4CL1 gene was negatively regulated by AtMYB4. There was no effect of AtMYB4 activity on transcript levels for CHS or 4CL3 genes.
Analysis of AtMYB4 regulation using transfection assays
To determine whether the effects of AtMYB4 on target gene expression involved direct interaction of the encoded transcription factor with the target gene promoters, transfection assays were undertaken in A.thaliana protoplasts. The promoters of the C4H, CHS, 4CL3 and CCoAOMT genes were fused to the GUS reporter gene. Each reporter gene construct was assayed with and without AtMYB4 in co-transfection assays. Expression from the C4H promoter was reduced 26-fold by co-transfection with AtMYB4, establishing that the effect of the transcription factor is to repress expression from the C4H promoter (Figure 8A). This relationship was confirmed by transforming wild-type and atmyb4– mutant lines with a C4H promoter–GUS reporter construct. GUS assays (three replicates each) from 10 independent transformants of the wild-type line gave 5.7 ± 1.2 nmol 4-methylumbelliferone (MU) mg/protein/min, whereas assays from 10 independent transformants of the mutant line gave 10.8 ± 1.0 nmol MU mg/protein/min, confirming the higher expression from the C4H promoter in the absence of AtMYB4 activity. Histochemical staining revealed that the C4H promoter gave GUS activity all over the leaf but particularly in the vascular tissue in the mutant, indicating that AtMYB4 is repressing expression of this gene all over the leaf (Figure 8B).
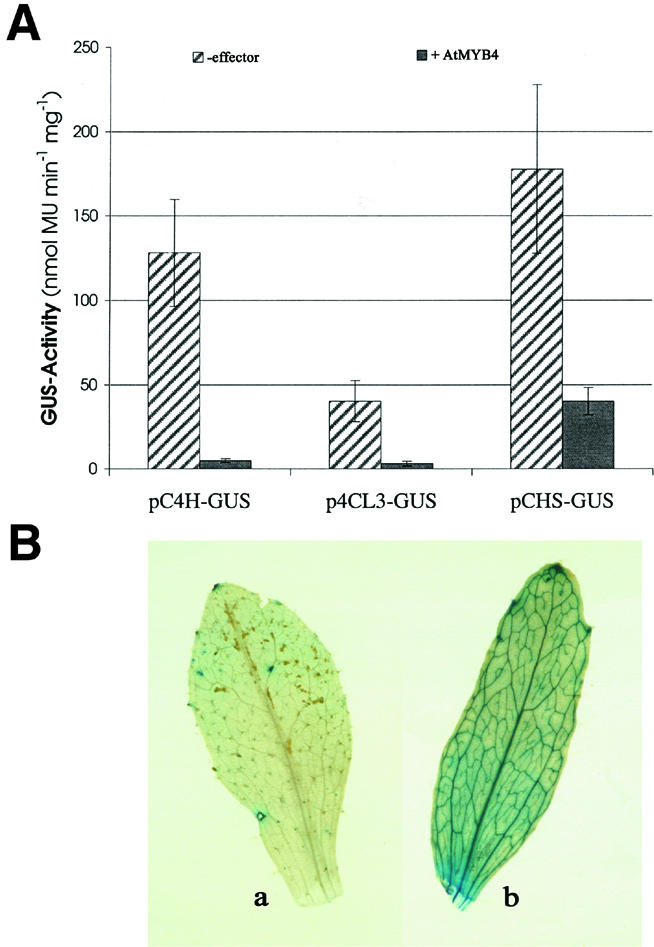
Fig. 8. (A) Transfection assays illustrating the effect of AtMYB4 on GUS expression driven by the C4H gene promoter, the CHS gene promoter and the 4CL3 gene promoter. (B) GUS staining of wild-type (a) and atmyb4– mutant plants (b) following transformation with the C4H promoter–GUS fusion. The activity driven by the C4H promoter was significantly enhanced in the mutant line, in the general tissue of the leaf and particularly in the vascular tissue, indicating, by comparison, the regions where AtMYB4 repressed C4H expression most strongly.
The transfection assays also revealed an inhibition by AtMYB4 of expression from the 4CL3 and the CHS promoters, although the downregulation of these reporter genes was not as great as for the C4H promoter (13- and 4-fold, respectively). These data confirmed that, at high concentrations, AtMYB4 can repress these genes, and that repression involves a direct interaction of the transcription factor with the promoters of its target genes. Interestingly, no activation or repression of the CCoAOMT promoter by AtMYB4 was observed in the transfection assays (data not shown), indicating that AtMYB4 does not act as a direct activator of the CCoAOMT gene.
Mutational analysis of AtMYB4
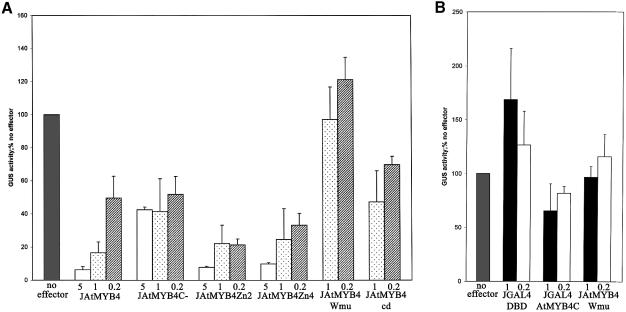
To investigate the mechanism of AtMYB4 repression, a series of mutations in the AtMYB4 gene construct were made and the effect of the altered proteins on the repression of the C4H promoter was monitored using transfection assays (Figure 9A). Mutagenesis of the tryptophan residue in the recognition helix of R3 should abolish DNA binding. This mutant (JAtMYB4Wmu) lost the ability to repress the C4H promoter, establishing that binding of DNA by AtMYB4 is essential for its function as a transcriptional repressor.
Fig. 9. (A) Analysis of the ability of AtMYB4 mutants to inhibit transcription from the C4H promoter in transfection assays. AtMYB4 repressedbasal expression of GUS from the C4H promoter between 20- and 40-fold depending on the concentration of effector supplied (5, 1 and 0.2 = 5, 1and 0.2 µg of effector plasmid, respectively). The repression exercised by AtMYB4 (JAtMYB4) was lifted by ∼50% by removal of the C-terminal domain of the protein (JAtMYB4C–). Mutation of either or both of the potential zinc finger domains had no effect on repression (JAtMYB4Zn2 and JAtMYB4Zn4). Mutation of the DNA binding domain of AtMYB4 (JAtMYB4Wmu) caused complete loss of all repression. Removal of the conserved domain (JAtMYB4cd) in the C-terminus of subgroup 4 family members reduced the ability of AtMYB4 to repress transcription, to a similar extent to complete removal of the C-terminus. Results are averages from at least four experiments. Error bars indicate standard deviations. (B) Analysis of the ability of the C-terminal domain of AtMYB4 to inhibit repression when linked to another DNA binding domain, using transfection assays. The GAL4 DNA binding domain slightly activated from the GAL4-90 promoter (JGAL4-DBD). Addition of the C-terminus of AtMYB4 to the GAL4 DNA binding domain (JGAL4AtMYB4C) reduced expression from the GAL4-90 promoter. This was not due to ‘squelching’ because the mutant of AtMYB4 with a defective DNA binding domain (JAtMYB4Wmu) had no effect on expression from the GAL4-90 promoter.
Deletion of the entire C-terminal domain reduced the repression, although it was not lost entirely at the concentration of effectors tested. Mutation of the cysteine residues within the region of the C-terminal domain suggested to form a zinc finger (Kranz et al., 1998) had no effect on the repression by AtMYB4. Deletion of a conserved motif found in AtMYB4 (JAtMYB4cd) did reduce repression to a similar extent to complete removal of the C-terminal domain (Figure 9A). These results were confirmed by multiple replicates and by assays conducted with varying amounts of effector DNA. In each case repression by AtMYB4 and mutated versions of the protein was reduced when lower concentrations of effector were used.
The results suggested that part of the repression effect could be attributed to the C-terminal domain of AtMYB4, specifically the conserved region found in subgroup 4 (Kranz et al., 1998). This motif is present in AmMYB308 and in AmMYB330, another MYB protein from A.majus that can repress hydroxycinnamic acid biosynthesis when overexpressed in tobacco (Tamagnone et al., 1998a).
It seemed likely that part of the repressing action of AtMYB4 stems directly from its recognition and binding of DNA. It is very likely that there is transcriptional activation of the C4H promoter during the preparation of protoplasts for the transfection assays. All the genes of phenylpropanoid metabolism are induced by wounding stimuli of this type (Bell-Lelong et al., 1997; Mizutani et al., 1997; Ehlting et al., 1999; Figure 4). Indeed, empirical evidence for such activation is provided by the background activity of the C4H promoter used to determine repression by AtMYB4 in the transfection assays. It is also likely that such activation results, in part, from the action of MYB-related transcriptional activators (Sablowski et al., 1994; Feldbrugge et al., 1997). Therefore, part of the repressing effect of AtMYB4 is likely to be due to competition with endogenous MYB activators for a common binding site in the C4H promoter.
Although part of the repressing activity of AtMYB4 may work through competition with activators, our mutational analysis showed that part of the repression exercised by AtMYB4 operates through its C-terminal domain. To determine whether repression by this domain operates indirectly on specific transcription factors associated with the C4H promoter, or whether repression operates directly on the basal transcriptional machinery, a construct encoding the C-terminal domain of AtMYB4 fused to the DNA binding domain of GAL4 was made. This was tested in transfection assays on a multimerized GAL4 motif fused upstream of the –90 sequence of the CaMV35S promoter, fused in turn to the GUS coding sequence. This reporter had appreciable activity in A.thaliana protoplasts in the absence of any effectors. This reporter also responded strongly to a GAL4–VP16 fusion (10- to 25-fold activation depending on the concentration of effector supplied). A construct expressing the GAL4 DNA binding domain alone showed a small stimulation of reporter activity in agreement with previous reports in plants (Schwechheimer et al., 1998). The GAL4 DNA binding domain–C-terminal AtMYB4 fusion reduced expression from the reporter gene by ∼50%. This was not the result of ‘squelching’, since inclusion of the version of AtMYB4 with the mutated DNA binding domain had no effect on expression of the reporter. The repression of transcription exercised by the C-terminal domain of AtMYB4 is, therefore, independent of the promoter on which it is working and likely to be a direct effect on the basal machinery.
Discussion
Analysis of A.thaliana lines mutant for the AtMYB4 gene has demonstrated that AtMYB4 acts as a negative regulator of hydroxycinnamic acid metabolism, principally through regulating the expression of the gene encoding cinnamate 4-hydroxylase. Expression of AtMYB4 is reduced by different environmental conditions including wounding and exposure to UV-B light. In this way it brings about a derepression of C4H gene expression, resulting in higher synthesis of protecting sinapate esters. A mutant of AtMYB4 that produces significantly higher levels of sinapate esters in its leaves also shows improved tolerance to UV-B treatment. It is interesting that the transcriptional response to UV-B that operates through AtMYB4 is focused on the regulation of expression of the C4H gene. There has been considerable debate as to which phenylpropanoids are involved in the response to UV-B and which provide the most effective sunscreens, and C4H is active in the synthesis of both sinapate esters and flavonoids. Recent evidence favours sinapate esters as being the primary source of protectant sunscreens in A.thaliana (Landry et al., 1995; Booij-James et al., 2000) and we observed a significant effect of the atmyb4– mutation on sinapoyl malate levels. Hydroxylation catalysed by C4H shifts the UV-absorbance spectrum of the hydroxycinnamic acid intermediates towards longer wavelengths to provide compounds that are effective in absorbing biologically relevant wavelengths of UV light (Landry et al., 1995; Bell-Lelong et al., 1997). Therefore, regulation of sinapate ester formation through control of C4H provides a biochemically effective strategy for plants to adopt in their acclimation to UV-B. C4H provides an effective rate-limiting step in the synthesis of sinapate esters because levels of sinapate esters are enhanced in the atmyb4– mutant, but, of the genes assayed, only the transcript levels of C4H also increase. Although it is possible that the control of flux along the hydroxycinnamic acid/monolignol pathway is vested in one of the genes not measured in our analysis, expression profiling of wild-type and mutant lines for 7000 expressed sequence tags on microarrays has not revealed additional responding genes (data not shown). The strong correlation between sinapate ester accumulation and C4H transcript levels in both mutant and AtMYB4-overexpressing lines supports the view that C4H is the principal rate-limiting step, at least in leaves of A.thaliana plants.
Analysis of atmyb4– mutants showed that they were also downregulated in CCoAOMT transcripts compared with wild type. However, transfection assays showed no evidence for direct activation of the CCoAOMT promoter by AtMYB4. One explanation for our results is that AtMYB4 represses a repressor of CCoAOMT. A simpler explanation is that expression of the CCoAOMT gene responds to feedback regulation by hydroxycinnamic acid derivatives. This could explain the elevated levels of CCoAOMT transcripts in lines overexpressing AtMYB4 (which have reduced hydroxycinnamic acid levels) and the lowered expression of the CCoAOMT gene in the atmyb4– mutant (where hydroxycinnamic acid intermediates are elevated). A similar type of metabolic regulation by transcinnamic acid has been reported for PAL in tobacco (Blount et al., 2000).
Our data reveal an important role for negative regulation in the synthesis of protecting sunscreens in plants. Such control has not been suggested by previous promoter analysis, although most of the detailed analysis of activation and silencing elements involved in UV response has been performed on the CHS gene promoter, for which negative regulation may not normally be operating. Activation of structural genes producing phenylpropanoid sunscreens by derepression is, however, unexpected. Our data suggest that AtMYB4 acts as both a direct repressor and as a competitive repressor through displacing activators binding to the MYB motifs common to the promoters of many genes in phenylpropanoid metabolism (P and L boxes: Sablowski et al., 1994). Such MYB-related activators of the C4H gene remain to be identified. However, there is evidence from tobacco to suggest that the direct repression exercised by the orthologue of AtMYB4, AmMYB308, is of primary significance in vivo, because overexpression of a truncated version of AmMYB308, comprising just the DNA binding domain, failed to give any phenotypic effects in transgenic plants (Tamagnone et al., 1998a).
Mutational analysis of AtMYB4 has shown the C-terminal region of the protein to be required for full repression. This domain can be transferred to a different DNA binding domain (GAL4) and confers silencing activity on the fusion protein. Although the direct silencing by the C-terminal domain was not strong, it was reproducible and was obtained with relatively low concentrations of effector plasmid. In fact, the GAL4 DNA binding domain retains some properties of a weak activator, as shown by our assays and those of others using this truncated protein (Schwechheimer et al., 1998). The silencing effect of the AtMYB4 C-terminal domain, therefore, may be somewhat greater than apparent from the transfection assays because it has to operate against a background of residual activation. Deletion and mutational analysis of AtMYB4 showed that the region involved in repression included the motif NLELRISLPDDV, which is also conserved in a number of R2R3 MYB proteins belonging to subgroup 4 (pdLNLD/ELxiG/S; Kranz et al., 1998). This subgroup includes AmMYB308 and AmMYB330, which have very similar effects to AtMYB4 when overexpressed in tobacco (Tamagnone et al., 1998a). Repressor functions have been described for MYB-related proteins before (Foos et al., 1992; Marhamati and Sonenshein, 1996), although often operating through interaction with negative regulatory proteins (Kaspar et al., 1999). No examples of R2R3 MYB-related repressors have previously been reported for plants.
The subgroup 4 MYB proteins include the products of four genes from A.thaliana, AtMYB4, AtMYB6, AtMYB7 and AtMYB32, which may be functionally redundant. It is likely that they serve similar roles in negatively regulating hydroxycinnamic acid metabolism. We have analysed expres sion of AtMYB32 and found transcripts to be significantly induced in the dark and in roots, unlike AtMYB4, which is constitutively expressed in most tissues. Therefore, AtMYB32 may play a particular role in negatively regulating hydroxycinnamic acid metabolism in roots, and in reducing expression in dark-grown plants, while AtMYB4 is particularly involved in the response to UV-B.
AtMYB4 appears to function rather specifically in the regulation of the C4H gene to control hydroxycinnamic acid metabolism. However, at high levels of AtMYB4 expression, additional genes fall under its negative transcriptional regulation. A similar reduction in target site selectivity has been reported for c-MYB activity in vivo (Andersson et al., 1999). The additional genes regulated by AtMYB4 are those encoding other steps in hydroxycinnamic acid metabolism and flavonoid biosynthesis. Indeed, the effect of high AtMYB4 activity can be seen as reinforcing the silencing of its primary target (C4H) by silencing other structural genes in the same pathway to ensure downregulation of the specific metabolic pathway.
These results have some important implications for experimental analysis of transcription factor action, suggesting caution in evaluating target genes by activation tagging or any other method involving high levels of transcription factor activity. Results based on inducible activation of MYB proteins may also be subject to reduced selectivity of target genes as a result of high cellular concentrations of transcription factor, which could explain reports of more diverse effects of the P and C1 genes from maize than originally expected from mutant analysis (Bruce et al., 2000).
Although our results suggest caution in deducing targets of MYB-transcription factors from overexpression analyses, they do provide some encouragement for the use of overexpression of regulatory genes to engineer metabolism. If high levels of specific regulatory genes are used, structural genes with similar regulatory motifs may fall under coordinate regulation. This reinforcement mechanism may be used normally by the plant, but could also be developed for biotechnological purposes to ensure effective metabolic regulation through coordinate control of several structural genes in a metabolic pathway.
Materials and methods
Screening for an insertion mutation of the AtMYB4 locus
A dSpm insertion was found amongst the Sainsbury Laboratory Arabidopsis thaliana population using gene-specific primers to amplify the gene (Meissner et al., 1999).
Constructs for plant transformation
To produce a construct for constitutive high level expression of AtMYB4, the full-length cDNA was cloned into pJIT60 (Guerineau and Mullineaux, 1993) between the double CaMV35S enhancer and the CaMV Terminator to create pJAtMYB4. The entire expression cassette was cloned into the HindIII site of pSMAK251. To produce a reporter construct for the promoter of the C4H gene of A.thaliana, two oligonucleotides, one containing the sequence from 942 bp upstream of the initiating ATG and the other the region just (39 bp) upstream of the initiating ATG were used. The promoter was amplified by PCR and cloned into pSMAK251 replacing the CaMV35S promoter, to drive expression of the downstream GUS gene.
Constructs for transfection assays
The reporter construct used for measuring expression from the C4H promoter consisted of a 1024 bp fragment upstream of the initiating ATG of the C4H gene, amplified by PCR and cloned XbaI–XhoI into pBT10GUS (Sprenger-Haussels and Weisshaar, 2000). The reporter for the CCoAOMT gene consisted of a 900 bp fragment upstream of the initiating ATG, amplified by PCR and cloned XbaI–XhoI into pBT10GUS. Reporter constructs for the 4CL3 and CHS genes of A.thaliana have been described by Ehlting et al. (1999) and Hartmann et al. (1998). The reporter construct used for assaying regulation specified by the GAL4 DNA binding domain involved four copies of the GAL4 binding site cloned immediately upstream of the –90 CaMV 35S promoter in pBT10GUS.
Effector constructs consisted of AtMYB4 in pJIT60 (pJAtMYB4) as described above, or modifications of the construct to change the amino acid sequence of the encoded protein. To produce pJAtMYB4C–, pJAtMYB4 was cut with BsaBI, which cuts at +593 bp in the cDNA coding sequence, and SmaI, which cuts in the polylinker at the 3′ end of the cDNA. Religation removed the intervening sequence to leave that encoding only the N-terminal 198 amino acids. The equivalent truncated version of AmMYB308 has been shown to form a stable protein in Escherichia coli (Tamagnone et al., 1998a).
A C-terminal region predicted to form a zinc finger motif was also mutated (Kranz et al., 1998), first by changing the sequence encoding amino acid 224 in pJAtMYB4 from TGT (cysteine) to TCT (serine) and that encoding amino acid 227 from TGC (cysteine) to TCC (serine) to form pJAtMYB4Zn2. pJAtMYB4Zn2 was mutagenized further, the sequence encoding amino acid 237 was changed from TGC (cysteine) to TCC (serine) and that encoding amino acid 239 was changed from TGC (cysteine) to TCC (serine) to form pJAtMYB4Zn4.
Digesting pJAtMYB4 with BsaBI, which cuts twice at +513 bp and +629 bp in the AtMYB4 cDNA coding sequence, created a construct expressing AtMYB4 with a deletion of the region encoding the conserved domain. The cut plasmid was religated to lose the sequence encoding NLELRISLPDDV (amino acids 199–210), and amino acid 198 was changed from leucine to phenylalanine. The ligation was in-frame so that the rest of the C-terminus of AtMYB4 was included in the protein expressed from this construct (pJAtMYB4cd).
A construct with a mutation in the AtMYB4 DNA binding domain was created by mutagenizing the sequence around the conserved tryptophan of the recognition helix of R3, changing amino acids 108–110 from NYW to IDP to form pJMYB4Wmu.
To fuse the sequence encoding the C-terminal domain of AtMYB4 to the DNA binding domain of GAL4, the sequences encoding amino acids 163–282 were amplified by PCR and cloned in-frame, BamHI–EcoRI, behind the sequences encoding the GAL4 DNA binding domain in pJIT60 (pGAL4; Schwechheimer et al., 1998). This plasmid was called pJGAL4AtMYB4C.
Co-transfection assays
The At7 cell culture from A.thaliana ecotype Columbia was used for the co-transfection assays as described by Hartmann et al. (1998). Following co-transfection, protoplasts were incubated for 20 h prior to harvest. GUS activity was measured in the extracts and normalized to luciferase (LUC) activity formed by co-transfection of a 35S LUC construct in each assay.
Stable transformation of Arabidopsis and tobacco
Tobacco (Nicotiana tabacum cv. Samsun NN) leaf discs were transformed using Agrobacterium tumifaciens strain LBA4404. Transformed plantlets were selected on Murashige and Skoog (MS) media containing 100 mg/ml kanamycin sulfate. More than 20 independent transformants were generated and analysed. Arabidopsis thaliana (ecotype Columbia) was transformed by vacuum infiltration with A.tumifaciens strain GV3101. To seeds were sown on germination medium (GM) containing 50 µg/ml kanamycin sulfate. At least 30 transformants carrying each construct were analysed.
UV treatment
Arabidopsis thaliana seedlings were grown on GM medium for 10 days (20°C, 10 h light/14 h dark). The lids of the Petri dishes were removed and the dishes were irradiated by a short wave transilluminator (UV-P 34003901 fitted with UV-B tubes F15T8 emitting 3.2 mW/cm2) for 10, 15, 20 or 25 min. After exposure the lids were replaced and the seedlings were grown for one week in the growth cabinet prior to scoring and photography.
Measurement of phenolics
Arabidopsis thaliana plants were grown for 12 days on MS agar or in soil. Leaves were frozen in liquid nitrogen and then phenolics were extracted in dry ice-cold methanol. Phenylpropanoids were analysed by HPLC as described in Tamagnone et al. (1998b). The identity of the major peaks was confirmed by LCMS.
RNA isolation and quantitative RT–PCR analysis
Plants were grown on agar for 12 days and total RNA was extracted from leaves according to Tamagnone et al. (1998a). RNA (5 µg) was treated with RNase-free DNase I to remove residual DNA. First strand cDNA was synthesized from 5 µg of RNA and amplified according to Frohman et al. (1988) using the dT17 adaptor as a primer and Superscript II RNase-H reverse transcriptase at 42°C for 1 h. In preliminary tests, samples were taken after 15, 20, 25 and 30 cycles, the products were separated by gel electrophoresis and then blotted onto nitrocellulose. These filters were probed with the appropriate labelled cDNA probes to confirm that amplification was linear and to establish the number of amplification cycles for cDNA detection within the linear range. Genes (see Table I) encoding actin (McDowell et al., 1996), C4H, 4CL, 4CL3, CHS, CAD1, F5H, OMT, PAL2, AtMYB4 (AF062860) and trpB (Berlyn et al., 1989) were amplified for between 20 and 25 cycles. CCoAOMT was amplified for 30 cycles. Samples were loaded onto a gel and separated by electrophoresis. Staining with ethidium bromide confirmed that no products could be visualized in the samples. The gels were blotted onto nitrocellulose and hybridized to gene-specific probes.
Light treatment and wounding expression analysis
Wild-type (ecotype Columbia) plants were grown on soil for 6 weeks (16 h light/8 h dark) and then dark adapted for 2 days. Plants were then transferred to continuous white, blue, UV-A or UV-B light, and RNA was extracted from rosette leaves. Light treatments were: white light, Sylvania cool white F48T12/CW/HO 60 48 µE/m2/s; blue light, GTE Sylvania fluorescent F48T12/246/HO 5R5459/ Blue filter 63 µE/m2/s; UV-A, Philips TLK40W/10R 0.7 mW/cm2; UV-B, Philips TLK20W/12RS 0.2 mW/cm2. For UV-C treatment no dark adaptation was given; plants were given a single dose of UV-C light (600 J/m2 from Philips TUV15W 2–75 mW/cm2).
Six-week-old plants were also used for wounding. Small cuts were made on half the rosette leaves and the other half was left intact. Wounded leaves were pooled separately from the non-wounded leaves that formed the sample exhibiting any systemic response. Control samples were from untreated plants of the same age.
Acknowledgments
Acknowledgements
Many thanks to Ruth Meissner and Mike Bevan for their roles in the ‘MYB consortium’; Ian Graham and Alan Crozier in the initial analysis of phenylpropanoids; Ute Tartler in the transfection assays and Steve Mackay in stable transformation; Javier Paz-Ares for critical and constructive reading of the manuscript; and Mandy Coventry for preparation of the manuscript. This work was supported by the European Union FPIV (BIO4-CT95-0129), the European Union FPV (QLG2-CT-1999-00876) in grants to C.M., B.W. and C.T., and by an EMBO short-term fellowship to H.J.
Note added in proof
During the review of this paper it was noticed that CAD1 (accession No. L37883) shows low similarity to CAD genes from other plant species. RT–PCR analysis of mutant and AtMYB4 over-expressing lines was repeated with another CAD gene from A.thaliana (accession No. Z31715), which is more similar to the CAD genes from other species. Exactly the same results were obtained for this gene as for CAD1 (Figure 5).
References
- Andersson K.B., Berge,T., Matre,V. and Gabrielsen,O.S. (1999) Sequence selectivity of c-Myb in vivo—resolution of a DNA target specificity paradox. J. Biol. Chem., 274, 21986–21994. [DOI] [PubMed] [Google Scholar]
- Ansieau S., Kowenz Leutz,E., Dechend,R. and Leutz,A. (1997) B-myb, a repressed trans-activating protein. J. Mol. Med., 75, 815–819. [DOI] [PubMed] [Google Scholar]
- Bell-Lelong D.A., Cusumano,J.C., Meyer,K. and Chapple,C. (1997) Cinnamate-4-hydroxylase expression in Arabidopsis—regulation in response to development and the environment. Plant Physiol., 113, 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M.B., Last,R.L. and Fink,G.R. (1989) A gene encoding the tryptophan synthase-β subunit of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 86, 4604–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount J.W., Korth,K.L., Masoud,S.A., Rasmussen,S., Lamb,C. and Dixon,R.A. (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol., 122, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij-James I.S., Dube,S.K., Jansen,M.A.K., Edelman,M. and Matoo,A.K. (2000) Ultraviolet-B radiation in plants: turnover of the PSII reaction center heterodimer in Arabidopsis mutants altered in phenolic metabolism. Plant Physiol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce W., Folkerts,O., Garnaat,C., Crasta,O., Roth,B. and Bowen,B. (2000) Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors, CRC and P. Plant Cell, 12, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J. and Hahlbrock,K. (1984) Transcription of plant defense genes in response to UV-light or fungal elicitor. Nature, 311, 76–78. [Google Scholar]
- Chapple C.C.S., Vogt,T., Ellis,B.E. and Somerville,C.R. (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell, 4, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C., Hoffmann,H., Schulz,W. and Hahlbrock,K. (1987) Structure and elicitor or UV-light-stimulated expression of 2 4-coumarate-CoA ligase genes in parsley. EMBO J., 6, 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorff J.W., Whittaker,L.J., Eltman,J.T. and Lipsick,J.S. (1992) Carboxy-terminal elements of c-myb negatively regulate transcriptional activation in cis and in trans. Genes Dev., 6, 2524–2535. [DOI] [PubMed] [Google Scholar]
- Ehlting J., Buttner,D., Wang,Q., Douglas,C.J., Somssich,I.E. and Kombrink,E. (1999) Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J., 19, 9–20. [DOI] [PubMed] [Google Scholar]
- Feldbrugge M., Sprenger,M., Dinkelbach,M., Yazaki,K., Harter,K. and Weisshaar,B. (1994) Functional-analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell, 6, 1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrugge M., Sprenger,M., Hahlbrock,K. and Weisshaar,B. (1997) PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J., 11, 1079–1093. [DOI] [PubMed] [Google Scholar]
- Foos G., Grimm,S. and Klempnauer,K.H. (1992) Functional antagonism between members of the MYB family-B-MYB inhibits V-MYB-induced gene activation. EMBO J., 11, 4619–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foos G., Grimm,S. and Klempnauer,K.H. (1994) The chicken A-myb protein is a transcriptional activator. Oncogene, 9, 2481–2488. [PubMed] [Google Scholar]
- Frohman M.A., Dush,M.K. and Martin,G.R. (1988) Rapid production of full-length cDNAs from rare transcripts—amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA, 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E., Drummond,B.J., Bowen,B. and Peterson,T. (1994) The MYB-homologous P-gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell, 76, 543–553. [DOI] [PubMed] [Google Scholar]
- Guerineau F. and Mullineaux,P. (1993) Plant transformation and expression vectors. In Croy,R.R.D. (ed.), Plant Molecular Biology Labfax, BIOS Scientific Publishers, Oxford, UK, pp. 121–147. [Google Scholar]
- Hartmann U., Valentine,W.J., Christie,J.M., Hays,J., Jenkins,G.I. and Weisshaar,B. (1998) Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol. Biol., 36, 741–754. [DOI] [PubMed] [Google Scholar]
- Kaspar P., Dvorakova,M., Kralova,J., Pajer,P., Kozmik,Z. and Dvorak,M. (1999) Myb-interacting protein, ATBF1, represses transcriptional activity of Myb oncoprotein. J. Biol. Chem., 274, 14422–14428. [DOI] [PubMed] [Google Scholar]
- Kranz H.D. et al. (1998) Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J., 16, 263–276. [DOI] [PubMed] [Google Scholar]
- Landry L.G., Chapple,C.C.S. and Last,R.L. (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol., 109, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Oulee,T.M., Raba,R., Amundson,R.G. and Last,R.L. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell, 5, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen M., Racicot,V., Strack,D. and Chapple,C. (1996) Sinapic acid ester metabolism in wild type and a sinapoylglucose-accumulating mutant of Arabidopsis. Plant Physiol., 112, 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhamati D.J. and Sonenshein,G.E. (1996) B-Myb expression in vascular smooth muscle cells occurs in a cell cycle-dependent fashion and down-regulates promoter activity of type I collagen genes. J. Biol. Chem., 271, 3359–3365. [DOI] [PubMed] [Google Scholar]
- Martin C. and Paz-Ares,J. (1997) MYB transcription factors in plants. Trends Genet., 13, 67–73. [DOI] [PubMed] [Google Scholar]
- McDowell J.M., An,Y.Q., Huang,S.R., McKinney,E.C. and Meagher,R.B. (1996) The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol., 111, 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R.C. et al. (1999) Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell, 11, 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M. Ohta,D. and Sato,R. (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol., 113, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano E., Martinez Garcia,J.F. and Martin,C. (1996) Apparent redundancy in Myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell, 8, 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero I., Fuertes,A., Benito,M.J., Malpica,J.M., Leyva,A. and Paz-Ares,J. (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J., 14, 273–284. [DOI] [PubMed] [Google Scholar]
- Sablowski R.W.M., Moyano,E., Culianez-Macia,F.A., Schuch,W., Martin,C. and Bevan,M. (1994) A flower-specific MYB protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J., 13, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz M.B., Grotewold,E. and Chandler,V.L. (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell, 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Dangl,J.L., Becker-Andre,M., Hahlbrock,K. and Schulz,W. (1989) Inducible in vivo DNA footprints define sequences necessary for UV-light activation of the parsley chalcone synthase gene. EMBO J., 8, 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Smith,C. and Bevan,M.W. (1998) The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol. Biol., 36, 195–204. [DOI] [PubMed] [Google Scholar]
- Sprenger-Haussels M. and Weisshaar,B. (2000) Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J., 22, 1–8. [DOI] [PubMed] [Google Scholar]
- Tamagnone L., Merida,A., Parr,A., Mackay,S., Culianez-Macia,F.A., Roberts,K. and Martin,C. (1998a) The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell, 10, 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L. et al. (1998b) Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell, 10, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.M., Dubendorff,J.W., Woo,C.H. and Lipsick,J.S. (1999) Functional analysis of carboxy-terminal deletion mutants of c-Myb. J. Virol., 73, 5875–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R.J., Robinson,C. and Lam,E.W.F. (1993) Transcription regulation by murine B-myb is distinct from that by c-myb. Nucleic Acids Res., 21, 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K. and Bishop,J.M. (1989) Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell, 58, 85–93. [DOI] [PubMed] [Google Scholar]