Abstract
Effects of the commercial drug zanamivir (Relenza™) covalently attached to poly-l-glutamine on the infectivity of influenza A viruses are examined using the plaque reduction assay and binding affinity to viral neuraminidase (NA). These multivalent drug conjugates exhibit (i) up to a 20,000-fold improvement in anti-influenza potency compared with the zanamivir parent against human and avian viral strains, including both wild-type and drug-resistant mutants, and (ii) superior neuraminidase (NA) inhibition constants, especially for the mutants. These findings provide a basis for exploring polymer-attached inhibitors as more efficacious therapeutics, particularly against drug-resistant influenza strains.
Keywords: drug resistance, polymeric drugs, zanamivir, influenza A, poly-l-glutamine, plaque reduction assay, neuraminidase inhibition assay
INTRODUCTION
In addition to yearly outbreaks affecting hundreds of millions of people worldwide,1 influenza pandemics of animal–human reassortment strains are increasingly common. Influenza infection is somewhat mitigated by prompt treatment with the neuraminidase (NA) inhibitors, oseltamivir (Tamiflu™), and zanamivir (Relenza™).1 Sialic acid cleavage from glycoproteins and glycolipids catalyzed by NA promotes both initiation of infection and multicycle infection by preventing viral aggregation, cleaving viral receptors from infected cells to allow release of newly formed virions, and degrading host’s mucins to enable access to epithelial cell surfaces.2-4
Point mutations in and around the active site of the NA enzyme can confer drug resistance and contribute to widespread infection.5,6 New therapeutic agents designed to circumvent drug resistance have been only moderately successful.6-9 An alternative strategy may be to covalently conjugate small-molecule drugs to biocompatible and resorbable water-soluble polymers. Although the resultant multivalent10 polymeric drugs have been shown to possess a greatly enhanced potency against wild-type human strains of influenza virus,11 they have not been explored against either mutant or nonhuman strains. In the present study, we fill this void and demonstrate that zanamivir covalently attached to poly-l-glutamine can overcome both oseltamivir and zanamivir resistance in not only human but also avian influenza A strains.
MATERIALS AND METHODS
Poly-l-glutamic acid (molecular weight = 50,000–100,000 Da) and all other chemicals, biochemicals, and solvents were purchased from Sigma-Aldrich Chemical Co (St. Louis, Missouri, USA). Influenza viruses [A/Wuhan/359/95 (H3N2) and A/turkey/MN/833/80 (H4N2), as well as their drug-resistant mutants] were obtained from the Centers for Disease Control and Prevention (Atlanta, Georgia, USA). Madin-Darby canine kidney (MDCK) cells were purchased from the ATCC. Zanamivir (1, Fig. 1) was obtained from BioDuro (Beijing, China).
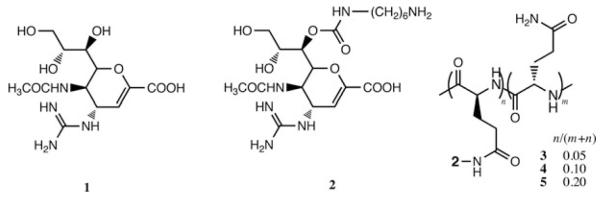
Figure 1.
Chemical structures of zanamivir (1), its derivative for attachment to poly-l-glutamic acid (2), and polymer-attached 2 at different degrees of loading (3–5).
Zanamivir derivative 2 (Fig. 1) was synthesized as described previously.12 To prepare polymer conjugates 3–5 (Fig. 1), 2 was reacted with the benzotriazole ester of poly-l-glutamic acid, followed by quenching with NH4OH (Fig. 2)11. Bare poly-l-glutamine was synthesized analogously.11 Zanamivir content in 3–5 was quantified by 1H NMR.
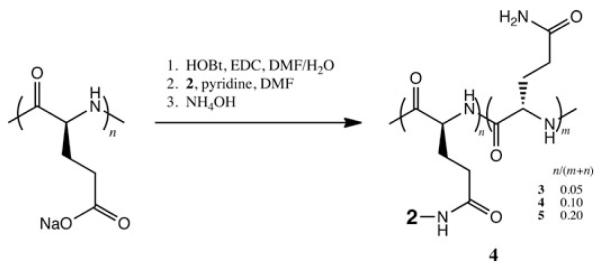
Figure 2.
Preparation of drug–polymer conjugates 3, 4, and 5. HOBt, 1-hydroxybenzotriazole; EDC, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide; DMF, N, N-dimethylformamide.
Plaque assays were performed in 12-well plates using a modified literature procedure.12,13 First, 55-μL aliquots of virus preparation (~800 pfu/mL in PBS) were incubated with equal volumes of inhibitor solutions (serially 10-fold diluted in PBS) for 1 h at room temperature. After washing, MDCK cells grown to confluency were infected at room temperature for 1 h with 100 μL of the virus–inhibitor mixture. The inoculum was removed by aspiration, and the cells were overlaid with 1 mL of plaque medium12 [2×F12 medium, 0.01% DEAE-dextran, 0.1% NaHCO3, 100 units/mL of penicillin G, 100 μg/mL of streptomycin, 4 μg/mL of trypsin, and 0.6% purified agar (L28; Oxoid Co., Basingstoke, UK)]. Plaques were counted after 3 to 4 days of incubation at 37°C.
Neuraminidase inhibition assays were performed using a modified literature procedure.14 Briefly, 20 μL of whole virus and 15 μL of inhibitor dilutions were incubated at room temperature for 1 h. Following addition of the fluorogenic NA substrate 4-methylumbelliferyl-α-d-N-acetylneuraminic acid (whose final concentration was 5- to 10-fold lower than the Km of the enzyme), generation of 4-methylumbelliferone was monitored for 1 h. Values of Ki were determined with nonlinear regression in KaleidaGraph.15
RESULTS AND DISCUSSION
To explore the benefits of multivalency10 (i.e., in the present case, covalent attachment of multiple copies of zanamivir to the same polymeric chain) with respect to drug-resistant strains, herein we have selected three influenza A viruses carrying NA mutations at position 119: two in vitro selected zanamivir-resistant16 avian A/turkey/MN/833/80 (turkey/MN; H4N2) E119D and E119G mutants and a clinically isolated oseltamivir-resistant A/Wuhan/359/95 (Wuhan; H3N2) E119V mutant, which is transmissible in ferrets.17
We have synthesized water-soluble polymeric derivatives of 1 in which the drug is conjugated to poly-l-glutamine through a flexible tether (Fig. 1).11,12 The introduction of the linker group did not drastically affect the anti-influenza or NA inhibitory activity. The inhibitory potency of 2 is within an order of magnitude of 1’s (Table 1), as well as of other zanamivir derivatives modified at the same position.18,19
Table 1.
Antiviral Activities of Zanamivir (1), As Well As of Its Monomeric (2) and Polymeric (3-5) Derivatives, Against Human Wuhan and Avian Turkey/MN Influenza Strains, As Determined by the Plaque Reduction Assay
| IC50 (nM of Zanamivir)a |
|||||
|---|---|---|---|---|---|
| Strain | 1 | 2b | 3 | 4 | 5 |
| A/Wuhan/359/95 | (2.3 ± 1.6) × 104 | (4.3 ± 0.2) × 105 | (1.8 ± 0.5) × 102 | 21 ±7 | (3.4 ± 1.0) × 102 |
| A/Wuhan/359/95 E119V | (4.8 ± 1.5) × 104 | (3.1 ± 0.1) × 105 | (7.7 ± 5.1) × 102 | 51 ±5 | (1.0 ± 0.6) × 103 |
| A/turkey/MN/833/80 | (2.1 ± 0.9) × 104 | (4.3 ± 1.1) × 104 | (1.2 ± 0.4) × 103 | (1.8 ± 0.2) × 102 | (9.6 ± 5.9) × 102 |
| A/turkey/MN/833/80 E119D | (1.8 ± 0.8) × 107 | >3.6 × 106 | (6.2 ± 4.0) × 103 | (1.7 ± 0.8) × 103 | (2.6 ± 1.5) × 103 |
| A/turkey/MN/833/80 E119G | n.d.c | >3.6 × 106 | (6.6 ± 3.3) × 103 | (3.6 ± 1.3) × 103 | (3.0 ± 1.2) × 104 |
All values were determined from experiments run at least in triplicate unless otherwise indicated. The IC50 values are expressed in concentrations of zanamavir, whether free or conjugated to poly-L-glutamine. To determine the IC50 values, zanamivir-based inhibitors and influenza viruses were incubated together prior to infection of confluent MDCK cells. Thus, the reported values reflect inhibition of infection. The IC50 values for bare poly-L-glutamine ranged from 2 to 10 mM (based on the monomer concentrations), as compared with 0.2 to 40 μM for 4, thus demonstrating that the polymer itself has no appreciable antiviral activity.
The A/turkey/MN mutant data stem from a single measurement each because of large quantities of 2 required to perform experiments with them.
Because of small quantities of 1 available, only one zanamivir-resistant mutant was assayed. Because the A/turkey/MN/833/80 E119D mutant exhibits the lowest sensitivity to 1, any observed effects with it should also be observed with the E119G mutant.
MDCK, Madin-Darby canine kidney.
Inhibition of influenza A/Yamagata (H1N1) is most effective with a 10% loading of zanamivir on poly-l-glutamine11; other multivalent systems reach a plateau in efficacy between 10% and 30% modification.20 Therefore, we have selected 10 mol% loading for our initial experiments as well. Using the plaque reduction assay, we have demonstrated that the poly-l-glutamine-attached drug 4 is not only about 20,000-fold more effective than 2 against another human influenza strain, Wuhan, but also some 200-fold more potent against an avian strain, turkey/MN.
Importantly, both oseltamivir- and zanamivir-resistant mutants of these viral strains are also far more susceptible to polymeric 4 than to small-molecule inhibitor 2: some 6000-fold improvement against oseltamivir-resistant Wuhan E119V mutant and a 2000-fold more enhancement against zanamivir-resistant turkey/MN mutants have been observed (Fig. 3). In comparison, maximal improvements for novel monomeric inhibitors to date have not exceeded 1000 times against oseltamivir-resistant influenza strains9 and 250 times against zanamivir-resistant ones.6
Figure 3.
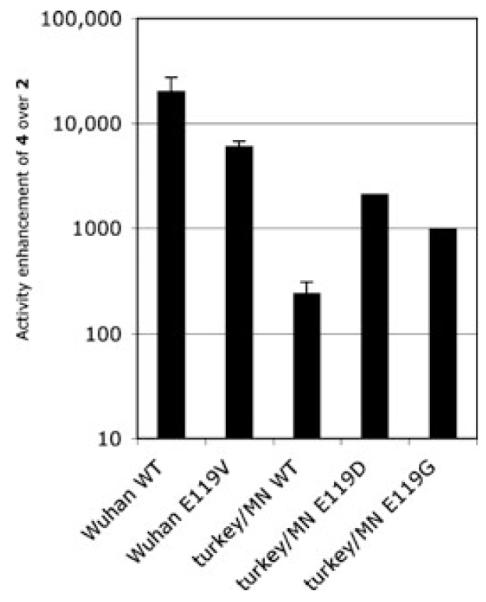
Antiviral activity enhancements of polymeric 4 over small-molecule 2 against wild-type and drug-resistant influenza strains. To determine the underlying IC50 values, zanamivir-based inhibitors and viruses were incubated together prior to infection of confluent MDCK cells. The antiviral activity enhancements shown were the results of experiments run at least in triplicate. The only exceptions were the A/turkey/MN mutants where a single measurement was run because of the large quantities of 2 required. An IC50 value for these zanamivir-resistant mutants was not reached event at millimolar concentrations of 2; thus, the actual enhancements are at least those indicated.
Next, we have examined how the amount of 2 conjugated to poly-l-glutamine affects inhibitory potency. Polymeric 4 has been found to be 10-fold more potent than either 3 or 5 against wild-type strains of both human Wuhan and avian turkey/MN viruses (Table 1). Importantly, 4 is also up to 20 times more potent than either 3 or 5 at inhibiting the drug-resistant strains (Table 1). The observation of the same optimal degree of loading for all the viral strains tested by us suggests that beyond a certain point the benefits of multivalency are counteracted by steric hindrances—presumably, too many ligand molecules attached to the same polymer chain interfere with each other’s action.
To gain further mechanistic insights, we have determined inhibition constants Ki for both 4 and 2, using the NA enzyme inhibition assay. The results presented in Table 2 afford several important conclusions. First, as seen in the last two lines of the table, the enzymes of zanamivir-resistant influenza strains expectedly bind 2 at least two orders of magnitude weaker than the wild-type and oseltamivir-resistant strains. Second, in all instances, the polymer-attached drug is a substantially more potent enzyme inhibitor than its small-molecule parent. Third, while the binding affinity enhancements caused by conjugation to poly-l-glutamine are relatively modest for wild-type human and avian strains, as well as for the oseltamivir-resistant human one, zanamivir-resistant turkey/MN mutants bind 4 2000 times more stronger than 2. As a result of this strengthened binding, the zanamivir-resistant mutants become as sensitive to 4 (in contrast to 2) as the others (Table 2). Thus, presentation of zanamivir on the polymer completely compensates for weakened binding in zanamivir-resistant strains.
Table 2.
Inhibition Constants (Ki) of Viral Neuraminidases by Small-Molecule (2) and Polymeric (4) Zanamivir Derivatives Against Both Wild-Type and Drug-Resistant Human and Avian Influenza Strains
|
Ki (nM of Zanamivira) |
||
|---|---|---|
| Strain | 2 | 4 |
| A/Wuhan/359/95 | 3.1 ± 1.0 | 0.93 ± 0.12 |
| A/Wuhan/359/95 E119V | 1.9 ± 0.8 | 0.90 ± 0.10 |
| A/turkey/MN/833/80 | 4.6 ± 0.9 | 0.27 ± 0.16 |
| A/turkey/MN/833/80 E119D | 800 ± 150 | 0.37 ± 0.12 |
| A/turkey/MN/833/80 E119G | 370 ± 20 | 0.13 ± 0.05 |
All values were determined from experiments run at least in triplicate unless otherwise indicated. The Ki values are expressed in concentrations of zanamavir, whether free or conjugated to poly-L-glutamine. For other conditions, see the Materials and Methods section.
Multivalent drug species, such as 4, exhibit better virus inhibition because of steric effects and increased affinity.10 Flexible linkers, such as that employed in this study, can promote improved ligand–protein binding by reducing steric obstacles and increasing effective ligand concentration21 (as is schematically illustrated in Fig. 4). Both free and polymer-bound 1 should have similar rotational and translational entropic costs for the first interaction with viral surface proteins but polymer-bound 1 should have lower costs for subsequent ones, thereby providing entropic benefits of multivalency.22 Herein, we have demonstrated that a multivalent presentation of 1 conjugated to a resorbable polymeric backbone overcomes binding deficiencies and leads to potent inhibition not only of wild-type but also of drug-resistant human and avian influenza strains, thus promising safe and more efficacious therapeutics.
Figure 4.
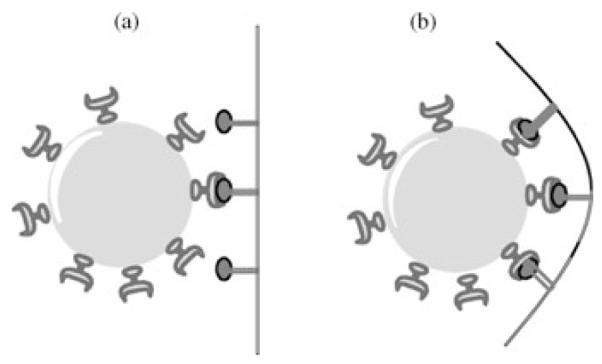
Schematic illustration of the proposed interactions of target viral surface proteins (gray claws) with complementary drugs (black ovals) in rigid (a) and flexible (b) drug–polymer conjugates. The large gray spheres represent influenza virus particles and black lines the polymeric chain and linkers.
ACKNOWLEDGMENTS
This work was partly supported by National Institutes of Health grant U01-AI074443. L.A.d.C. was supported by a postdoctoral fellowship from Fundación Ramón Areces of Spain.
REFERENCES
- 1.Luscher-Mattlie M. Influenza chemotherapy: A review of the present state of art and of new drugs in development. Arch Virol. 2000;145:2233–2248. doi: 10.1007/s007050070017. [DOI] [PubMed] [Google Scholar]
- 2.Greengard O, Poltoratskaia N, Leikina E, Zimmerberg J, Moscona A. The anti-influenza virus agent 4-GU-DANA (zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J Virol. 2000;74:11108–11114. doi: 10.1128/jvi.74.23.11108-11114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu C, Eichelberger MC, Compans RW, Air GM. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69:1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H-D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moscona A. Oseltamivir resistance—disabling our influenza defenses. N Engl J Med. 2005;353:2633–2636. doi: 10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- 6.Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother. 2005;49:4515–4520. doi: 10.1128/AAC.49.11.4515-4520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Sosa AT, Sild S, Maran U. Design of multibinding-site inhibitors, ligand efficiency, and consensus screening of avian influenza H5N1 wild-type neuraminidase and of the oseltamivir-resistant H274Y variant. J Chem Inf Model. 2008;48:2074–2080. doi: 10.1021/ci800242z. [DOI] [PubMed] [Google Scholar]
- 8.Koyama K, Takahashi M, Oitate M, Nakai N, Takakusa H, Miura S, Okazaki O. CS-8958, a prodrug of the novel neuraminidase inhibitor R-125489, demonstrates a favorable long-retention profile in the mouse respiratory tract. Antimicrob Agents Chemother. 2009;53:4845–4851. doi: 10.1128/AAC.00731-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita M, Tomozawa T, Kakuta M, Tokumitsu A, Nasu H, Kubo S. CS-8959, a prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrob Agents Chemother. 2009;53:186–192. doi: 10.1128/AAC.00333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mammen M, Choi S-K, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Masuda T, Yoshida S, Arai M, Kaneko S, Yamashita M, Honda T. Synthesis and anti-influenza evaluation of polyvalent sialidase inhibitors bearing 4-guanidino-Neu5Ac2en derivatives. Chem Pharm Bull. 2003;51:1386–1398. doi: 10.1248/cpb.51.1386. [DOI] [PubMed] [Google Scholar]
- 12.Haldar J, Álvarez de Cienfuegos L, Tumpey TM, Gubareva LV, Chen J, Klibanov AM. Bifunctional polymeric inhibitors of human influenza A viruses. Pharm Res. 2010;27:259–263. doi: 10.1007/s11095-009-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayden FG, Vote KM, Gordon DR., Jr Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob Agents Chemother. 1980;17:865–870. doi: 10.1128/aac.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wetherall NT, Trivedi T, Zeller J, Hodges-Savola C, McKimm-Breschkin JL, Zambon M, Hayden FG. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors. J Clin Microbiol. 2003;41:742–750. doi: 10.1128/JCM.41.2.742-750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kati WM, Montgomery D, Maring C, Stoll VS, Giranda V, Chen X, Graeme Laver W, Kohlbrenner W, Norbeck DW. Novel alpha and beta amino acid inhibitors of influenza virus neuraminidase. Antimicrob Agents Chemother. 2001;45:2563–2570. doi: 10.1128/AAC.45.9.2563-2570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubareva LV, Robinson MJ, Bethell RC, Webster RG. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol. 1997;71:3385–3390. doi: 10.1128/jvi.71.5.3385-3390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herlocher ML, Truscon R, Elias S, Yen H-L, Roberts NA, Ohmit SE, Monto AS. Influenza viruses resistant to the antiviral drug oseltamivir: Transmission studies in ferrets. J Infect Dis. 2004;190:1627–1630. doi: 10.1086/424572. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald SJF, Watson KG, Cameron R, Chalmers DK, Demaine DA, Fenton RJ, Gower D, Hamblin JN, Hamilton S, Hart GJ, Inglis GGA, Jin B, Jones HT, McConnell DB, Mason AM, Nguyen V, Owens IJ, Parry N, Reece PA, Shanahan SE, Smith D, Wu W-Y, Tucker SP. Potent and long-acting dimeric inhibitors of influenza virus neuraminidase are effective at a once-weekly dosing regimen. Antimicrob Agents Chemother. 2004;48:4542–4549. doi: 10.1128/AAC.48.12.4542-4549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews DM, Cherry PC, Humber DC, Jones PS, Keeling SP, Martin PF, Shaw CD, Swanson S. Synthesis and influenza sialidase inhibitory activity of analogues of 4-guanidino-Neu5Ac2en (zanamivir) modified in the glycerol side-chain. Eur J Med Chem. 1999;34:563–574. doi: 10.1016/s0223-5234(00)80026-4. [DOI] [PubMed] [Google Scholar]
- 20.Mammen M, Dahmann G, Whitesides GM. Effective inhibitors of hemagglutination by influenza virus synthesized from polymers having active ester groups. J Med Chem. 1995;38:4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 21.Kitov PI, Shimizu H, Homans SW, Bundle DR. Optimization of tether length in nonglycosidically linked bivalent ligands that target sites 2 and 1 of a shiga-like toxin. J Am Chem Soc. 2003;125:3284–3294. doi: 10.1021/ja0258529. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamurthy VM, Estroff LA, Whitesides GM. Multivalency in ligand design. In: Jahnke W, Erlanson DA, editors. Methods and principles in medicinal chemistry. Vol. 34. Wiley-VCH; Weinheim, Germany: 2006. pp. 11–53. [Google Scholar]