Abstract
Pentavalent organo-vanadates have been put forth as transition state analogues for a variety of phosphoryl transfer reactions. In particular, uridine 2′,3′-cyclic vanadate (U>v) has been proposed to resemble the transition state during catalysis by ribonuclease A (RNase A). Here, this hypothesis is tested. Lys41 of RNase A is known to donate a hydrogen bond to a nonbridging phosphoryl oxygen in the transition state during catalysis. Site-directed mutagenesis and semisynthesis were used to create enzymes with natural and nonnatural amino acid residues at position 41. These variants differ by 105-fold in their kcat/Km values for catalysis, but <40-fold in their Ki values for inhibition of catalysis by U>v. Plots of logKi vs log(Km/kcat) for three distinct substrates [poly(cytidylic acid), uridine 3′-(p-nitrophenyl phosphate), and cytidine 2′,3′-cyclic phosphate] have slopes that range from 0.25 and 0.36. These plots would have a slope of unity if U>v were a perfect transition state analogue. Values of Ki for U>v correlate weakly with the equilibrium dissociation constant for the enzymic complexes with substrate or product, indicating that U>v bears some resemblance to the substrate and product as well as the transition state. Thus, U>v is a transition state analogue for RNase A, but only a marginal one. This finding indicates that a pentavalent organo-vanadate cannot necessarily be the basis for a rigorous analysis of the transition state for a phosphoryl transfer reaction.
Introduction
Understanding the basis of enzymatic catalysis entails describing how forces acting between an enzyme and its substrate(s) increase as the transition state is formed.1–3 The transition state, the most energetic species along a reaction pathway, is fleeting. Having a lifetime near 10−12 s,4 the transition state is not readily amenable to direct study. Hence, molecules whose ground state structures mimic the transition state for a particular enzymatic reaction can be useful in probing the means by which the enzyme lowers the activation barrier of chemical transformation. How, then, is a molecule judged to be an analogue of a transition state?
A criterion for judging transition state analogy is suggested by the equation:5,6
| (1) |
where Ktx refers to the equilibrium dissociation constant of the enzyme•transition state complex. If a molecule is a perfect mimic of the transition state, then its equilibrium dissociation constant from the enzyme (Kd = Ki) should equal Ktx. Still, knowing the value of Ki for a particular inhibitor and the corresponding value of Ktx does not allow for an evaluation of the inhibitor as a transition state analogue. For example, a bound inhibitor could participate in favorable or unfavorable interactions with little resemblance to those in the enzyme•transition state complex. Bartlett has therefore argued that a more appropriate criteria for assessing transition state analogy is that the relative affinity of an enzyme for the chemical transition state (given by kcat/Km) and for a putative transition state analogue (given by 1/Ki) be constant over a range of values.7,8 In other words, Δlog(Km/kcat) = ΔlogKi.
Two methods exist for systematically altering the variables kcat/Km and Ki. One method employs a range of inhibitors that mirror a range of substrates. This first method has been applied extensively to proteases, using an aldehyde, phosphonate, or phosphonamidate group in place of the scissile amide in a peptidyl analogue.7,9–12 The other method involves systematic alteration of the active site. This second method is more generally applicable, as it does not require the enzyme to accommodate a variety of substrates.13–15 It does, however, require structural knowledge of the active site and the ability to alter that active site systematically. Meaningful correlations have also been obtained between kcat/Km and the conformational stability of the related enzyme•inhibitor complexes.16
Pentavalent organo-vanadates have been proposed as transition state analogues for phosphoryl transfer reactions. These proposals arise from V(V) being able to form stable pentoxy complexes, some of which adopt the trigonal bipyramidal geometry that occurs during phosphoryl transfer reactions.17 The first evidence18 of an organo-vanadate being an inhibitor of catalysis by an enzyme was obtained with ribonuclease A (RNase A;19 EC 3.1.27.5), which catalyzes two phosphoryl transfer reactions: the transphosphorylation (and hence cleavage) of RNA, and the hydrolysis of nucleoside 2′,3′-cyclic phosphates (Figure 1).20 Uridine and vanadate, in combination, inhibit catalysis by RNase A more strongly than would be predicted from their individual Ki values.18 Vanadate was presumed to combine with uridine and water to form a 1:1:1 complex, uridine 2′,3′-cyclic vanadate monohydrate (U>v), and this complex was presumed to mimic the transition state. X-Ray diffraction analyses have confirmed the existence of U>v in the active site, where the observed geometry is somewhat distorted from the predicted trigonal bipyramid (Figure 1C)21,22. A recent structural study indicates that the degree of distortion is greater than was appreciated previously.23
Figure 1.
(A) Putative mechanism of the transphosphorylation reaction (top) and hydrolysis reaction (bottom) catalyzed by ribonuclease A. “B” is His12, and “A” is His119. (B) Putative structure of the transition state during the transphosphorylation reaction (HOR = nucleoside) or hydrolysis reaction (HOR = H2O) catalyzed by RNase A. (C) Stereoview of the structure of the active site in the RNase A•U>v complex. The structure was refined to 2.0 Å from X-ray and neutron diffraction data collected from crystals grown at pH 5.3 (Protein Data Bank entry 6RSA).22 The distance from Lys41 to the 2′ oxygen is 2.8 Å, and to the nearest nonbridging oxygen is 3.5 Å. The distance from His12 (right) to the nearest oxygen is 2.7 Å, and to the 2′ oxygen is 3.0 Å. The other enzymic residue is His119.
The degree to which the RNase A•U>v complex is analogous to the enzymic transition state is uncertain. In the crystalline RNase A•U>v complex, active-site residues are found in positions not congruent with their commonly accepted mechanistic roles.22–24 For example, we have used semisynthesis to show that Lys41 donates a hydrogen bond to a non-bridging phosphoryl oxygen in the transition state for RNA cleavage.25 Yet, the side-chain amino group of Lys41 is much closer to the 2′ oxygen than to a non-bridging vanadyl oxygen in the RNase A•U>v complex (Figure 1C). This observation calls into question at least one of the following: the commonly-held conception of the catalytic mechanism (Figures 1A and 1B),20 the details of the crytallographic models (Figure 1C),21,22 or the validity of U>v as a transition state mimic. Studies have used the structure of U>v bound to RNase A as a basis for computational investigations.26,27 The relevance of these studies depends in great measure on the ability of U>v to mimic the transition state.
Here, we assess the relevance of the RNase A•U>v complex to catalysis by the enzyme. We do so by assessing catalysis and its inhibition in enzymes in which Lys41 has been altered systematically by site-directed mutagenesis and semisynthesis. Thus, our data report specifically on the interaction between U>v and residue 41, and the degree to which this interaction mimics that in the enzyme•transition state complex. From this perspective, we find that U>v is only a marginal analogue of the transition state for catalysis by RNase A.
Experimental Section
Materials
Wild-type RNase A and the K41A, K41R, and K41C variants were produced and purified as described.27–29 The Cys41 sulfhydryl group of K41C RNase A was modified by alkylation with bromoethylamine to yield a semisynthetic variant with an S-(aminoethyl)cysteine residue at position 41 (K41CEA RNase A) as described.27 Uridine 3′-(p-nitrophenyl phosphate) [Up(OC6H4-p-NO2)] was synthesized by J. E. Thompson and T. G. Kutateladze as the 2′,5′-O-tetrahydropyranyl-protected material,30 and handled as described.31 Sodium vanadate was obtained from Aldrich Chemical (Milwaukee, WI). All other chemicals were obtained from Sigma Chemical (St. Louis, MO).
Preparation of Uridine- and Vanadate-Containing Buffers
Concentrated solutions of uridine (1 M) were prepared in 0.010 M sodium succinate buffer (pH 6.0) containing NaCl (0.10 M). The concentration of these stocks was determined spectrophotometrically in H2O with ε = 9.9 × 103 M−1cm−1 at 260 nm.32 Sodium vanadate was dried for 1 h at 115 °C and cooled in a dessicator before weighing and dissolution in water with gentle heating (to 0.06 M). The pH after dissolution was 9.2 and no visible color was present, indicating the absence of appreciable amounts of decavanadate species.33,29 The uridine and vanadate stock solutions were added to 10 mM sodium succinate buffer (pH 6.0) containing NaCl (0.10 M). After preparing each solution, the pH was re-adjusted to 6.0 with dilute HCl, and the solutions were passed through a 0.22-μm filter. To insure that all species were at equilibrium, buffers were incubated at room temperature overnight and the pH was checked again prior to use in enzyme assays.
Assays of Enzymatic Catalysis
The substrate chosen for uridine and vanadate inhibition studies was Up(OC6H4-p-NO2), which undergoes RNase A catalyzed transphosphorylation to release p-nitrophenol. Even though Up(OC6H4-p-NO2) is a relatively poor substrate, the large change in extinction coefficient associated with this reaction makes for a sensitive assay. More importantly, p-nitrophenol can be monitored at a wavelength far from the peak absorbance of uridine, allowing for use of high uridine concentrations.
2′,5′-O-Tetrahydropyranyl-protected Up(OC6H4-p-NO2) was deprotected and handled as described.31 The initial rate of release of p-nitrophenol from Up(OC6H4-p-NO2) was followed by the increase in absorbance at 330 nm in 10 mM sodium succinate buffer (pH 6.0) containing NaCl (0.10 M), Up(OC6H4-p-NO2) (30 μM – 1.0 mM), and enzyme (7 nM – 7 μM). The uncatalyzed rate is not negligible compared to the lower enzyme-catalyzed rates measured herein; the rate constant for the nonenzymatic transphosphorylation of Up(OC6H4-p-NO2) at pH 6.0 has been reported to be k ≅ 1.5 × 10−5 s−1.34 To interpret properly the changes in absorbance at 330 nm, measurements of uncatalyzed Up(OC6H4-p-NO2) cleavage were made in 10 mM sodium succinate buffer (pH 6.0) absent any enzyme. These rates were in agreement with the reported rate constant,34 and were subtracted from the total rate to give the enzyme-catalyzed rate.
Inhibition of Catalysis by Uridine 3′-Phosphate
Uridine 3′-phosphate (3′-UMP) is the product of the transphosphorylation and hydrolysis of Up(OC6H4-p-NO2) by RNase A. Inhibition by 3′-UMP was measured by determining the initial velocities of enzyme-catalyzed transphosphorylation of Up(OC6H4-p-NO2). Reactions were performed in 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M), Up(OC6H4-p-NO2) (30 μM – 1.0 mM), 3′-UMP (60 μM – 4.3 mM), and enzyme (7 nM – 13 μM). Assays were otherwise identical to those used for inhibition by U>v (vide supra). Because the initial velocity was only measured for 2% or less of the total reaction, the amount of 3′-UMP generated enzymatically was negligible.
Previously, we and others had used isothermal titration calorimetery to determine the value of the equilibrium dissociation constant (Kd) for the RNase A•3′-UMP complex.35,36 Here, we performed the same experiment with the K41A variant. Aliquots (2.5 μL) of a solution of 3′-UMP (12.8 mM) were injected into a solution of K41A RNase A (0.36 mM) until the heat generated upon each individual injection, resulting from the exothermic nature of protein ligand binding, had approached a constant level. This titration was done with a MicroCal MCS system (MicroCal Software, Northhampton, MA) Data were fitted with the program ORIGIN (MicroCal) to obtain the value of Kd.35
Data Analyses
Initial velocity data were obtained for four enzyme variants (wild-type, K41CEA, K41R, or K41A) and two sodium vanadate concentrations (0.1 or 0.4 mM). For each enzyme variant, initial velocities were measured not only with uridine plus vanadate, but also with uridine alone or vanadate alone, as well as in the absence of inhibitors. The measured inhibition by vanadate, in the absence of uridine, was probably the result of inhibitory effects of one or several minor, multimeric vanadate species, not VO3− itself.37,38 No Ki value for vanadate can be derived because the nature of the actual inhibitor moiety and its concentration relative to [VO3−] remain unclear.29 Consequently, each concentration of vanadate was treated separately, and the derived inhibition parameter was considered to be an apparent one that applied only to that particular vanadate concentration. Data from a set were fitted by multiple regression analysis to simultaneous equations, sharing the parameters Km, Vmax, KiU, , and Ki/[U>v], where KiU is the equilibrium dissociation constant for competitive inhibition by uridine, , the apparent equilibrium dissociation constant for competitive inhibition by vanadate, and Ki/[U>v] is the ratio of the concentration of U>v to its apparent equilibrium dissociation constant for competitive inhibition. Analyses were performed on unweighted data with the program GRAFIT (Erithicus Software, Palo Alto, CA) and the equations:
| (2) |
if neither uridine nor vanadate was present,
| (3) |
if uridine was present alone,
| (4) |
if vanadate was present alone, and
| (5) |
if both uridine and vanadate were present. Reported errors are standard deviations of the average Ki/[U>v] value from replicate determinations. Relationships between kinetic parameters of the enzyme variants were determined with the program GRAFIT.
Results and Discussion
Synergism of Inhibition by Uridine and Vanadate
To test U>v as an analogue of the transition state during catalysis by RNase A, we first determined whether uridine and vanadate inhibit catalysis synergistically. Practical considerations severely limit the amount of uridine and vanadate that can be used to observe the inhibition due to U>v. Uridine is itself a weak inhibitor (Ki = 15 mM under the current assay conditions), and large uridine concentrations overwhelm measurements of inhibition. The predominant monomeric vanadate species at pH 6.0 is H2VO4− (pKa 8.15 ref 39), which has been reported to be non-inhibitory.37 Our experience indicates, however, that the inhibition by H2VO4− at concentrations above 0.4 mM is not negligible.38 Still, all of the RNase A variants studied here were inhibited to an extent greater than would be expected from uridine alone and vanadate alone. This synergy of inhibition was apparent at both concentrations of vanadate tested (0.1 and 0.4 mM). It was therefore always possible to determine a meaningful value of Ki/[U>v] by using eq 5. The values of Ki/[U>v] are listed in Table 1.
Table 1.
Values of Ki/[Uridine 2′,3′-Cyclic Vanadate] for Inhibition of Catalysis by Wild-Type Ribonuclease A and the K41CEA, K41R, and K41A Variants a
| Ribonuclease A | [Uridine] (mM) | [NaVO3] (mM) | Ki/[U>v] |
|---|---|---|---|
| Wild-type | 15 | 0.1 | 0.15 ± 0.4 |
| Wild-type | 15 | 0.4 | 0.061 ± 0.011 |
| K41CEA | 15 | 0.1 | 1.1 ± 0.1 |
| K41CEA | 15 | 0.4 | 0.37 ± 0.04 |
| K41R | 15 | 0.1 | 1.8 ± 0.7 |
| K41R | 15 | 0.4 | 0.36 ± 0.04 |
| K41A | 15 | 0.1 | 2.7 ± 0.8 |
| K41A | 15 | 0.4 | 2.3 ± 1.5 |
Data were obtained at 25 °C in 10 mM sodium succinate buffer (pH 6.0) containing NaCl (0.10 M) and Up(OC6H4-p-NO2).
What is the Value of Ki for Uridine 2′,3′-Cyclic Vanadate?
The true value of Ki for U>v is somewhat elusive. Converting the parameter determined here, Ki/[U>v], into an explicit value for Ki would require knowing how much U>v is actually present in solution. The original characterization of the equilibrium constant for U>v formation, done by means of ultraviolet/visible spectroscopy, rested on the faulty assumption that there was a single major species, possessing 1:1 stoichiometry.18 The authors noted that irregularities in their data suggested the existence of a minor 2:1 uridine:vanadate species as well. Recent work by Tracey and coworkers used 1H, 13C, and 51V NMR spectroscopy to demonstrate that a 2:2 nucleoside:vanadate dimer predominates in solution under most conditions.40 A 2:2 species has also been observed by X-ray diffraction analysis.41 Tetrahedral and pentacoordinate 1:1 complexes do occur in solution, but only in minor proportions.40 The formation constant for U>v has been estimated to be (1.8 ± 1.5) M−1 at pH 7.42 The value estimated for the cyclic tetrahedral complex (i.e., U>v minus H2O) is (4.5 ± 1.1) M−1. In addition, acyclic 1:1 nucleoside:vanadate complexes may occur at low levels.40
Although some uncertainty persists in the quantitative description of minor 1:1 uridine:vanadate species in solution, we can estimate a value of Ki with some confidence. Tracey and coworkers have reported that Ki = 0.45 μM for wild-type RNase A and U>v at pH 7.37 This value was calculated based on their prior estimates of the abundance of the 1:1 and 2:2 complexes.40 We measured a value of Ki/[U>v] = 0.15 for wild-type RNase A at 15 mM uridine and 0.1 mM vanadate (Table 1). Using the analysis of Tracey and coworkers,37 we expect the concentration of the 1:1 uridine:vanadate complex to be 2.9 μM.38 The resulting value of Ki = 2.9 μM/6.5 = 0.45 μM is in complete agreement with that published for pH 7.37 At 15 mM uridine and 0.4 mM vanadate, we measured Ki/[U>v] = 0.061 (Table 1). Under these conditions, we expect the concentration of the 1:1 complex to be 5.9 μM.38 The resulting value of Ki = 5.9 μM/16.4 = 0.36 μM is likewise in close agreement with the published value.37 Thus, we believe that Ki ≅ 0.4 μM for inhibition of RNase A by U>v.
A comparison of the affinity of U>v for RNase A with that of related molecules is noteworthy. 2′-Deoxy-2′-fluorouridylyl(3′→5′) adenosine (UFpA) is an analogue of UpA, a substrate for the transphosphorylation step. The reported dissociation constant for the RNase A•UFpA complex is 0.4 mM at pH 5.5.43 The Km for U>p, which is a substrate for the hydrolysis step, is probably reflective of its dissociation constant,44 and is 2.2 mM at pH 6.0 and 2.1 mM at pH 5.5.45 3′-UMP, which is the product of the hydrolysis of U>p, binds somewhat more tightly than does U>p but still has a Ki of only 72 μM at pH 6.0. Thus, the affinity of RNase A for U>v at pH 6.0 is >102-fold greater than that for any substrate, product, or analogue thereof. Although a Ki value of 0.4 μM is much larger than the value of Ktx (<2× 10−15 M for the transphosphorylation of UpA28), it is nonetheless the lowest Ki value of any known molecule related to the reaction coordinate of catalysis by RNase A. The Ki of U>v is also lower than that of decavanadate (V10O286−; Ki = 1 μM).29
Fortunately, we need only relative measurements of binding to detect an inverse correlation of Ki with kcat/Km. The quandary of poorly characterized equilibria of multiple complexes can be side-stepped. Because we can measure inhibition for all the RNase A variants under a constant set of conditions, the U>v concentration, though unknown, will be constant. If kcat/Km exhibits an inverse correlation with Ki, then it also does so with Ki/[U>v]. Such correlations, which were determined over a range of kcat/Km values spanning 105-fold, are presented on a logarithmic scale in Figure 2. Increases in Ki correspond to decreases in kcat/Km for three substrates, which undergo either the transphosphorylation or hydrolysis reactions catalyzed by RNase A. The results are similar for the two different vanadate concentrations, as the lines in Figure 2 are essentially parallel.
Figure 2.
Plot of log(Ki/[U>v]) vs log(Km/kcat) for (A) cleavage of poly(cytidylic acid), (B) cleavage of Up(OC6H4-p-NO2), and (C) hydrolysis of cytidine 2′,3′-cyclic phosphate by wild-type ribonuclease A and the K41CEA, K41R, and K41A variants. Values of kcat/Km were obtained at 25 °C in 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M). Values of Ki/[U>v] (Table 1) were obtained at 25 °C in 0.010 M sodium succinate buffer (pH 6.0) containing NaCl (0.10 M) and poly(C). Open circles (○) denote 15 mM uridine and 0.1 mM NaVO3 and have (A) slope = 0.25 ± 0.12, (B) slope = 0.31 ± 0.15, and (C) slope = 0.34 ± 0.12. Closed circles (●) denote 15 mM uridine and 0.4 mM NaVO3 and have (A) slope = 0.29 ± 0.16, (B) slope = 0.36 ± 0.18, and (C) slope = 0.36 ± 0.15. The dashed lines have a slope of unity.
Is Uridine 2′,3′-Cyclic Vanadate a Transition State Analogue?
The slope of the line in a plot of logKi vs log(Km/kcat) is meaningful. If Ki were to mirror Ktx perfectly, the value of the slope would be unity.7,9 In practice, the transition state and its imperfect analogue can show differential levels of sensitivity to ligand or active-site changes. Peptide aldehydes as analogues for the serine protease elastase exhibit a slope of 0.74.9 Peptide analogues containing a phosphonate linkage, when compared with substrates for the zinc protease thermolysin, show a slope of 1.05.7 A peptide analogue with a phosphonamidate linkage yield a slope of 1.03 when alterations were made at a catalytic arginine in carboxypeptidase A.13 The linear correlations of U>v inhibition and catalytic changes, resulting from alterations made to RNase A at position 41, show slopes that are much more shallow, between 0.25 and 0.36 (Figure 2).
Lys41 plays a similar role in catalysis of both the transphosphorylation of RNA and the hydrolysis of nucleoside 2′,3′-cyclic phosphates.19b,38 Comparing the correlation of U>v inhibition with catalysis of these two distinct reactions can reveal whether U>v more closely mimics the transition state for transphosphorylation or that for hydrolysis. A priori, the transition state for hydrolysis would appear to be the species that U>v resembles more closely, as both U>v and a nucleoside 2′,3′-cyclic phosphate lack the ability to access the enzymic subsites that interact with the nucleobases and phosphoryl groups on the 3′ side of the scissile bond.46 Yet, this resemblance only applies to ground state features and is not relevant to spatial or electronic similarities specific to the transition state. Because the slopes of the lines in Figures 2A and 2B (where kcat/Km is for the transphosphorylation of two different substrates) are similar to those in Figure 2C (where kcat/Km is for a hydrolysis reaction), we conclude that U>v bears a similar resemblance to the transition state for transphosphorylation and hydrolysis.
The transphosphorylation of poly(C) by wild-type RNase A appears to be limited by a step other than the chemical transformation.28 The value of Ktx pertains to the chemical transition state. Thus, measured values of kcat/Km for the more active enzymes, in contrast to those for sluggish variants, might not fully reflect the value of Ktx. The slope in a plot of log(Ki/[U>v]) vs logKtx could therefore be even more shallow than the slope in the plot of log(Ki/[U>v]) vs log(Km/kcat) (Figures 2A and 2B).
The substrate and the product can also be considered to be mimics of the transition state, as they are inevitably related. By extension, a mimic of the substrate or product is in some sense also a mimic of the transition state. Does U>v mimic the transition state simply by trivial mimicry of the substrate or product? To compare binding of the product to the binding of U>v, the inhibition constant for 3′-UMP (Kp) was measured for all four enzymes. These values are listed in Table 2. For wild-type RNase A and the K41A variant, the value of Kd for the enzyme•3′-UMP complex was ascertained by using isothermal titration calorimetry. The binding of another ground state, C>p, is reflected in the value of Km, as the forward commitment of the RNase A•C>p complex is low (Cf < 1).44,47
Table 2.
Values of Kp for Uridine 3′-Phosphate Inhibition of Catalysis by Wild-type Ribonuclease A and the K41CEA, K41R, and K41A Variants a
| Ribonuclease A | Kp (mM) |
|---|---|
| Wild-type | 0.072 ± 0.012 |
| Wild-type | 0.054 ± 0.005 b,c |
| K41CEA | 0.057 ± 0.005 |
| K41R | 0.046 ± 0.003 |
| K41A | 0.26 ± 0.12 |
| K41A | 0.41 ± 0.07 b |
Values of Kp were determined at 25 °C in 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M), Up(OC6H4-p-NO2), and 3′-UMP.
Value of Kd for the complex with 3′-UMP determined by isothermal titration calorimetry at 25 °C in 0.10 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M).
From ref 36.
The binding of U>v, like that of the transition state, is more sensitive to changes at position 41 than is the binding of the substrate or product. Values of Ki change by ≤ 38-fold (Table 1), but values of Km and Kp change by ≤ 9-fold (Table 2 and Figure 3). As shown in Figure 3, the slope in plots of log(Ki/[U>v]) vs logKm and log(Ki/[U>v]) vs logKp is near 1.5. The proximity of the slopes in Figure 3 to unity indicates that U>v does bear some resemblance to the substrate and product. This conclusion is made somewhat tentative, however, by the small variation in Km and Kp.
Figure 3.
Plot of log(Ki/[U>v]) vs logKm and logKp for wild-type ribonuclease A and the K41CEA, K41R, and K41A variants. Values of Ki/[U>v] (Table 1) were obtained at 25 °C in 0.010 M sodium succinate buffer, (pH 6.0) containing NaCl (0.10 M) and Up(OC6H4-p-NO2). Values of Km (●) were obtained from initial velocity data at 25 °C in 0.025 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M) and C>p. Values of Kp (○; Table 2) were obtained from initial velocity data measured at 25 °C in 0.10 M MES-NaOH buffer, pH 6.0, containing NaCl (0.10 M), Up(OC6H4-p-NO2), and 3′-UMP. Values of Kp (□; Table 2) for the wild-type and K41A enzymes were also obtained by isothermal titration calorimetry of enzyme•3′-UMP complex formation. The solid line has slope ≈ 1.5; the dashed line has a slope of unity.
We had shown previously that Lys41 interacts extensively with the transition state during catalysis by RNase A.27 This analysis is extended in Figure 4, which is a plot of logKp vs log(Km/kcat). The shallow slope of this plot (near 0.2) indicates that Lys41 interacts far more extensively with the transition state than the product during catalysis of C>p hydrolysis.
Figure 4.
Plot of logKp vs log(kcat/Km) for wild-type ribonuclease A and the K41CEA, K41R, and K41A variants. Values of Kp (○; Table 2) were obtained from initial velocity data measured at 25 °C in 0.10 M MES-NaOH buffer, pH 6.0, containing NaCl (0.10 M), Up(OC6H4-p-NO2), and 3′-UMP. Values of kcat/Km were obtained at 25 °C in 0.025 M MES-NaOH buffer (pH 6.0) containing NaCl (0.10 M) and C>p. Values of Kp (□; Table 2) for the wild-type and K41A enzymes were also obtained by isothermal titration calorimetry of enzyme•3′-UMP complex formation. The solid line has slope ≈ 0.2; the dashed line has a slope of unity.
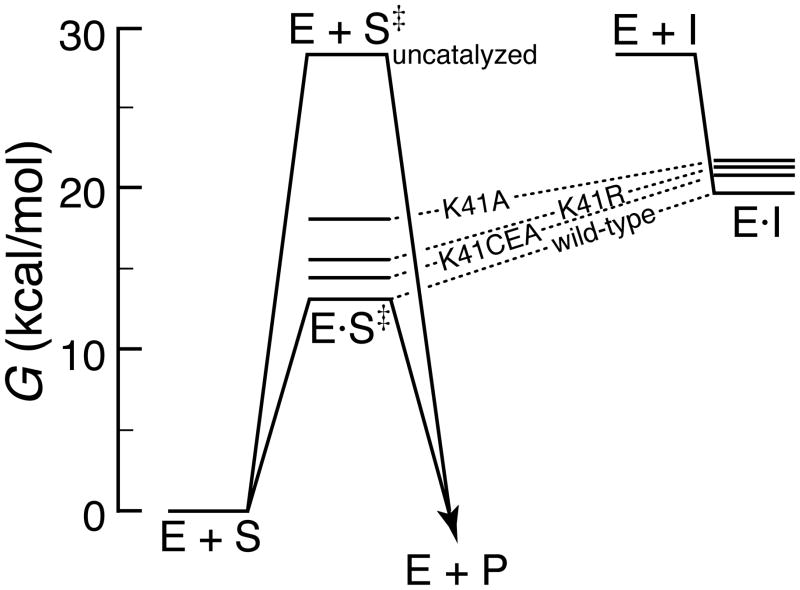
The mimicry of U>v to the transition state can be summarized in two related free energy profiles. In Figure 5, the effect of changing residue 41 on catalysis (that is, binding to the rate-limiting transition state) is depicted on the left, and the effect of changing residue 41 on inhibition (that is, binding to U>v) is depicted on the right. If U>v were a true analogue of the transition state, then the four dotted lines would be parallel. They are not.
Figure 5.
Free energy profile of catalysis by ribonuclease A and its inhibition by uridine 2′,3′-cyclic vanadate. The left profile depicts the free energy barrier for the uncatalyzed hydrolysis of C>p [ΔGuncat‡ = −RTln(kuncath/kbT) ref 28] and the free energy barriers for hydrolysis of C>p catalyzed by the wild-type enzyme and three variants [ΔGtx° = − RTln(kcat/Km)/kuncat]. The right profile depicts the binding of these variants to the uridine vanadate complex [ΔGi° = −RTln(M/Ki)]. For wild-type RNase A, Ki = 0.45 μM.37 For the variant enzymes, Ki is calculated from the values of Ki/[U>v] (Table 1). To ease comparisons, the free energy of the inhibitor ‘I’ in the right profile is set equal to that of the transition state ,S‡, in the left profile.
Organo-Vanadates as Mimics of Transition States
A pentavalent organo-vanadate is likely to be a less accurate mimic of the transition state for a phosphoryl transfer reaction than is, say, a phosphonate or phosphonamidate for an amide hydrolysis reaction.7,10,12,13 The failure of pentavalent organo-vanadates as transition state analogues for phosphoryl transfer reactions is explicable. The location and charge of the vanadyl oxygens differ significantly from those of the corresponding phosphoryl oxygens in the enzymic transition state. For example, the O2′···P···O5″ bond angle in the transition state is likely to be 180° (Figure 1B), whereas the analogous O3′ –V–O1V bond angle in the RNase A•U>v complex is only 150.5° (Figure 1C).23 Moreover, V–O bonds in vanadates exhibit a wide spectrum of strengths and polarities.24,48 Likewise, P–O bonds in the transition states of phosphoryl transfer reactions exhibit a variety of bond orders.17 In general, however, a V–O bond is far less polar than a P–O bond.24 Consequently, the non-bridging oxygens in a pentavalent vanadate form weaker hydrogen bonds than do those in a phosphorane.24 This distinction could explain why Lys41, which donates a hydrogen bond to a non-bridging phosphoryl oxygen in the transition state during catalysis by RNase A,25 does not donate a hydrogen bond to a non-bridging vanadyl oxygen in the crystalline RNase A•U>v complex (Figure 2C).24 Hence, we argue for skepticism about detailed or quantitative conclusions regarding a phosphoryl transfer reaction based solely on analyses of a complex with a pentavalent organo-vanadate.
Acknowledgments
This work was supported by Grant GM44783 (NIH). Isothermal titration calorimetry data were collected at the University of Wisconsin–Madison Biophysics Instrumentation Facility, which is supported by the University of Wisconsin–Madison and was established by Grant BIR-9512577 (NSF).
References
- 1.Pauling L. Chem Eng News. 1946;24:1375. [Google Scholar]
- 2.Pauling L. Nature. 1948;161:707–709. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 3.Jencks WP. In: Current Aspects of Biochemical Energetics. Kaplan NO, Kennedy EP, editors. Academic Press; New York: 1966. pp. 273–298. [Google Scholar]
- 4.Lolis E, Petsko GA. Annu Rev Biochem. 1990;59:597–630. doi: 10.1146/annurev.bi.59.070190.003121. [DOI] [PubMed] [Google Scholar]
- 5.Wolfenden R. Nature. 1969;223:704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 6.Radzicka A, Wolfenden R. Methods Enzymol. 1995;249:284–312. doi: 10.1016/0076-6879(95)49039-6. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett PA, Marlowe CK. Biochemistry. 1983;22:4618–4624. doi: 10.1021/bi00289a002. [DOI] [PubMed] [Google Scholar]
- 8.Mader MM, Bartlett PA. Chem Rev. 1997;97:1281–1301. doi: 10.1021/cr960435y. [DOI] [PubMed] [Google Scholar]
- 9.Thompson RC. Biochemistry. 1973;12:47–51. doi: 10.1021/bi00725a009. [DOI] [PubMed] [Google Scholar]
- 10.Hanson JE, Kaplan AP, Bartlett PA. Biochemistry. 1989;28:6294–6305. doi: 10.1021/bi00441a022. [DOI] [PubMed] [Google Scholar]
- 11.Smith AA, Carlow DC, Wolfenden R, Short SA. Biochemistry. 1994;33:6468–6474. doi: 10.1021/bi00187a012. [DOI] [PubMed] [Google Scholar]
- 12.Bartlett PA, Biangiordano MA. J Org Chem. 1996;61:3433–3438. [Google Scholar]
- 13.Phillips MA, Kaplan AP, Rutter WJ, Bartlett PA. Biochemistry. 1992;31:959–963. doi: 10.1021/bi00119a003. [DOI] [PubMed] [Google Scholar]
- 14.Kerr KM, Hedstrom L. Biochemistry. 1997;36:13365–13373. doi: 10.1021/bi9714161. [DOI] [PubMed] [Google Scholar]
- 15.Mosi R, Sham H, Uitdehaag JCM, Ruiterkamp R, Dijkstra BW, Withers SG. Biochemistry. 1998;37:17192–17198. doi: 10.1021/bi981109a. [DOI] [PubMed] [Google Scholar]
- 16.Rahil J, Pratt RF. Biochemistry. 1994;33:116–125. doi: 10.1021/bi00167a015. [DOI] [PubMed] [Google Scholar]
- 17.Cleland WW, Hengge AC. FASEB J. 1995;9:1585–1594. doi: 10.1096/fasebj.9.15.8529838. [DOI] [PubMed] [Google Scholar]
- 18.Lindquist RN, Lynn JL, Jr, Lienhard GE. J Am Chem Soc. 1973;95:8762–8768. doi: 10.1021/ja00807a043. [DOI] [PubMed] [Google Scholar]
- 19.(a) D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1987. [Google Scholar]; (b) Raines RT. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 20.For other proposed mechanisms, see: Witzel H. Progr Nucleic Acid Res. 1963;2:221–258.Hammes GG. Adv Protein Chem. 1968;23:1–57. doi: 10.1016/s0065-3233(08)60399-x.Wang JH. Science. 1968;161:328–334. doi: 10.1126/science.161.3839.328.Anslyn E, Breslow R. J Am Chem Soc. 1989;111:4473–4482.
- 21.Alber T, Gilbert W, Ponzi D, Petsko G. In: The Role of Mobility in Substrate Binding and Catalytic Machinery of Enzymes. Porter R, O’Connor MD, Whelan J, editors. Pitman; London: 1982. pp. 4–24. [Google Scholar]
- 22.Wlodawer A, Miller M, Sjölin L. Proc Natl Acad Sci USA. 1983;80:3628–3631. doi: 10.1073/pnas.80.12.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wladkowski BD, Svensson LA, Sjolin L, Ladner JE, Gilliland GL. J Am Chem Soc. 1998;120:5488–5498. [Google Scholar]
- 24.Krauss M, Basch H. J Am Chem Soc. 1992;114:3630–3634. [Google Scholar]
- 25.Messmore JM, Fuchs DN, Raines RT. J Am Chem Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001.For other analyses of catalysis by RNase A variants at Lys41, see: Trautwein K, Holliger P, Stackhouse J, Benner SA. FEBS Lett. 1991;281:275–277. doi: 10.1016/0014-5793(91)80410-5.Messmore JM, Holmgren SK, Grilley JE, Raines RT. Bioconjug Chem. 2000;11:408–413. doi: 10.1021/bc990142m.
- 26.Bruenger A, Brooks C, III, Karplus M. Proc Natl Acad Sci USA. 1985;82:8458–8462. doi: 10.1073/pnas.82.24.8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wladkowski BD, Krauss M, Stevens WJ. J Am Chem Soc. 1995;117:10537–10545. [Google Scholar]
- 28.Thompson JE, Kutateladze TG, Schuster MC, Venegas FD, Messmore JM, Raines RT. Bioorg Chem. 1995;23:471–481. doi: 10.1006/bioo.1995.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messmore JM, Raines RT. Arch Biochem Biophys. 2000;381:25–30. doi: 10.1006/abbi.2000.1951. [DOI] [PubMed] [Google Scholar]
- 30.Davis AM, Regan AC, Williams A. Biochemistry. 1988;27:9042–9047. doi: 10.1021/bi00425a024. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JE, Venegas FD, Raines RT. Biochemistry. 1994;33:7408–7414. doi: 10.1021/bi00189a047. [DOI] [PubMed] [Google Scholar]
- 32.Beaven GH, Holiday ER, Johnson EA. In: Optical Properties of Nucleic Acids and Their Components. Chargraff E, Davidson JN, editors. Vol. 1. Academic Press; New York: 1955. pp. 493–553. [Google Scholar]
- 33.Goddard JB, Gonas AM. Inorg Chem. 1973;12:574–579. [Google Scholar]
- 34.Dantzman CL, Kiessling LL. J Am Chem Soc. 1996;118:11715–11719. [Google Scholar]
- 35.Wiseman T, Williston S, Brandts JF, Lin LN. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 36.Fisher BM, Schultz LW, Raines RT. Biochemistry. 1998;37:17386–17401. doi: 10.1021/bi981369s. [DOI] [PubMed] [Google Scholar]
- 37.Leon-Lai CH, Gresser MJ, Tracey AS. Can J Chem. 1996;74:38–48. [Google Scholar]
- 38.Messmore JM. PhD Thesis. University of Wisconsin; Madison: 1999. [Google Scholar]
- 39.Bailey NA, Carrington A, Lott KAK, Symons MCR. J Chem Soc. 1960:290–297. [Google Scholar]
- 40.Tracey AS, Jaswal JS, Gresser MJ, Rehder D. Inorg Chem. 1990;29:4283–4288. [Google Scholar]
- 41.Angus-Dunne SJ, Batchelor RJ, Tracey AS, Einstein FWB. J Am Chem Soc. 1995;117:5292–5296. [Google Scholar]
- 42.Tracey AS, Leon-Lai CH. Inorg Chem. 1991;30:3200–3204. [Google Scholar]
- 43.Anonov IV, Gurevich AZ, Dudkin SM, Karpiesky MA, Sakhorovsky VG. Eur J Biochem. 1978;87:45–54. doi: 10.1111/j.1432-1033.1978.tb12350.x. [DOI] [PubMed] [Google Scholar]
- 44.Eftink MR, Biltonen RL. Biochemistry. 1983;22:5123–5134. doi: 10.1021/bi00291a011. [DOI] [PubMed] [Google Scholar]
- 45.Schultz LW, Quirk DJ, Raines RT. Biochemistry. 1998;37:8886–8898. doi: 10.1021/bi972766q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.(a) Nogués MV, Vilanova M, Cuchillo CM. Biochim Biophys Acta. 1995;1253:16–24. doi: 10.1016/0167-4838(95)00138-k. [DOI] [PubMed] [Google Scholar]; (b) Nogués MV, Moussaoui M, Boix E, Vilanova M, Ribó M, Cuchillo CM. Cell Mol Life Sci. 1998;54:766–774. doi: 10.1007/s000180050205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.del Rosario EJ, Hammes GG. J Am Chem Soc. 1970;92:1750. doi: 10.1021/ja00709a056. [DOI] [PubMed] [Google Scholar]
- 48.Ray WJ, Crans DC, Zheng J, Burgner JW, II, Deng H, Mahroof-Tahir M. J Am Chem Soc. 1995;117:6015–6026. [Google Scholar]